ABSTRACT
Influenza viral infection represents a serious public health problem that causes contagious respiratory disease, which is most effectively prevented through vaccination to reduce transmission and future infection. The nonstructural (NS) gene of influenza A virus encodes an mRNA transcript that is alternatively spliced to express two viral proteins, the nonstructural protein 1 (NS1) and the nuclear export protein (NEP). The importance of the NS gene of influenza A virus for viral replication and virulence has been well described and represents an attractive target to generate live attenuated influenza viruses with vaccine potential. Considering that most amino acids can be synthesized from several synonymous codons, this study employed the use of misrepresented mammalian codons (codon deoptimization) for the de novo synthesis of a viral NS RNA segment based on influenza A/Puerto Rico/8/1934 (H1N1) (PR8) virus. We generated three different recombinant influenza PR8 viruses containing codon-deoptimized synonymous mutations in coding regions comprising the entire NS gene or the mRNA corresponding to the individual viral protein NS1 or NEP, without modifying the respective splicing and packaging signals of the viral segment. The fitness of these synthetic viruses was attenuated in vivo, while they retained immunogenicity, conferring both homologous and heterologous protection against influenza A virus challenges. These results indicate that influenza viruses can be effectively attenuated by synonymous codon deoptimization of the NS gene and open the possibility of their use as a safe vaccine to prevent infections with these important human pathogens.
IMPORTANCE Vaccination serves as the best therapeutic option to protect humans against influenza viral infections. However, the efficacy of current influenza vaccines is suboptimal, and novel approaches are necessary for the prevention of disease cause by this important human respiratory pathogen. The nonstructural (NS) gene of influenza virus encodes both the multifunctional nonstructural protein 1 (NS1), essential for innate immune evasion, and the nuclear export protein (NEP), required for the nuclear export of viral ribonucleoproteins and for timing of the virus life cycle. Here, we have generated a recombinant influenza A/Puerto Rico/8/1934 (H1N1) (PR8) virus containing a codon-deoptimized NS segment that is attenuated in vivo yet retains immunogenicity and protection efficacy against homologous and heterologous influenza virus challenges. These results open the exciting possibility of using this NS codon deoptimization methodology alone or in combination with other approaches for the future development of vaccine candidates to prevent influenza viral infections.
INTRODUCTION
Influenza viruses are enveloped viruses that belong to the Orthomyxoviridae family and contain a segmented genome of eight, single-stranded RNA molecules with negative polarity (1). Each year, influenza viruses are responsible for upwards of 40,000 deaths and over 200,000 hospitalizations in the United States, making influenza the most common cause of vaccine-preventable morbidity and mortality (2, 3). Infection with influenza virus poses a threat to health and results in significant negative economic impacts on society every year (4). Additionally, influenza viruses can lead to sporadic pandemics when novel viruses are introduced into humans (5). The first influenza pandemic of this century was declared in 2009 with the emergence of a quadruple-reassortant swine-origin H1N1 virus (6). By the end of 2009, the virus was detected in approximately 200 countries, infecting more than 600,000 individuals and being associated with nearly 8,000 deaths (7). Public health concerns over influenza viruses are aggravated by their efficient transmission and the limited antiviral therapeutic options (8). Hence, vaccination remains our best medical intervention to protect humans against influenza virus (9), even though the effectiveness of current vaccines is suboptimal (10). The first option for vaccination is the injectable inactivated influenza vaccine (IIV), which requires a large quantity of virus and cannot induce a significant cytolytic cellular response, which is important for generating memory against subsequent infections (11–13). Additionally, recombinant influenza vaccine (RIV), recently approved by the U.S. Food and Drug Administration (FDA), induces antibody-mediated protection similar to that induced by IIV (14). The remaining option is the live attenuated influenza vaccine (LAIV), which induces better cellular and humoral immunity but is recommended only for immunocompetent 2- to 49-year-old persons (15). Thus, new vaccination strategies that overcome the limitations associated with current vaccination approaches are required for the prevention of influenza viral infections in humans.
Influenza virus RNA segment 8, or the nonstructural (NS) gene, encodes two fundamentally distinct proteins through an alternative splicing mechanism (16). Influenza virus produces segment 8 as a continuous primary transcript. Standard processing of this NS mRNA then generates nonstructural protein 1 (NS1), whereas alternative processing using a weak 5′ splice site results in a second, less abundant splice product, nuclear export protein (NEP) (16), which accounts for ∼10 to 15% of NS-derived mRNA (17). Although both polypeptides are ultimately translated from different open reading frames (ORFs), they share the first 10 N-terminal amino acids (16). NS1 is a multifunctional protein that most notably antagonizes the type I interferon (IFN-I) response and represents the main virus countermeasure against innate immune activation (18, 19). In addition, NS1 inhibits several IFN-I-induced genes (20, 21), limits the processing and nuclear export of host mRNA (22), and contributes to virulence (23). Because of the ability of influenza virus NS1 to counteract the host immune response, a variety of potential vaccine strategies rely on modified NS1 protein as a means for virus attenuation. Typically, these recombinant viruses showed significant growth attenuation in vivo (24–28). NEP, the underrepresented product for NS mRNA, is necessary for the production of replication-competent virus (29) and has several biologically important functions (30–32). NEP is mainly involved in the nuclear export of viral ribonucleoproteins (vRNPs) (31), the end product of genome replication that consists of each viral RNA segment encapsidated by the nucleoprotein (NP), and the heterotrimeric polymerase complex of polymerase basic 1 (PB1), PB2, and polymerase acidic (PA). NEP has also been reported to regulate the accumulation of viral RNA species, which leads to a switch in viral polymerase activity from a replicase (viral RNA [vRNA] to cRNA) to a transcriptase (vRNA to mRNA) (30). Lastly, NEP has been postulated to be responsible for controlling the timing of influenza viral infection through protein abundance (33). Hence, the multifunctional roles of the influenza virus NS gene products make for an ideal target for attenuation to create novel live attenuated influenza vaccines (19–21, 34–37).
Codon usage bias refers to differences in the frequency of occurrence of synonymous codons in coding DNA/RNA sequences, where all mammals share essentially the same codon bias (38). Due to the degeneracy of the genetic code, most amino acids are encoded by more than one codon (synonymous codons), and such codons have been observed to be unequally represented both within and between genomes. Importantly, recombinant viruses containing suboptimal codon organization could be used as potential live attenuated vaccines with the following advantages: (i) novel gene synthesis technologies allow rapid virus generation within a few weeks, (ii) vaccine candidates express identical protein sequences conserving an intact antigenic repertoire, and (iii) due to the large number of mutations introduced, the development of virulent revertants of RNA viruses through gradual nucleotide sequence mutations is unlikely. To date, two studies with influenza A virus attenuated by codon rearrangement of the PB1, hemagglutinin (HA), NP, and NA genes or combinations thereof have been described (39, 40). In those studies, the authors made use of codon pair bias to engineer codon-deoptimized viruses through an algorithm that creates a suboptimal organization of codon pairs (39, 40).
In the present work, we have reengineered the NS gene of influenza A/Puerto Rico/8/1934 (H1N1) (PR8) virus by introducing, through de novo gene synthesis, the least-used human synonymous codons in a way that perfectly preserves the wild-type (WT) amino acid sequence. In contrast to prior studies where genes involved in the synthesis of viral RNA or the spread of virus were modified, we have instead deoptimized the coding sequence of the NS gene by modifying each individual codon not involved in cis-acting regulation. Three viruses containing synonymous codon-deoptimized mutations in the separate splice products of NS (NS1 or NEP) or the coding region comprising the entire NS gene were generated. In vitro characterization of the recombinant codon-deoptimized influenza viruses showed altered protein expression levels for NS1, NEP, or both compared with those for the WT, reflecting the degree of deoptimization and the number of mutations introduced. However, all viruses grew with similar plaque phenotypes and to similar titers in Madin-Darby canine kidney (MDCK) cells, a recently FDA-approved cell line for influenza vaccine production (41). Unlike their in vitro phenotype, NS codon-deoptimized influenza viruses were attenuated in vivo and were also shown to conserve the immunogenicity properties of WT virus. Interestingly, immunization of mice by a single intranasal dose of codon-deoptimized virus conferred homologous and heterologous protection against subsequent influenza viral challenge, suggesting that this approach can provide protection against antigenically distinct influenza viruses.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney 293T (293T; ATCC CRL-11268) cells, human lung epithelial carcinoma (A549; ATCC CCL-185) cells, African green monkey kidney epithelial (Vero; ATCC CCL-81) cells, and MDCK (ATCC CCL-34) cells were grown at 37°C in air enriched with 5% CO2 and Dulbecco's modified Eagle's medium (DMEM; Mediatech, Inc.) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals) and 1% penicillin (100 units/ml)–streptomycin (100 μg/ml)–2 mM l-glutamine (P-S-G; Mediatech, Inc.). Human 293T ISRE-mRFP/ISRE-Fluc cells, encoding the monomeric red fluorescent protein (mRFP) and the firefly luciferase (Fluc) reporter genes under the control of an IFN-I-stimulated response element (ISRE), have been previously described (42). Influenza A/Puerto Rico/8/1934 (H1N1) (PR8) virus (43) and a recombinant virus containing the HA and NA segments of A/Hong Kong/1/1968 (H3N2) virus in the background of PR8 virus (X31 virus) (44) were grown in MDCK cells as previously described (45). Viral titers were determined by immunofocus assay (fluorescent forming units [FFU]) or standard plaque assay (PFU) in MDCK cells as previously described (45). The recombinant Newcastle disease virus (rNDV) expressing the green fluorescent protein (GFP), rNDV-GFP, was described previously (46, 47).
Construction of plasmids containing codon-deoptimized viral NS sequences.
To engineer a set of attenuated PR8 viruses through genome-scale changes in codon bias, a PR8 virus NS cDNA that contained the codon-deoptimized sequence comprising nucleotides 89 to 488 and 727 to 811 (NScd) was synthesized de novo (Biomatik) with appropriate flanking SapI restriction sites for subcloning into ambisense plasmid pDZ (48) to generate pDZ-NScd. Sequences including the previously described NS packaging signals (49) and the splicing donor and acceptor sites (16) were not altered (GenBank accession number AF389122.1). In addition, nucleotides surrounded the packaging signals or the splicing donor and acceptor sites that could interfere with their functions or virus rescue were not modified (Fig. 1). An ambisense plasmid containing the NS1 ORF codon-deoptimized sequence (pDZ-NS1cd) was generated by subcloning the codon-deoptimized NS1 region (NS1cd; nucleotides 89 to 488) from pDZ-NScd into the pDZ-NS plasmid. Plasmid pDZ-NEPcd was generated using a similar cloning strategy and contains the codon-deoptimized NEP region (NEPcd; nucleotides 727 to 811). In this case, most of the NEP nucleotide sequence was not modified because it either overlaps the NS1 ORF (16) or is part of the packaging signals required for efficient incorporation of the NS segment during viral infection (49). Polymerase II expression pCAGGS plasmids (50) containing NS1 or NEP codon-deoptimized sequences fused to an HA epitope tag (YPYDVPDYA) (46) or GFP (35) at the C terminus were obtained by subcloning the ORFs from pDZ-NS1cd and pDZ-NEP using standard molecular biology techniques. All pDZ and pCAGGS plasmids containing a codon-deoptimized NS, NS1, or NEP sequence were confirmed by sequencing (ACGT, Inc.).
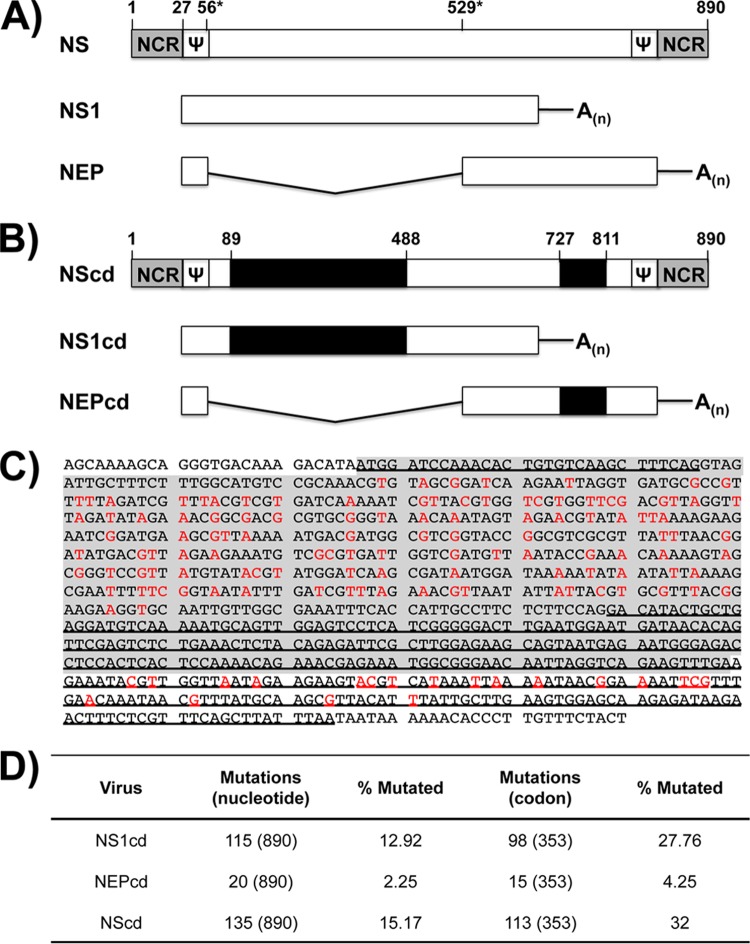
FIG 1.
Schematic representation of PR8 (H1N1) virus NS gene and gene transcripts. Wild-type (A) and codon-deoptimized (B) influenza PR8 virus NS genes are indicated by white boxes (WT sequence) or black boxes (codon-deoptimized sequence). Noncoding regions (NCR) are indicated with gray boxes. Packaging signals and splicing donor or acceptor regions are indicated by Ψ and asterisks, respectively. Nucleotide length is indicated with a number above the gene segments. (C) Replacement of nucleotides (red) for NS1 (gray shading) and NEP (underline). (D) Number and percentage of synonymous nucleotide and codon mutations in NS1cd, NEPcd, and NScd.
Virus rescue.
Three different viruses with codon-deoptimized synonymous mutations were generated: virus with codon deoptimization of the entire NS gene (NScd virus), which includes 113 (32%) codon substitutions, and viruses with codon deoptimization of the individual gene products NS1 (NS1cd virus), which incorporates 98 (27.76%) codon substitutions, and NEP (NEPcd virus), which incorporates 15 (4.25%) codon substitutions. Virus rescues were performed as previously described (51, 52). Briefly, cocultures (1:1) of 293T/MDCK cells (6-well plate format, 106 cells/well) in suspension were cotransfected, using Lipofectamine 2000 (Invitrogen), with 1 μg each of the seven ambisense wild-type (WT) plasmids (pDZ-PB2, -PB1, -PA, -NP, -NA, -M, -HA) plus the ambisense WT NS plasmid (pDZ-NS) or codon-deoptimized plasmid pDZ-NScd, -NS1cd, or -NEPcd. At 12 h posttransfection, the medium was replaced with DMEM containing 0.3% bovine serum albumin (BSA), 1% P-S-G, and 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma). At 48 h posttransfection, tissue culture supernatants were collected, clarified, and used to infect fresh MDCK cells (6-well plate format, 106 cells/well). At 3 days postinfection, recombinant viruses were plaque purified and scaled up in MDCK cells (45). Virus stocks were generated by infecting confluent 10-cm dishes of MDCK cells at a low multiplicity of infection (MOI; 0.001) (45). Stocks were titrated by plaque assay on MDCK cells (51), and the identity of the segment 8 vRNA was confirmed by restriction analysis and sequencing.
Semiquantitative RT-PCR.
Total RNA from infected MDCK cells was purified using the TRIzol reagent (Invitrogen) according to the manufacturer's specifications. The cDNAs were synthesized with SuperScript II reverse transcriptase (Invitrogen), using 1 μg of total RNA as the template and oligo(dT) to amplify total mRNAs or a primer specific for the NS noncoding region (5′-AGTAGAAACAAGGGTGTTTTTTA-3′) to amplify the vRNA. The cDNAs were used as the templates for semiquantitative reverse transcription-PCR (RT-PCR) with primers specific for segment 8 (forward primer 5′-AGCAAAAGCAGGGTGAACAAAGAC-3′; reverse primer 5′-TAATAATAAAAAACACCCTTGTTTCTACT-3′), the coding region of NS1 and NEP (forward primer NS-1/24 [5′-ATGGATCCAAACACTGTGTCA-3′]; reverse primer NS-684/707 [5′-CCTAATTGTTCCCGCCATTTCTCG-3′]), NP (forward primer 5′-ATGGCGTCCCAAGGCACCAAACGG-3′; reverse primer 5′-TTAATTGTCGTACTCCTCTGCATT-3′), and canine GAPDH (glyceraldehyde-3-phosphate dehydrogenase; forward primer 5′-AACATCATCCCTGCTTCCAC-3′; reverse primer 5′-GACCACCTGGTCCTCAGTGT-3′).
Transient cell transfection.
To evaluate WT and codon-deoptimized NS1 and NEP expression levels, human 293T cells were transfected for analysis by epifluorescence (GFP-tagged pCAGGS plasmids) and Western blotting (HA-tagged pCAGGS plasmids). Briefly, 106 human 293T cells in 6-well plates were transfected in suspension with the indicated concentrations of pCAGGS protein expression plasmids or empty (E) pCAGGS plasmid as a negative control. At 24 h posttransfection, cells were collected by centrifugation at 14,000 rpm for 5 min at 4°C and lysed with 400 μl of lysis buffer (20 mM Tris-HCl [pH 7.4], 5 mM EDTA, 100 mM NaCl, 1% NP-40, complete protease inhibitor cocktail; Roche) for 30 min on ice. Cell lysates were clarified by centrifugation at 14,000 rpm for 30 min at 4°C (53).
Protein gel electrophoresis and Western blot analysis.
Proteins from transfected or infected cell lysates were separated by denaturing electrophoresis in 10% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane (Bio-Rad) with a Bio-Rad Mini Protean II electroblotting apparatus at 175 mA for 2 h at 4°C. Membranes were blocked for 1 h with 10% dried skim milk in phosphate-buffered saline (PBS), 0.1% Tween 20 (T-PBS) and incubated overnight at 4°C with specific primary monoclonal antibodies (MAbs) or primary polyclonal antibodies (pAbs): HA (mouse MAb; catalog no. H9658; Sigma), GFP (rabbit pAb; catalog no. SC8334; Santa Cruz), NS1 (mouse MAb 1A7) (19), NEP (rabbit pAb) (31), or NP (mouse MAb HT103) (31). Mouse MAb against actin (catalog no. A1978; Sigma) was used as an internal loading control. Bound primary antibodies were detected with horseradish peroxidase (HRP)-conjugated antibodies against the different species, and proteins were detected by chemiluminescence (Hyglo; Dennville Scientific), following the manufacturer's recommendations. Western blots were quantified by densitometry using the software ImageJ (v1.46). The bands were normalized to the level of actin expression, and then the level of expression of the WT protein was considered 100% for all amounts of transfected plasmids for comparison with the levels of expression of the corresponding codon-deoptimized constructs.
Virus growth curve analysis.
To determine in vitro virus growth rates, triplicate wells of confluent MDCK or A549 cells (6-well plate format, 106 cells/well) were infected at an MOI of 0.001. After 1 h of virus adsorption at room temperature, cells were washed and overlaid with DMEM containing 0.3% BSA and TPCK-treated trypsin. At the indicated times postinfection (12, 24, 48, and 72 h), tissue culture supernatants were collected and viral titers were determined by immunofocus assay (FFU/ml) as previously described (45). Briefly, triplicate confluent wells of MDCK cells (96-well format, 104 cells/well) were infected with 10-fold serial dilutions of tissue culture supernatants. At 8 h postinfection, cells were fixed and permeabilized (4% formaldehyde, 0.5% Triton X-100 in PBS) for 15 min at room temperature. After washing with PBS, the cells were incubated in blocking solution (2.5% BSA in PBS) for 1 h at room temperature, washed with PBS, and incubated with 1 μg/ml of influenza virus NP MAb HT103 for 1 h at 37°C. After washing with PBS, the cells were incubated with a fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG secondary antibody (Dako) and 1 mg/ml of 4′,6′-diamidino-2-phenylindole (DAPI; Research Organics) for 1 h at 37°C. Cells were visualized using a fluorescence microscope and photographed (Cooke Sensicam QE) at ×20 magnification, and the images were processed using Adobe Photoshop (CS4) software. NP-expressing cells were enumerated to determine the virus titer (FFU/ml). The mean value and standard deviation (SD) were calculated using Microsoft Excel software.
Bioassay to evaluate IFN-I production.
The levels of IFN-I secreted by virus-infected cells were evaluated as previously described (46, 54). Briefly, monolayers of A549 cells (12-well plate format, 5 × 105 cells/well) were mock infected or infected (MOI, 3) with WT NS or NScd PR8 viruses. At the indicated times postinfection (0, 6, 12, 24, and 48 h), tissue culture supernatants were collected, and viruses were UV inactivated by exposure to shortwave (254 nm) UV radiation (Mineralight UV lamp; UV S-68; Ultra-Violet Products) for 40 min at a distance of 6 cm (46, 54). Vero cells that had been seeded in 96-well plates (1 × 104 cells/well) the day before were treated with the UV-inactivated tissue culture supernatants for 24 h and then infected with rNDV-GFP (MOI, 1) for 1 h at room temperature and examined at 24 h postinfection for GFP expression under a fluorescence microscope. GFP intensity was measured with a microplate reader (DTX880; Beckman Coulter). For controls, Vero cells were mock treated or treated with 1, 10, or 100 units (U) of universal IFN-I (PBL Assay Science). Human 293T cells expressing mRFP and Fluc under the control of an ISRE promoter (42) were seeded in 96-well culture plates (1 × 104 cells/well) on the day before treatment with the UV-inactivated tissue culture supernatants and visualized at 24 h posttreatment under a fluorescence microscope (mRFP). Luciferase activities in cell lysates were determined using a Promega (Fitchburg, WA) luciferase reporter assay and a Lumicount luminometer. As internal controls, human 293T ISRE-mRFP/ISRE-Fluc cells were mock treated or treated with 1, 10, or 100 U of universal IFN-I.
Mouse experiments.
Female 6- to 8-week-old C57BL/6 mice were purchased from the National Cancer Institute (NCI) and maintained in the animal care facility at the University of Rochester under specific-pathogen-free conditions. All animal protocols were approved by the University of Rochester Committee of Animal Resources and complied with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council (55). Mice were anesthetized intraperitoneally (i.p.) with 2,2,2-tribromoethanol (Avertin; 240 mg/kg of body weight) and then inoculated intranasally (i.n.) with 30 μl of a virus preparation containing 10 or 100 PFU of WT NS, NScd, NS1cd, or NEPcd PR8 virus and monitored daily for morbidity (body weight loss) and mortality (survival). Mice showing a more than 25% loss of body weight were considered to have reached the experimental endpoint and were humanely euthanized. Virus replication was evaluated by determination of viral titers in the lungs at days 3 and 6 postinfection (45). To that end, three mice in each group were sacrificed and lungs were surgically extracted and homogenized. Virus titers were determined by immunofocus assay (FFU/ml) as indicated above. Geometric mean titers (GMTs) and statistical analyses (Mann-Whitney test) were performed using GraphPad Prism software. The 50% mouse lethal dose (MLD50) for WT or codon-deoptimized recombinant influenza viruses was determined using the method of Reed and Muench (56) in mice infected with 10, 100, and 1,000 PFU (n = 4) of WT, NScd, NS1cd, or NEPcd PR8 virus.
Vaccination and challenge of mice.
Female 6- to 8-week-old C57BL/6 mice were i.n. inoculated with PBS or 100 PFU of NScd virus. Two weeks after priming, the mice were challenged i.n. with 10 or 100 50% lethal doses (LD50s) of PR8 or X31 virus to determine homologous and heterologous protection, respectively. After challenge, the mice were monitored daily for morbidity and mortality, and viral replication in the lungs was evaluated at days 3 and 6 postinfection, as described above. Mouse sera were collected by submandibular bleeding 24 h prior to viral vaccinations and challenges and evaluated for the presence of influenza virus antibodies by an enzyme-linked immunosorbent assay (ELISA).
ELISA.
To assess the levels of virus-specific antibodies present in immunized mice, ELISAs were performed as previously described (45). Briefly, 96-well plates were coated for 18 h at 4°C with lysates from mock-, PR8 virus-, or X31 virus-infected MDCK cells. Alternatively, plates were coated with PR8 virus (H1N1) or A/Uruguay/716/2007 (H3N2) virus HA (100 ng per well; NR-19240 and NR-15168, respectively; BEI Resources). After washing with PBS, the coated wells were blocked with PBS containing 1% BSA, and then the plates were incubated with 1:2 dilutions of mouse serum (starting dilution, 1:64) for 1 h at 37°C, washed with H2O, and incubated with HRP-conjugated goat anti-mouse IgG (1:2,000; Southern Biotech) for 30 min at 37°C. The reactions were developed with 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (BioLegend) for 5 min at room temperature, quenched with 2 N H2SO4, and read at 450 nm (Vmax kinetic microplate reader; Molecular Devices).
HAI assays.
Hemagglutination inhibition (HAI) assays were used to evaluate the presence of neutralizing antibodies. To that end, mouse sera were treated with receptor-destroying enzyme (RDE; Sigma) prior to heat inactivation for 30 min at 56°C. Sera were then serially diluted 2-fold beginning at a 1:16 dilution and mixed 1:1 with 8 hemagglutination units (HAU) of PR8 or X31 virus for 30 min at room temperature. HAI was visualized by adding 0.5% turkey red blood cells (RBCs) to the virus-antibody mixtures for 30 min on ice (57, 58). The GMT ± SD from individual mice (n = 4) was calculated from the last well where hemagglutination was inhibited, using Microsoft Excel software.
RESULTS
Codon deoptimization of PR8 virus NS.
Codon deoptimization of the influenza virus NS gene should result in lower levels of NS1 and NEP protein expression, which would likely attenuate the virus so that it could then be used as a prototypic vaccine. To engineer a codon-deoptimized NS gene, we introduced underrepresented codons through genome-scale changes to the NS cDNA based on influenza A/Puerto Rico/8/1934 (H1N1) (PR8) virus, which was synthesized de novo. The amino acid sequence was completely conserved, and previously described NS packaging signals (49) and sequences involved in splicing (16) were excluded from the deoptimization for the successful generation of recombinant viruses (Fig. 1A and B). The NS codon-deoptimized gene included 113 (32%) codon changes through 135 (15.17%) nucleotide substitutions. Each mRNA processed from the NS codon-deoptimized gene incorporated 98 (27.76%) or 15 (4.25%) codons through 115 (12.92%) or 20 (2.25%) nucleotide changes in NS1 and NEP, respectively (Fig. 1C and D). These three NS gene deoptimization strategies were used to evaluate independently or in combination the effect of each codon-deoptimized viral product in the context of both protein expression and virus replication.
Downregulation of protein synthesis by the codon-deoptimized NS gene.
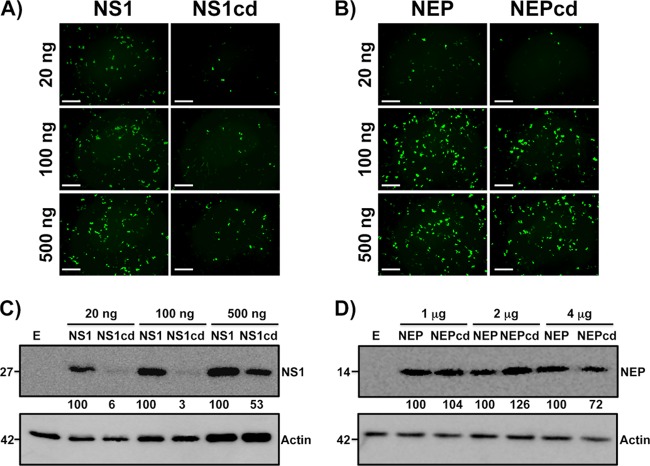
To determine whether the codon deoptimization of NS1 or NEP ORFs could lead to a reduction in protein expression levels in the absence of other viral factors, human 293T cells were transfected with 20, 100, and 500 ng of plasmids pCAGGS-NS1-GFP and pCAGGS-NS1cd-GFP (Fig. 2A) or pCAGGS-NEP-GFP and pCAGGS-NEPcd-GFP (Fig. 2B) and characterized for protein expression 24 h later by epifluorescence microscopy. A large reduction in the fluorescent signal and the number of fluorescent cells was observed in cells transfected with NS1cd constructs compared with the results for cells transfected with the WT, while no differences were observed with the NEP constructs.
FIG 2.
Analysis of the transitory expression of NS1cd and NEPcd in human 293T cells. (A and B) Fluorescence microscopy. Human 293T cells were transiently transfected with the indicated amounts of pCAGGS-NS1-GFP (NS1) and pCAGGS-NS1cd-GFP (NS1cd) (A) or pCAGGS-NEP-GFP (NEP) and pCAGGS-NEPcd-GFP (NEPcd) (B). Protein synthesis was evaluated 24 h posttransfection by determination of the level of GFP expression using fluorescence microscopy. Representative images are shown. Magnification, ×10. Bars, 500 μm. (C and D) Western blot analysis. Extracts from human 293T cells transiently transfected for 24 h with different amounts of pCAGGS-NS1-HA (lanes NS1) and pCAGGS-NS1cd-HA (lanes NS1cd) (C) or pCAGGS-NEP-HA (lanes NEP) and pCAGGS-NEPcd-HA (lanes NEPcd) (D) and empty pCAGGS (lanes E) were probed with anti-HA (top) or anti-actin (bottom) antibodies as described in Materials and Methods. Protein molecular sizes (in kDa) are indicated to the left. Western blots were quantified by densitometry using the software ImageJ (v1.46). Relative band intensities (as described in Materials and Methods) are indicated.
The effects of codon deoptimization on protein expression were also tested by Western blotting. For that, the C termini of the WT, the NS1cd construct (Fig. 2C), and the NEPcd construct (Fig. 2D) were fused to an HA epitope tag and used to transfect human 293T cells. Protein expression was evaluated at 24 h posttransfection using an anti-HA MAb. In this case, higher concentrations of pCAGGS NEP-HA expression plasmids were required for protein detection due to the small size of the viral protein (12 kDa) (32). The pattern of expression of both proteins correlated with that previously observed for GFP-tagged constructs. Whereas abundant polypeptide corresponding to the expected size of NS1 WT was clearly detected, the polypeptide corresponding to NS1cd was drastically reduced (Fig. 2C), while no differences between NEP WT and NEPcd polypeptides were observed (Fig. 2D). Protein densitometry of Western blot bands confirmed this observation. Overall, these data indicate that codon deoptimization of NS1 reduces protein expression, while codon deoptimization of NEP does not reduce protein levels, which may be attributed to differences in the percentage of codon changes introduced into the two viral proteins, the relative length of the mRNAs, or a combination thereof.
Generation of recombinant codon-deoptimized PR8 virus NS.
Since NS1 and NEP constructs were efficiently expressed in transfected cells, we wanted to ascertain whether recombinant influenza viruses encoding codon-deoptimized NS products could be rescued, as well as assess the effect of the deoptimization of viral NS products individually or in combination in the context of viral replication. To this end, codon-deoptimized viral NS RNA segments were incorporated into plasmid-based reverse genetics techniques (52) in order to generate recombinant, codon-deoptimized viruses. We generated three different viruses containing codon-deoptimized synonymous mutations in coding regions comprising the entire NS gene (NScd) or the individual viral proteins (NS1cd and NEPcd). The identity of the recombinant viruses was confirmed by RT-PCR using restriction analysis and sequencing of the NS segments. Sequence data revealed that the NS segment in all recombinant viruses did not contain additional changes (data not shown). To ascertain whether the splicing of NS mRNA was affected upon infection, MDCK cells were infected with WT NS or codon-deoptimized recombinant viruses, and total RNA was harvested and used to amplify the mRNAs by semiquantitative RT-PCR (Fig. 3). Using primers specific for the mRNAs of NS1 and NEP, we observed comparable levels of both segment 8 products for all viruses, suggesting that the splicing was not affected as a consequence of the nucleotide modifications introduced during codon deoptimization. To control for levels of viral infection and cellular transcription, influenza viral NP and canine GAPDH mRNA levels, respectively, were also evaluated by RT-PCR in the same cell samples.
FIG 3.
Analysis of segment 8 splicing in MDCK cells infected with wild-type and codon-deoptimized viruses by semiquantitative RT-PCR. MDCK cells were mock infected (mock) or infected with WT NS or codon-deoptimized viruses (NS1cd, NEPcd, and NScd) at an MOI of 5, and total RNA was harvested at 12 h postinfection. RT-PCRs were performed as described in Materials and Methods. NS vRNA or NS1 and NEP mRNAs were analyzed using specific primers. Primers for viral NP or cellular GAPDH mRNAs were included as internal controls.
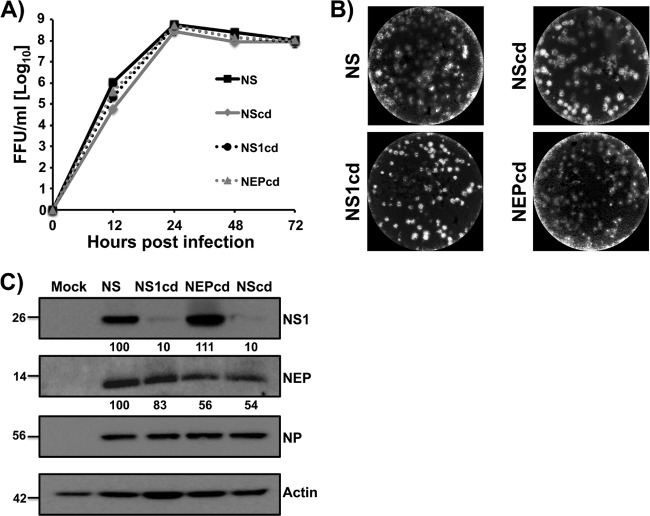
Growth properties of codon-deoptimized recombinant influenza viruses in tissue culture.
To analyze the replicative properties of recombinant codon-deoptimized viruses, multicycle growth curves of viruses with WT NS, NScd, NS1cd, or NEPcd in MDCK cells (MOI, 0.001) were assessed at 12, 24, 48, and 72 h postinfection (Fig. 4A). Despite the codon alterations in the NS gene, codon-deoptimized viruses displayed replication kinetics comparable to that of WT NS virus. Only at 12 h postinfection did the NScd show a slightly delayed kinetics, suggesting that reduced protein levels did not grossly impair virus growth in MDCK cells. Moreover, comparable plaque phenotypes in MDCK cells were observed (Fig. 4B), suggesting that the viruses with codon-deoptimized NS could propagate to levels comparable to those of viruses with WT NS in vitro. Next, we evaluated NS1 and NEP expression levels in the context of viral infection by Western blotting (Fig. 4C). Extracts from either mock-infected or infected (MOI, 0.001) MDCK cells were examined at 24 h postinfection using antibodies specific for NS1 (MAb 1A7), NEP (pAb NEP), or NP (MAb HT103). Actin was used as a protein loading control. Western blot analysis demonstrated that the levels of NP were not altered significantly in any codon-deoptimized virus infections, which was expected on the basis of virus growth kinetics. The amount of NS1 was not altered for NEPcd virus; however, it was significantly reduced in NS1cd and NScd viruses, correlating with codon deoptimization. Interestingly, under these experimental conditions, the level of NEP expression was slightly reduced for all recombinant viruses compared with that for WT NS virus.
FIG 4.
Analysis of wild-type and codon-deoptimized viruses in vitro in MDCK cells. (A) Multicycle growth kinetics. Virus titers in tissue culture supernatants of MDCK cells infected (MOI, 0.001) with WT NS or codon-deoptimized (NS1cd, NEPcd, and NScd) viruses were analyzed at the indicated times postinfection (12, 24, 48, and 72 h) by immunofocus assay (FFU/ml) using the anti-NP MAb HT103. Data represent the means ± SDs of the results determined for triplicate wells. (B) Plaque phenotype. MDCK cells were infected with WT NS or codon-deoptimized (NS1cd, NEPcd, and NScd) viruses, and at 2 days postinfection, monolayers were stained with crystal violet. (C) Analysis of protein expression levels in infected cells by Western blotting. MDCK cells were mock infected (mock) or infected with WT NS or codon-deoptimized (NS1cd, NEPcd, and NScd) viruses at an MOI of 0.001, and NS1, NEP, and NP expression levels were analyzed (24 h postinfection) by Western blotting with specific antibodies. Actin was used as a loading control. The protein molecular sizes (in kDa) are indicated to the left. Western blots were quantified by densitometry using the software ImageJ (v1.46). The bands were normalized to the level of actin expression for protein quantitation and then to the level of viral NP expression for determination of viral infection. The expression of WT protein was considered 100% for comparison with the level of expression by the corresponding codon-deoptimized viruses.
To evaluate if the influenza viruses carrying codon-deoptimized NS showed different replication kinetics in a cell line that more accurately represents the cells targeted in a natural human infection, A549 human lung epithelial cells were used (59, 60). To that end, human A549 cells were infected with WT NS or codon-deoptimized NS viruses (MOI, 0.001), and at 12, 24, 48, and 72 h postinfection, viral titers were evaluated (Fig. 5A). Contrary to observations in MDCK cells, viruses in which NS1 was codon deoptimized did not achieve levels of replication similar to those of WT NS viruses. However, NEPcd viruses replicated similarly to WT NS viruses. These data suggest that the influenza viruses with codon-deoptimized NS have an attenuated phenotype when tested in cells targeted during natural human infection. To evaluate if infection with codon-deoptimized viruses led to differential expression of NS1 and NEP proteins, lysates of infected A549 human cells were evaluated by Western blotting (Fig. 5B). For this purpose, A549 cells were either mock infected or infected at a high MOI (3) to achieve similar levels of infection. Then, cell extracts were collected at 24 h postinfection and probed using the antibodies indicated above. The levels of NP were not altered significantly in any codon-deoptimized virus infections, suggesting similar levels of infection. The amount of NS1 was notably reduced in NS1cd and NScd viruses, and the amount of NEP detected was markedly reduced in NEPcd virus, correlating with codon deoptimization. Altogether, these data suggest that the influenza viruses with codon-deoptimized NS have an attenuated phenotype and an expected decrease in protein expression levels when tested in cells targeted during natural human infection (i.e., human airway epithelial [A549] cells).
FIG 5.
Analysis of wild-type and codon-deoptimized viruses in vitro in A549 cells. (A) Multicycle growth kinetics. Human A549 cells were infected (MOI, 0.001) with WT NS or codon-deoptimized (NS1cd, NEPcd, and NScd) viruses. At the indicated times postinfection (12, 24, 48, and 72 h), tissue culture cell supernatants were collected and analyzed for virus titers by immunofocus assay (FFU/ml) using the anti-NP MAb HT103. Data represent the means ± SDs of the results determined for triplicate wells. *, P < 0.05 by an unpaired two-tailed Student's t test (n = 3 per time point). (B) Analysis of protein expression levels in infected cells by Western blotting. A549 cells were mock infected (mock) or infected with WT NS or codon-deoptimized (NS1cd, NEPcd, and NScd) viruses at an MOI of 3, and NS1, NEP, and NP expression levels were analyzed (24 h postinfection) by Western blotting with specific antibodies. Actin was used as a loading control. The protein molecular sizes (in kDa) are indicated to the left. Western blots were quantified by densitometry using the software ImageJ (v1.46). The bands were normalized to the level of actin expression for protein quantitation and then to the level of viral NP expression for determination of viral infection. The expression of WT protein was considered 100% for comparison with the level of expression by the corresponding codon-deoptimized viruses.
In vivo characterization of codon-deoptimized viruses.
Due to the delayed growth in human A549 cells, we postulated that NScd and NS1cd viruses would be attenuated in vivo, as A549 cells more accurately reflect the target cells in the respiratory airway (59, 60). Thus, we compared the virulence of WT NS and codon-deoptimized viruses in mice. To ascertain whether the loss of NS1, NEP, or both NS1 and NEP expression impacts the course of an in vivo virus infection, groups of mice (n = 4) were inoculated i.n. with 10 or 100 PFU of WT NS, NS1cd, NEPcd, or NScd virus and monitored for 14 days for signs of illness, weight loss, and mortality. As expected, codon-deoptimized viruses showed levels of attenuation and pathogenicity different from those for WT NS viruses. Animals infected with 10 PFU of NScd or NS1cd virus lost less than 5% of their initial body weight (Fig. 6Ai), and all survived (Fig. 6Bi). In spite of the limited attenuation observed in vitro, mice infected with NEPcd virus showed less weight loss and mortality than mice infected with WT NS virus (Fig. 6Ai and Bi). In mice infected with 100 PFU, all mice infected with NScd virus survived, whereas only 50% of mice infected with NS1cd virus survived. In contrast, all mice infected with the same dose of NEPcd or WT NS virus succumbed to viral infection by day 8 (Fig. 6Aii and Bii). The MLD50 (56) for each virus was ∼300 PFU for NScd virus, ∼200 PFU for NS1cd virus, ∼20 PFU for NEPcd virus, and ∼10 PFU for WT NS virus (Table 1). We also evaluated the viral titers in lungs at days 3 and 6 postinfection (Fig. 6Ci and Cii). Animals infected with NS1cd virus and, more markedly, those infected with NScd virus showed significantly lower viral titers at day 3 postinfection than animals infected with NEPcd and WT NS viruses, regardless of whether 10 (Fig. 6Ci) or 100 (Fig. 6Cii) PFU was used. However, at day 6 postinfection, differences in viral titers were significant only with 10 PFU of NScd virus. Overall, viral titers correlated with the virus dose and the degree of morbidity and mortality, indicating that NS codon deoptimization of influenza virus results in viral attenuation in vivo that correlates with the amount of codon changes introduced in the viral segment (NScd > NS1cd > NEPcd > WT NS).
FIG 6.
Attenuation of codon-deoptimized viruses in mice. Female 6- to-8-week-old C57BL/6 mice were infected i.n. with 10 (Ai, Bi, and Ci) or 100 (Aii, Bii, and Cii) PFU of WT NS, NScd, NS1cd, or NEPcd virus and then monitored daily for 2 weeks for body weight loss (Ai and Aii) and survival (Bi and Bii). Mice that lost 25% of their body weight were sacrificed. Data represent the means ± SDs of the results determined for individual mice (n = 4). To evaluate viral lung replication (Ci and Cii), mice were sacrificed at 3 and 6 days postinfection and whole lungs were harvested, homogenized, and used to quantify viral titers by immunofocus assay (FFU/ml). Symbols represent data from individual mice. Bars, geometric mean lung virus titers; dotted lines, limit of detection (20 FFU/ml). Statistical analyses were conducted using the Mann-Whitney test. *, P <0.05 (n = 3).
TABLE 1.
Recombinant influenza virus MLD50s
| Virus NSa | MLD50 (no. of PFU/mouse) |
|---|---|
| WT | 10 |
| NScd | 316.2 |
| NS1cd | 215.4 |
| NEPcd | 21.54 |
Ten, 100, and 1,000 PFU (n = 4) of each recombinant WT NS virus and codon-deoptimized virus were inoculated i.n., and mortality was determined over 2 weeks.
NS codon-deoptimized influenza virus protects mice from homologous and heterologous lethal viral challenges.
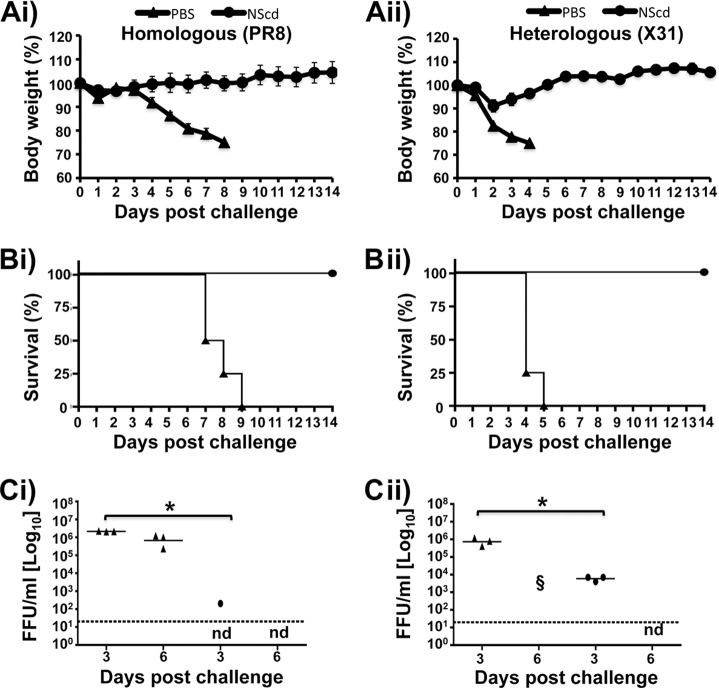
Given that NScd virus was attenuated in mice, we hypothesized that NScd virus could potentially be used as a vaccine. To evaluate this possibility, mice were vaccinated i.n. with 100 PFU of the NScd virus or mock vaccinated with PBS. Mice were then rested for 2 weeks to clear any residual vaccine virus. At 14 days postvaccination, either homologous or heterologous protection was evaluated by challenging vaccinated mice with 10 (Fig. 7) or 100 (Fig. 8) MLD50s of PR8 (H1N1) or X31 (H3N2) virus. All eight segments of PR8 virus should carry proteins identical to NScd, whereas X31 virus carries the same six internal segments as PR8 virus, but the HA and NA segments are derived from A/Hong Kong/1/1968 (H3N2) virus. None of vaccinated animals showed any weight loss (Fig. 7Ai and Aii), and all vaccinated animals survived both homologous and heterologous challenge with 10 LD50s (Fig. 7Bi and Bii), while all mock-vaccinated mice drastically lost weight and died after challenge. In addition, virus replication levels in the lungs from vaccinated mice were significantly lower than those in the lungs from mock-vaccinated mice at day 3 following homologous and heterologous challenge (Fig. 7Ci and Cii).
FIG 7.
Protection efficacy of virus with codon-deoptimized NS. Female 6- to-8-week-old C57BL/6 mice were immunized with 100 PFU of the NScd virus or mock vaccinated with PBS. At 2 weeks postvaccination, animals were challenged with 10 LD50s of PR8 (H1N1) virus (Ai, Bi, and Ci) or X31 (H3N2) virus (Aii, Bii, and Cii) to evaluate homologous and heterologous protection, respectively. Relative body weight (Ai and Aii) and survival (Bi and Bii) were evaluated for 14 days after the challenge. Mice that lost 25% of their body weight were euthanized. Data represent the means ± SDs of the results determined for individual mice (n = 4). To evaluate viral lung replication at 3 and 6 days postchallenge (Ci and Cii), mouse lungs were prepared as described in the legend to Fig. 5 and used to quantify viral titers by immunofocus assay (FFU/ml). Symbols represent data from individual mice. Bars, geometric mean lung virus titers; dotted line, limit of detection (20 FFU/ml). Statistical analyses were conducted using the Mann-Whitney test. *, P < 0.05 (n = 3). Statistics could not be calculated for day 6, because most mock-vaccinated mice died from infection and vaccinated mice showed no signs of virus replication. §, dead animals; nd, not determined.
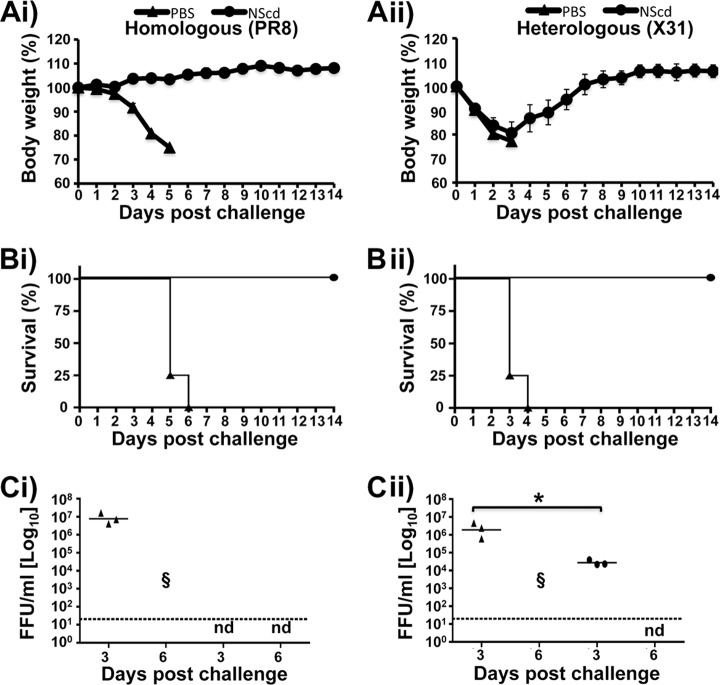
FIG 8.
NScd-immunized mice are protected after a lethal challenge (100 LD50s) of wild-type viruses. Female 6- to-8-week-old C57BL/6 mice were immunized with 100 PFU of NScd or mock vaccinated with PBS. Fourteen days after the immunization, animals were challenged with 100 LD50s of PR8 (H1N1) virus (Ai, Bi, and Ci) or X31 (H3N2) virus (Aii, Bii, and Cii) to evaluate homologous and heterologous protection, respectively. The relative body weight (Ai and Aii) and survival (Bi and Bii) were evaluated daily for 2 weeks, and mice that lost 25% of their body weight were sacrificed. Data represent the means ± SDs of the results determined for individual mice (n = 4). To evaluate viral lung replication at days 3 and 6 postchallenge (Ci and Cii), an immunofocus assay was performed as described in the legend to Fig. 5 (FFU/ml). Symbols represent data from individual mice. Bars, geometric mean lung virus titers; dotted lines, limit of detection (20 FFU/ml). Statistical analyses were conducted using the Mann-Whitney test. *, P <0.05 (n = 3).
When the animals were challenged with 100 LD50s, all immunized mice survived challenge with either PR8 or X31 virus. In contrast, all mock-vaccinated mice showed a progressive loss of body weight (Fig. 8Ai and Aii) and succumbed to viral infection (Fig. 8Bi and Bii). Although immunized animals challenged with X31 virus suffered severe body weight loss (20%), all recovered and survived. To determine the inhibition of viral replication in lungs, viral titers were analyzed at days 3 and 6 postchallenge (Fig. 8Ci and Cii). Mock-vaccinated mice showed high viral titers at day 3, while immunized mice showed no detectable virus with homologous challenge (Fig. 8Ci) or a 100-fold decrease with heterologous challenge (Fig. 8Cii). Strikingly, no detectable levels of challenge virus were observed in vaccinated animals at day 6 postchallenge with homologous or heterologous viruses, in direct contrast to the results for mock-vaccinated animals, in which all animals succumbed to challenge (Fig. 8Ci and Cii). These results indicate that NScd virus can induce protective immune responses against both homologous (PR8 virus) and heterologous (X31 virus) lethal challenges.
Induction of humoral responses.
Humoral immune responses induced by vaccination with 0, 10, or 100 PFU of NScd virus were evaluated in sera collected 2 weeks after immunization. The responses were characterized by ELISA using cell lysates from mock-, PR8 virus-, or X31 virus-infected MDCK cells (Fig. 9A and B) or using H1 and H3 purified proteins (Fig. 9C and D). As expected, mice responded to NScd vaccination by eliciting higher serum IgG titers against PR8 virus antigens (Fig. 9A) and H1 protein (Fig. 9C) but not H3 protein (Fig. 9D). Antibodies against X31 virus antigens were, however, detected (Fig. 9B), although the levels were less than those in convalescent-phase sera from animals infected with X31 virus, presumably due to the lack of anti-H3 antibodies induced by NScd (H1) vaccination. In addition, protective HAI titers against PR8 virus but not X31 virus were observed in NScd-vaccinated animals (Table 2). These data suggest that vaccination with NScd virus, besides its humoral activation, likely induces a cellular response that is responsible for the observed heterologous protection against X31 virus. Overall, this provides evidence that immunization with NScd safely confers protection from both homologous and heterologous virus infections.
FIG 9.
Humoral responses to NS codon-deoptimized vaccination. Female 6- to-8-week-old C56BL/6 mice were mock infected with PBS or infected with 10 or 100 PFU of NScd virus. At 14 days postinfection, mice were bled and the sera were collected and evaluated by ELISA for IgG antibodies against total influenza virus protein using cell extracts of MDCK cells infected with PR8 virus (A) or X31 virus (B) or against recombinant protein H1 (C) or H3 (D). Serum obtained from a mouse infected with X31 virus was used as an internal control. OD, optical density. Data represent the means ± SDs of the results for 5 individual mice.
TABLE 2.
Immunogenicity of NS codon-deoptimized virusa
| Immunization and dose (PFU) | Mean (SD) HAI titer |
|
|---|---|---|
| PR8 virus | X31 virus | |
| PBS | ≤16 (ND) | ≤16 (ND) |
| NScd | ||
| 10 | 49 (41.72) | ≤16 (ND) |
| 100 | 147 (57.24) | ≤16 (ND) |
Data were calculated from 5 immunized or 2 mock-immunized individual animals. ND, not determined.
Reduced interferon antagonism may contribute to attenuation of NS codon-deoptimized virus.
One of the main functions of the viral NS1 protein is to counteract the IFN-I response during infection (18, 19). Thus, we sought to test whether attenuation of NScd virus correlated with a diminished antagonism of IFN-I. To that end, tissue culture supernatants of A549 cells infected with either NS or NScd viruses were collected at various times postinfection (0, 6, 12, 24, and 48 h). Comparable levels of infection in A549 cells were verified by NP expression (Fig. 10B), and the levels of secreted IFN-I were then analyzed using two complementary bioassays (Fig. 10A). In the virus-based assay, inhibition of rNDV-GFP infection is dependent on IFN-I pretreatment (46, 54), which was recapitulated by the dose-dependent addition of recombinant IFN-I (Fig. 10C and D). Although tissue culture supernatants from cells infected with WT NS at 24 and 48 h postinfection led to the modest inhibition of rNDV-GFP, a significant decrease of rNDV-GFP infection was observed when cells were pretreated with supernatants from cells infected with NScd virus, as determined by GFP expression using a fluorescence microscope (Fig. 10C) or a GFP microplate reader (Fig. 10D). In the cell-based assay, reporter gene (mRFP and Fluc) expression is driven by the ISRE promoter, which is activated with the presence of IFN-I in tissue culture supernatants (42). The assay was validated by detection of a dose-dependent increase in reporter gene expression upon exposure of cells to recombinant IFN-I at various concentrations (Fig. 10E and F). Similar to the virus-based assay, supernatants from cells infected with NScd virus collected at 24 and 48 h postinfection induced higher levels of reporter gene expression than supernatants from WT NS virus-infected cells, as determined by mRFP (Fig. 10E) or luciferase (Fig. 10F) expression. Interestingly, tissue culture supernatants from NScd virus-infected cells contained less IFN-I at 48 h than at 24 h postinfection, probably due to the accumulation of NS1 protein. Altogether, these results suggest that the mechanism of NScd PR8 virus attenuation in vitro is due, at least in part, to reduced IFN-I antagonism, which may also account for its attenuation in vivo.
FIG 10.
Differences in induction of IFN-I with wild-type virus NS and viruses with codon-deoptimized NS. (A) Schematic representation of the IFN-I bioassays. Human A549 cells were infected with WT NS or NScd PR8 viruses, and tissue culture supernatants (TCS) were collected at the indicated times postinfection (0, 6, 12, 24, and 48 h) and UV inactivated before they were used to treat Vero or 293T ISRE mRFP/ISRE Fluc cells. (B) Indirect immunofluorescence. WT NS and NScd PR8 virus infections of A549 cells were evaluated at 12 h postinfection using the anti-NP MAb HT103. Mock-infected cells were included as a negative control. (C and D) Analysis of IFN-I induction using a virus-based bioassay. Vero cells were treated with UV-inactivated tissue culture supernatants collected from WT NS and NScd PR8 virus-infected A549 cells at different times postinfection (0, 6, 12, 24, and 48 h) and infected with rNDV-GFP (MOI, 1). The levels of IFN-I were evaluated by determination of the ability to inhibit rNDV-GFP infection under a fluorescence microscope (C) and quantified by using a microplate reader (D). Data are represented in relative GFP units, where mock-treated Vero cells infected with rNDV-GFP were considered to have a value of 100%. Vero cells treated with 1, 10, and 100 U of IFN-I were used as internal controls. (E and F) Analysis of IFN-I using a cell-based assay. Human 293T cells expressing mRFP and firefly luciferase under the control of an ISRE promoter were treated with the UV-inactivated tissue culture supernatants from WT NS and NScd PR8 virus-infected A549 cells. Activation of the ISRE promoter was evaluated by determination of mRFP expression under a fluorescence microscope (E) and quantified by determination of firefly luciferase activity (F). Representative fields are shown in panels C and E. Data represented in panels D and F show the means ± SDs of the results determined for triplicate wells. *, P <0.05. hpi, hours postinfection; RLU, relative light units.
DISCUSSION
Using plasmid-based reverse genetics techniques and de novo cDNA synthesis, we have engineered recombinant influenza PR8 viruses carrying a synonymous codon-deoptimized NS segment. Recombinant viruses were designed under the hypothesis that introduction of many underrepresented codons into the NS gene should lead to a reduction of protein expression and thus an attenuated phenotype. Viruses harboring deoptimized codons in the NS gene (NScd viruses) or the ORFs for NS1 or NEP (NS1cd and NEPcd viruses, respectively) were evaluated in vitro and in vivo and showed levels of attenuation that correlated with the amount of codon changes introduced in the viral segment. Additionally, the most attenuated NScd virus demonstrated applicability as a safe, live attenuated vaccine that produced immunogenicity and provided protection against both homologous and heterologous influenza virus lethal challenges in mice.
In MDCK cells, protein expression by NScd and NS1cd viruses was decreased in relation to that by WT virus, while protein expression by NEPcd virus was only slightly decreased. These results were expected because only 4.25% of the NEP ORF codons were altered (Fig. 1). Generally, an optimal coadaptation of codon usage and tRNA concentration evolves within a species to obtain a highly efficient rate of translation (61–63). Our data suggest that the phenotypes of codon-deoptimized viruses may be determined in part by the rate of gene translation, as also shown by a reduction in protein expression in transiently transfected human 293T cells (Fig. 2). However, other mechanistic explanations for the attenuation of codon-deoptimized viruses may also be possible. First, polypeptide elongation is determined in part by the concentration of aminoacyl-tRNA; therefore, the dependence of an mRNA on many underrepresented tRNAs could increase the time of translation (64). This would result in ribosomal stalling, premature dissociation of the translation complex from the RNA, and subsequent protein truncation and degradation. Second, changes in the mRNA secondary structure could contribute to the observed decreases in protein synthesis because the mRNA secondary structure is an important element for codon selection (65, 66).
In this study, codon deoptimization of the NS gene of influenza PR8 virus revealed interesting insights into the biology of the virus. Virus replication in vitro was not affected in MDCK cells (Fig. 4), despite protein expression changes, determined by Western blotting, in transfected human 293T cells (Fig. 2) and infected MDCK cells (Fig. 4). Unlike in MDCK cells, the replication capabilities of codon-deoptimized influenza viruses were altered in human A549 cells (compare Fig. 4A and 5A), perhaps due to signaling pathways in human A549 cells that are different from those in MDCK cells or absent in MDCK cells (59, 60). For instance, since NS1 is an IFN-I antagonist protein, the differences in replication rates observed could correspond with species-specific IFN-I production or be correlated with IFN-I-stimulated gene (ISG) expression (20, 21, 54). Also, differences in preferred tRNA usage between canine and human cells or kidney and lung tissues could contribute to the differences in growth kinetics of the codon-deoptimized viruses.
Codon deoptimization had no observable effect on NS splicing in virus-infected MDCK cells, as assessed using semiquantitative PCR (Fig. 3). This is important because inefficient splicing of influenza A virus NS controls the accumulation of NEP, which could serve as an important switch from viral transcription to replication (17, 33). In accord with the results reported here, Robb et al. showed that similar levels of spliced NEP mRNA were produced in the presence and absence of the NS1 protein (17). In contrast, Garaigorta and Ortin showed that segment 8 splicing is controlled, in part, by the viral NS1 protein itself, since NS1 appears to inhibit the splicing and nuclear export of NS1 mRNA transcripts within infected cells (67). However, in our system, after decreasing the expression of NS1 by codon deoptimization, there was no diminution of either NEP protein or mRNA.
Recently, Chua et al. showed that limiting NS1 accumulation does not impact virus replication in vitro (68), which is similar to our findings (Fig. 4). The authors also evaluated virus replication in model systems that either decreased or enhanced NEP levels and showed that deregulation of NEP abrogates viral replication, likely due to NEP-mediated vRNP export. In our system, infection of MDCK and A549 cells with NEPcd virus showed replication kinetics similar to that obtained by infection with WT NS virus, suggesting that the reduction of NEP levels by codon deoptimization was not enough to significantly disrupt viral replication. These observations correlated with the results of experiments performed in vivo, where NEPcd virus produced levels of morbidity similar to those produced by WT NS virus, whereas NScd and NS1cd viruses were notably attenuated. However, since the NScd virus was slightly more attenuated than the NS1cd virus in vitro and in vivo, it remains possible that codon deoptimization of NEP may modestly contribute to the higher attenuation observed with NScd virus.
The in vitro results translated to mice, where codon-deoptimized NS and NS1 viruses also showed attenuated replication compared to the replication of WT NS virus (Fig. 6). Mice infected with NEPcd virus showed body weight loss and survival similar to those of mice infected with WT NS virus. Notably, both NS1cd viruses and, mainly, NScd viruses were significantly attenuated. NScd virus-infected mice showed minimal body weight loss and 100% survival when animals were infected with both 10 and 100 PFU of virus. Furthermore, the virus titers recovered from the lungs of NS1cd and NScd virus-inoculated mice were significantly lower than those recovered from the lungs of NEPcd or WT NS virus-infected mice, suggesting a more rapid viral clearance and a shorter duration of viral shedding.
Vaccine potential was further evaluated by homologous or heterologous challenge of immunized mice (Fig. 7 and 8). We found that although the NScd virus is attenuated in mice, the virus can still induce immune responses capable of providing significant protection against homologous and heterologous viral challenges. We hypothesize that the induction of neutralizing antibodies protects against homologous challenge (Fig. 9), and the induction of a T cell response directed against conserved internal proteins might likely protect against heterologous challenge, although this hypothesis was not directly evaluated.
Among the first battles between the host and the virus, IFN-Is play an important role in controlling viral infection (69). Therefore, the attenuated phenotype observed with NScd virus could be the consequence of the reduced production of the IFN-I antagonist NS1, which would hasten virus clearance from the respiratory tract and lungs due to increased IFN-I antiviral activity and activation of the adaptive immune response (22, 23, 25–27). This hypothesis was assessed using two complementary bioassays aimed at the evaluation of differences in IFN-I production during WT NS or NScd viral infections (Fig. 10). Our results indicate a correlation between viral attenuation in vivo and the larger amounts of IFN-I produced during NScd viral infection than during WT NS viral infection, suggesting that lower NS1 protein expression levels at early times postinfection result in higher levels of IFN-I production and in virus attenuation in vivo. However, our conclusions are based on in vitro experiments, and in vivo approaches are needed to confirm the mechanism of viral attenuation in an animal model. Additionally, we cannot rule out the possibility that differences in NEP expression levels could also contribute to viral attenuation.
Successful vaccine candidates must show attenuation in the host while retaining immunogenicity and must also grow well in manufacturing-suitable tissue culture platforms (70, 71). Importantly, we showed that NS codon-deoptimized viruses were attenuated in mice but replicated efficiently in the FDA-approved MDCK cell line used for human vaccine production (41). Therefore, our viruses may prove useful as safe alternatives in the generation of LAIV. Because the influenza virus NS1 protein can antagonize IFN-I, LAIVs based on mutated or truncated NS1 proteins have been previously proposed (24–26, 28, 72, 73). However, this is the first time that influenza viruses have been attenuated due to deoptimized codon use in the influenza viral NS segment, resulting in decreased levels of NS1 protein expression. Our approach has several advantages over previously reported systems that rely on mutated or truncated NS1 for virus attenuation. In recombinant viruses containing mutant NS1, the possibility that the virus will revert to the WT is high because the number of changes in the NS1 sequence is limited. However, in our codon-deoptimized NS virus, the possibility of reversion is diminished due to the large number of modifications introduced. Moreover, unlike other systems, we have also included changes in NEP. On the other hand, compared to deltaNS1 virus (24–28, 35, 36, 72, 73), our codon-deoptimized NS virus is able to replicate, bringing an optimal balance between viral attenuation and immunogenicity, similar to the situation observed with current LAIV approaches.
In summary, we have shown that vaccination with a single dose of the NScd LAIV can induce protective homo- and heterotypic immunity in mice. These results suggest that codon-deoptimized influenza viruses could be considered for use alone or in combination with current and/or experimental vaccine approaches in the development of safe, immunogenic, and protective vaccine approaches against influenza (39, 40, 74). Live attenuated vaccine viruses engineered by codon deoptimization express the intact viral repertoire of antigenic sites because the amino acid sequence is conserved, inducing both cellular and humoral immunity against all viral immunogenic epitopes. To date, two similar approaches have been used to generate recombinant attenuated influenza viruses using codon permutations to modify the expression of different genes involved in viral replication or viral spread (39, 40). In these studies, the authors made use of codon pair bias to engineer codon-deoptimized viruses by creating a suboptimal organization of pairs of codons, instead of modifying each codon individually. Unlike these prior studies, where genes involved in the synthesis of viral RNA and the spread of virus were modified, we have deoptimized the coding sequence of the NS gene involved in counteracting the antiviral host response. Although similar approaches modifying a gene involved in innate antagonism have not yet been described, our results correlate well with those of previous studies where viral IFN-I antagonists were affected (35, 37, 46, 75). Moreover, our studies provide the first evidence that a viral segment undergoing splicing could be used for viral attenuation and that the attenuation induced by codon deoptimization is most probably regulated by the number of nucleotide changes introduced in the viral genome. In fact, recombinant virus containing several codon-deoptimized genes involved in different steps of the virus life cycle could be used to generate controlled and novel live attenuated vaccines.
ACKNOWLEDGMENTS
We thank Adolfo Garcia-Sastre for providing the influenza PR8 virus reverse genetics plasmid-based techniques, B. Paige Lawrence for the X31 virus, Tom Moran for the NP monoclonal antibody (HT103), and Peter Palese for providing the NEP polyclonal antibody and pDZ ambisense plasmid. We thank BEI Resources for the influenza virus H1 (NR-19240) and H3 (NR-15168) purified proteins.
A.N. is a recipient of the University of Rochester Vaccine Fellowship (2013) and a Centers for Excellence in Influenza Research & Surveillance (CEIRS) intercollaborative training grant (2013). S.F.B. is currently supported by a University of Rochester immunology training grant (AI 007285-26). E.O.-R. is supported by a Postdoctoral Fellowship for Diversity and Academic Excellence from the Office for Faculty Development & Diversity at the University of Rochester. Research in the L.M.-S. laboratory is funded by NIH grants RO1 AI077719 and R03AI099681-01A1, the NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS HHSN266200700008C), and the University of Rochester Center for Biodefense and Immune Modeling (CBIM HHSN272201000055C).
Footnotes
Published ahead of print 25 June 2014
REFERENCES
- 1.Wright PF, Neumann G, Kawaoka Y. 2007. Orthomyxoviruses. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Strauss SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Luckhaupt SE, Sweeney MH, Funk R, Calvert GM, Nowell M, D'Mello T, Reingold A, Meek J, Yousey-Hindes K, Arnold KE, Ryan P, Lynfield R, Morin C, Baumbach J, Zansky S, Bennett NM, Thomas A, Schaffner W, Jones T. 2012. Influenza-associated hospitalizations by industry, 2009-10 influenza season, United States. Emerg. Infect. Dis. 18:556–562. 10.3201/eid1804.110337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. 2004. Influenza-associated hospitalizations in the United States. JAMA 292:1333–1340. 10.1001/jama.292.11.1333 [DOI] [PubMed] [Google Scholar]
- 4.Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. 2007. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25:5086–5096. 10.1016/j.vaccine.2007.03.046 [DOI] [PubMed] [Google Scholar]
- 5.Li KS, Guan Y, Wang J, Smith GJ, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, Estoepangestie AT, Chaisingh A, Auewarakul P, Long HT, Hanh NT, Webby RJ, Poon LL, Chen H, Shortridge KF, Yuen KY, Webster RG, Peiris JS. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209–213. 10.1038/nature02746 [DOI] [PubMed] [Google Scholar]
- 6.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. 10.1126/science.1176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. 10.1126/science.1177238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch JP, III, Walsh EE. 2007. Influenza: evolving strategies in treatment and prevention. Semin. Respir. Crit. Care Med. 28:144–158. 10.1055/s-2007-976487 [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2012. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP)—United States, 2012-13 influenza season. MMWR Morb. Mortal. Wkly. Rep. 61:613–618 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm6132a3.htm [PubMed] [Google Scholar]
- 10.Osterholm MT, Kelley NS, Sommer A, Belongia EA. 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis. 12:36–44. 10.1016/S1473-3099(11)70295-X [DOI] [PubMed] [Google Scholar]
- 11.Belshe RB, Newman FK, Wilkins K, Graham IL, Babusis E, Ewell M, Frey SE. 2007. Comparative immunogenicity of trivalent influenza vaccine administered by intradermal or intramuscular route in healthy adults. Vaccine 25:6755–6763. 10.1016/j.vaccine.2007.06.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 2010. Licensure of a high-dose inactivated influenza vaccine for persons aged ≥ 65 years (Fluzone high-dose) and guidance for use—United States, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:485–486 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5916a2.htm [PubMed] [Google Scholar]
- 13.Rimmelzwaan GF, Fouchier RA, Osterhaus AD. 2007. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr. Opin. Biotechnol. 18:529–536. 10.1016/j.copbio.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 14.Cox MM, Patriarca PA, Treanor J. 2008. FluBlok, a recombinant hemagglutinin influenza vaccine. Influenza Other Respir. Viruses 2:211–219. 10.1111/j.1750-2659.2008.00053.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Villiers PJ, Steele AD, Hiemstra LA, Rappaport R, Dunning AJ, Gruber WC, Forrest BD. 2009. Efficacy and safety of a live attenuated influenza vaccine in adults 60 years of age and older. Vaccine 28:228–234. 10.1016/j.vaccine.2009.09.092 [DOI] [PubMed] [Google Scholar]
- 16.Lamb RA, Lai CJ. 1980. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell 21:475–485. 10.1016/0092-8674(80)90484-5 [DOI] [PubMed] [Google Scholar]
- 17.Robb NC, Jackson D, Vreede FT, Fodor E. 2010. Splicing of influenza A virus NS1 mRNA is independent of the viral NS1 protein. J. Gen. Virol. 91:2331–2340. 10.1099/vir.0.022004-0 [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. 10.1006/viro.1998.9508 [DOI] [PubMed] [Google Scholar]
- 19.Steidle S, Martinez-Sobrido L, Mordstein M, Lienenklaus S, Garcia-Sastre A, Staheli P, Kochs G. 2010. Glycine 184 in nonstructural protein NS1 determines the virulence of influenza A virus strain PR8 without affecting the host interferon response. J. Virol. 84:12761–12770. 10.1128/JVI.00701-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr, Garcia-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514–524. 10.1128/JVI.01265-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989–7996. 10.1128/JVI.74.17.7989-7996.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376. 10.1099/vir.0.2008/004606-0 [DOI] [PubMed] [Google Scholar]
- 23.Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang X, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG, Garcia-Sastre A. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. U. S. A. 99:10736–10741. 10.1073/pnas.112338099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baskin CR, Bielefeldt-Ohmann H, Garcia-Sastre A, Tumpey TM, Van Hoeven N, Carter VS, Thomas MJ, Proll S, Solorzano A, Billharz R, Fornek JL, Thomas S, Chen CH, Clark EA, Murali-Krishna K, Katze MG. 2007. Functional genomic and serological analysis of the protective immune response resulting from vaccination of macaques with an NS1-truncated influenza virus. J. Virol. 81:11817–11827. 10.1128/JVI.00590-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falcon AM, Fernandez-Sesma A, Nakaya Y, Moran TM, Ortin J, Garcia-Sastre A. 2005. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J. Gen. Virol. 86:2817–2821. 10.1099/vir.0.80991-0 [DOI] [PubMed] [Google Scholar]
- 26.Ferko B, Stasakova J, Romanova J, Kittel C, Sereinig S, Katinger H, Egorov A. 2004. Immunogenicity and protection efficacy of replication-deficient influenza A viruses with altered NS1 genes. J. Virol. 78:13037–13045. 10.1128/JVI.78.23.13037-13045.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solorzano A, Webby RJ, Lager KM, Janke BH, Garcia-Sastre A, Richt JA. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535–7543. 10.1128/JVI.79.12.7535-7543.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talon J, Salvatore M, O'Neill RE, Nakaya Y, Zheng H, Muster T, Garcia-Sastre A, Palese P. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. U. S. A. 97:4309–4314. 10.1073/pnas.070525997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann G, Hughes MT, Kawaoka Y. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751–6758. 10.1093/emboj/19.24.6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bullido R, Gomez-Puertas P, Saiz MJ, Portela A. 2001. Influenza A virus NEP (NS2 protein) downregulates RNA synthesis of model template RNAs. J. Virol. 75:4912–4917. 10.1128/JVI.75.10.4912-4917.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Neill RE, Talon J, Palese P. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288–296. 10.1093/emboj/17.1.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paterson D, Fodor E. 2012. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog. 8:e1003019. 10.1371/journal.ppat.1003019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, Garcia-Sastre A, tenOever BR. 2010. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl. Acad. Sci. U. S. A. 107:11525–11530. 10.1073/pnas.1001984107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerra S, Abaitua F, Martinez-Sobrido L, Esteban M, Garcia-Sastre A, Rodriguez D. 2011. Host-range restriction of vaccinia virus E3L deletion mutant can be overcome in vitro, but not in vivo, by expression of the influenza virus NS1 protein. PLoS One 6:e28677. 10.1371/journal.pone.0028677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hai R, Martinez-Sobrido L, Fraser KA, Ayllon J, Garcia-Sastre A, Palese P. 2008. Influenza B virus NS1-truncated mutants: live-attenuated vaccine approach. J. Virol. 82:10580–10590. 10.1128/JVI.01213-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011–7021. 10.1128/JVI.02581-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kochs G, Martinez-Sobrido L, Lienenklaus S, Weiss S, Garcia-Sastre A, Staeheli P. 2009. Strong interferon-inducing capacity of a highly virulent variant of influenza A virus strain PR8 with deletions in the NS1 gene. J. Gen. Virol. 90:2990–2994. 10.1099/vir.0.015727-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura Y, Gojobori T, Ikemura T. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. 10.1093/nar/28.1.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller S, Coleman JR, Papamichail D, Ward CB, Nimnual A, Futcher B, Skiena S, Wimmer E. 2010. Live attenuated influenza virus vaccines by computer-aided rational design. Nat. Biotechnol. 28:723–726. 10.1038/nbt.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang C, Skiena S, Futcher B, Mueller S, Wimmer E. 2013. Deliberate reduction of hemagglutinin and neuraminidase expression of influenza virus leads to an ultraprotective live vaccine in mice. Proc. Natl. Acad. Sci. U. S. A. 110:9481–9486. 10.1073/pnas.1307473110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun W. 2012. Approval letter—FluMist® quadrivalent (BL 125020/1668). U.S. Food and Drug Administration, Rockville, MD [Google Scholar]
- 42.Nguyen DN, Kim P, Martinez-Sobrido L, Beitzel B, Garcia-Sastre A, Langer R, Anderson DG. 2009. A novel high-throughput cell-based method for integrated quantification of type I interferons and in vitro screening of immunostimulatory RNA drug delivery. Biotechnol. Bioeng. 103:664–675. 10.1002/bit.22312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schickli JH, Flandorfer A, Nakaya T, Martinez-Sobrido L, Garcia-Sastre A, Palese P. 2001. Plasmid-only rescue of influenza A virus vaccine candidates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1965–1973. 10.1098/rstb.2001.0979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kilbourne ED. 1969. Future influenza vaccines and the use of genetic recombinants. Bull. World Health Organ. 41:643–645 [PMC free article] [PubMed] [Google Scholar]
- 45.Baker SF, Guo H, Albrecht RA, Garcia-Sastre A, Topham DJ, Martinez-Sobrido L. 2013. Protection against lethal influenza with a viral mimic. J. Virol. 87:8591–8605. 10.1128/JVI.01081-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Sobrido L, Zuniga EI, Rosario D, Garcia-Sastre A, de la Torre JC. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 80:9192–9199. 10.1128/JVI.00555-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501–1511. 10.1128/JVI.77.2.1501-1511.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlivan M, Zamarin D, Garcia-Sastre A, Cullinane A, Chambers T, Palese P. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431–8439. 10.1128/JVI.79.13.8431-8439.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozawa M, Kawaoka Y. 2011. Taming influenza viruses. Virus Res. 162:8–11. 10.1016/j.virusres.2011.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. 10.1016/0378-1119(91)90434-D [DOI] [PubMed] [Google Scholar]
- 51.Martinez-Sobrido L, Cadagan R, Steel J, Basler CF, Palese P, Moran TM, Garcia-Sastre A. 2010. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 84:2157–2163. 10.1128/JVI.01433-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez-Sobrido L, Garcia-Sastre A. 2010. Generation of recombinant influenza virus from plasmid DNA. J. Vis. Exp. 2010:2057. 10.3791/2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ortiz-Riano E, Cheng BY, de la Torre JC, Martinez-Sobrido L. 2012. Self-association of lymphocytic choriomeningitis virus nucleoprotein is mediated by its N-terminal region and is not required for its anti-interferon function. J. Virol. 86:3307–3317. 10.1128/JVI.05503-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Donelan NR, Basler CF, Garcia-Sastre A. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257–13266. 10.1128/JVI.77.24.13257-13266.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]
- 56.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 57.Guo H, Santiago F, Lambert K, Takimoto T, Topham DJ. 2011. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. J. Virol. 85:448–455. 10.1128/JVI.01812-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steel J, Lowen AC, Pena L, Angel M, Solorzano A, Albrecht R, Perez DR, Garcia-Sastre A, Palese P. 2009. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J. Virol. 83:1742–1753. 10.1128/JVI.01920-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dove BK, Surtees R, Bean TJ, Munday D, Wise HM, Digard P, Carroll MW, Ajuh P, Barr JN, Hiscox JA. 2012. A quantitative proteomic analysis of lung epithelial (A549) cells infected with 2009 pandemic influenza A virus using stable isotope labelling with amino acids in cell culture. Proteomics 12:1431–1436. 10.1002/pmic.201100470 [DOI] [PubMed] [Google Scholar]
- 60.Sutejo R, Yeo DS, Myaing MZ, Hui C, Xia J, Ko D, Cheung PC, Tan BH, Sugrue RJ. 2012. Activation of type I and III interferon signalling pathways occurs in lung epithelial cells infected with low pathogenic avian influenza viruses. PLoS One 7:e33732. 10.1371/journal.pone.0033732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chamary JV, Hurst LD. 2005. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 6:R75. 10.1186/gb-2005-6-9-r75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. 2001. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 53:290–298. 10.1007/s002390010219 [DOI] [PubMed] [Google Scholar]
- 63.Lavner Y, Kotlar D. 2005. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene 345:127–138. 10.1016/j.gene.2004.11.035 [DOI] [PubMed] [Google Scholar]
- 64.Gustafsson C, Govindarajan S, Minshull J. 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22:346–353. 10.1016/j.tibtech.2004.04.006 [DOI] [PubMed] [Google Scholar]
- 65.Kozak M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1–34. 10.1016/S0378-1119(02)01056-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shabalina SA, Ogurtsov AY, Spiridonov NA. 2006. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 34:2428–2437. 10.1093/nar/gkl287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garaigorta U, Ortin J. 2007. Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res. 35:4573–4582. 10.1093/nar/gkm230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chua MA, Schmid S, Perez JT, Langlois RA, Tenoever BR. 2013. Influenza A virus utilizes suboptimal splicing to coordinate the timing of infection. Cell Rep. 3:23–29. 10.1016/j.celrep.2012.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47. 10.1099/vir.0.83391-0 [DOI] [PubMed] [Google Scholar]
- 70.Hussain AI, Cordeiro M, Sevilla E, Liu J. 2010. Comparison of egg and high yielding MDCK cell-derived live attenuated influenza virus for commercial production of trivalent influenza vaccine: in vitro cell susceptibility and influenza virus replication kinetics in permissive and semi-permissive cells. Vaccine 28:3848–3855. 10.1016/j.vaccine.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pica N, Palese P. 2013. Toward a universal influenza virus vaccine: prospects and challenges. Annu. Rev. Med. 64:189–202. 10.1146/annurev-med-120611-145115 [DOI] [PubMed] [Google Scholar]
- 72.Richt JA, Lekcharoensuk P, Lager KM, Vincent AL, Loiacono CM, Janke BH, Wu WH, Yoon KJ, Webby RJ, Solorzano A, Garcia-Sastre A. 2006. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J. Virol. 80:11009–11018. 10.1128/JVI.00787-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vincent AL, Ma W, Lager KM, Janke BH, Webby RJ, Garcia-Sastre A, Richt JA. 2007. Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 25:7999–8009. 10.1016/j.vaccine.2007.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wimmer E, Mueller S, Tumpey TM, Taubenberger JK. 2009. Synthetic viruses: a new opportunity to understand and prevent viral disease. Nat. Biotechnol. 27:1163–1172. 10.1038/nbt.1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Versteeg GA, Garcia-Sastre A. 2010. Viral tricks to grid-lock the type I interferon system. Curr. Opin. Microbiol. 13:508–516. 10.1016/j.mib.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]