ABSTRACT
Cytotoxic T lymphocytes recognizing conserved peptide epitopes are crucial for protection against influenza A virus (IAV) infection. The CD8 T cell response against the M158–66 (GILGFVFTL) matrix protein epitope is immunodominant when restricted by HLA-A*02, a major histocompatibility complex (MHC) molecule expressed by approximately half of the human population. Here we report that the GILGFVFTL peptide is restricted by multiple HLA-C*08 alleles as well. We observed that M158–66 was able to elicit cytotoxic T lymphocyte (CTL) responses in both HLA-A*02- and HLA-C*08-positive individuals and that GILGFVFTL-specific CTLs in individuals expressing both restriction elements were distinct and not cross-reactive. The crystal structure of GILGFVFTL–HLA-C*08:01 was solved at 1.84 Å, and comparison with the known GILGFVFTL–HLA-A*02:01 structure revealed that the antigen bound both complexes in near-identical conformations, accommodated by binding pockets shaped from shared as well as unique residues. This discovery of degenerate peptide presentation by both HLA-A and HLA-C allelic variants eliciting unique CTL responses to IAV infection contributes fundamental knowledge with important implications for vaccine development strategies.
IMPORTANCE The presentation of influenza A virus peptides to elicit immunity is thought to be narrowly restricted, with a single peptide presented by a specific HLA molecule. In this study, we show that the same influenza A virus peptide can be more broadly presented by both HLA-A and HLA-C molecules. This discovery may help to explain the differences in immunity to influenza A virus between individuals and populations and may also aid in the design of vaccines.
INTRODUCTION
Influenza A viruses (IAVs) cause seasonal epidemics that affect millions of individuals, resulting in substantial morbidity and mortality each year. The viruses are also responsible for recurring pandemics due to the emergence of highly pathogenic strains from spontaneous reassortment when the same cell is coinfected (1). In addition to humoral immunity, cellular immunity against influenza involving both CD8+ and CD4+ T lymphocytes has been shown to be important (2–5). Cross-protection against heterosubtypic IAV strains is mediated predominantly by cross-reactive T cells recognizing epitopes that are conserved across viral subtypes (6–9). Within each individual, only a small subset of peptides can stimulate virus-specific cytotoxic T lymphocytes (CTLs). This epitope repertoire uniquely corresponds to the HLA haplotype of that person, while the high degree of HLA polymorphism within the general population is believed to prevent viral escape from immune pressure by epitope mutation. Degenerate binding of peptides to microvariants of the same HLA allele has been demonstrated in the context of infectious agents such as Plasmodium falciparum (10), HIV (11), cytomegalovirus (CMV) (12), Epstein-Barr virus (EBV) (13), and influenza A viruses (14, 15). However, degenerate peptide binding and subsequent stimulation of an immune response by two alternative HLA loci involving HLA-C are rare (12, 16–19).
We have identified novel restrictions to multiple HLA-C*08 alleles for a highly conserved immunodominant epitope derived from the influenza A virus M1 protein, M158–66 (GILGFVFTL), known to be restricted by HLA-A*02 (20, 21). This is the first peptide epitope reported to be presented degenerately by alleles from both HLA-A and HLA-C in the context of IAV infection. Using major histocompatibility complex (MHC) tetramers, we demonstrate that CTL recognition of the GILGFVFTL peptide presented by both HLA-A*02 and HLA-C*08 is nonredundant, giving rise to two mutually exclusive CTL populations within the same individual's T cell repertoire. By comparing the crystal structures of GILGFVFTL-bound HLA-A*02:01 and GILGFVFTL-bound HLA-C*08:01, we observed that the epitope binds to the two complexes similarly at the anchor pockets but that peptide residue positions p4 to p6 are oriented differently, which could impact T cell receptor interaction. These enhanced virus-specific CTL responses elicited by a single epitope presented by multiple restriction elements suggest that vaccine strategies employing T cell epitopes are probably more broadly effective than previously appreciated. This novel HLA-C*08:01 crystal structure provides new insight into the role and peptide binding properties of the less-studied HLA-C-restricted T cell responses. In addition, our findings could provide guidance for the future development and evaluation of IAV and other vaccines.
MATERIALS AND METHODS
HLA typing.
This study was performed according to the ethical principles stated in the World Medical Association Declaration of Helsinki. The protocols were approved by the Institutional Review Board of the National University of Singapore. With informed consent, 100 μl of blood was collected from each donor, and genomic DNA (gDNA) from these blood samples was extracted using the PureLink genomic DNA kit (Invitrogen). The exon-2-to-exon-3 regions of the class I HLA genes were amplified via PCR using the following primer pairs: HLA-A1 (TGGCCCCYGGTACCCGT)/HLA-A2 (GAAACSGCCTCTGYGGGGAGAAGCAA), HLA-B1 (GGGTCCCAGTTCTAAAGTCCCCACG)/HLA-B2 (CCATCCCSGGCGAYCTAT), and HLA-C1 (AGCGAGGKGCCCGCCCGGCGA)/HLA-C2 (GGAGATGGGGAAGGCTCCCCACT) (where K indicates G or T, S indicates G or C, and Y indicates T or C.). Each of the PCR products was then sequenced, and the sequences were compared with the HLA sequence alignment from the IMGT/HLA database to determine the HLA genotypes.
Donors and in vitro stimulation of human PBMCs.
Fresh whole blood was collected from 15 healthy donors after written informed consent was obtained, and peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Paque Plus (GE Healthcare). PBMCs were stimulated in vitro at a cell density of 106/ml with 10 μg/ml GILGFVFTL peptide (GenScript, USA). The culture medium used was RPMI 1640 medium supplemented with 5% pooled AB serum (Invitrogen), 2.05 mM l-glutamine (Invitrogen), 100 IU/ml penicillin-streptomycin (Invitrogen), and 40 μM β-mercaptoethanol (Gibco). For a 14-day culture, the PBMCs were incubated at 37°C under 5% CO2 and were supplemented with 25 U/ml recombinant interleukin 2 (rIL-2) (R&D Systems) every 2 to 3 days from day 2 onward. Also, a half-medium change was carried out every 2 to 3 days from day 5 onward.
Preparation of MHC tetramers.
Recombinant MHC molecules were prepared using a protocol described previously (22). pET-28a(+) encoding the HLA-A*02:01 and -C*08:01 heavy chains and human β2 microglobulin was synthesized (GenScript, USA), transformed into Escherichia coli BL21 cells, and overexpressed by triggering with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Inclusion bodies from E. coli BL21 were extracted, purified under reducing conditions, and solubilized in 8 M urea buffer. The MHC proteins were refolded for 36 h with a MHC heavy chain/β2-microglobulin/peptide ratio of 1:2:10. The product was then dialyzed three times in 20 mM Tris (pH 8.0) prior to biotinylation by recombinant BirA enzymes. The MHC proteins were purified by using gel filtration chromatography (S200). MHC tetramers were assembled by mixing purified HLA-A*02:01 and -C*08:01 molecules with streptavidin-phycoerythrin (PE) (Invitrogen) and streptavidin-Brilliant Violet 421 (BioLegend) at a molar ratio of 4:1.
Tetramer staining.
HLA-A*02:01 and -C*08:01 tetramers were titrated using PBMCs isolated from an HLA-A*02:01- and HLA-C*08:01-positive donor that were stimulated with the GILGFVFTL peptide to obtain the concentration of each tetramer that gives half-maximal staining intensity. Subsequent tetramer staining was performed using the titrated concentrations. Briefly, cells were stained with a LIVE/DEAD fixable near-infrared (near-IR) stain (Molecular Probes), washed with cold phosphate-buffered saline (PBS), and coincubated with 47.72 nM HLA-A*02:01 tetramers and 8.267 nM HLA-C*08:01 tetramers on ice for 20 min. After incubation, the cells were stained with peridinin chlorophyll protein (PerCP)-conjugated anti-CD8 antibodies (BD Biosciences), thoroughly washed with cold PBS, and fixed with 1% paraformaldehyde–PBS. Data were acquired by flow cytometry on a BD LSR II flow cytometer, and analysis was performed using FlowJo (TreeStar).
Intracellular cytokine staining.
At day 14 after the first peptide stimulation, T cell lines were resuspended in fresh culture medium at 106 cells/ml and were stimulated with 1 μg/ml GILGFVFTL for 5 h at 37°C under 5% CO2 in the presence of GolgiPlug (containing brefeldin A) (BD Biosciences) and fluorescein isothiocyanate (FITC)-conjugated anti-CD107a antibodies (eBioscience). For positive controls, cells were stimulated with 10 ng/ml phorbol myristate acetate (PMA) and 400 ng/ml ionomycin (Sigma-Aldrich) instead of the antigenic peptide. After stimulation, the cells were tetramer and surface stained as described above and were fixed and permeabilized with Cytofix/Cytoperm solution (BD Biosciences). Intracellular staining was carried out using allophycocyanin (APC)-conjugated antibodies against gamma interferon (IFN-γ) (BD Biosciences).
Protein crystallization.
Crystal-growing conditions were first screened by Rigaku's CrystalMation system using Hampton Research and Molecular Dimensions screening kits. Each drop contained 0.2 μl protein and 0.2 μl screening buffer. To generate peptide-MHC crystals, crystallization by the hanging drop method was set up manually under six selected conditions. Each drop contained 2 μl protein and 2 μl screening solution and was further equilibrated at 18°C against the respective reservoir solutions. GILGFVFTL–HLA-C*08:01 crystals were subsequently observed under the following condition: 0.05 M calcium chloride dehydrate, 0.1 M Bis-Tris (pH 6.5), 30% (vol/vol) polyethylene glycol monomethyl ether 550 (Hampton Research). A single crystal was further obtained by the streak-seeding method (23). Perfluoropolyether Cryo Oil was used as a cryoprotectant solution.
X-ray diffraction data collection, structure determination, and structure analysis.
Single-wavelength native data (1.54178 A°) were collected from a single crystal of HLA-C*08:01-influenza A virus with the CCD Proteum X-ray diffraction system (Institute of Molecular and Cell Biology, A*STAR, Singapore). The crystal belongs to space group C2. The data collection strategy was created by Proteum2, and the data were indexed, integrated, and scaled by Proteum2 (see Table 2). All data were further processed by CCP4 (24). The structure of GILGFVFTL–HLA-C*08:01 was determined by the molecular replacement method using the crystal structure of HLA-Cw4 (PDB code 1QQD) as the search model. The rotation and translation function and restrained refinement were carried out using the MOLREP and REFMAC5 programs in CCP4, respectively. Simple mutations were applied to initial models in the Coot program after molecular replacement in CCP4. These models were further refined by REFMAC5, during which the parameters R-factor (Rf) and Rfree that assesses the agreement between the calculated model and observed data were carefully monitored. In the final stage, water molecules were added, and the model was subjected to additional refinement using REFMAC5. This was then checked by Coot, and the water molecules were removed if either the B-factor was >80 Å2, the map sigma level was <1.00 electron/Å3, or the closest contact was not in the range from 2.3 to 3.5 Å. Following water refinement, restrained individual B-factor refinement was performed. Model geometry was verified using the PROCHECK program (25) with all dihedral angles in the favored or allowed regions. The HLA-peptide complex was displayed and analyzed by the PyMOL molecular graphics system (26). The PDBePISA (Protein Interfaces, Structures and Assemblies) service (27) was used to generate interface data, energetic data, and hydrogen bond counts. Structural coordinates from the GILGFVFTL–HLA-A*02:01 complex (PDB code 2VLL) were used for performing comparison analysis.
TABLE 2.
Data collection and refinement statistics for GILGFVFTL–HLA-C*08:01
| Parameter | Value(s) |
|---|---|
| Data collection statistics | |
| Space group | C2 |
| No. of molecules/AU | 1 |
| Cell parameters | |
| a, b, c (Å) | 94.4, 75.7, 61.9 |
| α = γ (°) | 90.0 |
| β (°) | 120.3 |
| Resolution range (Å)a | 55.4–1.84 (1.88–1.84) |
| No. of reflections | |
| Total | 445,969 |
| Unique | 35,257 |
| Completeness (%)a | 97.5 (89.90) |
| Rmergea | 0.041 (0.302) |
| I/σ(I)a | 12.11 (3.30) |
| Multiplicity | 12.65 (2.64) |
| Refinement statistics | |
| Rcrysta,b | 0.180 (0.23) |
| Rfreea,c | 0.229 (0.26) |
| No. of: | |
| Reflections | 32,697 |
| Atoms | 3,559 |
| Waters | 419 |
| Stereochemistry | |
| Bond length (Å) | 0.012 |
| Bond angle (°) | 1.5 |
| Ramachandran plot (%) | |
| Most favored region | 97.07 |
| Allowed region | 2.93 |
| Disallowed region | 0 |
Values in parentheses refer to the highest-resolution shell; I/σ(I) is a measurement of signal-to-noise ratio; Rmerge indicates agreement among multiple measurements of data sets.
Calculated as Σ||Fo| − |Fc||/Σ Fo.
Same as Rcryst but calculated on the 5% of the data excluded from refinement.
CD spectra and thermal unfolding.
Circular dichroism (CD) experiments for the GILGFVFTL–HLA-C*08:01 and GILGFVFTL–HLA-A*02:01 complexes were performed on a Jasco J-810 spectropolarimeter equipped with a thermal controller. The far-UV CD spectra (200 to 260 nm) were collected at a protein concentration of between 15 and 20 μM in 20 mM Tris (pH 8.0) buffer, using a 1-mm-path-length cuvette with 0.1-nm spectral resolution. The ellipticity at 218 nm was recorded continuously during heating. The water jacket cell containing the sample was heated at a linear rate of 2°C/min. Thermal unfolding spectra were normalized and were smoothened by the adjacent average method. The basal line values before and after unfolding were calculated by averaging the ellipticity values in the ranges of 25 to 35°C and 80 to 90°C, respectively. The denaturation temperatures (Td) at the midpoint were determined, assuming that the unfolding equilibrium follows a two-state mechanism. The characteristic wavelength of 218 nm was selected for the detection of secondary-structure change during thermal unfolding.
Protein structure accession number.
The coordinates for the GILGFVFTL–HLA-C*08:01 complex have been deposited in the Protein Data Bank under accession code 4NT6.
RESULTS
Immunogenicity of M158–66 (GILGFVFTL) in CTL recall responses.
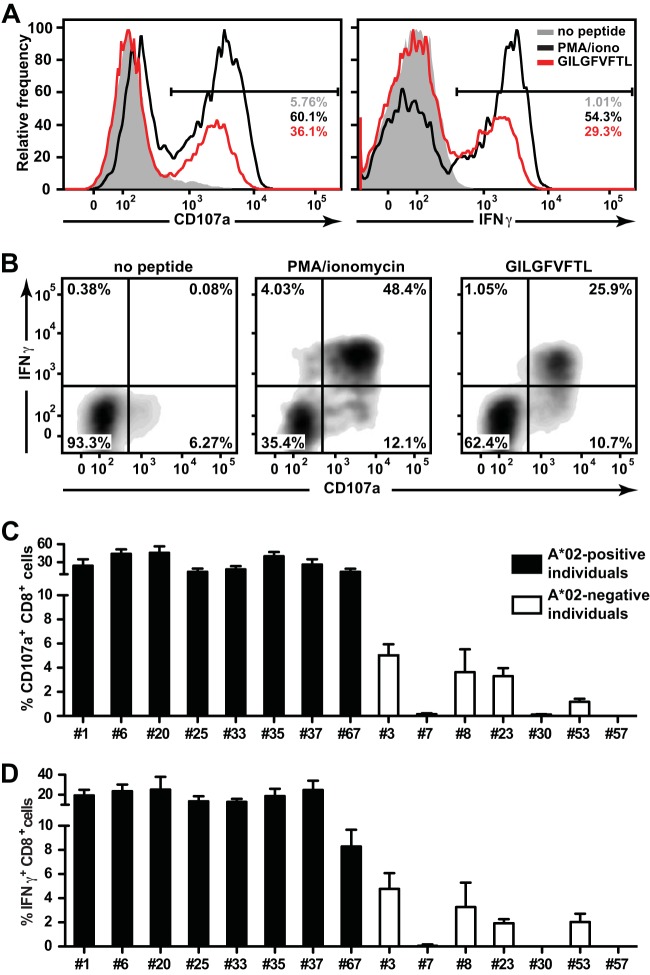
Cytotoxic T cell responses are often directed against a limited number of determinants encoded in complex antigens of a particular pathogen. In influenza A virus infection, the paradigm in immunodominant human CTL responses among IAV antigens concerns the HLA-A*02-restricted M158–66 peptide (GILGFVFTL). CTLs specific for GILGFVFTL can be observed in almost all HLA-A*02-positive (A*02+) donors who have been naturally exposed to influenza A virus (28, 29). In a cohort of recruited individuals, we aimed to confirm adequate CTL responses against influenza A virus, and therefore, we first sought to characterize the GILGFVFTL-specific responses in these individuals. PBMCs from 15 individuals (8 HLA-A*02-positive and 7 HLA-A*02-negative [A*02−] individuals) were collected, pulsed with the GILGFVFTL peptide, and cultured in vitro for 14 days. The cells were then restimulated with GILGFVFTL, and CD107a surface expression and gamma interferon (IFN-γ) secretion (Fig. 1A and B) were measured. CTL responses against the M158–66 peptide were detected in 100% (8/8) of the HLA-A*02-positive individuals. A good proportion of the CTLs from these individuals expressed both CD107a and IFN-γ in response to the stimulation (Fig. 1B). The frequencies of CD107a-expressing CTLs ranged from 14.3% to 45.7% of the total CD8+ population (Fig. 1C). The frequencies of IFN-γ secretion were slightly lower, between 8.3% and 25.3% of the total CD8+ population (Fig. 1D).
FIG 1.
Immunogenicity of M158–66 in the CTL recall responses of HLA-A2-positive and -negative individuals. (A) Representative histograms of CD107a and IFN-γ expression in the total CD8+ population derived from an A2-positive individual (donor 25) after stimulation with the GILGFVFTL peptide (red). Unstimulated CTLs (shaded histograms) and CTLs stimulated with PMA-ionomycin (black curves) were included as negative and positive controls, respectively. The frequencies of positive cells among total CD8+ populations are given. (B) Representative plot of IFN-γ versus CD107a expression in the CD8+ cells from panel A. Cells were either left unstimulated (left) or stimulated with PMA-ionomycin (center) or GILGFVFTL (right). (C and D) Summaries of the frequencies of cells expressing CD107a (C) and IFN-γ (D) among cells from each of the 15 individuals recruited in this study.
Surprisingly, GILGFVFTL-specific CTL responses were also observed in 57% (4/7) of the HLA-A*02-negative donors (i.e., donors 3, 8, 23, and 53). The frequencies of the responding CTLs were lower than those for the HLA-A*02-positive individuals, with CD107a expression and IFN-γ secretion ranging from 1.2% to 5% and from 1.9% to 4.8% of the total CD8+ population, respectively. These data suggested the existence of degenerate antigenicity for the GILGFVFTL peptide involving MHC restriction elements other than the well-studied HLA-A2.
Degenerate presentation of the GILGFVFTL peptide by non-HLA-A2 restriction elements.
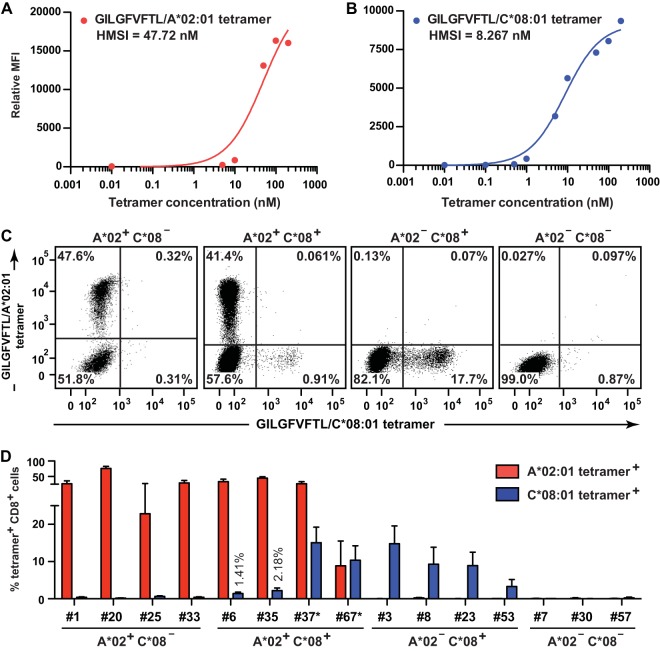
Which alternative HLA products could be responsible for the presentation of GILGFVFTL in the HLA-A*02-negative individuals? We first compared the HLA profiles of all 15 individuals (Table 1). All four GILGFVFTL-responding individuals from the HLA-A*02-negative group were noted to carry HLA-C*08 alleles; among these, three individuals (donors 3, 8, and 23) expressed HLA-A*11:01. This finding highlighted HLA-C*08 and HLA-A*11:01 as potential allelic variants for degenerate peptide binding and presentation. HLA-A*11:01 could reasonably be excluded from consideration, because an equal number of individuals (3/3 [donors 7, 30, and 57]) displayed no reactivity toward the GILGFVFTL peptide (Fig. 1C and D). To determine whether there were CTLs restricted by HLA-C*08:01, donor PBMCs were stained with GILGFVFTL–HLA-A*02:01 and GILGFVFTL–HLA-C*08:01 tetramers labeled with alternative fluorophores (Fig. 2). The 15 donors were divided into the following groups: A*02+ C*08− (n = 4), A*02+ C*08+ (n = 4), A*02− C*08+ (n = 4), and A*02− C*08− (n = 3). Representative staining of the GILGFVFTL-specific CTLs is shown in Fig. 2C. CTLs from A*02+ C*08− and A*02− C*08+ individuals could be stained with the GILGFVFTL–HLA-A*02:01 and GILGFVFTL–HLA-C*08:01 tetramers, respectively, whereas CTLs from A*02− C*08− individuals did not bind either of the tetramers. This finding confirms the authenticity of GILGFVFTL-specific CTL restriction by HLA-C*08. More importantly, GILGFVFTL-specific CTLs in A*02+ C*08+ donors bound independently to either HLA-A*02:01 or HLA-C*08:01 tetramers, forming two mutually exclusive CTL populations. This showed that the CTLs were not cross-reactive in T cell recognition when GILGFVFTL was presented in the context of an alternative HLA product within the same individual. In general, the frequencies of GILGFVFTL-specific CTLs measured by tetramer staining show the same hierarchy as that of the recalled GILGFVFTL responses measured in the total CD8+ population (Fig. 1C and D and 2D).
TABLE 1.
HLA allele profiles of the 15 donors recruited in this studya
| Donor ID | HLA-A | HLA-B | HLA-C | |||
|---|---|---|---|---|---|---|
| 1 | A*02:01 | A*02:06 | B*27:05 | B*40:01 | C*03:03 | C*03:04 |
| 3 | A*11:01 | A*11:01 | B*13:06 | B*48:03 | C*03:04 | C*08:06 |
| 6 | A*02:01 | A*02:06 | B*56:01 | B*95:21 | C*07:02 | C*08:01 |
| 7 | A*11:01 | A*32:01 | B*46:01 | B*51:07 | C*01:02 | C*14:02 |
| 8 | A*11:01 | A*33:03 | B*15:02 | B*51:01 | C*08:01 | C*14:02 |
| 20 | A*02:01 | A*11:01 | B*13:01 | B*40:01 | C*03:04 | C*07:02 |
| 23 | A*11:01 | A*24:02 | B*40:53 | B*40:75 | C*01:02 | C*08:03 |
| 25 | A*02:01 | A*33:03 | B*35:03 | B*40:06 | C*01:02 | C*04:01 |
| 30 | A*11:01 | A*33:03 | B*13:01 | B*40:01 | C*03:04 | C*07:02 |
| 33 | A*02:01 | A*02:01 | B*40:01 | B*46:01 | C*01:02 | C*15:02 |
| 35 | A*02:01 | A*02:06 | B*15:02 | B*51:01 | C*08:01 | C*14:02 |
| 37 | A*02:06 | A*02:06 | B*15:02 | B*55:02 | C*01:02 | C*08:01 |
| 53 | A*24:02 | A*30:01 | B*13:02 | B*15:02 | C*06:02 | C*08:01 |
| 57 | A*11:01 | A*11:01 | B*39:01 | B*40:01 | C*03:04 | C*07:02 |
| 67 | A*02:06 | A*02:07 | B*15:02 | B*46:01 | C*08:01 | C*08:03 |
Alleles from HLA-A2 and HLA-C8 are shown in boldface.
FIG 2.
Tetramer-staining M158–66-specific CTLs. (A and B) Titration of GILGFVFTL–HLA-A*02:01 and GILGFVFTL–HLA-C*08:01 tetramers used for the detection of GILGFVFTL-specific T cells. MFI, mean fluorescence intensity; HMSI, half-maximal staining intensity. (C and D) A mixture of the two tetramers with the concentration of each tetramer giving HMSI was used for dual-tetramer staining of GILGFVFTL-specific T cells from donors. (C) PBMCs were stained with the GILGFVFTL–HLA-A*02:01 and GILGFVFTL–HLA-C*08:01 tetramers. The four plots represent the staining of PBMCs from an individual of each group: HLA-A*02+ HLA-C*08− (donor 33), HLA-A*02+ HLA-C*08+ (donor 6), HLA-A*02− HLA-C*08+ (donor 3), and HLA-A*02− HLA-C*08− (donor 57). The frequency within the total CD8+ population is given in each quadrant of each plot. (D) Summary of frequencies of HLA-A*02:01 and HLA-C*08:01 tetramer-positive cells in the total CD8+ population from each of the 15 individuals studied. For donors 37* and 67*, antigen-specific T cells were quantified with GILGFVFTL–HLA-A*02:06 tetramers instead of GILGFVFTL–HLA-A*02:01 tetramers.
HLA-C*08:01 tetramer-positive but not HLA-A*02:01 tetramer-positive GILGFVFTL-specific CTLs were detected in two A*02+ C*08+ donors (donors 37 and 67) (Table 1). Previous studies have shown that micropolymorphisms for HLA-A*02 can cause functionally distinct peptide presentation (14, 30). To determine whether this phenomenon was the root cause of our failed tetramer staining, we repeated the dual-tetramer staining with four-digit-matched GILGFVFTL–HLA-A*02:06 and GILGFVFTL–HLA-C*08:01 tetramers for donors 37 (HLA-A*02:06) and 67 (HLA-A*02:06 and -A*02:07) (Fig. 2D). Indeed, the HLA-A*02-restricted GILGFVFTL-specific CTLs that could not be detected previously by HLA-A*02:01 tetramers were readily quantified with HLA-A*02:06 tetramers. This agrees with the observation that minor sequence variations between HLA-A*02:01, -A*02:06, and -A*02:07 can profoundly affect antigen-specific CD8 T cell detection with MHC multimers (31). HLA-A*02:01 differs from HLA-A*02:06 at F9Y and also from HLA-A*02:07 at Y99C. Since both positions 9 and 99 are located within the peptide-binding groove, we assume that these substitutions induce subtle changes in peptide orientation and therefore activate different clonal T cell populations. The GILGFVFTL–HLA-C*08:01 tetramers, on the other hand, were able to stain CTLs from HLA-C*08:01-, -C*08:03-, and -C*08:06-positive individuals (Fig. 2D). This promiscuity can be rationalized by the fact that the three HLA-C*08 alleles share identical amino acid sequences within the peptide-binding groove, with a single amino acid difference located at the edge of the α2 helix (G175 in HLA-C*08:01 and R175 in HLA-C*08:03 and -C*08:06).
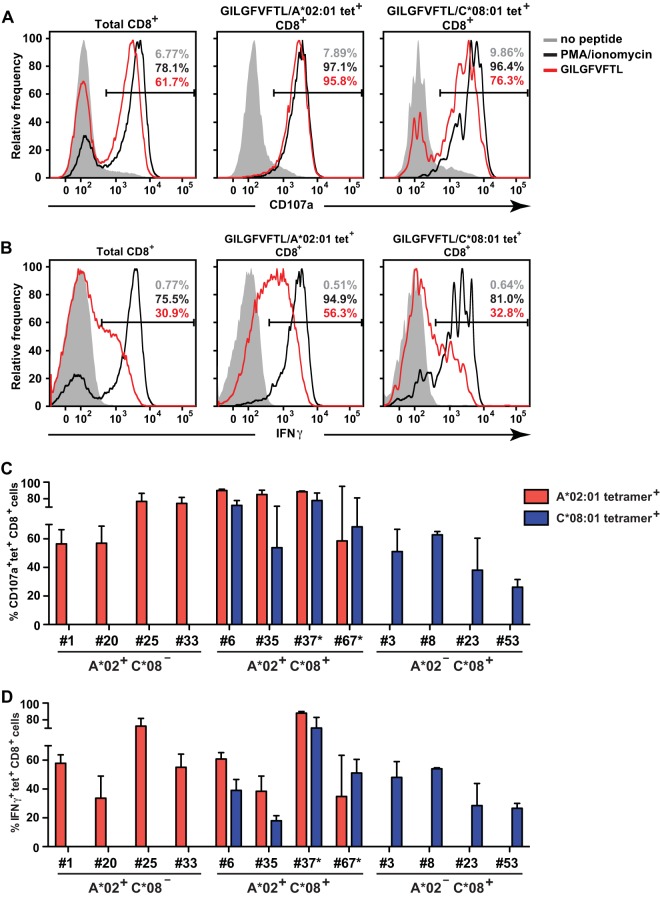
Next, we examined the functional responses of the HLA-A*02:01 and HLA-C*08:01 tetramer-positive CTLs to GILGFVFTL stimulation. Both HLA-A*02:01 and HLA-C*08:01 tetramer-positive CTLs expressed CD107a and IFN-γ after peptide stimulation (Fig. 3A and B, respectively). The frequencies of CD107a and IFN-γ expression among HLA-C*08:01 tetramer-positive T cells, however, were generally lower than those for HLA-A*02:01 tetramer-positive T cells (Fig. 3C and D). In agreement with the data in Fig. 2D, HLA-A2-restricted CTLs from individuals 23 and 67 did not stain with GILGFVFTL–HLA-A*02:01 tetramers, and thus, the proportions of CD107a and IFN-γ expression by HLA-A*02:01 tetramer-positive CTLs were not quantified. Instead, the frequencies of CD107a- and IFN-γ-expressing HLA-A*02:06 tetramer-positive CTLs in these individuals were measured (Fig. 3C and D). These data suggest that HLA-C*08-restricted CTLs are functional yet expand in vitro to lower frequencies than HLA-A*02-restricted CTLs.
FIG 3.
Functional characterization of HLA-A*02:01 and HLA-C*08:01 tetramer-positive CTLs. (A) Representative histograms of CD107a expression by CTLs derived from an A*02:01+ C*08:01+ individual (donor 6) after stimulation with the GILGFVFTL peptide (red). Unstimulated CTLs (shaded histograms) and CTLs stimulated with PMA-ionomycin (black curves) were included as negative and positive controls, respectively. The plots shown were gated on total (left), HLA-A*02:01 tetramer-positive (center), and HLA-C*08:01 tetramer-positive (right) CD8+ cells. The frequencies of positive cells within those populations are given. (B) Representative histograms of IFN-γ expression by CTLs, depicted as for CD107a expression in panel A. (C) Summary of the frequencies of CD107a-expressing tetramer-positive CTLs from individuals possessing HLA-A2 and/or HLA-C8 alleles. (D) Summary of the frequencies of IFN-γ-expressing tetramer-positive CTLs, represented as for CD107a-expressing tetramer-positive CTLs in panel C. For donors 37* and 67* in panels C and D, antigen-specific T cells were quantified with GILGFVFTL–HLA-A*02:06 tetramers instead of GILGFVFTL–HLA-A*02:01 tetramers.
Structural comparison of GILGFVFTL presented by HLA-C*08:01 and HLA-A*02:01.
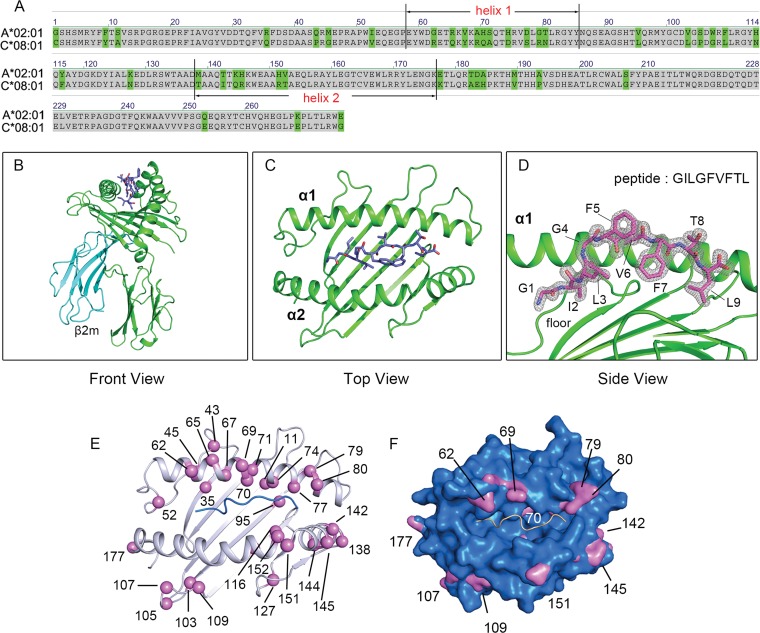
To elucidate the structural basis of the presentation of the same peptide by both HLA-A*02:01 and HLA-C*08:01, with 41 amino acid differences between them (Fig. 4A), we proceeded to generate protein crystals of GILGFVFTL–HLA-C*08:01, and the structure was successfully solved to 1.84 Å resolution (Table 2) (PDB code 4NT6). The typical global structure and features of an HLA molecule are immediately apparent (Fig. 4B and C), with the GILGFVFTL peptide residing in the binding groove surrounded by the α1 helix, the α2 helix, and the β-pleated floor (Fig. 4D). The characteristic HLA-C-specific KYRV motif around pocket B of the binding groove, the polymorphic amino acids residing along the α1 and α2 helices (amino acids 140 to 149), and the Cα atom positions are shown in Fig. 4E. To understand the difference in surface occupancy between HLA-C*08:01 and HLA-A*02:01, we overlapped their surfaces and pinpointed the contour differences (Fig. 4F). The portions of HLA-C*08:01 protruding above the HLA-A*02:01 surface gathered in three regions: the α1 helix solvent-exposed surface (R62, R69, R79, and N80), the α2 helix solvent-exposed surface (I142, R145, R151, and K177), and the loop region following the α2 helix (G107 and L109).
FIG 4.
Structural comparison between HLA-A*02:01 and HLA-C*08:01. (A) Protein sequence alignment of HLA-A*02:01 and HLA-C*08:01. Amino acid differences are highlighted in green. (B) Overview of the crystal structure of GILGFVFTL–HLA-C*08:01. The HLA-C*08:01 heavy chain, β2 microglobulin, and peptide are highlighted in green, cyan, and blue, respectively. (C) Top view of the peptide-binding groove. (D) Peptide conformation and electron density of the peptide. Electron density maps with the Fourier coefficients (2Fo − Fc) of the loops, hinge, peptide, and contact residues were contoured at 1.5 σ (standard deviation). (E) Positions of polymorphic residues around the binding groove. The Cα atoms of these residues are represented as purple spheres. (F) Surface comparison between HLA-A*02:01 and HLA-C*08:01. The HLA-A*02:01 surface is shown in blue. Portions of HLA-C*08:01 protruding above the HLA-A*02:01 surface are highlighted in purple.
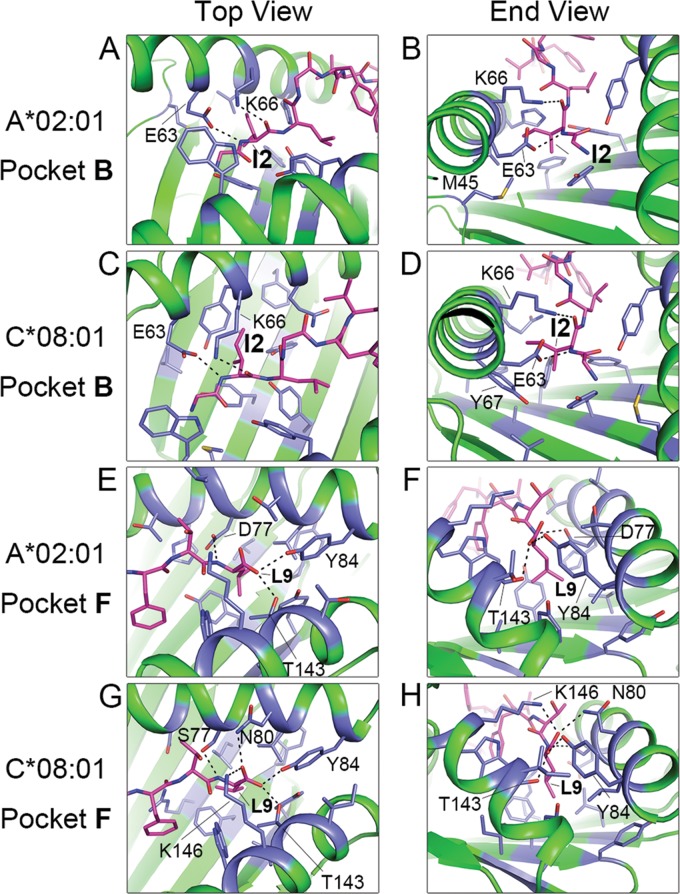
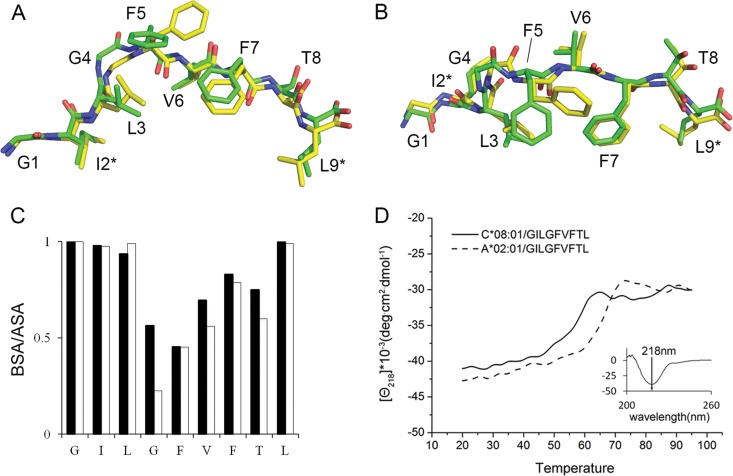
Comparison of the structures of the GILGFVFTL–HLA-A*02:01 and -C*08:01 complexes revealed that the peptide is stably bound in the anchor pockets B and F (Fig. 5). In both HLA-A*02:01 and HLA-C*08:01, the peptide at pocket B is held in position by the formation of hydrogen bond contacts by its isoleucine at peptide position 2 (p2) with E63 and K66 (Fig. 5A to D and Table 3). Unique features involving the long side chains of M45 (HLA-A*02:01) and Y7 (HLA-C*08:01) additionally contribute to engaging I2 of the peptide, though not at a level sufficient for the formation of hydrogen bonds. Aside from the involvement of Y7 and Y9, no significant bond formation is noted from residues along the floor of the β-plated sheet. Pocket F is described by positions 77, 80, 84, 143, and 146 of HLA-A*02:01 and -C*08:01 (Fig. 5E to H). Due to amino acid differences (D77, T80, V95, and Y116 for HLA-A*02:01; S77, N80, L95, and F116 for HLA-C*08:01), HLA-A*02:01 and -C*08:01 form multiple but different contacts with the p9 (Leu) of the peptide (Table 3). The p9 of the peptide appears to be well accommodated in the F pockets of both HLAs, and as with pocket B, the contribution from the binding groove floor is in the form of hydrophobic interactions via Y116 (HLA-A*02:01) and F116 (HLA-C*08:01). Because more HLA-peptide interactions are seen in pocket F, we assign to it a more significant role for providing binding stability than to pocket B. Torsional rotation of the bound peptide results in prominent differences in the peptide contour, particularly at p5, which contribute to eliciting very different T cell responses against the HLA-A*02- and HLA-C*08-presented epitopes (Fig. 6A to C). The overall stability of the MHC-peptide complex is influenced not only by the different amino acids in the critical residue positions of the HLA binding pockets but also by secondary interactions with the backbone and amino acid side chains over the entire length of the peptide. To assess the comparative stabilities of GILGFVFTL binding to HLA-A*02:01 and -C*08:01, a circular dichroism thermal stability assay was performed using in vitro-refolded GILGFVFTL–HLA-A*02:01 and GILGFVFTL–HLA-C*08:01 complexes. The circular dichroism experiment revealed that GILGFVFTL–HLA-A*02:01 has a midpoint denaturation temperature (Td) of 64.5°C, compared to 56°C for GILGFVFTL–HLA-C*08:01 (Fig. 6D). Together with the generally less dense expression of HLA-C on the cell surface, this may explain the lower level of T cell responses that could be recalled toward GILGFVFTL–HLA-C*08:01 than toward GILGFVFTL–HLA-A*02:01, as seen in Fig. 1C and D.
FIG 5.
Comparison of anchor contacts for the GILGFVFTL–HLA-A*02:01 and GILGFVFTL–HLA-C*08:01 complexes. (A through D) Interactions between residues in pocket B of the MHC and the N-terminal anchor (Ile2) of the peptide in the GILGFVFTL–HLA-A*02:01 (A and B) and GILGFVFTL–HLA-C*08:01 (C and D) structures. (E through H) Interactions between residues in pocket F of the MHC and the C-terminal anchor (Leu9) of the peptide in the GILGFVFTL–HLA-A*02:01 (E and F) and GILGFVFTL–HLA-C*08:01 (G and H) structures. The GILGFVFTL peptide is shown in pink. Residues of the MHC interacting with the anchor residues are highlighted in blue. Dashed lines indicate hydrogen bonds between anchor residues and the HLA heavy chain.
TABLE 3.
Interactions between anchor residues (p2 and p9) and HLA in pockets B and F
| GILGFVFTL–HLA-A*02:01 |
GILGFVFTL–HLA-C*08:01 |
|||||
|---|---|---|---|---|---|---|
| Interacting residue/atom in: |
Distance (Å)a | Interacting residue/atom in: |
Distance (Å)a | |||
| Peptide | HLA-A*02:01 | Peptide | HLA-C*08:01 | |||
| B | I2/CG2 | Y7/CE2 | 3.31 | |||
| I2/CB | F9/CZ | 4.90 | I2/CB | Y9/OH | 3.43 | |
| I2/CD1 | M45/CE | 4.10 | ||||
| I2/N | E63/OE1 | 2.94b | I2/N | E63/OE2 | 2.85b | |
| I2/O | K66/NZ | 2.81b | I2/O | K66/NZ | 2.83b | |
| I2/CD1 | V67/N | 3.41 | I2/CD1 | Y67/N | 3.55 | |
| I2/CD1 | H70/CE1 | 3.80 | I2/CD1 | Q70/CB | 4.97 | |
| I2/CB | Y99/OH | 3.26 | I2/CB | Y99/OH | 3.28 | |
| I2/CA | Y159/OH | 3.77 | I2/C | Y159/OH | 3.86 | |
| I2/N | W167/NE1 | 4.98 | I2/N | W167/NE1 | 4.88 | |
| F | L9/N | D77/OD1 | 2.91b | L9/N | S77/OG | 3.01b |
| L9/OXT | T80/CG2 | 3.63 | L9/O | N80/ND2 | 2.94b | |
| L9/CD1 | L81/CD1 | 3.64 | L9/CD1 | L81/CD1 | 4.08 | |
| L9/O | Y84/OH | 2.97b | L9/OXT | Y84/OH | 2.65b | |
| L9/CD1 | V95/CG1 | 4.85 | L9/CD1 | L95/CD1 | 3.72 | |
| L9/CD2 | Y116/CD2 | 3.95 | L9/CD2 | F116/CE2 | 4.00 | |
| L9/CD2 | Y123/CE2 | 3.54 | L9/CD1 | Y123/CE2 | 3.71 | |
| L9/CD2 | I124/CG2 | 4.29 | ||||
| L9/O | T143/OG1 | 2.79b | L9/OXT | T143/OG1 | 2.69b | |
| L9/OXT | K146/NZ | 3.36 | L9/O | K146/NZ | 2.86 | |
| L9/CD2 | W147/CH2 | 3.42b | L9/CG | W147/CZ2 | 3.59b | |
Distance (atom-to-atom) in peptide-HLA contact.
Hydrogen bond.
FIG 6.
Peptide orientation of GILGFVFTL within the HLA-A*02:01 and -C*08:01 complexes and thermal stabilities of the peptide-MHC molecules. (A) Side view of the conformations of the GILGFVFTL peptide within the HLA-A*02:01 and -C*08:01 complexes. The structural conformation of GILGFVFTL within the HLA-C*08:01 complex (shown in yellow) is superimposed on that of GILGFVFTL within the HLA-A*02:01 complex (shown in green). (B) Top view of the peptide conformations (90° rotation) shown in panel A. (C) Ratios of the buried surface area (BSA) to the accessible surface area (ASA) for GILGFVFTL bound to HLA-C*08:01 (filled bars) or to HLA-A*02:01 (open bars). (D) Circular dichroism plot. The ellipticities of GILGFVFTL–C*08:01 (solid line) and GILGFVFTL–A*02:01 (dashed line) at 218 nm are plotted as a function of temperature. (Inset) The far-UV CD spectrum of HLA-C*08:01 expressed as a function of wavelength (200 to 260 nm) was determined, and 218 nm was chosen for monitoring of the secondary-structure change during thermal unfolding.
DISCUSSION
The M158–66 epitope is highly conserved among seasonal and pandemic influenza A virus strains, with 93% conservation reported for 69 strains tested (29). The immunogenicity of this peptide in the context of HLA-A*02-restricted CTL responses has been well characterized. Although T cell-mediated immunity against influenza A virus infection is broad, with multiple specificities, studies have shown that GILGFVFTL is clearly the immunodominant epitope among HLA-A*02 individuals; the magnitude of memory responses has been shown to be one of the highest among the known conserved HLA-A*02-restricted IAV epitopes (9, 28, 29, 32). The allele frequency of HLA-C*08:01 is high among Asian populations and very low in other parts of the world. Among Singapore Chinese and Han Chinese populations, the allele frequencies are 8.6% and 13.3%, respectively (33, 34), as opposed to 0.09% in Caucasians (35). In contrast, the prevalence of HLA-A*02:01 is relatively high globally; it is found in 9% to 19% of Chinese individuals and 27% to 29% of Caucasians. Micropolymorphisms of HLA-A2, such as HLA-A*02:03, -A*02:06, -A*02:07, and -A*02:11, are more commonplace among Asians (33–35). In this study, we observed that GILGFVFTL-specific responses were present in all the HLA-A*02-positive individuals recruited and that as many as 93% and 88% of the GILGFVFTL–HLA-A*02 tetramer-positive CTLs could express CD107a and IFN-γ, respectively. CTL responses measured by CD107a and IFN-γ expression were observed to be slightly higher with HLA-A*02 restriction than with HLA-C*08 restriction, although this may not necessarily be a property of the CTLs themselves, since influences such as the density of surface HLA molecule expression and the nature of peptide binding can shape the overall CTL response. Degenerate binding of this epitope, described previously, was known to occur only within HLA-A2 alleles (14, 28–30), including HLA-A*02:01, -A*02:02, -A*02:03, -A*02:06, -A*68:02, and -A*69:01. In this study, we report the unexpected finding of novel restrictions of the T cell epitope to three HLA-C8 alleles: HLA-C*08:01, -C*08:03, and -C*08:06. This finding, for the first time in the context of IAV infection, demonstrates degenerate peptide binding and cellular immune responses comprising two different HLA products: HLA-A and HLA-C molecules. In addition, we also detected CTL responses in all HLA-C*08-positive individuals, suggesting that the immunoprevalence of HLA-C*08-restricted GILGFVFTL responses in the human population may be comparable to that of HLA-A*02-restricted responses. Interestingly, among HLA-C molecules, HLA-C*08:01 appears to have broad peptide binding capacity, as shown in reports of binding to hepatitis B virus (HBV) envelope residues 171 to 180 (36, 37) and CMV pp65 residues 198 to 206 (12).
Whether the CTLs recognizing an identical epitope presented by unrelated HLA allelic variants were derived from the same CTL clones, resulting in T cell cross-reactivity, remains to be elucidated. By using GILGFVFTL–HLA-A*02:01, -A*02:06, or -C*08:01 tetramers, it was demonstrated that GILGFVFTL-specific CTLs stained positive with both the HLA-A*02 and -C*08 tetramers yet formed two mutually exclusive populations in all four A*02+ C*08+ individuals. Our data therefore demonstrate that CTLs recognizing HLA-A*02- or -C*08-restricted GILGFVFTL are clonally distinct and that there is no cross-reactivity in antigen recognition. This suggests that epitope-based IAV vaccine strategies involving GILGFVFTL may be more effective than we would have expected, since the newly identified novel nondegenerate HLA-C*08-restricted CTL populations could potentially confer wider protection on HLA-A*02-negative individuals expressing HLA-C*08-based HLAs.
Since T cell recognition of GILGFVFTL presented by HLA-A2 is distinct from that of GILGFVFTL presented by HLA-C8, it should be noted that these HLA-A2- and -C8-restricted CTLs could possibly recognize different epitopes nested within the same peptide. One example of this phenomenon is the degenerate peptide binding of an HLA-A*02-restricted HBV core antigen, HBcAg18–27, to several alleles from the HLA-A2 and HLA-B7 supertypes (38, 39). Peptide-binding motifs for both the A2 and B7 supertypes were found to nest within the same peptide. The minimum epitope for A2-supertype molecules is HBcAg18–27, whereas the minimum epitope for B7-supertype alleles, such as HLA-B35 and -B51, is HBcAg19–27.
Another interesting observation is that for the allelic variants explored in this study, HLA-A2-restricted but not HLA-C8-restricted GILGFVFTL-specific CTLs are sensitive to micropolymorphisms among the various HLA subtypes. The GILGFVFTL-specific CTLs from HLA-A*02:06- and/or -A*02:07-positive individuals (donors 37 and 67) did not recognize or bind to GILGFVFTL–HLA-A*02:01 tetramers, whereas detection of these CTLs using GILGFVFTL–HLA-A*02:06 tetramers yielded 8.89% to 26.4% antigen-specific CTLs (Fig. 2D). We have shown recently that HLA-A*02:01-restricted GILGFVFTL T cell clones could bind only to HLA-A*02:01, -A*02:71, and -A*02:77 tetramers, whereas other HLA-A2 subtype tetramers, including HLA-A*02:06 tetramers, could not be recognized by the T cells (31). Therefore, it can be concluded that small differences among HLA-A*02 products can lead to functionally different antigen presentations, highlighting the fine specificity of these CTLs. In individuals carrying both HLA-A*02 and HLA-C*08 alleles, the CTL response with HLA-A*02 restriction appears to be higher than that with HLA-C*08 restriction. This raises the possibility that there is competition for the same peptide between the two HLA molecules within the endoplasmic reticulum.
In conclusion, our present study characterized novel HLA-C*08-restricted GILGFVFTL-specific CTL populations, and using MHC tetramers, we demonstrated that degenerate CTL recognition of the epitope occurs within the HLA-C*08 family but not when the epitope is presented by various HLA-A*02 subtypes. More importantly, our findings provided evidence for degenerate antigenicity between HLA products derived from separate loci in the context of influenza A virus. This information will help address important knowledge gaps in the understanding of HLA-C-mediated immune responses against influenza A virus and will provide new insights for the future development of polyvalent IAV vaccines.
ACKNOWLEDGMENTS
J.A.L.C. performed the cellular and tetramer studies. J.L. collected the X-ray data and solved the structure. J.L. and X.T. generated the protein crystals. G.M.G. and E.C.R. designed the study. J.A.L.C., G.M.G., and E.C.R. wrote the manuscript.
This project was supported by the Singapore Immunology Network (A*STAR) and by a Singapore National Research Foundation (NRF) Research Fellowship (NRF2007NRF-RF001-226). J.A.L.C. is a National University of Singapore Yong Loo Lin School of Medicine Graduate Scholarship recipient.
We declare no conflict of interest.
Footnotes
Published ahead of print 2 July 2014
REFERENCES
- 1.Webster RG, Wright SM, Castrucci MR, Bean WJ, Kawaoka Y. 1993. Influenza—a model of an emerging virus disease. Intervirology 35:16–25 [DOI] [PubMed] [Google Scholar]
- 2.Yap KL, Ada GL, McKenzie IF. 1978. Transfer of specific cytotoxic T lymphocytes protects mice inoculated with influenza virus. Nature 273:238–239. 10.1038/273238a0 [DOI] [PubMed] [Google Scholar]
- 3.McMichael AJ, Gotch FM, Noble GR, Beare PA. 1983. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 309:13–17. 10.1056/NEJM198307073090103 [DOI] [PubMed] [Google Scholar]
- 4.Guo H, Santiago F, Lambert K, Takimoto T, Topham DJ. 2011. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. J. Virol. 85:448–455. 10.1128/JVI.01812-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, Bean T, Barclay W, Deeks JJ, Lalvani A. 2013. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 19:1305–1312. 10.1038/nm.3350 [DOI] [PubMed] [Google Scholar]
- 6.Lee LY-H, Ha DLA, Simmons C, de Jong MD, Chau NVV, Schumacher R, Peng Y-C, McMichael AJ, Farrar JJ, Smith GL, Townsend ARM, Askonas BA, Rowland-Jones S, Dong T. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118:3478–3490. 10.1172/JCI32460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gras S, Kedzierski L, Valkenburg SA, Laurie K, Liu YC, Denholm JT, Richards MJ, Rimmelzwaan GF, Kelso A, Doherty PC, Turner SJ, Rossjohn J, Kedzierska K. 2010. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1–2009 and H1N1–1918 influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 107:12599–12604. 10.1073/pnas.1007270107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurie KL, Carolan LA, Middleton D, Lowther S, Kelso A, Barr IG. 2010. Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J. Infect. Dis. 202:1011–1020. 10.1086/656188 [DOI] [PubMed] [Google Scholar]
- 9.Quiñones-Parra S, Grant E, Loh L, Nguyen THO, Campbell K-A, Tong SYC, Miller A, Doherty PC, Vijaykrishna D, Rossjohn J, Gras S, Kedzierska K. 2014. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc. Natl. Acad. Sci. U. S. A. 111:1049–1054. 10.1073/pnas.1322229111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doolan DL, Hoffman SL, Southwood S, Wentworth PA, Sidney J, Chesnut RW, Keogh E, Appella E, Nutman TB, Lal AA, Gordon DM, Oloo A, Sette A. 1997. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity 7:97–112. 10.1016/S1074-7613(00)80513-0 [DOI] [PubMed] [Google Scholar]
- 11.Threlkeld SC, Wentworth PA, Kalams SA, Wilkes BM, Ruhl DJ, Keogh E, Sidney J, Southwood S, Walker BD, Sette A. 1997. Degenerate and promiscuous recognition by CTL of peptides presented by the MHC class I A3-like superfamily: implications for vaccine development. J. Immunol. 159:1648–1657 [PubMed] [Google Scholar]
- 12.Kondo E, Akatsuka Y, Kuzushima K, Tsujimura K, Asakura S, Tajima K, Kagami Y, Kodera Y, Tanimoto M, Morishima Y, Takahashi T. 2004. Identification of novel CTL epitopes of CMV-pp65 presented by a variety of HLA alleles. Blood 103:630–638. 10.1182/blood-2003-03-0824 [DOI] [PubMed] [Google Scholar]
- 13.Burrows SR, Elkington RA, Miles JJ, Green KJ, Walker S, Haryana SM, Moss DJ, Dunckley H, Burrows JM, Khanna R. 2003. Promiscuous CTL recognition of viral epitopes on multiple human leukocyte antigens: biological validation of the proposed HLA A24 supertype. J. Immunol. 171:1407–1412. 10.4049/jimmunol.171.3.1407 [DOI] [PubMed] [Google Scholar]
- 14.Zhang H-G, Pang X-W, Shang X-Y, Xing Q, Chen W-F. 2003. Functional supertype of HLA-A2 in the presentation of Flu matrix p58–66 to induce CD8+ T-cell response in a Northern Chinese population. Tissue Antigens 62:285–295. 10.1034/j.1399-0039.2003.00102.x [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Zhang S, Tan S, Yi Y, Wu B, Cao B, Zhu F, Wang C, Wang H, Qi J, Gao GF. 2012. Cross-allele cytotoxic T lymphocyte responses against 2009 pandemic H1N1 influenza A virus among HLA-A24 and HLA-A3 supertype-positive individuals. J. Virol. 86:13281–13294. 10.1128/JVI.01841-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Currier JR, deSouza M, Chanbancherd P, Bernstein W, Birx DL, Cox JH. 2002. Comprehensive screening for human immunodeficiency virus type 1 subtype-specific CD8 cytotoxic T lymphocytes and definition of degenerate epitopes restricted by HLA-A0207 and -C(W)0304 alleles. J. Virol. 76:4971–4986. 10.1128/JVI.76.10.4971-4986.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabbaj S, Bansal A, Ritter GD, Perkins C, Edwards BH, Gough E, Tang J, Szinger JJ, Korber B, Wilson CM, Kaslow RA, Mulligan MJ, Goepfert PA. 2003. Cross-reactive CD8+ T cell epitopes identified in US adolescent minorities. J. Acquir. Immune Defic. Syndr. 33:426–438. 10.1097/00126334-200308010-00003 [DOI] [PubMed] [Google Scholar]
- 18.Masemola AM, Mashishi TN, Khoury G, Bredell H, Paximadis M, Mathebula T, Barkhan D, Puren A, Vardas E, Colvin M, Zijenah L, Katzenstein D, Musonda R, Allen S, Kumwenda N, Taha T, Gray G, McIntyre J, Karim SA, Sheppard HW, Gray CM, HIVNET 028 Study Team 2004. Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from southern Africa. J. Immunol. 173:4607–4617. 10.4049/jimmunol.173.7.4607 [DOI] [PubMed] [Google Scholar]
- 19.Frahm N, Yusim K, Suscovich TJ, Adams S, Sidney J, Hraber P, Hewitt HS, Linde CH, Kavanagh DG, Woodberry T, Henry LM, Faircloth K, Listgarten J, Kadie C, Jojic N, Sango K, Brown NV, Pae E, Zaman MT, Bihl F, Khatri A, John M, Mallal S, Marincola FM, Walker BD, Sette A, Heckerman D, Korber BT, Brander C. 2007. Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur. J. Immunol. 37:2419–2433. 10.1002/eji.200737365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotch F, Rothbard J, Howland K, Townsend A, McMichael A. 1987. Cytotoxic T lymphocytes recognize a fragment of influenza virus matrix protein in association with HLA-A2. Nature 326:881–882. 10.1038/326881a0 [DOI] [PubMed] [Google Scholar]
- 21.Morrison J, Elvin J, Latron F, Gotch F, Moots R, Strominger JL, McMichael A. 1992. Identification of the nonamer peptide from influenza A matrix protein and the role of pockets of HLA-A2 in its recognition by cytotoxic T lymphocytes. Eur. J. Immunol. 22:903–907. 10.1002/eji.1830220404 [DOI] [PubMed] [Google Scholar]
- 22.Garboczi DN, Hung DT, Wiley DC. 1992. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. U. S. A. 89:3429–3433. 10.1073/pnas.89.8.3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stura EA, Wilson IA. 1991. Applications of the streak seeding technique in protein crystallization. J. Crystal Growth 110:270–282. 10.1016/0022-0248(91)90896-D [DOI] [Google Scholar]
- 24.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laskowski RA, Moss DS, Thornton JM. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049–1067. 10.1006/jmbi.1993.1351 [DOI] [PubMed] [Google Scholar]
- 26.Delano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 27.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 28.Gianfrani C, Oseroff C, Sidney J, Chesnut RW, Sette A. 2000. Human memory CTL response specific for influenza A virus is broad and multispecific. Hum. Immunol. 61:438–452. 10.1016/S0198-8859(00)00105-1 [DOI] [PubMed] [Google Scholar]
- 29.Alexander J, Bilsel P, del Guercio M-F, Marinkovic-Petrovic A, Southwood S, Stewart S, Ishioka G, Kotturi MF, Botten J, Sidney J, Newman M, Sette A. 2010. Identification of broad binding class I HLA supertype epitopes to provide universal coverage of influenza A virus. Hum. Immunol. 71:468–474. 10.1016/j.humimm.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barouch D, Friede T, Stevanović S, Tussey L, Smith K, Rowland-Jones S, Braud V, McMichael A, Rammensee HG. 1995. HLA-A2 subtypes are functionally distinct in peptide binding and presentation. J. Exp. Med. 182:1847–1856. 10.1084/jem.182.6.1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Buuren MM, Dijkgraaf FE, Linnemann C, Toebes M, Chang CXL, Mok JY, Nguyen M, van Esch WJE, Kvistborg P, Grotenbreg GM, Schumacher TNM. 2014. HLA micropolymorphisms strongly affect peptide-MHC multimer-based monitoring of antigen-specific CD8+ T cell responses. J. Immunol. 192:641–648. 10.4049/jimmunol.1301770 [DOI] [PubMed] [Google Scholar]
- 32.Boon ACM, de Mutsert G, Graus YMF, Fouchier RAM, Sintnicolaas K, Osterhaus ADME, Rimmelzwaan GF. 2002. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J. Virol. 76:582–590. 10.1128/JVI.76.2.582-590.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam TH, Shen M, Chia J-M, Chan SH, Ren EC. 2013. Population-specific recombination sites within the human MHC region. Heredity (Edinb.) 111:131–138. 10.1038/hdy.2013.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trachtenberg E, Vinson M, Hayes E, Hsu Y-M, Houtchens K, Erlich H, Klitz W, Hsia Y, Hollenbach J. 2007. HLA class I (A, B, C) and class II (DRB1, DQA1, DQB1, DPB1) alleles and haplotypes in the Han from southern China. Tissue Antigens 70:455–463. 10.1111/j.1399-0039.2007.00932.x [DOI] [PubMed] [Google Scholar]
- 35.Wang SS, Abdou AM, Morton LM, Thomas R, Cerhan JR, Gao X, Cozen W, Rothman N, Davis S, Severson RK, Bernstein L, Hartge P, Carrington M. 2010. Human leukocyte antigen class I and II alleles in non-Hodgkin lymphoma etiology. Blood 115:4820–4823. 10.1182/blood-2010-01-266775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CXL, Tan AT, Or MY, Toh KY, Lim PY, Chia ASE, Froesig TM, Nadua KD, Oh H-LJ, Leong HN, Hadrup SR, Gehring AJ, Tan Y-J, Bertoletti A, Grotenbreg GM. 2013. Conditional ligands for Asian HLA variants facilitate the definition of CD8+ T-cell responses in acute and chronic viral diseases. Eur. J. Immunol. 43:1109–1120. 10.1002/eji.201243088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan AT, Sodsai P, Chia A, Moreau E, Chng MHY, Tham CYL, Ho ZZ, Banu N, Hirankarn N, Bertoletti A. 2014. Immunoprevalence and immunodominance of HLA-Cw*0801-restricted T cell response targeting the hepatitis B virus envelope transmembrane region. J. Virol. 88:1332–1341. 10.1128/JVI.02600-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertoni R, Sidney J, Fowler P, Chesnut RW, Chisari FV, Sette A. 1997. Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J. Clin. Invest. 100:503–513. 10.1172/JCI119559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thimme R, Chang KM, Pemberton J, Sette A, Chisari FV. 2001. Degenerate immunogenicity of an HLA-A2-restricted hepatitis B virus nucleocapsid cytotoxic T-lymphocyte epitope that is also presented by HLA-B51. J. Virol. 75:3984–3987. 10.1128/JVI.75.8.3984-3987.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]