ABSTRACT
Dengue virus (DENV), composed of four distinct serotypes, is the most important and rapidly emerging arthropod-borne pathogen and imposes substantial economic and public health burdens. We constructed candidate vaccines containing the DNA of five of the genotypes of dengue virus serotype 2 (DENV-2) and evaluated the immunogenicity, the neutralizing (Nt) activity of the elicited antibodies, and the protective efficacy elicited in mice immunized with the vaccine candidates. We observed a significant correlation between the level of in vitro virus-like particle secretion, the elicited antibody response, and the protective efficacy of the vaccines containing the DNA of the different DENV genotypes in immunized mice. However, higher total IgG antibody levels did not always translate into higher Nt antibodies against homologous and heterologous viruses. We also found that, in contrast to previous reports, more than 50% of total IgG targeted ectodomain III (EDIII) of the E protein, and a substantial fraction of this population was interdomain highly neutralizing flavivirus subgroup-cross-reactive antibodies, such as monoclonal antibody 1B7-5. In addition, the lack of a critical epitope(s) in the Sylvatic genotype virus recognized by interdomain antibodies could be the major cause of the poor protection of mice vaccinated with the Asian 1 genotype vaccine (pVD2-Asian 1) from lethal challenge with virus of the Sylvatic genotype. In conclusion, although the pVD2-Asian 1 vaccine was immunogenic, elicited sufficient titers of Nt antibodies against all DENV-2 genotypes, and provided 100% protection against challenge with virus of the homologous Asian 1 genotype and virus of the heterologous Cosmopolitan genotype, it is critical to monitor the potential emergence of Sylvatic genotype viruses, since vaccine candidates under development may not protect vaccinated humans from these viruses.
IMPORTANCE Five genotype-specific dengue virus serotype 2 (DENV-2) DNA vaccine candidates were evaluated for their immunogenicity, homologous and heterologous neutralizing (Nt) antibody titers, and cross-genotype protection in a murine model. The immunity elicited by our prototype vaccine candidate (Asian 1 genotype strain 16681) in mice was protective against viruses of other genotypes but not against virus of the Sylvatic genotype, whose emergence and potential risk after introduction into the human population have previously been demonstrated. The underlying mechanism of a lack of protection elicited by the prototype vaccine may at least be contributed by the absence of a flavivirus subgroup-cross-reactive, highly neutralizing monoclonal antibody 1B7-5-like epitope in DENV-2 of the Sylvatic genotype. The DENV DNA vaccine directs the synthesis and assembly of virus-like particles (VLPs) and induces immune responses similar to those elicited by live-attenuated vaccines, and its flexibility permits the fast deployment of vaccine to combat emerging viruses, such as Sylvatic genotype viruses. The enhanced VLP secretion obtained by replacement of ectodomain I-II (EDI-II) of the Cosmopolitan genotype vaccine construct (VD2-Cosmopolitan) with the Asian 1 EDI-II elicited significantly higher total IgG and Nt antibody titers and suggests a novel approach to enhance the immunogenicity of the DNA vaccine. A DENV vaccine capable of eliciting protective immunity against viruses of existing and emerging genotypes should be the focus of future DENV vaccine development.
INTRODUCTION
Dengue virus (DENV) is the most important and rapidly emerging arthropod-borne pathogen and imposes substantial economic and public health burdens, especially in tropical and subtropical countries (1, 2). It is transmitted to humans through the bite of Aedes mosquitoes. A recent study estimated that 390 million DENV infections occur annually worldwide, with 500,000 of these cases being severe and with 25,000 cases resulting in death, mostly among children (3). Despite the impact of this disease, neither a licensed vaccine nor a specific antiviral drug is available; vector control is the only control measure available (4, 5). There are four antigenically distinct serotypes of the virus (DENV serotype 1 [DENV-1] to DENV-4), and each can cause a wide spectrum of clinical manifestations, including asymptomatic infection, self-limited flu-like dengue fever (DF), and the severe life-threatening dengue hemorrhagic fever and dengue shock syndrome (DHF/DSS) (2, 6).
Dengue virus belongs to the genus Flavivirus of the family Flaviviridae. It is an enveloped, single-stranded, positive-sense RNA virus, and the genome, which is approximately 11 kb long, encodes a polyprotein precursor of about 3,400 amino acid residues (7). Co- and posttranslational processing by cellular and viral proteases generates three structural proteins (the capsid [C], premembrane [prM], and envelope [E] proteins) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (7, 8). The E protein, which is approximately 500 amino acids in length, is folded into three structurally distinct ectodomains (EDs), termed EDI, EDII, and EDIII (9). The E glycoprotein mediates two crucial functions in flavivirus replication: it is responsible for virus attachment and fusion to susceptible cells (10). It also possesses important flavivirus epitopes that elicit protective neutralizing (Nt) antibody responses (11, 12). Antibodies against prM and the tip of EDII, which contains a highly conserved sequence (i.e., the fusion peptide loop), are immunodominant and believed to constitute a major portion of the total IgG population in the sera of humans naturally infected by dengue viruses (13–15).
The most advanced dengue vaccine in development is the chimeric yellow fever 17D-dengue (CYD) vaccine, and the CYD tetravalent dengue vaccine has progressed to late-phase human clinical trials (16). However, recent clinical trial data showed that this vaccine failed to protect against infection by DENV-2, probably due to viral replication and immune interference in human hosts who received this vaccine (17). Use of the DNA vaccine platform is a unique approach which elicits immune responses after immunization that closely resemble those seen in natural infections with intracellular pathogens (18). An important feature of DNA vaccines is that antigen processing occurs both intracellularly, allowing the processed antigen to enter the major histocompatibility complex (MHC) class I pathway, and extracellularly, allowing the secreted antigen to be processed through the MHC class II pathway (18, 19). This feature of cross presentation of endogenous and exogenous antigens can also induce higher antigen-specific T- and B-cell immune responses, similar to live-attenuated vaccines (19). A phase I clinical trial of a monovalent DENV-1 DNA vaccine candidate, the design of which was different from that used in this study, has been completed (20). The results demonstrated that the vaccine has favorable reactogenicity and safety but is poorly immunogenic. Despite this shortcoming, the tetravalent DENV DNA vaccine study was still initiated in 2011 (21).
Each DENV serotype is further grouped into genotypes on the basis of the genetic diversity of the E-protein gene sequences (22). DENV-2 has been classified into six genotypes, including Asian 1, Asian 2, American, American/Asian, Cosmopolitan, and Sylvatic (23). The most advanced chimeric yellow fever 17D–DENV-2 vaccine candidate is developed from an Asian 1 genotype strain (strain PUO-218), and experimental results suggested that the vaccine can neutralize a broad spectrum of DENV strains of the same serotype, including representative strains from each of the six DENV-2 genotypes (24). However, the immunogenicity and the cross-neutralization potency of a dengue vaccine prepared with other strains from different genotypes have not been extensively directly compared. In the present study, we sought to evaluate the immunogenicity of DENV-2 recombinant plasmid DNA by selecting one representative strain from each genotype and constructing a panel of plasmid constructs containing genotypically unique prM and E genes. The focus of our current study is (i) to determine the immunogenicity of vaccines containing the DNA of the different DENV genotypes, (ii) to determine the cross-neutralizing activities of antibodies elicited by vaccines containing the DNA of the different DENV genotypes against strains of different genotypes, and (iii) to investigate the protection against challenge with heterologous viruses of different genotypes afforded by the current DNA vaccine.
MATERIALS AND METHODS
Amino acid sequence analyses.
Seventy-six unique full-length E-protein sequences of representative strains of the different DENV-2 genotypes were retrieved from the NCBI dengue virus database and aligned using the BioEdit software package suite (version 7.2.0). Variable amino acid positions were identified to be informative sites when substitutions occurred in at least three independent sequences of strains of each of the recognized DENV-2 genotypes (23). Localization and surface exposure of the variable amino acid positions were analyzed by use of PyMOL molecular graphics system software (version 1.6) on the basis of the published crystal structures of mature and immature dengue virus serotype 2 virions (PDB accession numbers 3J27 and 3C6D) (25, 26). Calculations of the stable free energy (ΔΔG) of the variable amino acid substitutions were performed using the PoPMuSiC (version 2.1) server (http://babylone.ulb.ac.be/popmusic/) on the basis of the structural coordinates of the E protein of the mature dengue virus serotype 2 particle (25) and E soluble protein (27).
Cells and viruses.
COS-1, Vero, and C6/36 cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone, Logan, UT), 2 mM l-glutamine, 110 mg/liter sodium pyruvate, 0.1 mM nonessential amino acids, 20 ml/liter 7.5% NaHCO3, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were maintained at 37°C with 5% CO2, except for C6/36 cells, which were maintained at 28°C without CO2.
The DENV-2 strains used in this study (Table 1) included viruses belonging to the following genotypes: Asian 1 (strain 16681), Asian 2 (strains PL046 and BC27/96), American (strains S14635 and 1340), American/Asian (strains BC141/96 and BC100/98), Cosmopolitan (strains BC145/97 and 1222), and Sylvatic (strains DakHD 10674, DakAr A1247, and DakAr A2022 from Africa and strain P8-1407 from Asia). All viruses were provided by one of us (G.-J.J.C.), except for strains 16681 and 1222 (DENV-2), which were obtained from H. C. Wu (Academia Sinica, Taiwan) and C. C. King (National Taiwan University), respectively. Virus stocks were propagated by infecting C6/36 cells in DMEM with 2% FBS for 7 days. Culture supernatants were harvested, clarified by centrifugation, and stored in aliquots at −80°C.
TABLE 1.
Characteristics of representative DENV-2 strains used in this study
| Straina | Genotype | Yr isolated | Host | Country of origin |
|---|---|---|---|---|
| 16681 | Asian 1 | 1964 | Human | Thailand |
| BC27/96 | Asian 2 | 1995 | Human | Vietnam |
| PL046 | Asian 2 | ? | Human | Taiwan |
| S14635 | American | 1974 | Human | Tonga |
| 1340 | American | 1977 | Human | Puerto Rico |
| BC141/96 | American/Asian | 1944 | Human | Puerto Rico |
| BC100/98 | American/Asian | 1998 | Human | Bolivia |
| BC145/97 | Cosmopolitan | 1997 | Human | Malaysia |
| 1222 | Cosmopolitan | 2002 | Human | Taiwan |
| DakHD 10674 | Sylvatic | 1970 | Human | Senegal |
| DakAr A1247 | Sylvatic | 1980 | Mosquito | Ivory Coast |
| DakAr A2022 | Sylvatic | 1980 | Mosquito | Burkina Faso |
| P8-1407 | Sylvatic | 1970 | Monkey | Malaysia |
Strains in boldface were used to construct plasmids carrying the DNA of the different genotypes.
Antibodies.
A panel of murine monoclonal antibodies (MAbs) with distinct flavivirus E-protein reactivity was provided by one of us (G.-J.J.C.). These included group-cross-reactive antibody (6B6C-1) recognizing viruses of the four major pathogenic flavivirus serocomplexes, subgroup-cross-reactive antibodies (1B7-5 and T5-1) recognizing all or some members of two or more different flavivirus serocomplexes, subcomplex-cross-reactive antibodies (1A1D-2 and 9D12-16) recognizing a subset of the four DENV complex viruses, and a type-specific antibody (3H5-1) recognizing DENV-2 only. MAb 155-49 recognizing DENV-2 prM was obtained from H. Y. Lei (National Cheng Kung University, Taiwan). Anti-DENV-1 and -2 rabbit sera and mouse hyperimmune ascitic fluid (MHIAF) were also provided by one of us (G.-J.J.C.). MAbs 155-49, 3H5-1, 1B7-5, and 6B6C-1 were also labeled with horseradish peroxidase (HRP) using an EZ-Link Plus activated peroxidase kit (Thermo Fisher Scientific Inc., Rockford, IL) according to the manufacturer's instruction.
Construction of plasmids.
The previously constructed and characterized recombinant plasmid pVD2i (14, 28, 29) expressing the prM and E proteins of DENV-2 Asian 1 genotype strain 16681 was used in this study and herein is termed pVD2-Asian 1.
To construct plasmids expressing proteins of the other DENV-2 genotypes, genomic RNA was extracted from 150 μl of C6/36 cell culture medium infected with a strain of each DENV-2 genotype using a QIAmp viral RNA kit (Qiagen, Santa Clarita, CA) and was used as the template in reverse transcription-PCR for the amplification of the prM gene and 80% of envelope gene (amino acids [aa] 1 to 396). The primer sequences are listed in Table 2. AfeI and StuI restriction enzyme sites were incorporated at the 5′ and 3′ termini of the cDNA amplicons, respectively. cDNA amplicons were digested with the AfeI and StuI enzymes and inserted into the AfeI- and StuI-cutting sites of the pVD1i expression vector plasmid (30) to obtain plasmids pVD2-American, pVD2-American/Asian, pVD2-Cosmopolitan, and pVD2-Sylvatic using DENV-2 strain S14635, BC100/98, BC145/97, and DakHD 10674 as the templates, respectively. A plasmid containing Asian 2 genotype DNA was not constructed because of the sequence similarity between the Asian 1 and Asian 2 genotypes, which have only three commonly shared nonconservative amino acid substitutions when the E-protein sequences are compared. The furin cleavage site of prM of the pVD2-Asian 1 plasmid was further mutated from amino acid residues 87-RREKR-91 to 87-RREST-91 to produce an immature form of virus-like particles (VLPs) using a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA).
TABLE 2.
Oligonucleotides used in plasmid constructions
| Primer | Primer sequence (5′–3′)a | Amplified region |
|---|---|---|
| 102 forward | AATTATAGCGCTTTTCATCTGACCACACGCAAC | S14635 prM/E (aa 1–396) |
| 102 reverse | AATTATAGGCCTTGCCCAGCGTGCTTCCTTTCTTGAACCAGTCCAG | |
| 70 forward | AATTATAGCGCTTTCCATTTAACCACACGTAAT | BC100/98 prM/E (aa 1–396) |
| 70 reverse | AATTATAGGCCTTGCCCAGCGTACTTCCCTTCTTAAACCAATTGAG | |
| 85 forward | AATTATAGCGCTTTTCATTTGACCACACGCAAC | BC145/97 prM/E (aa 1–396) |
| 85 reverse | AATTATAGGCCTTGCCCAGCGTGCTTCCTTTTTTGAACCAGTTTAA | |
| 68 forward | AATTATAGCGCTTTTCATTTGACCACACGCAAC | DakHD 10674 prM/E (aa 1–396) |
| 68 reverse | AATTATAGGCCTTGCCCAGCGTGCTTCCTTTTTTGAACCAGTTTAA | |
| 16681 EDI-II forward | AATTAACGTCTCACAATGCGTTGCATAGGAATG | 16681 EDI-II (aa 1–294) |
| 16681 EDI-II reverse | AATTAACGTCTCAGCTGTAGCTTGTCCATTCTC | |
| 85 vector forward | AATTAACGTCTCGCAGCTCAAAGGAATGTCATAC | pVD2- Cosmopolitan excluding EDI-II |
| 85 vector reverse | AATTAACGTCTCCATTGTCATTGAAGGAGCGAC | |
| 68 EDI-II forward-a | AATTAACGTCTCACAATGCGATGTATAGGCATG | DakHD 10674 EDI-II (aa 1–294) |
| 68 EDI-II reverse-a | AATTAACGTCTCTTGAGTTGCAGCTTGTCCATC | |
| 16681 vector forward-a | AATTAACGTCTCGCTCAAAGGAATGTCATACTC | 16681/pVD2 excluding EDI-II |
| 16681 vector reverse-a | AATTAACGTCTCCATTGTCATTGAAGGAGTGAC | |
| 68 EDIII forward-b | AATTAACGTCTCAAAGGGATGTCGTACTCCATG | DakHD 10674 EDIII (aa 294–396) |
| 68 EDIII reverse-b | AATTAACGTCTCAGCGTGCTTCCTTTTTTGAAC | |
| 16681 vector forward-b | AATTAACGTCTCCACGCTGGGCAAGGCCTTTTC | 16681/pVD2 excluding EDIII |
| 16681 vector reverse-b | AATTAACGTCTCTCCTTTGAGCTGTAGCTTGTC | |
| pVD1i EDIII forward | AATCGTCTCAAAGGGATGTCATATGTGATGTGC | pVD1i EDIII (aa 294–396) |
| pVD1i EDIII reverse | AATTAACGTCTCAGCGTGCTTCCTTTCTTGAAC | |
| 16681 EDIII forward | AATCGTCTCAAAGGAATGTCATACTCTATGTGCAC | pVD2-Asian 1 EDIII (aa 294–396) |
| 16681 EDIII reverse | AATCGTCTCAGCGTGCTTCCTTTCTTAAACCAG | |
| pVD1i vector forward | AATTAACGTCTCCACGCTGGGCAAGGCCTTTTC | pVD1i excluding EDIII |
| pVD1i vector reverse | AATGAACGTCTCCCCTTTTAGAGTCAGTTTGTC |
Nucleotides in boldface are AfeI (AGCGCT), StuI (AGGCCT), and BsmBI (CGTCTC) enzyme restriction sites.
To create chimeric plasmids, replacement of the E-protein EDI-II of pVD2-Asian 1 with that of pVD2-Sylvatic or EDI-II of pVD2-Cosmopolitan with that of pVD2-Asian 1, replacement of EDIII of pVD2-Asian 1 with that of pVD2-Sylvatic or pVD1i, or replacement of EDIII of pVD1i with that of pVD2-Asian 1 was carried out using the Golden Gate assembly method (31). The ectodomains of interest and the expression vector backbones were PCR amplified from wild-type (WT) plasmids as the templates. Primer sets (Table 2) were designed to introduce BsmBI restriction enzyme sites flanking the 5′ and 3′ termini of the resulting PCR products. The DNA fragments were then digested with BsmBI and ligated with T4 DNA ligase to obtain new plasmids pVD2-Asian 1/Sylvatic EDI-II, pVD2-Cosmopolitan/Asian 1 EDI-II, pVD2-Asian 1/Sylvatic EDIII, pVD2-Asian 1/D1 EDIII, and pVD1/D2-Asian 1 EDIII.
Automated DNA sequencing was performed on an ABI Prism 3730 sequencer (Applied Biosystems, Foster City, CA), and recombinant plasmids with correct prM and E sequences were identified using Lasergene software (DNAStar, Madison, WI).
Plasmids for in vitro transfection and mouse immunizations were purified from Escherichia coli TOP10 cells using a PureLink HiPure plasmid midiprep kit (Invitrogen, Life Technologies Corp., Carlsbad, CA) and reconstituted in diethyl pyrocarbonate-treated water.
Immunofluorescence assay (IFA).
COS-1 cells were transfected with plasmids containing the prM and E genes of strains representing each of the DENV-2 genotypes to evaluate intracellular protein expression. Briefly, 8 × 104 cells per well were seeded into 24-well Costar cell culture plates (Corning Inc., Corning, NY) containing coverslips and transfected with 1.2 μg of DNA and 3 μl of Lipofectamine 2000 (Invitrogen, Life Technologies Corp., Carlsbad, CA) following the manufacturer's recommendations. Cells were then incubated at 37°C with 5% CO2. After 48 h, the cells were washed twice with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS at room temperature for 20 min. Cells were then blocked with 3% bovine serum albumin (BSA) in PBS for 1 h at 37°C and incubated with anti-DENV-2 MHIAF at 1:200 for 1 h at 37°C. The cells were washed three times and incubated with a mixture of fluorescein isothiocyanate-conjugated goat anti-mouse IgG (1:200; Jackson ImmunoResearch, West Grove, PA) and DAPI (4′,6-diamidino-2-phenylindole dihydrochloride; 1:500; Invitrogen, Molecular Probes, Inc., Eugene, OR) for 1 h at 37°C. After washing 5 times, coverslips were mounted on a glass slide and cells were visualized under a fluorescence microscope (CKX41; Olympus).
Ag production and secretion level characterization.
To produce VLP antigens (Ags), COS-1 cells at a density of 1.5 × 107 cells/ml were electroporated with 30 μg of a plasmid containing the genes of each DENV-2 genotype following the previously described protocol (32). After electroporation, cells were seeded into a 75-cm2 culture flasks containing 15 ml growth medium and allowed to recover overnight at 37°C. The growth medium was replaced on the next day with a maintenance medium containing 2% FBS, and cells were continuously incubated at 28°C with 5% CO2 for VLP secretion. Tissue culture media were harvested 5 days after transfection and clarified by centrifugation at 8,000 × g for 30 min at 4°C.
An antigen-capture enzyme-linked immunosorbent assay (ELISA) was performed as previously described (14) to detect and quantify the level of secretion of VLP antigens harvested from COS-1 cells transfected with plasmids containing the genes of each DENV-2 genotype. Briefly, flat-bottom 96-well MaxiSorp Nunc-Immuno plates (Nunc, Roskilde, Denmark) were coated with 50 μl of rabbit anti-DENV-2 VLP serum at 1:500 in carbonate buffer (0.015 M Na2CO3, 0.035 M NaHCO3, pH 9.6) and incubated overnight at 4°C, and the wells were blocked with 200 μl of 1% BSA in PBS for 1 h at 37°C. Harvested culture medium and normal COS-1 cell culture fluid were titrated 2-fold in PBS, 50 μl was added to wells in duplicate, and the plates were incubated for 2 h at 37°C and washed 5 times with 200 μl of PBS with 0.1% Tween 20 (0.1% PBST). Captured antigens were detected by adding 50 μl of anti-DENV-2 MHIAF at 1:2,000 in blocking buffer, and the plates were incubated for 1 h at 37°C and washed 5 times. Fifty microliters of HRP-conjugated goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) at 1:6,000 in blocking buffer was added to the wells, and the plates were incubated for 1 h at 37°C to detect MHIAF. Subsequently, the plates were washed 10 times. Bound conjugate was detected with 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Enhanced K-Blue TMB; Neogen Corp., Lexington, KY), the plates were incubated at room temperature for 10 min, and the reaction was stopped with 2 N H2SO4. The A450s of the reactions were measured using a Tecan Sunrise microplate reader (Tecan, Grödig, Austria). Endpoint antigen secretion titers from two independent experiments were determined from twice the average optical density (OD) of the negative-control antigen by using a sigmoidal dose-response equation in GraphPad Prism (version 6.0) software (GraphPad Software, Inc., La Jolla, CA).
Mouse experiments.
This study was carried out in compliance with the guidelines for the care and use of laboratory animals of the National Laboratory Animal Center, Taiwan. The animal use protocol has been reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of National Chung Hsing University (approval number 97-69). All efforts were made to minimize the suffering of the mice. At weeks 0, 4, and 8, groups of four 4-week-old female BALB/c mice were injected intramuscularly with plasmids containing the DNA of each DENV-2 genotype at a dose of 100 μg/100 μl in PBS, and administration of the dose was divided between the right and left quadriceps muscles. The mice were bled from the retro-orbital sinus before immunization and 4 weeks following the third injection. Due to the similarity of the genetic backgrounds of the inbred BALB/c mice used in this study and to increase the volume of sera, pooled sera from each group of mice were evaluated for DENV-2-specific antibodies by ELISA and a focus-reduction microneutralization test (FRμNT), as described below.
For the evaluation of passive protection by maternal antibody, ICR pups from the mating of nonimmunized males with immunized females 9 weeks following an initial vaccination at 4 weeks of age were obtained. Immunized females were also boosted twice at 4-week intervals after priming. Immune sera were collected from immunized females 1 week prior to mating to confirm the presence of the total IgG titer and the virus neutralization response. Pups from unvaccinated females were used as the challenge control. Groups of pups were challenged intracranially at 2 days after birth with 2,000, 1,000, and 300 50% lethal doses (LD50s) of DENV-2 strains 16681, DakArHD 10674, and 1222, respectively. The percent survival of the mice was evaluated daily for up to 21 days.
ELISAs.
Postvaccination mouse sera were assayed for the presence of DENV-2-specific total IgG with the same Ag-capture ELISA protocol described above with minor modifications. The concentrations of the VLP antigens of the different genotypes were standardized at a single concentration producing an OD of 1.4 to determine the total IgG titer after appropriate dilutions. Pooled sera from mice with the same immunization schedule were initially diluted 1:1,000, titrated 2-fold, added to wells in duplicate, and incubated for 1 h at 37°C. Prevaccination mouse sera were included as negative controls. Incubations with conjugate and substrate were carried out according to the standard Ag-capture ELISA, as described above. Values of the OD at 450 nm were modeled as nonlinear functions of the log10 serum dilutions using a sigmoidal dose-response (variable-slope) equation, and the endpoint antibody titers from two independent experiments were determined to be the dilutions where the OD value was twice the average OD for the negative control. Domain-swapped VLPs (pVD2-Asian 1/D1 EDIII and pVD1/D2-Asian 1 EDIII) were also used in parallel with wild-type VLP (pVD2-Asian 1) to determine the domain-specific antibody response from sera from mice vaccinated with DNA of the different genotypes. The proportion of non-EDIII (EDI-II- and prM-specific) IgG from vaccinated mouse serum was calculated by dividing the endpoint titer obtained from the specific knockout antigen (pVD2-Asian 1/D1 EDIII) by that obtained from WT antigen with the same serum sample. EDIII-specific IgG proportions were calculated from the equation 1 minus the proportion of non-EDIII-specific IgG, as previously described (14, 30).
Binding ELISA was done by use of a setup similar to that described above to determine the activity of binding to wild-type or mutant VLPs, except that serial 2-fold dilutions of anti-DENV-2 MHIAF were replaced by the MAbs indicated below or pVD2-Asian 1 postvaccination sera to determine the binding reactivity to DENV-2 wild-type VLPs (pVD2-Asian 1), mutant VLPs (pVD2-Asian 1/D1 EDIII, and pVD1/D2-Asian 1 EDIII, pVD2-Asian 1/Sylvatic EDIII, pVD2-Asian 1/Sylvatic EDI-II), and VLPs of the other genotypes.
An epitope-blocking ELISA was performed to determine the genotype differences of the vaccinated mouse antibody responses to prM protein and cross-reactive or serotype-specific epitopes by the well-characterized prM MAb 155-49, the EDII-cross-reactive MAbs 6B6C-1 and 1B7-5, and DENV-2 EDIII type-specific MAb 3H5-1, respectively. The setup was similar to that of the total IgG ELISA, wherein plates were coated with rabbit anti-DENV-2 VLP serum and blocked with 1% BSA in PBS and DENV-2 VLP antigen was captured. After washing, pooled sera were diluted 1:40 in blocking buffer and incubated for 1 h at 37°C. After incubation and washing of the sera, a 1:2,000 dilution of HRP-conjugated MAbs was added to each well and the plates were incubated for 1 h at 37°C so that the MAbs could compete with the already bound antibody to the DENV-2 VLP antigen from the immune mouse sera. Bound conjugate was detected with TMB substrate, the plates were incubated for 10 min, and the reaction was stopped with 2 N H2SO4. The A450 values of the reactions were measured. Percent blocking was determined by comparing the results for replicate wells with HRP-conjugated MAb competing against preadsorbed naive mouse serum using the following formula: {100 – [(OD of immune serum on DENV-2 VLPs – OD of immune serum on normal Ag)/(OD of naive serum on DENV-2 VLPs – OD of naive serum on normal Ag)]} × 100.
To determine the prM/E ratio of the VLPs of the different DENV-2 genotypes, the same Ag-capture ELISA described above was performed, except that antigens were detected with anti-prM or anti-E MAbs (155-49 or 3H5-1) following a previously described protocol (13). prM/E ratios were calculated from the ratio of the OD of prM/OD of E and expressed as a percentage relative to the ratio of the OD of prM/OD of E of pVD2-Asian 1 immature VLPs.
Virus neutralization.
The ability of the immune mouse sera to neutralize a number of representative strains of the different DENV-2 genotypes was measured by FRμNT, as previously described (14). Briefly, 2.475 × 104 Vero cells per well were seeded into flat-bottom 96-well Costar cell culture plates (Corning Inc., Corning, NY) and incubated for 16 h overnight at 37°C with 5% CO2. Pooled sera were initially diluted 1:10, heat inactivated for 30 min at 56°C, and titrated 2-fold to a 40-μl volume, and 320 PFU/40 μl of DENV-2 was added to each dilution. The mixtures were then incubated for 1 h at 37°C. After incubation, 25 μl of the immune complexes was inoculated in duplicate into plates containing Vero cell monolayers. The plates were incubated for 1 h at 37°C with 5% CO2 and rocked every 10 min to allow infection. Overlay medium containing 1% methylcellulose (Sigma-Aldrich Inc., St. Louis, MO) in DMEM with 2% FBS was added, and the plates were incubated at 37°C with 5% CO2. Forty-eight hours later, the plates were washed, fixed with 75% acetone in PBS, and air dried. Immunostaining was performed by adding anti-DENV-2 MHIAF at 1:600 in 5% milk in 0.1% PBST. The plates were incubated for 60 min at 37°C and washed, goat anti-mouse IgG–HRP at 1:100 in 5% milk in 0.1% PBST was added, and the plates were incubated for 45 min at 37°C. Infection foci were visualized using a peroxidase substrate kit (Vector VIP SK-4600; Vector Laboratories, Inc., Burlingame, CA) following the manufacturer's instructions. FRμNT titers were calculated for each virus relative to that for a back-titration with a virus-only control. The titers for the exact 50% reduction of infectious foci (Nt50 titers) were modeled using a sigmoidal dose-response (variable slope) formula. All values were taken from the averages of two independent experiments. The cross-neutralization of the different DENV-2 strains by antisera from the vaccines containing the DNA of the different genotypes is presented by use of a heatmap generated using the web tool of the HIV sequence database (http://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html).
Statistical analyses.
All data are represented as means ± standard errors and were analyzed using GraphPad Prism (version 6.0) software. One-way analysis of variance (ANOVA), followed by Holm-Sidak's posttest, was used to analyze the ELISA and neutralization titers across multiple groups. An unpaired t test was used to analyze sets of data between two groups. Kaplan-Meier survival curves were analyzed by the log-rank test. P values of <0.05 were considered significant.
RESULTS
Variable epitopes in E proteins of different DENV-2 genotypes.
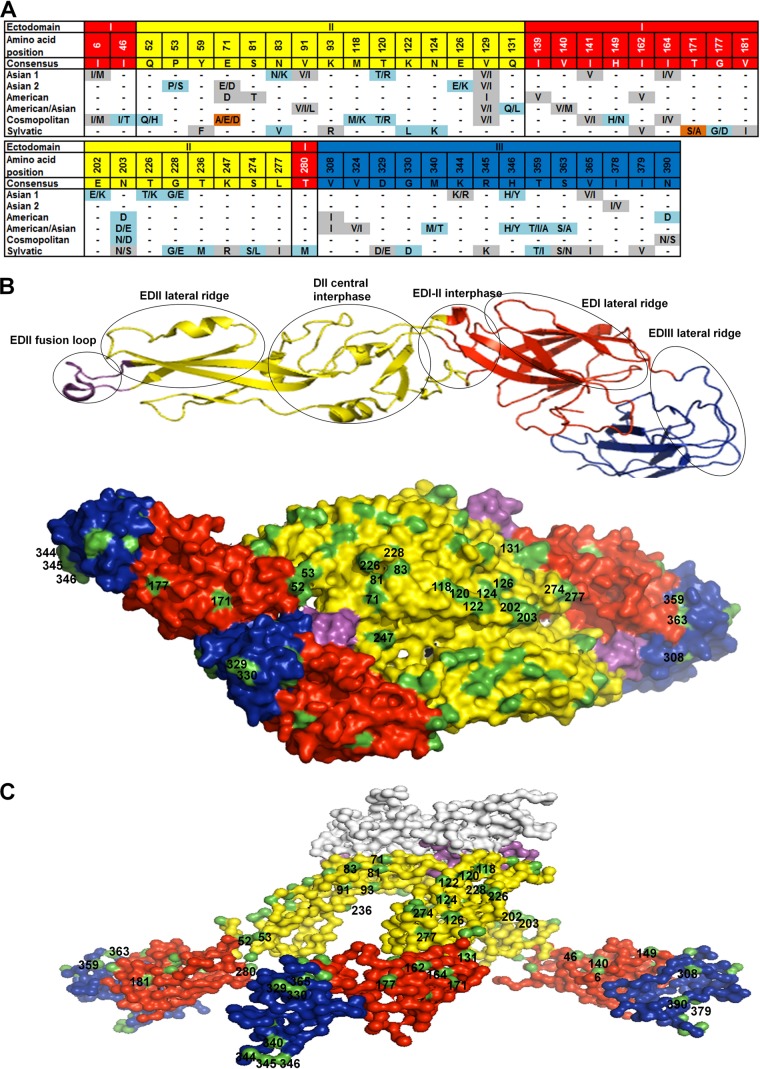
Full-length ectodomain E-protein sequences from 76 representative strains of known DENV-2 genotype (23) were downloaded from the NCBI dengue virus database and aligned to examine the genetic variability among different genotypes (complete alignments are available upon request). Variable amino acid positions occurring in at least 3 independent sequences of strains of a particular genotype were defined to be informative sites. On the basis of the number of amino acid differences compared to the consensus sequence (Fig. 1A), the Sylvatic genotype showed the greatest sequence diversity, followed by the Asian 1, Cosmopolitan, American/Asian, American, and Asian 2 genotypes, with the six genotypes showing 23, 13, 12, 11, 8, and 5 variable residues compared to the consensus sequence, respectively. Among the six genotypes, Asian 1 and Asian 2 shared three nonconservative amino acid changes at E-protein amino acid 53 (E53), E126, and E346. The other genotypes had at least 8 amino acid differences compared to the sequence of Asian 1. Further analysis revealed a total of 49 variable amino acids in the E protein that were scattered along EDI, EDII, and EDIII. Structural analysis using the coordinates of a mature dengue virus serotype 2 strain (25) showed that among these variable residues, 25 are surface exposed (Fig. 1B), whereas on the basis of the resolved crystal structures of an immature dengue virus serotype 2 strain (26), 42 variable residues are surfaced exposed (Fig. 1C). The amino acids at positions 344, 345, and 346 are buried in the mature virus but appear to be exposed during the viral thermodynamic transitional states (25). These variable residues are located within or adjacent to the known major E-protein antigenic sites (Fig. 1B) and include 1 conservative substitution and 10 nonconservative substitutions at either critical antibody recognition sites or neutralization epitopes (33–36). These important substitutions are located in the EDI lateral ridge (G177D), the EDII lateral ridge (E7D/A, N83K/V), the EDII central dimer interphase (K122L and N124K), the EDI-II interphase (S274L), and the EDIII lateral ridge (D329E, G330D, M340T, H346Y, and T359I).
FIG 1.
Informative sites in the envelope protein of DENV-2 genotypes. (A) Alignment showing the variable amino acid positions and consensus sequence in the E-protein EDs. Gray shading, conservative amino acid substitutions; light blue shading, nonconservative substitutions; brown shading, nonconservative substitutions unique to certain genotypes. (B) Structural locations of surface-exposed informative sites. (Top) Ribbon diagram of the monomeric E protein with several of the known antigenic sites marked in circles; (bottom) top view of the dimeric E-protein crystal model of the mature DENV-2 particle (PDB accession number 3J27) depicted as a surface representation. (C) Side view surface representation of the model of the trimeric E-protein crystal structure of the immature DENV-2 particle (PDB accession number 3C6D). In panels A to C, EDI, EDII, EDIII, and the fusion loop are colored red, yellow, blue, and violet, respectively. Surface-exposed residues (B and C) are highlighted in green. The prM protein (C) is white.
DNA vaccine construction.
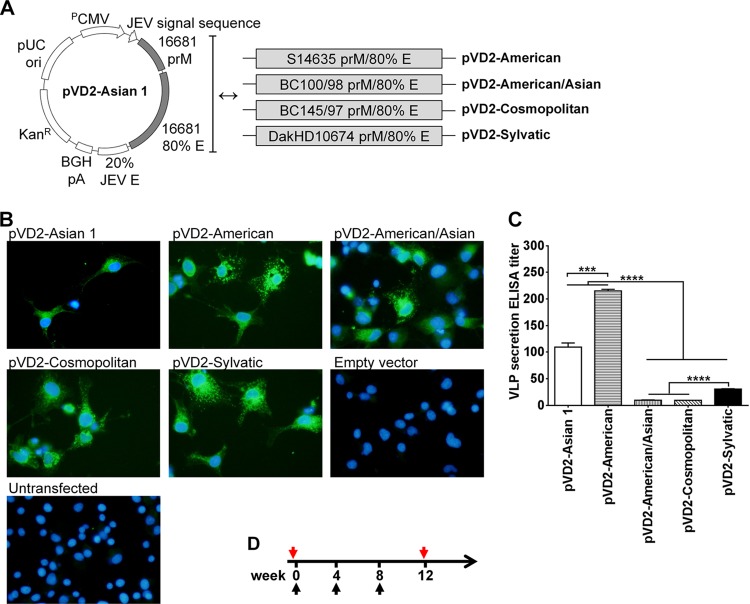
To gain better biological insights into how E-protein sequence diversity could possibly affect the immunogenicity of a dengue virus DNA vaccine, we constructed plasmids containing the prM and E genes from strains representing different DENV-2 genotypes for use in our subsequent immunization studies. In addition to our previously developed pVD2-Asian 1 genotype vaccine (28), DENV-2 strains representing the American, American/Asian, Cosmopolitan, and Sylvatic genotypes (Table 1) were chosen to create a genetically diverse set of DENV-2 DNA vaccines. The Asian 2 genotype was not included in the construction since the amino acid sequences from virus strains of this genotype contained substitutions already represented by the Asian 1 strain used in this study. The design of the recombinant plasmid constructs (Fig. 2A) includes the human cytomegalovirus early gene promoter, the Japanese encephalitis virus (JEV) signal sequence, the prM gene of each DENV-2 genotype, 80% of the envelope (E) gene region (amino acids 1 to 396) of each DENV-2 genotype, 20% of the E gene (amino acids 397 to 495) of the homologous region of JEV, and a bovine growth hormone poly(A) signal. This replacement of the C-terminal 20% of the DENV-2 E gene, which was found to contain a membrane retention sequence, with the homologous region of the JEV E gene enhances the biosynthesis and extracellular secretion of the assembled VLPs without altering the native DENV-2 E-glycoprotein conformation.
FIG 2.
Vaccine design, analysis of recombinant antigen expression and secretion, and immunization schedule. (A) Schematic representation of plasmid vectors containing DNA of the different DENV-2 genotypes: pVD2-Asian 1, pVD2-American, pVD2-American/Asian, pVD2-Cosmopolitan, and pVD2-Sylvatic. These plasmid constructs include the human cytomegalovirus early gene promoter (PCMV), the JEV signal sequence, prM, 80% of the E-gene region (amino acids 1 to 396) of each DENV-2 genotype, 20% of the homologous E-gene region of JEV (amino acids 397 to 495), and a bovine growth hormone poly(A) signal (BGH pA). (B) Immunofluorescence analysis of recombinant protein expression in COS-1 cells transfected with plasmids containing DNA of the different DENV-2 genotypes. After fixation and permeabilization, recombinant proteins were detected with anti-DENV-2 MHIAF, followed by incubation with goat anti-mouse IgG-fluorescein isothiocyanate and DAPI to counterstain the nucleus. The characteristic green fluorescence in the cells indicates positivity for the intracellular expression of recombinant proteins. (C) Detection and quantification by an Ag-capture ELISA of secreted VLPs in culture supernatants harvested from transiently transformed COS-1 cells. Data are presented as means ± SEMs of two independent repeat transfection experiments. Means were analyzed by one-way ANOVA (P < 0.05), and Holm-Sidak's posttest significance is indicated with asterisks (***, P = 0.0002; ****, P < 0.0001). (D) Schedule of immunization with different plasmid vectors containing DNA of the different DENV-2 genotypes. Groups of 4-week-old BALB/c mice were immunized with three doses of 100 μg DNA intramuscularly. At 4 weeks following the last immunization, blood samples from each group of mice were taken and pooled for further serum antibody analyses. Black arrows, immunization times; red arrows, blood sampling times.
Distinct levels of antigen secretion by different genotype DNA vaccines.
To ascertain that all constructs can drive recombinant protein expression in vitro, an immunofluorescence assay was conducted in COS-1 cells transiently transformed with plasmids carrying the genes of the different DENV-2 genotypes. A positive reaction to anti-DENV-2 MHIAF, which recognizes multiple epitopes of DENV-2, was observed (Fig. 2B) in all plasmid-transfected cells, indicating that our plasmid constructs can direct the proper intracellular expression of recombinant proteins of the different DENV-2 genotypes. As expected, no expression was observed in either empty vector-transfected cells or untransfected cells.
Extracellular secretion of the recombinant proteins in harvested culture supernatants was next assessed by an Ag-capture ELISA. VLPs of all genotypes were secreted, but the levels of secretion were statistically significantly different (P < 0.0001) (Fig. 2C). pVD2-American and pVD2-Cosmopolitan had the highest and lowest secretion levels, respectively. pVD2-American and pVD2-Asian 1 had higher secretion titers (1:215 and 1:109, respectively) than pVD2-Sylvatic, pVD2-American/Asian, and pVD2-Cosmopolitan (1:31, 1:10, and 1:9, respectively). Since DNA vaccines stimulate an immune response through endogenous and exogenous pathways, the various levels of extracellular VLP secretion hint that these plasmid constructs could also vary in their immunogenicity when inoculated in vivo as DNA vaccines.
Distinct antigenicity of vaccines containing the DNA of the different genotypes.
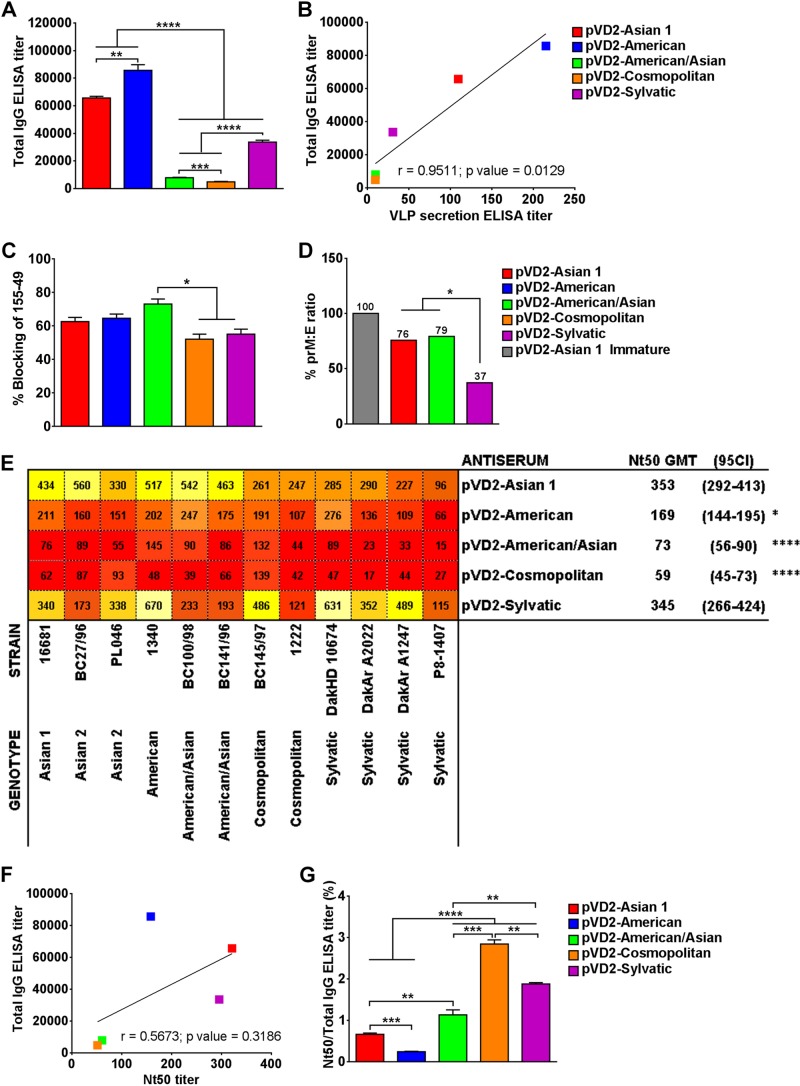
To define the influence of secretion level on the immunogenicity of our vaccines containing the DNA of the different DENV-2 genotypes, we immunized groups of 4-week-old BALB/c mice (see the immunization schedule in Fig. 2D) at weeks 0, 4, and 8 with plasmids carrying DNA of the different genotypes. At 4 weeks after the third immunization, blood samples were collected and pooled sera were analyzed for an antibody response by ELISA. All vaccinated mice produced anti-DENV-2-specific antibodies, with remarkable differences being seen among groups (Fig. 3A). The IgG titers in mice vaccinated with the pVD2-American (1:85,678), pVD2-Asian 1 (1:65,690), and pVD2-Sylvatic (1:33,652) vaccines were significantly greater than those in mice vaccinated with the pVD2-American/Asian (1:7,941) and pVD2-Cosmopolitan (1:4,892) vaccines (P < 0.0001). Additionally, the antibody response was highly correlated to the in vitro level of VLP secretion (P = 0.0129) (Fig. 3B). Our results suggest that the DNA of the different DENV-2 genotypes affected not only VLP secretion but also the antigenic properties presented by the amino acid variation of the VLPs of the different genotypes.
FIG 3.
Naturally occurring amino acid variation affects the immunogenicity of DENV-2 DNA vaccines. (A) Total IgG antibody responses in mice injected with different DENV-2 DNA vaccines were determined by a standard Ag-capture ELISA 4 weeks following the last immunization. **, P = 0.0057; ***, P = 0.0007, ****, P < 0.0001. (B) The total IgG ELISA titer is positively and significantly correlated with the VLP secretion titer, as determined by a regression correlation analysis (r = 0.9511; P = 0.0129). (C) Percent blocking of an HRP-labeled anti-prM MAb (155-49) by sera from mice vaccinated with different DENV-2 genotypes containing antibodies to VLPs of the different DENV-2 genotypes determined by an epitope-blocking ELISA. *, P < 0.05. (D) The ratio of uncleaved prM and E among the VLPs of the different DENV-2 genotypes was measured by ELISA and is expressed as a percentage relative to that for immature pVD2-Asian 1 VLPs. *, P = 0.0366. (E) Cross-neutralization activity of antisera elicited by vaccines containing the different DENV-2 genotypes against a set of viruses of different DENV-2 genotypes determined by FRμNT. Nt50 titers are presented as a heatmap generated using the web tool of the HIV sequence database. Lower Nt50 values are red, and the colors progress to orange and then yellow to bright yellow at higher values. GMT, geometric mean titer. *, P < 0.05; ****, P < 0.0001. (F) The total IgG ELISA titer is negatively correlated with the Nt50 titer upon regression correlation analysis (r = 0.5673; P = 0.3186). (G) Ratio of Nt50 titers to DENV-2-specific ELISA titers as a measure of the functional neutralizing activity of the elicited immune sera. ELISA data are means ± SEMs of two independent repeat experiments. **, P = 0.0038; ***, P = 0.003; ****, P < 0.0001. Nt50 titers are expressed as the mean ± 95% CI. Means were analyzed by one-way ANOVA (P < 0.05), and Holm-Sidak's posttest significance is indicated with asterisks.
Maturation status of VLPs of the different genotypes.
Previous studies suggested that sera from humans with natural dengue virus infection contain significant amounts of anti-prM antibodies (13) because of the partially mature state of dengue virion particles. However, whether a similar situation occurs among the different genotypes of DENV has not been explored before. Therefore, an epitope-blocking ELISA was performed to characterize the ability of the antibodies elicited by the vaccines containing the DNA of the different genotypes to block MAb 155-49 against the DENV-2 prM epitope. This was used as an indirect measure of the maturation status of the secreted VLP antigens. As indicated in Fig. 3C, the maturity of the VLPs expressed by the different DNA vaccines did not vary significantly, except for the VLPs expressed by the pVD2-American/Asian vaccine, which induced a significantly higher proportion of prM-targeting antibodies than the pVD2-Cosmopolitan and pVD2-Sylvatic vaccines (P < 0.05). The antibodies elicited by the different vaccines could block 50 to 70% of the ability of MAb 155-49 to bind to prM epitopes. Since the prM antibodies could also be induced when the injected plasmid DNA was processed intracellularly, we further measured the relative density of the uncleaved prM on selected VLPs of the different genotypes to demonstrate the maturation status of the VLPs. Consistent with the prM epitope blocking by immune sera produced from the different vaccines, the VLPs of the different genotypes exhibited differences in the densities of uncleaved prM, expressed as a percentage of the prM/E ratio (Fig. 3D). The higher the prM/E ratio is, the less efficient the prM cleavage is and the more immature VLPs are generated. Our data suggest that pVD2-Asian 1 and pVD2-American/Asian secreted VLPs that have significantly higher prM/E ratios than pVD2-Sylvatic VLPs (P = 0.0366). pVD2-Asian 1 and pVD2-American/Asian VLPs were about 80% immature, whereas pVD2-Sylvatic VLPs were only 40% immature. Together, these results suggest that the efficiency of prM cleavage varies among VLPs of the different DENV-2 genotypes, which were neither completely mature nor immature but were partially mature in form.
The immunogenicity of the DNA vaccine is genotype dependent.
To rigorously analyze the influence of the secretion level on the immunogenicity of the vaccines containing the DNA of the different genotypes, a FRμNT against a panel of diverse DENV-2 strains was conducted using pooled vaccinated mouse sera. The vaccines exhibited significant differences in immunogenicity in mice (P < 0.0001). Both the pVD2-Asian 1 and pVD2-Sylvatic vaccines neutralized representative DENV-2 strains tested, with the neutralization titers being higher than 1:80, whereas the pVD2-American, pVD2-American/Asian, and pVD2-Comopolitan vaccines had comparatively lower neutralization titers (Fig. 3E; see also Table S1 in the supplemental material). The immune sera elicited by pVD2-Asian 1 and pVD2-Sylvatic vaccines had, on average, a greater ability to neutralize (Nt50 titers, 1:353 and 1:345, respectively) homologous and heterologous viruses representing the six DENV-2 genotypes than the immune sera elicited by the pVD2-American (Nt50 titer, 1:169) (P < 0.05), pVD2-American/Asian (Nt50 titer, 1:73), and pVD2-Cosmopolitan (Nt50 titer, 1:59) (P < 0.0001) vaccines.
Although the anti-DENV-2 IgG antibody response was highly correlated to the level of VLP secretion, the neutralization activity of the vaccinated mouse sera was not consistently correlated with the IgG response (Fig. 3F). An obvious discrepancy between the sera from the groups of mice immunized with pVD2-American and pVD2-Sylvatic vaccines was observed (Fig. 3A and E). The low neutralization titer of the sera from pVD2-American-vaccinated mice was inversely proportional to the high total IgG titer of those sera, which is in contrast to the findings for the sera from the pVD2-Sylvatic-vaccinated mice. This discrepancy provided evidence that the different antibody compositions induced by the different DNA vaccines might affect their functional activities in virus neutralization, as suggested by previous investigations (37, 38). We therefore calculated the ratio of the Nt50 titer to the total IgG titer to measure the DENV-2-specific neutralizing activity of the elicited antibodies. Significant differences in the compositions of the functional activities of the neutralizing antibodies from each group of vaccinated mice were found (P < 0.0001) (Fig. 3G). Interestingly, the pVD2-Cosmopolitan, pVD2-Sylvatic, and pVD2-American/Asian vaccines induced a higher proportion of functional antibodies than the pVD2-Asian 1 and pVD2-American vaccines (P < 0.0001). Even though sera from pVD2-Cosmopolitan- and pVD2-American/Asian-vaccinated mice presented a higher proportion of functional antibodies, a low total IgG antibody titer might partially explain the overall low level of neutralization activity (Fig. 3A and G). Conversely, sera from pVD2-American- and pVD2-Asian 1-vaccinated mice had relatively larger total antibody populations but contained the smallest neutralizing antibody composition compared to the sera from pVD2-Cosmopolitan- and pVD2-American/Asian-vaccinated mice. Taken together, these results show that the amino acid variability in the E-protein ectodomain of the various genotypic DNA vaccines affects not only the magnitude of antibody production but also, ultimately, the proportion of neutralizing antibodies elicited.
Characterization of domain specificity and type-specific and cross-reactive responses of polyclonal sera.
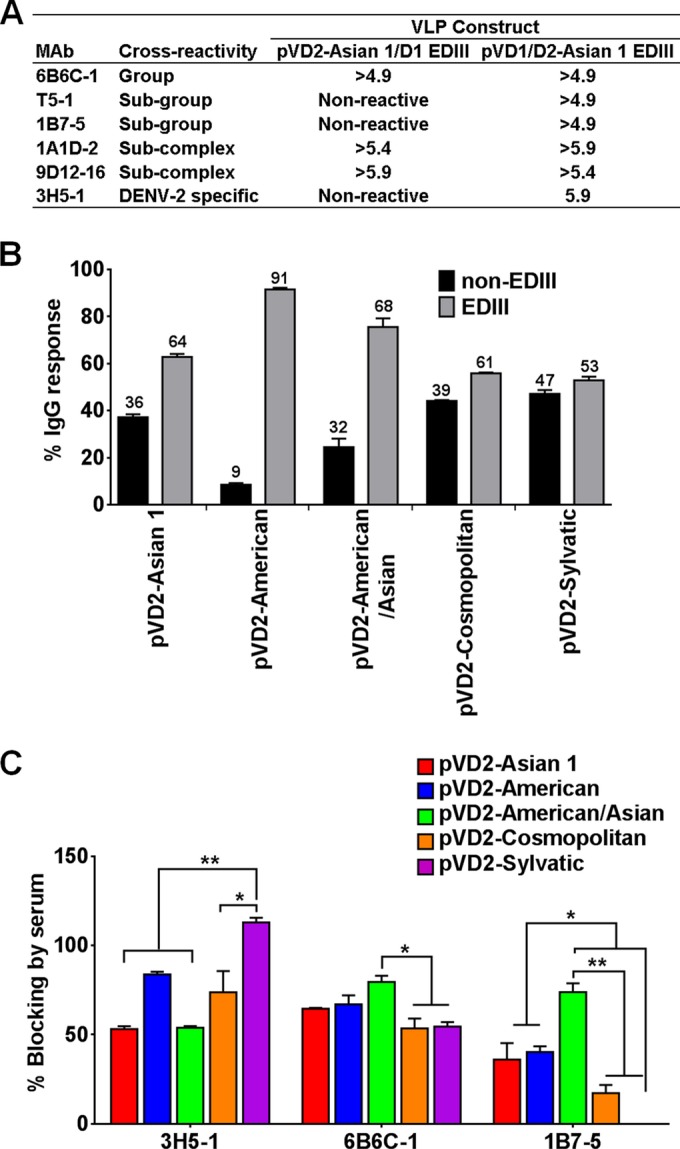
To examine the domain-specific proportions of the polyclonal E-protein-specific IgG response, mutant DENV-2 VLPs (pVD2-Asian 1/D1 EDIII or pVD1/D2-Asian 1 EDIII) produced from mutant pVD2i-Asian 1 plasmids, where EDIII or EDI-II was replaced with DENV-1 EDIII or EDI-II, respectively, were used. In order to confirm if the EDIII-specific antibodies failed to bind to the mutant VLP (pVD2-Asian 1/D1 EDIII), a monoclonal antibody binding ELISA was performed. Using mutant VLPs, our MAb mapping (Fig. 4A) not only showed that MAb 3H5-1 (EDIII serotype-specific antibody) failed to bind to the DENV-1 EDIII-containing VLPs but also showed the presence of interdomain antibodies, as evidenced by the loss of reactivity of MAbs 1B7-5 and T5-1. The subgroup-cross-reactive MAb 1B7-5 lost reactivity to pVD2-Asian 1/D1 EDIII VLPs but not to pVD1/D2-Asian 1 EDIII VLPs. This result provided more evidence to confirm previous observations that epitopes defined by 1B7-5 might be composed of discontinuous parts extending not only across EDI-II but also across EDIII (39, 40). Another subgroup-cross-reactive MAb, T5-1, whose epitope is not yet known, also displayed reactivity similar to that of 1B7-5.
FIG 4.
Characterization of type-specific and cross-reactive activity and domain-specific responses of antibodies from sera from mice vaccinated with the different genotypes. (A) ELISA reactivities of MAbs against E-protein EDIII mutant DENV-1 and DENV-2 VLPs; (B) domain-specific IgG responses to the EDIII and non-EDIII epitopes of sera from mice vaccinated with the different genotypes; (C) blocking of an HRP-labeled DENV-2-specific MAb (3H5-1) and cross-reactive MAbs (6B6C-1 and 1B7-5) by sera from mice vaccinated with the different DENV-2 genotypes containing antibodies to VLPs of the different DENV-2 genotypes determined by an epitope-blocking ELISA. ELISA data are the means ± SEMs of two independent repeat experiments. Means were analyzed by one-way ANOVA (P < 0.05), and Holm-Sidak's posttest significance is indicated with asterisks (*, P < 0.05; **, P < 0.01).
By comparing the endpoint titers between the mutant (pVD2-Asian 1/D1 EDIII) and wild-type (pVD2-Asian I) VLPs, our results demonstrate that a major proportion of the total IgG population of different polyclonal sera targets the EDIII region and a minor fraction is directed against non-EDIII regions (Fig. 4B). The EDIII antibodies elicited by the different vaccines constitute a substantial proportion of the total IgG population: 50% for pVD2-Sylvatic, 60% for pVD2-Asian 1 and pVD2-Cosmopolitan, and 70% for pVD2-American/Asian. pVD2-American produced an even larger amount of EDIII antibodies, which constituted 90% of the total IgG population.
Epitope-blocking ELISAs were performed to further characterize the proportion of type-specific and cross-reactive antibodies in sera from mice receiving the vaccines with DNA from the different genotypes. As shown in Fig. 4C, the IgGs in the polyclonal sera produced after vaccination with the different DNA vaccines had variable proportions of type-specific and cross-reactive activity. In terms of the type-specific response, sera from all mice vaccinated with the DNA vaccines of the various genotypes had more than 50% MAb 3H5-1-blocking activity; however, sera from pVD2-Sylvatic-vaccinated mice had significantly higher levels of activity than sera from pVD2-Cosmopolitan (P < 0.05)-, pVD2-Asian 1-, and pVD2-American/Asian-vaccinated mice (P < 0.01). There was no significant variation in the percent inhibition of group-cross-reactive MAb 6B6C-1 by antibodies elicited by the different vaccines except for pVD2-American/Asian, which induced a significantly larger amount of anti-fusion loop antibodies (blocking activity, about 80%) than the pVD2-Cosmopolitan and pVD2-Sylvatic vaccines (P < 0.01). The subgroup-cross-reactive antibodies in the different polyclonal sera varied in their ability to inhibit MAb 1B7-5 (P < 0.05). Sera from pVD2-Asian 1- and pVD2-American-vaccinated mice exhibited 40% blocking activity, while sera from pVD2-American/Asian-vaccinated mice had more than 70% blocking activity. Sera from pVD2-Cosmopolitan-vaccinated mice had only less than 20% blocking activity. Interestingly, antibodies elicited by the pVD2-Sylvatic vaccine had no blocking activity at all against MAb 1B7-5, which suggests that VLPs secreted by the pVD2-Sylvatic vaccine do not properly present the epitopes that are targeted by this particular MAb.
Naturally occurring mutations alter the binding potential of neutralizing antibodies.
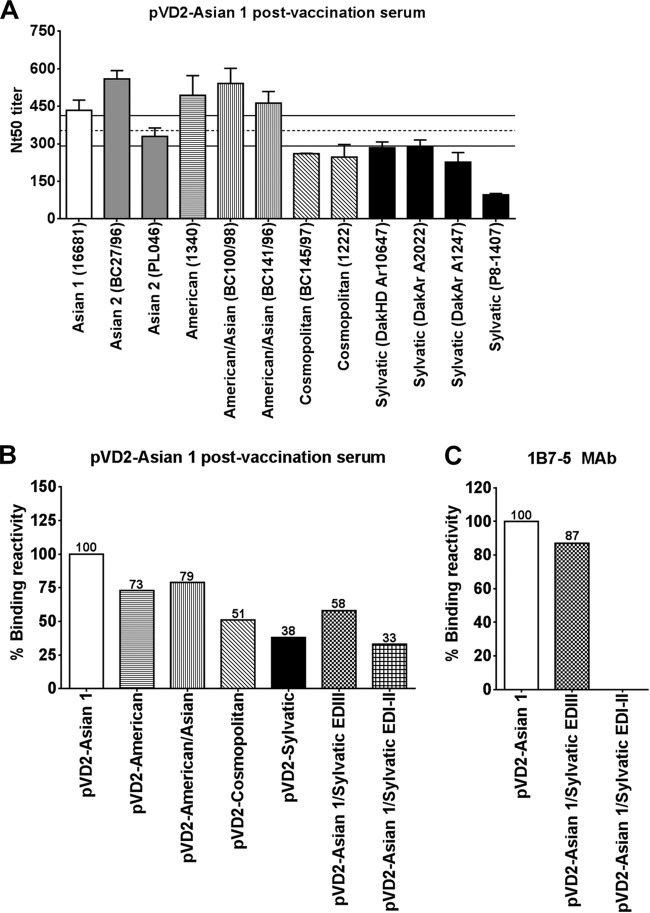
The results of an in-depth analysis of the virus neutralization activities of sera from pVD2-Asian 1-vaccinated mice are shown in Fig. 5A. Although sera from pVD2-Asian 1-vaccinated mice strongly neutralized multiple strains of the different DENV-2 genotypes, we found that the Nt50 titers for strains BC145/97 and 1222 (Cosmopolitan genotype) and strains DakHD 10674, DakAr A1247, DakAr A2022, and P8-1407 (Sylvatic genotype) were below the range of the reciprocal geometric mean Nt50 titer (95% confidence interval [CI], 292 to 413). We speculated that the amino acid variations in these viruses might affect the binding affinity of the elicited antibodies, which is constitutively correlated with the neutralization of virus infectivity (41). In order to evaluate the influence of the naturally occurring amino acid variations among strains of the different DENV-2 genotypes, we first performed a binding assay using antigens from VLPs of the different genotypes to determine if the observed genotype-specific neutralization of sera from pVD2-Asian 1-vaccinated mice was due to variations in the binding affinities of antibodies to the different genotypes. The percent binding reactivity of sera from pVD2-Asian 1-vaccinated mice to VLPs of the different genotypes relative to that to wild-type pVD2-Asian 1 VLPs was calculated. Indeed, the differences in the neutralization activity observed were consistent with the differences in binding reactivity (Fig. 5B). The strong binding reactivity to pVD2-American and pVD2-American/Asian VLPs of more than 70% translated to efficient neutralization of viruses of the American and American/Asian genotypes. Remarkably, the more than 40 and 60% reductions in reactivity to pVD2-Cosmopolitan and pVD2-Sylvatic VLPs, respectively, were consistent with the poor neutralization of viruses of these genotypes.
FIG 5.
Naturally occurring mutations affect the neutralizing ability and physical binding of antibodies. (A) Cross-FRμNT titers of pVD2-Asian 1 postvaccination serum against a set of strains of different DENV-2 genotypes. Dashed line, geometric mean Nt50 titer; solid lines, 95% confidence interval of the titer of the serum pool. Error bars represent SEMs. (B) Binding ELISA reactivity of pVD2-Asian 1 postvaccination serum to different VLP antigens. The percentage of the reactivity of sera with antibodies to heterologous and mutant VLP antigens remaining relative to that for the wild-type Asian 1 genotype VLPs was calculated. (C) The percentage of the reactivity of MAb 1B7-5 to the chimeric Asian 1 VLP antigens remaining relative to that for the wild-type Asian 1 genotype VLPs was calculated. All data represent the means ± SEMs of two independent experiments.
Second, to confirm the finding described above, we used two chimeric pVD2-Asian 1 VLPs where EDI-II and EDIII were replaced by pVD2-Sylvatic EDI-II and EDIII, respectively. The reason was that strains of the Sylvatic genotype showed the greatest sequence diversity among the six DENV-2 genotypes. It was therefore a logical choice in the construction of a chimeric plasmid for a loss-of-function analysis. We found that sera from pVD2-Asian 1-vaccinated mice still retained substantial reactivity (about 60%) to chimeric pVD2-Asian1/Sylvatic EDIII VLPs (Fig. 5B). However, replacement of EDI-II of wild-type pVD2-Asian 1 with EDI-II of pVD2-Sylvatic resulted in a dramatic reduction in reactivity of more than 60%, suggesting that the epitopes located in EDI-II are the determinants highly targeted by antibodies elicited by the pVD2-Asian 1 vaccine.
We also screened our VLPs of various genotypes for reactivity to a panel of well-characterized neutralizing murine MAbs (14, 30, 40) recognizing flavivirus group-, subgroup-, subcomplex-, and type-specific epitopes to investigate whether the VLPs expressed in vivo from DNA vaccines of the different DENV-2 genotypes contain B-cell epitopes that elicit antibodies with reactivities similar to the reactivities of these MAbs. As shown in Table 3, most of the MAbs were highly reactive to antigens on the VLPs of the different genotypes. However, it is interesting to note that only one specific MAb (1B7-5) demonstrated a striking loss of reactivity to pVD2-Sylvatic VLPs, indicating that a certain antigenic epitope(s) mainly targeted by 1B7-5-like antibodies is not represented in these expressed Sylvatic genotype VLPs. MAb 1B7-5 is flavivirus subgroup cross-reactive and a potently neutralizing MAb, and the exact docking epitope recognized by this antibody is currently unknown, although several previous studies have suggested that the location might be in the E-protein EDI-II (39, 40, 42). As expected, the binding of this MAb to the chimeric pVD2-Asian 1/Sylvatic EDI-II VLP was abrogated (Fig. 5C).
TABLE 3.
MAb reactivities for DENV-2 genotype VLPsa
| MAb | CRb | Virusc | MAb reactivityd for the following VLP construct: |
||||
|---|---|---|---|---|---|---|---|
| pVD2-Asian 1 | pVD2-American | pVD2-American/Asian | pVD2-Cosmopolitan | pVD2-Sylvatic | |||
| 6B6C-1 | Group | SLEV | >6.0 | >6.0 | >6.0 | >6.0 | >5.1 |
| 1B7-5 | Subgroup | DENV-2 | >5.1 | >5.4 | 6 | >6.0 | NRe |
| 1A1D-2 | Subcomplex | DENV-2 | >5.1 | >5.1 | >5.1 | >5.1 | >5.1 |
| 9D12-16 | Subcomplex | DENV-2 | >5.1 | >5.4 | >5.4 | >5.1 | >5.4 |
| 3H5-1 | Type specific | DENV-2 | >6.0 | >5.7 | >5.7 | >6.0 | >5.7 |
Reactivities of MAbs that had various cross-reactivities and that were selected from different flaviviruses. All MAbs had neutralizing activity (30, 40, 42).
CR, cross-reactivity. Group-cross-reactive MAbs recognize all viruses of the four major pathogenic flavivirus serocomplexes, subgroup-cross-reactive MAbs recognize all or some members of two or more different flavivirus serocomplexes, subcomplex-cross-reactive MAbs recognize a subset of the DENV complex, and type-specific MAbs recognize DENV-2 only.
Virus against which the MAb was raised. DENV-2, Dengue virus serotype 2; SLEV, St. Louis encephalitis virus.
MAb reactivities of VLPs of the different DENV-2 genotypes are presented as inverse log10 Ag-capture ELISA endpoint values.
NR, nonreactive.
Genotype-specific in vivo protective efficacy of the DNA vaccines.
Next, we assessed if the immunogenicities of the different DNA vaccines influenced by their genotypic characteristics could be translated into protection in vivo. We focused our attention on the protective efficacy of the pVD2-Asian-1 DNA vaccine. Initially, BALB/c mice with the same genetic background as the mice used to obtain the immunogenicity data were used to evaluate protection. Unfortunately, the number of pups produced was too few and variable for us to compare the results in a consistent manner. Therefore, 4-week-old female ICR mice were used instead. The mice were vaccinated with 100 μg/100 μl of plasmid DNA and boosted twice at 4-week intervals. For evaluation of passive protection by maternal antibody, pups were obtained after mating of the vaccinated females, which had an Nt50 titer of >1:200 at 9 weeks following the initial vaccination, with nonimmunized male mice. Pups were infected intracranially at 2 days after birth with 2,000 LD50s of virus strain 16681. Pups from unvaccinated females were used as unvaccinated controls. As expected, all pups from vaccinated mothers survived the homologous viral infection, whereas pups from unvaccinated mothers did not (P < 0.0001) (Fig. 6A). We further investigated if this protective efficacy could be sustained against strains of the other DENV-2 genotypes, particularly virus strains belonging to the Cosmopolitan and Sylvatic genotypes (strains 1222 and DakHD 10674, respectively) since sera from pVD2-Asian 1-vaccinated mice displayed lower neutralization activity against these two genotypes, as shown in Fig. 4A. Pups were infected intracranially 2 days after birth with 1,000 and 300 LD50s of virus strains DakHD 10674 and 1222, respectively. Similarly, pups from vaccinated mothers were completely protected from heterologous strain 1222 infection, whereas pups from unvaccinated mothers were not (P < 0.0001) (Fig. 6B). In contrast, all pups from both vaccinated and unvaccinated mothers succumbed to death following challenge with strain DakHD 10674 (Fig. 6C). Even though none of the pups survived the DakHD 10674 infection, the time to death was significantly prolonged in pups nursed by vaccinated mothers compared to that for pups from control mothers (P = 0.0232). We speculate that the absence of protection in pups from vaccinated mothers could be due to the absence of MAb 1B7-5-like binding against the virus strain of the Sylvatic genotype and that 1B7-5-like antibodies play a major role in this mouse model of passive challenge.
FIG 6.
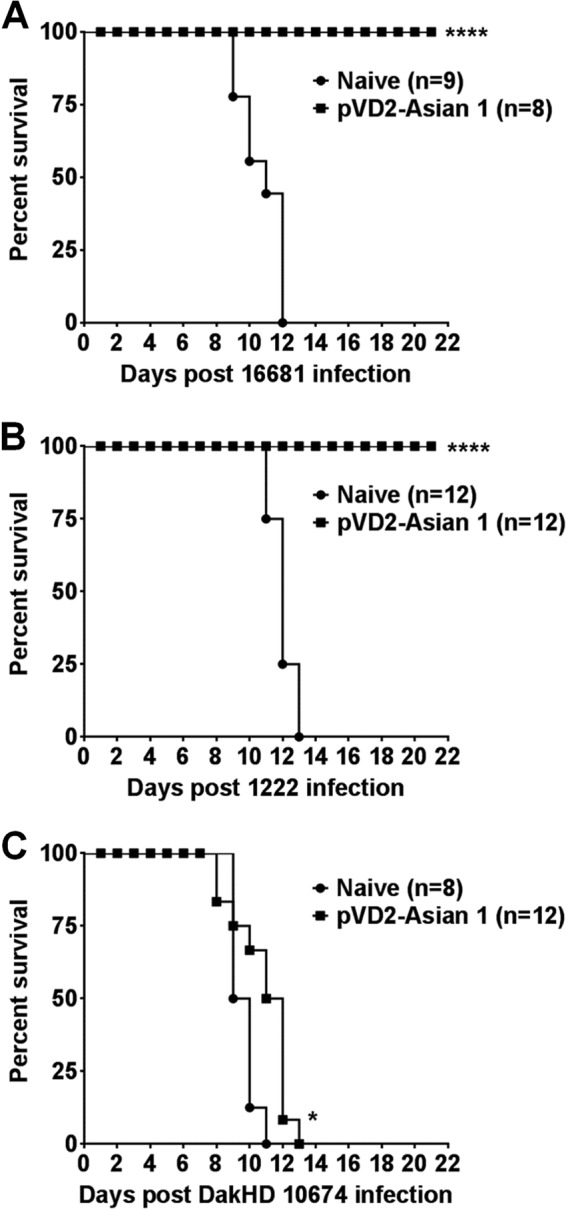
The in vivo protective efficacy of the DENV-2 DNA vaccine is genotype dependent. (A to C) Groups of 2-day-old suckling mice obtained from pVD2-Asian 1-vaccinated ICR female mice (Nt50 titer, >1:200) were infected intracranially with 2,000 LD50s of DENV-2 strain 16681 (A), 300 LD50s of DENV-2 strain 1222 (B), and 1,000 LD50s of DENV-2 strain DakHD 10674 (C). The survival of the mice was monitored for 21 days after infection. Kaplan-Meier survival curves were analyzed by the log-rank test. P values of <0.05 were considered significant. The statistical significance of the difference between mice from vaccinated mothers and mice from naive mothers is indicated with asterisks. *, P = 0.0232; ****, P < 0.0001.
Ectodomain manipulation improves the poorly immunogenic Cosmopolitan genotype DNA vaccine.
To prove that the immunogenicity of our DNA vaccine is dependent on the level of secretion of the expressed recombinant proteins, we manipulated the E-protein ectodomain of the poorly immunogenic Cosmopolitan genotype vaccine to enhance VLP secretion. The pVD2-Cosmopolitan vaccine has the same EDIII amino acid sequence as the highly immunogenic pVD2-Asian 1 vaccine but a different EDI-II sequence (see Fig. S1 in the supplemental material). Thus, we engineered a chimeric pVD2-Cosmopolitan vaccine wherein its EDI-II was replaced by the EDI-II of pVD2-Asian 1. After replacement of the EDI-II of pVD2-Cosmopolitan with that of pVD2-Asian 1, VLP secretion in transiently transformed COS-1 cells was dramatically augmented, showing a significant 15-fold increase (Fig. 7A) compared to that achieved with the wild-type pVD2-Cosmopolitan plasmid DNA vaccine (P < 0.0001). In parallel with this was a significant 4-fold increase in antibody production (Fig. 7B) in mice following immunization with the chimeric pVD2-Cosmopolitan/Asian 1 EDI-II vaccine compared to that following immunization with the wild-type pVD2-Cosmopolitan vaccine (P = 0.0017). FRμNT results further revealed an increase in the neutralizing activities of the immune sera from pVD2-Cosmopolitan/Asian 1 EDI-II-vaccinated mice concomitant with the increase in the IgG titer (Fig. 7C). Compared to the neutralization potency of sera from wild-type pVD2-Cosmopolitan-vaccinated mice, a significant gain in the neutralization potency of sera from pVD2-Cosmopolitan/Asian 1 EDI-II-vaccinated mice against virus strains 16681 (P = 0.0031), 1340 (P = 0.0195), and BC100/98 (P = 0.0004) was observed. These results demonstrate that replacement of the E-protein ectodomain of one genotype with that of another can be exploited as a strategy to improve the immunogenicity of a poorly immunogenic DNA vaccine.
FIG 7.
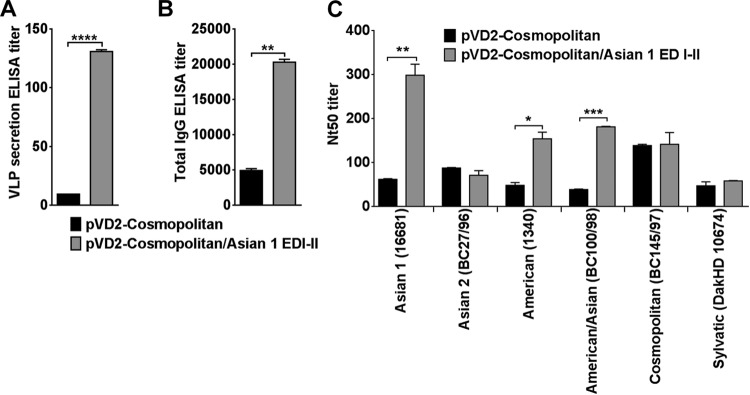
Enhancement of VLP secretion and immunogenicity by manipulation of ectodomain of E protein. (A) An increase in the VLP secretion titer in pVD2-Cosmopolitan/Asian 1 EDI-II-transfected COS-1 cells after replacement of the EDI-II gene of the WT pVD2-Cosmopolitan plasmid vector with the corresponding gene segment from the pVD2-Asian plasmid was determined by ELISA. ****, P < 0.0001. (B) pVD2-Cosmopolitan/Asian 1 EDI-II-vaccinated mice exhibit a larger total IgG antibody response. **, P = 0.0017. (C) Improvement of FRμNT titers against the different DENV-2 genotypes by sera from pVD2-Cosmopolitan/Asian 1 EDI-II-vaccinated mice. *, P = 0.0195; **, P = 0.0031; ***, P = 0.0004. All data represent the means ± SEMs of two independent experiments. Means were analyzed by an unpaired t test, and significance (P < 0.05) is indicated with asterisks.
Destabilizing mutations in the poorly immunogenic Cosmopolitan genotype DNA vaccine.
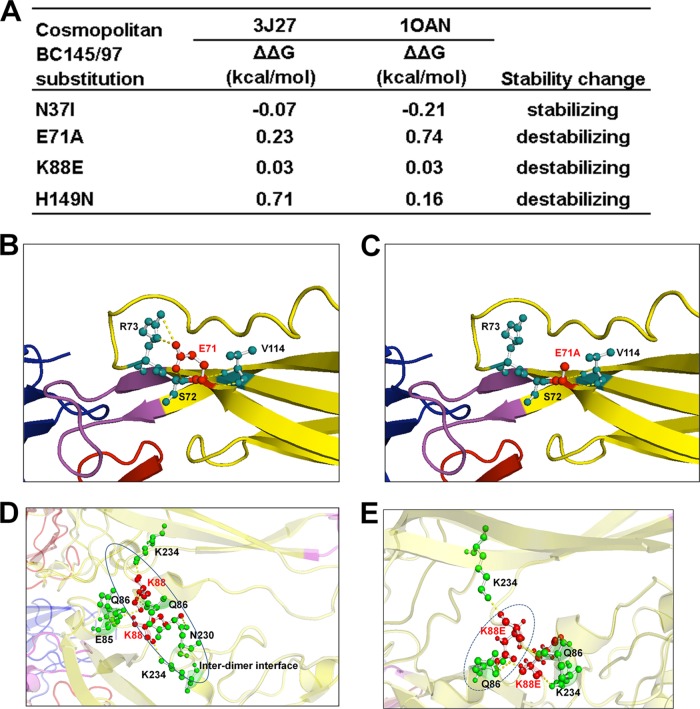
Mutations to the amino acid sequence of E protein could potentially alter the proper glycoprotein structural conformation, could disrupt inter- or intradimer interactions, and might lead to impaired particle formation, maturation, or secretion and, thus, impaired immunogenicity when administered as a vaccine. We analyzed if the naturally occurring mutations present in the poorly immunogenic pVD2-Cosmopolitan vaccine (see Fig. S1 in the supplemental material) affect the stability of VLP expression. Using the atomic coordinates of both the mature dengue virus particle and soluble E protein, analyses revealed that the nonconservative substitutions E71A, K88E, and H149N have destabilizing effects on the E-protein structure on the basis of the high values of the predicted changes in free energy (Fig. 8A) for all substitutions except N37I, which does not entail an energy cost. These destabilizing effects are brought about by disruption of the local hydrogen bonding network and charged interactions with neighboring amino acids (Fig. 8B to E) that could disrupt the structural integrity of E glycoprotein.
FIG 8.
Naturally occurring mutations affect the structural stability of the E protein of pVD2-Cosmopolitan VLPs. (A) Stable free energy (ΔΔG) calculations for E-protein substitutions in Cosmopolitan genotype strain BC145/97 based upon the Protein Data Bank structural coordinates for the mature dengue virus particle (PDB accession number 3J27) and soluble E protein (PDB accession number 1OAN) (25, 27). (B to E) Amino acid residues involved in E-protein intra- and interdimer polar contacts. (B) E71 (in a red ball-and-stick representation) and its intradimeric contact with amino acid residues S72, R73, and V114 (in green ball-and-stick representations), connected by yellow dashed lines. (C) The construct with the E71A substitution shows a loss of intradimeric contact with R73. (D) K88 (in a red ball-and-stick representation) and its contact with various amino acid residues (in green ball-and-stick representations), connected by yellow dashed lines. K88 of chain A of the E-protein asymmetric unit has intradimeric contacts within chain A at positions Q86 and K234 and has an interdimeric contact with E85 of chain C. K88 of chain C has intradimeric contacts within chain C at positions Q86, N230, and K234. (E) K88E of chain A shows a loss of interdimeric contact with E85 of chain C and a steric clash with Q86 of chain C. K88E of chain C shows a loss of intradimeric contact with N230 of chain C.
DISCUSSION
The leading candidates for a dengue vaccine are chimeric live-attenuated viruses (cLAV), but it has been proved difficult to produce cLAV vaccines that are satisfactorily attenuated and at the same time sufficiently immunogenic (43). Clearly, there is a reason for exploring alternatives. DNA immunization is thought to elicit immune responses that closely resemble those seen in natural infections with intracellular pathogens. We previously reported that a DENV-2 DNA vaccine consisting of prM and 80% E (aa 1 to 396) of DENV-2, followed by 20% E from the homologous JEV membrane-anchoring region, enhanced VLP secretion after intracellular expression and was able to generate high-titer antibodies in mice which persisted for more than 6 months (28, 44). DENV-neutralizing antibodies directed against the E protein are thought to play a key role in protection against the disease, an idea supported directly by passive antibody transfer experiments in animal models and indirectly by epidemiological data from prospective studies in areas where dengue is endemic (45, 46). However, the levels of antibody induction and protection for a dengue vaccine have not been established under conditions of different antigenic backbones, and these might differ depending on the phylogenetic genotype differences. In the present study, we constructed DNA vaccines with the different DENV-2 genotypes and further evaluated their immunogenicities, the neutralization activities of the elicited antibodies, and the protection provided in vaccinated mice against a set of representative DENV-2 strains that has circulated worldwide.
First, to explore if different antigenic properties due to amino acid variation in the E-protein ectodomains of different DENV-2 genotypes affected the immunogenicity of our DNA vaccines, we immunized groups of 4-week-old BALB/c mice with plasmids carrying the DNA of the different genotypes. It was interesting to find that the antibody response measured by a total IgG ELISA was highly correlated to the level of in vitro VLP secretion (P = 0.0129) (Fig. 3B). Although only one haplotype of mice was tested in this study, the different immunogenicities induced by the different DNA vaccines could be consistent when different inbred or outbred mice are used, as suggested by a previous study (47). To further confirm that the level of secretion of the expressed recombinant proteins of our DNA vaccines affects their immunogenicity, the E-protein ectodomain of the poorly immunogenic Cosmopolitan genotype vaccine was manipulated to enhance its VLP secretion by engineering a chimeric pVD2-Cosmopolitan vaccine wherein its EDI-II sequence was transplanted from pVD2-Asian 1. This ectodomain manipulation indeed brought a significant improvement to the secretion of VLPs compared to that for the wild-type pVD2-Cosmopolitan plasmid (Fig. 7A), and when used as a DNA vaccine in mice, it led to the increased production of antibodies (Fig. 7B) and a concomitant enhancement of the potential of the immune sera to neutralize those heterologous DENV-2 genotype strains that were previously poorly neutralized (Fig. 7C). This gain-of-function phenomenon observed after transplanting the EDI-II sequence of pVD2-Asian 1 into a pVD2-Cosmopolitan backbone could be attributed to the effects of E71, K88, and H149, contained in Asian 1 EDI-II, in reestablishing a stable inter- and intradimer local network of amino acid interactions that provide a proper E-glycoprotein structural conformation. On the basis of the structural analyses, the nonconservative mutations found in Cosmopolitan genotype strain BC145/97 are predicted to have greater changes in free energy (Fig. 8A) and, thus, decrease the stability of the E protein. The E71A substitution caused structural disturbances within the E-protein homodimer by disrupting a hydrogen bond with S72 and V114 and a charged interaction with R73 (Fig. 8B and C). E71 is also involved in a charged interaction with K52 of the pr peptide within the prominent complementary electrostatic patches of the pr and E contact area (26), and a loss of this interaction by an alanine substitution may contribute to the disruption of the prM and E interaction during VLP maturation. A K88E substitution in one dimer pair could potentially destabilize the E-protein interdimer interaction by creating an interdimeric steric clash with Q86 of the other dimer pair and a loss of interdimeric polar contacts with E85 and N230 (Fig. 8D and E). H149 is believed to function as a switch in the dissolution of a network of local interactions (EDI residues 142 to 157, including the glycan linked on N153) around the fusion loop during maturation (48). This rigid conformation of the fusion loop tethering network is disrupted during the protonation of H149 at low pH to trigger fusion loop exposure. An H149N substitution could possibly block this event and somehow entail a high energy cost for this dynamic change to proceed.
Since each of the four DENV serotypes has multiple DENV genotypes and frequent shifts of the predominantly circulating genotype occur in many different geographic regions, the ability of a vaccine to successfully overcome this diversity is crucial, and creation of a vaccine with this ability was the second objective of our study. Consistent with a previous report (24), our results suggest that use of the Asian 1 genotype (strain 16681 in this study) as the backbone for a DENV-2 vaccine can neutralize representative strains of different genotypes of DENV-2. Similarly, we observed variable neutralization activities of the elicited immune sera (Fig. 5A). Lower neutralization titers against strains of the Cosmopolitan and Sylvatic genotypes were particularly observed. Several studies on the four serotypes of DENV extensively examined how genotypic sequence variation affects the potency of antibody neutralization (33, 37, 38, 49–53). Observations from those studies analogously showed that a number of antibodies inefficiently neutralized heterologous virus strains of a distinct genotype. In addition, a MAb mapping study carried out with different DENV-2 genotypes showed conservation of the DENV-2 type-specific and DENV complex-reactive antigenic sites in EDIII (54) and that substitutions in these regions did not significantly affect antibody binding affinity or neutralization, findings which further suggest that DENV-2 EDIII-specific neutralizing antibodies will likely be effective against DENV-2 strains of all six genotypes. Surprisingly, a novel finding in the current study demonstrated that the Sylvatic genotype of DENV-2 escaped binding to a critical flavivirus subgroup-cross-reactive, highly potent neutralizing MAb, 1B7-5. The exact binding epitope of 1B7-5 is currently unknown, and our ongoing study mapped the possible binding epitope in EDI-II, which is consistent with the findings reported in previous publications (39, 40). Whether the binding escape of a single MAb (1B7-5) would result in the loss of protection from challenge by the current vaccine strain of the Sylvatic genotype needs further study. Detailed mapping to find the exact location of this epitope and its contribution to neutralization and protection against Sylvatic genotype viruses is under way. A study using human monoclonal antibodies also demonstrated that EDI-II contains potent cross-neutralization epitopes (55) and suggested its potential for use in future vaccine design (56, 57). Furthermore, the finding in this study that MAb 1B7-5 is an interdomain antibody which recognizes not only EDI-II but also EDIII emphasizes the important role of neutralizing antibodies recognizing quaternary epitopes. Mapping of these cross-neutralization epitopes will facilitate the future development of a universal dengue vaccine with broad neutralization activities.
Previous reports suggested that MAbs with strongly neutralizing activity against DENV are serotype specific and bind to EDIII (10, 36, 58). Recent data suggest that humans develop low levels of EDIII-reactive antibodies during DENV infection (less than 10% of the total virus-reactive antibodies) and that they make only a minor contribution (5 to 15%) to the neutralizing activity of human immune sera (59). On the other hand, mice infected with DENV develop anti-EDIII antibodies that account for one-third (∼34%) of the in vitro serotype-specific neutralization, and it was shown that these are not critical for in vivo protection (60). These studies seem to suggest that epitopes present in E ectodomains other than EDIII are primarily responsible for DENV neutralization. However, we found in our study that mice immunized with DENV-2 DNA vaccines develop EDIII antibodies that constitute at least more than 50% of the total IgG population. This inconsistency with the results of previous studies could be due to the different nature of the antigen that we employed in our experiment. We estimated the proportions of IgG antibodies recognizing EDIII and non-EDIII using a chimeric VLP system, while the other group used an EDIII–maltose-binding protein fusion expressed in E. coli. One advantage of using VLPs, aside from the fact that they display an E protein with proper folding and the proper conformation, is their usefulness in confirming the presence of interdomain antibodies in polyclonal sera. Interdomain antibodies could not possibly be determined by using EDIII domain-specific proteins since this type of antibody binds only to complex, quaternary epitopes displayed only on an intact particle, as suggested by previous studies (35, 61, 62). These epitopes span either EDII and EDIII, EDI and EDIII, or EDI-II and EDIII (63–65). Using this new approach in determining the domain specificity of antibodies in polyclonal sera, we have shown that EDIII interdomain antibodies contributed a substantial amount to the total abundance of EDIII-reactive antibodies, and these interdomain antibodies present in the polyclonal sera elicited from DENV-2 DNA-immunized mice are T5-1- and 1B7-5-like antibodies. One limitation of this study is that the calculation of the amount of domain-specific antibodies in the immune mouse sera other than those elicited from the pVD2-Asian 1 vaccine might not be totally accurate since the chimeric VLP used in the binding assay was based on the pVD2-Asian 1 VLP backbone.
Neutralization activity has been used during the development of dengue vaccine candidates and provided useful information in terms of immunogenicity (seroneutralizing antibodies in particular) and protection against infection. In this regard, protection against the morbidity/mortality induced by infection with wild-type dengue viruses could be assessed with this model. Indeed, it has previously been demonstrated that neutralizing antibodies elicited by a single injection of the CYD vaccine protected against virulent virus challenge (66). However, the recent failure to correlate the neutralization activity with protection from DENV-2 infection in a CYD vaccine phase IIb clinical trial raised a concern (17). In our study, a DNA vaccine prepared with the strain 16681 backbone did not protect the mice from challenge with DENV-2 of the Sylvatic genotype. Several reports have discussed the potential of Sylvatic genotype viruses to reemerge after the worldwide implementation of a DENV vaccine (67, 68). Although previous studies (24, 69) using infected human sera provided sufficient neutralization activity against Sylvatic genotype viruses, a finding which was consistent with the results of our in vitro study, our in vivo study in mice suggested otherwise. Development of a reliable assay to measure the correlate of protection in vitro and in vivo will be crucial for vaccine development efforts.
There are limitations to this study and the broader applicability of our conclusions. First, it is essential to obtain serum from an individual mouse to reveal individual variations in the antibody response after vaccination. Instead, we pooled sera from each treatment group to obtain a sufficient volume of serum and used these pooled group sera to conduct multiple FRμNTs against 12 DENV-2 strains of different genotypes (one American, one Asian 1, two Asian 2, two American/Asian, two Cosmopolitan, and four Sylvatic genotype viruses). Second, the applicability of the results obtained in this study with mice to humans requires further studies. Third, recombinant subviral particles (RSPs; VLPs in our terminology) produced by tick-borne encephalitis virus (TBE) are heterogeneous in size, but the majority of TBE RSPs are about 30 nm in diameter and consist of 60 copies of E protein packed as dimers arranged in a T=1 icosahedral lattice, an assemblage that is different from that for the 90 E-protein dimers containing virion particles (70, 71). Using DENV-2 VLPs, MAb 1B7-5 was found to be an interdomain antibody, but whether it is similarly expressed in virion particles requires further investigation, as does the contribution of this antibody to human protection.
In summary, our study suggests that DENV-2 DNA vaccines with backbones from different genotypes differed in their antigenicities and immunogenicities and the protection from challenge that they offered in mice. We also suggest a strategy to enhance the level of VLP secretion and thereby increase the immunogenicity of VLPs by swapping the E-protein ectodomain from one genotype to the other. Furthermore, recent structure studies using humanized monoclonal antibodies suggested that potent neutralizing antibodies recognize a spatial region which exists only on virion particles and not on subunit or monomeric E protein (61). Therefore, using live-attenuated, inactivated whole-virus vaccines or vaccines that form virus-like particles should be a better and more efficacious approach for dengue vaccines.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Science Council of Taiwan (96-3111-B-005-001- and 98-2320-B-005-003-MY3).
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 9 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00810-14.
REFERENCES
- 1.Kyle J, Harris E. 2008. Global spread and persistence of dengue. Annu. Rev. Microbiol. 62:71–92. 10.1146/annurev.micro.62.081307.163005 [DOI] [PubMed] [Google Scholar]
- 2.Guzman M, Halstead S, Artsob H, Buchy P, Farrar J, Gubler D, Hunsperger E, Kroeger A, Margolis H, Martínez E, Nathan M, Pelegrino J, Simmons C, Yoksan S, Peeling R. 2010. Dengue: a continuing global threat. Nat. Rev. Microbiol. 8(12 Suppl):S7–S16. 10.1038/nrmicro2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt S, Gething P, Brady O, Messina J, Farlow A, Moyes C, Drake J, Brownstein J, Hoen A, Sankoh O, Myers M, George D, Jaenisch T, Wint G, Simmons C, Scott T, Farrar J, Hay S. 2013. The global distribution and burden of dengue. Nature 496:504–507. 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilder-Smith A, Ooi E, Vasudevan S, Gubler D. 2010. Update on dengue: epidemiology, virus evolution, antiviral drugs, and vaccine development. Curr. Infect. Dis. Rep. 12:157–164. 10.1007/s11908-010-0102-7 [DOI] [PubMed] [Google Scholar]
- 5.Morrison A, Zielinski-Gutierrez E, Scott T, Rosenberg R. 2008. Defining challenges and proposing solutions for control of the virus vector Aedes aegypti. PLoS Med. 5:e68. 10.1371/journal.pmed.0050068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halstead SB. 2007. Dengue. Lancet 370:1644–1652. 10.1016/S0140-6736(07)61687-0 [DOI] [PubMed] [Google Scholar]
- 7.Chambers TJ, Hahn CS, Galler R, Rice CM. 1990. Flavivirus genome: organization, expression and replication. Annu. Rev. Microbiol. 44:649–688. 10.1146/annurev.mi.44.100190.003245 [DOI] [PubMed] [Google Scholar]
- 8.Lindenbach BD, Rice CM. 2001. Flaviviridae: the viruses and their replication, p 991–1110 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 9.Mukhopadhyay S, Kuhn R, Rossmann M. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13–22. 10.1038/nrmicro1067 [DOI] [PubMed] [Google Scholar]
- 10.Crill WD, Roehrig JT. 2001. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 75:7769–7773. 10.1128/JVI.75.16.7769-7773.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierson T, Fremont D, Kuhn R, Diamond M. 2008. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe 4:229–238. 10.1016/j.chom.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinz F, Stiasny K. 2012. Flaviviruses and their antigenic structure. J. Clin. Virol. 55:289–295. 10.1016/j.jcv.2012.08.024 [DOI] [PubMed] [Google Scholar]
- 13.Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, Chawansuntati K, Malasit P, Mongkolsapaya J, Screaton G. 2010. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328:745–748. 10.1126/science.1185181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crill W, Hughes H, Delorey M, Chang G. 2009. Humoral immune responses of dengue fever patients using epitope-specific serotype-2 virus-like particle antigens. PLoS One 4:e4991. 10.1371/journal.pone.0004991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai CY, Williams KL, Wu YC, Knight S, Balmaseda A, Harris E, Wang WK. 2013. Analysis of cross-reactive antibodies recognizing the fusion loop of envelope protein and correlation with neutralizing antibody titers in Nicaraguan dengue cases. PLoS Negl. Trop. Dis. 7:e2451. 10.1371/journal.pntd.0002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinz F, Stiasny K. 2012. Flaviviruses and flavivirus vaccines. Vaccine 30:4301–4306. 10.1016/j.vaccine.2011.09.114 [DOI] [PubMed] [Google Scholar]
- 17.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel T, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth N, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567. 10.1016/S0140-6736(12)61428-7 [DOI] [PubMed] [Google Scholar]
- 18.Robinson HL, Torres C. 1997. DNA vaccines. Semin. Immunol. 9:271–283. 10.1006/smim.1997.0083 [DOI] [PubMed] [Google Scholar]
- 19.Shedlock D, Weiner D. 2000. DNA vaccination: antigen presentation and the induction of immunity. J. Leukoc. Biol. 68:793–806. 10.1189/jlb.1938-3673 [DOI] [PubMed] [Google Scholar]
- 20.Beckett CG, Tjaden J, Burgess T, Danko JR, Tamminga C, Simmons M, Wu SJ, Sun P, Kochel T, Raviprakash K, Hayes CG, Porter KR. 2011. Evaluation of a prototype dengue-1 DNA vaccine in a phase 1 clinical trial. Vaccine 29:960–968. 10.1016/j.vaccine.2010.11.050 [DOI] [PubMed] [Google Scholar]
- 21.Yauch LE, Shresta S. 2014. Dengue virus vaccine development. Adv. Virus Res. 88:315–372. 10.1016/B978-0-12-800098-4.00007-6 [DOI] [PubMed] [Google Scholar]
- 22.Holmes EC, Twiddy SS. 2003. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 3:19–28. 10.1016/S1567-1348(03)00004-2 [DOI] [PubMed] [Google Scholar]
- 23.Twiddy SS, Farrar JJ, Chau NV, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. 2002. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology 298:63–72. 10.1006/viro.2002.1447 [DOI] [PubMed] [Google Scholar]
- 24.Barban V, Munoz-Jordan J, Santiago G, Mantel N, Girerd Y. 2012. Broad neutralization of wild-type dengue virus isolates following immunization in monkeys with a tetravalent dengue vaccine based on chimeric yellow fever 17D/dengue viruses. Virology 429:91–98. 10.1016/j.virol.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Ge P, Yu X, Brannan J, Bi G, Zhang Q, Schein S, Zhou Z. 2013. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20:105–110. 10.1038/nsmb.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. 2008. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319:1830–1834. 10.1126/science.1153263 [DOI] [PubMed] [Google Scholar]
- 27.Modis Y, Ogata S, Clements D, Harrison SC. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 100:6986–6991. 10.1073/pnas.0832193100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang G-JJ, Hunt AR, Holmes DA, Springfield T, Chiueh TS, Roehrig JT, Gubler DJ. 2003. Enhancing biosynthesis and secretion of premembrane and envelope proteins by the chimeric plasmid of dengue virus type 2 and Japanese encephalitis virus. Virology 306:170–180. 10.1016/S0042-6822(02)00028-4 [DOI] [PubMed] [Google Scholar]
- 29.Hughes H, Crill W, Chang G. 2012. Manipulation of immunodominant dengue virus E protein epitopes reduces potential antibody-dependent enhancement. Virol. J. 9:115. 10.1186/1743-422X-9-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crill WD, Hughes HR, Trainor NB, Davis BS, Whitney MT, Chang GJ. 2012. Sculpting humoral immunity through dengue vaccination to enhance protective immunity. Front. Immunol. 3:334. 10.3389/fimmu.2012.00334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS One 3:e3647. 10.1371/journal.pone.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang G, Hunt A, Davis B. 2000. A single intramuscular injection of recombinant plasmid DNA induces protective immunity and prevents Japanese encephalitis in mice. J. Virol. 74:4244–4252. 10.1128/JVI.74.9.4244-4252.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sukupolvi-Petty S, Austin S, Engle M, Brien J, Dowd K, Williams K, Johnson S, Rico-Hesse R, Harris E, Pierson T, Fremont D, Diamond M. 2010. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J. Virol. 84:9227–9239. 10.1128/JVI.01087-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lok S, Ng M, Aaskov J. 2001. Amino acid and phenotypic changes in dengue 2 virus associated with escape from neutralisation by IgM antibody. J. Med. Virol. 65:315–323. 10.1002/jmv.2036 [DOI] [PubMed] [Google Scholar]
- 35.de Alwis R, Smith S, Olivarez N, Messer W, Huynh J, Wahala W, White L, Diamond M, Baric R, Crowe JJ, de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl. Acad. Sci. U. S. A. 109:7439–7444. 10.1073/pnas.1200566109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gromowski G, Barrett A. 2007. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 366:349–360. 10.1016/j.virol.2007.05.042 [DOI] [PubMed] [Google Scholar]
- 37.Bernardo L, Yndart A, Vazquez S, Morier L, Guzman MG. 2005. Antibody responses to Asian and American genotypes of dengue 2 virus in immunized mice. Clin. Diagn. Lab. Immunol. 12:361–362. 10.1128/CDLI.12.2.361-362.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zulueta A, Martin J, Hermida L, Alvarez M, Valdes I, Prado I, Chinea G, Rosario D, Guillen G, Guzman MG. 2006. Amino acid changes in the recombinant dengue 3 envelope domain III determine its antigenicity and immunogenicity in mice. Virus Res. 121:65–73. 10.1016/j.virusres.2006.04.003 [DOI] [PubMed] [Google Scholar]
- 39.Aaskov JG, Geysen HM, Mason TJ. 1989. Serologically defined linear epitopes in the envelope protein of dengue 2 (Jamaica strain 1409). Arch. Virol. 105:209–221. 10.1007/BF01311358 [DOI] [PubMed] [Google Scholar]
- 40.Roehrig JT, Bolin RA, Kelly RG. 1998. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246:317–328. 10.1006/viro.1998.9200 [DOI] [PubMed] [Google Scholar]
- 41.Gromowski G, Barrett N, Barrett A. 2008. Characterization of dengue virus complex-specific neutralizing epitopes on envelope protein domain III of dengue 2 virus. J. Virol. 82:8828–8837. 10.1128/JVI.00606-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henchal EA, McCown JM, Burke DS, Seguin MC, Brandt WE. 1985. Epitopic analysis of antigenic determinants on the surface of dengue 2 virions using monoclonal antibodies. Am. J. Trop. Med. Hyg. 34:162–169 [DOI] [PubMed] [Google Scholar]
- 43.Webster D, Farrar J, Rowland-Jones S. 2009. Progress towards a dengue vaccine. Lancet Infect. Dis. 9:678–687. 10.1016/S1473-3099(09)70254-3 [DOI] [PubMed] [Google Scholar]
- 44.Men RH, Bray M, Lai CJ. 1991. Carboxy-terminally truncated dengue virus envelope glycoproteins expressed on the cell surface and secreted extracellularly exhibit increased immunogenicity in mice. J. Virol. 65:1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kyle J, Balsitis S, Zhang L, Beatty P, Harris E. 2008. Antibodies play a greater role than immune cells in heterologous protection against secondary dengue virus infection in a mouse model. Virology 380:296–303. 10.1016/j.virol.2008.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Endy T, Nisalak A, Chunsuttitwat S, Vaughn DW, Green S, Ennis FA, Rothman AL, Libraty DH. 2004. Relationship of preexisting dengue virus (DV) neutralizing antibody levels to viremia and severity of disease in a prospective cohort study of DV infection in Thailand. J. Infect. Dis. 189:990–1000. 10.1086/382280 [DOI] [PubMed] [Google Scholar]
- 47.Jan LR, Yang CS, Henchal LS, Sumiyoshi H, Summers PL, Dubois DR, Lai CJ. 1993. Increased immunogenicity and protective efficacy in outbred and inbred mice by strategic carboxyl-terminal truncation of Japanese encephalitis virus envelope glycoprotein. Am. J. Trop. Med. Hyg. 48:412–423 [DOI] [PubMed] [Google Scholar]
- 48.Christian EA, Kahle KM, Mattia K, Puffer BA, Pfaff JM, Miller A, Paes C, Davidson E, Doranz BJ. 2013. Atomic-level functional model of dengue virus envelope protein infectivity. Proc. Natl. Acad. Sci. U. S. A. 110:18662–18667. 10.1073/pnas.1310962110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blaney JE, Matro JM, Murphy BR, Whitehead SS. 2005. Recombinant, live-attenuated tetravalent dengue virus vaccine formulations induce a balanced, broad, and protective neutralizing antibody response against each of the four serotypes in rhesus monkeys. J. Virol. 79:5516–5528. 10.1128/JVI.79.9.5516-5528.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O'Brien KM, Nelson CA, Johnson S, Fremont DH, Diamond MS. 2010. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of denguevirus type 1. PLoS Pathog. 6:e1000823. 10.1371/journal.ppat.1000823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brien JD, Austin SK, Sukupolvi-Petty S, O'Brien KM, Johnson S, Fremont DH, Diamond MS. 2010. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J. Virol. 84:10630–10643. 10.1128/JVI.01190-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wahala WM, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS, de Silva AM. 2010. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog. 6:e1000821. 10.1371/journal.ppat.1000821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sukupolvi-Petty S, Brien JD, Austin SK, Shrestha B, Swayne S, Kahle K, Doranz BJ, Johnson S, Pierson TC, Fremont DH, Diamond MS. 2013. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J. Virol. 87:8826–8842. 10.1128/JVI.01314-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pitcher TJ, Gromowski GD, Beasley DW, Barrett AD. 2012. Conservation of the DENV-2 type-specific and DEN complex-reactive antigenic sites among DENV-2 genotypes. Virology 422:386–392. 10.1016/j.virol.2011.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F. 2010. The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8:271–283. 10.1016/j.chom.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azevedo A, Yamamura A, Freire M, Trindade G, Bonaldo M, Galler R, Alves A. 2011. DNA vaccines against dengue virus type 2 based on truncate envelope protein or its domain III. PLoS One 6:e20528. 10.1371/journal.pone.0020528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai WY, Lai CY, Wu YC, Lin HE, Edwards C, Jumnainsong A, Kliks S, Halstead S, Mongkolsapaya J, Screaton GR, Wang WK. 2013. High-avidity and potently neutralizing cross-reactive human monoclonal antibodies derived from secondary dengue virus infection. J. Virol. 87:12562–12575. 10.1128/JVI.00871-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS. 2007. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J. Virol. 81:12816–12826. 10.1128/JVI.00432-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wahala WM, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM. 2009. Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology 392:103–113. 10.1016/j.virol.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams KL, Wahala WM, Orozco S, de Silva AM, Harris E. 2012. Antibodies targeting dengue virus envelope domain III are not required for serotype-specific protection or prevention of enhancement in vivo. Virology 429:12–20. 10.1016/j.virol.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teoh E, Kukkaro P, Teo E, Lim A, Tan T, Yip A, Schul W, Aung M, Kostyuchenko V, Leo Y, Chan S, Smith K, Chan A, Zou G, Ooi E, Kemeny D, Tan G, Ng J, Ng M, Alonso S, Fisher D, Shi P, Hanson B, Lok S, MacAry P. 2012. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci. Transl. Med. 4:139–183. 10.1126/scitranslmed.3003888 [DOI] [PubMed] [Google Scholar]
- 62.Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A, Crowe JE, Jr, Lok SM. 2014. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol. Med. 6:358–371. 10.1002/emmm.201303404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goncalvez AP, Men R, Wernly C, Purcell RH, Lai CJ. 2004. Chimpanzee Fab fragments and a derived humanized immunoglobulin G1 antibody that efficiently cross-neutralize dengue type 1 and type 2 viruses. J. Virol. 78:12910–12918. 10.1128/JVI.78.23.12910-12918.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cockburn JJ, Navarro Sanchez ME, Goncalvez AP, Zaitseva E, Stura EA, Kikuti CM, Duquerroy S, Dussart P, Chernomordik LV, Lai CJ, Rey FA. 2012. Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus. EMBO J. 31:767–779. 10.1038/emboj.2011.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG. 2010. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc. Natl. Acad. Sci. U. S. A. 107:18950–18955. 10.1073/pnas.1011036107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guy B, Barrere B, Malinowski C, Saville M, Teyssou R, Lang J. 2011. From research to phase III: preclinical, industrial and clinical development of the Sanofi Pasteur tetravalent dengue vaccine. Vaccine 29:7229–7241. 10.1016/j.vaccine.2011.06.094 [DOI] [PubMed] [Google Scholar]
- 67.Vasilakis N, Shell E, Fokam E, Mason P, Hanley K, Estes D, Weaver S. 2007. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology 358:402–412. 10.1016/j.virol.2006.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vasilakis N, Cardosa J, Hanley K, Holmes E, Weaver S. 2011. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat. Rev. Microbiol. 9:532–541. 10.1038/nrmicro2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Durbin A, Mayer S, Rossi S, Amaya-Larios I, Ramos-Castaneda J, Eong Ooi E, Jane Cardosa M, Munoz-Jordan J, Tesh R, Messer W, Weaver S, Vasilakis N. 2013. Emergence potential of sylvatic dengue virus type 4 in the urban transmission cycle is restrained by vaccination and homotypic immunity. Virology 439:34–41. 10.1016/j.virol.2013.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferlenghi I, Clarke M, Ruttan T, Allison SL, Schalich J, Heinz FX, Harrison SC, Rey FA, Fuller SD. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593–602. 10.1016/S1097-2765(01)00206-4 [DOI] [PubMed] [Google Scholar]
- 71.Lorenz IC, Kartenbeck J, Mezzacasa A, Allison SL, Heinz FX, Helenius A. 2003. Intracellular assembly and secretion of recombinant subviral particles from tick-borne encephalitis virus. J. Virol. 77:4370–4382. 10.1128/JVI.77.7.4370-4382.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.