Abstract
Myosins are a large superfamily of actin-dependent molecule motors that carry out many functions in cells. Some myosins are cargo carriers that move processively along actin which means that a single molecule of myosin can take many ATP-dependent steps on actin per initial encounter. Other myosins are designed to work in large ensembles such as myosin thick filaments. In vitro motility assays are a powerful method for studying the function of myosins. These assays in general use small amounts of protein, are simple to implement, and can be done on microscopes commonly found in many laboratories. There are two basic versions of the assay which involve different geometries. In the sliding actin in vitro motility assay, myosin molecules are bound to a coverslip surface in a simply constructed microscopic flow chamber. Fluorescently labeled actin filaments are added to the flow chamber in the presence of ATP, and the movement of these actin filaments powered by the surface-bound myosins is observed. This assay has been used widely for a variety of myosins including both processive and nonprocessive ones. From this assay, one can easily measure the rate at which myosin is translocating actin. The single-molecule motility assay uses an inverted geometry compared to the sliding actin in vitro motility assay. It is most useful for processive myosins. Here, actin filaments are affixed to the coverslip surface. Fluorescently labeled single molecules of myosins (usually ones with processive kinetics) are introduced, and the movement of single molecules along the actin filaments is observed. This assay typically uses total internal reflection fluorescent (TIRF) microscopy to reduce the background signal arising from myosins in solution. From this assay, one can measure the velocity of movement, the frequency of movement, and the run length. If sufficient photons can be collected, one can use Gaussian fitting of the point spread function to determine the position of the labeled myosin to within a few nanometers which allows for measurement of the step size and the stepping kinetics. Together, these two assays are powerful tools to elucidate myosin function.
Keywords: In vitro motility, Myosin, Actin, Fluorescence, TIRF
9.1 Introduction
Myosins are a superfamily of molecular motors that use the energy from ATP to interact with actin filaments in a variety of manners in muscle or nonmuscle cells [1]. The classic example of myosin function is its role in skeletal muscle fibers where myosin forms thick filaments which are interdigitated with actin filaments in the almost crystalline lattice of the sarcomere. These myosins are members of class 2, but now more than 35 classes of myosins present throughout phylogeny have been defined which are evolutionarily adapted to perform a plethora of intracellular tasks [2]. There are two simplistic methods of classifying members of the myosin superfamily. One historic classification system terms all class 2 myosins to be “conventional” and all other myosin “unconventional.” Another functional classification system of the myosin superfamily divides the members into two categories based on their duty ratios. The “duty ratio” is defined as the fraction of time myosin is bound tightly to actin during a cycle of ATP hydrolysis. Low-duty ratio myosins work as large ensembles and only interact with actin strongly for a small percentage of their kinetic cycles, examples of which include skeletal muscle myosins. High-duty ratio myosins such as myosin 5a spend the majority of their kinetic cycle in a strongly actin-bound state. This type of kinetics is suitable for myosins that transport cargo and need to move along actin for several microns without detaching [3]. Prior to the early 1980s, in vitro assays applied to myosins measured the ability of actin to bind myosin and to accelerate its MgATPase activity in vitro. Transient kinetic studies revealed the rate and equilibrium constants of the individual steps involved in their enzymatic cycle. However, an in vitro assay to measure myosin’s ability to translocate actin was missing until Sheetz and Spudich [4] developed the first functional in vitro motility assay. This assay used long actin cables exposed by dissection of cylindrical, multinucleated intermodal cells of the green alga, Nitella axillaris, as a “railroad” track to support the movement of micron-sized beads coated with myosin. This was a major innovation to myosin research, which yielded some very interesting insights. However, the assay had several limitations. Nitella had to be cultured in the lab, the dissection was not trivial, the preparations were typically only stable for an hour or so, and myosin had to be attached to a bead. The beads sometimes picked up the very fast endogenous myosin motors from the alga, and the movement was occurring on bundles of plant actins, with unknown specificity for mammalian myosins. Thus, while this assay was very useful, a truly in vitro assay that only used purified proteins was still needed.
Such an assay became available in 1986 when Kron and Spudich [5] introduced the sliding actin in vitro motility assay which required only two highly purified proteins, myosin and actin, to reconstitute the essence of a muscle contraction, the ability for myosin to translocate actin. This assay was made possible by the observations that single F-actin filaments were readily visible in solution by fluorescence microscopy, if labeled with rhodamine phalloidin [6]. Kron and Spudich described the movement of fluorescent rhodamine phalloidin labeled actin by myosin filaments from skeletal muscle or the amoeba, Dictyostelium discoideum, which had been bound to a glass coverslip surface [5]. This assay was easy to implement, used microscopy equipment already present in many labs, and was fast, reproducible, and quantitative. Furthermore, it could be performed with very small amounts of purified myosin and actin. The methodologies for this assay were further refined by Toyoshima et al. [7] who used a nitrocellulose-coated coverslip surface and demonstrated that not only myosin filaments but even the tailless proteolytic fragments of myosin, heavy meromyosin (HMM), and subfragment-1 (S1) moved actin. The motility assay provided a new way to classify and assess the characteristics of different classes of myosin by quantifying how fast they could move actin. The effects of varying ionic strength, magnesium, calcium, pH, and temperature on the speed of movement could be easily studied [8]. Below, we will describe this assay, highlight the evolution of this assay into versions that can measure mechanical properties of single molecules of myosin, and cover some of the major findings that have derived from the use of these elegant assays.
9.2 Discoveries Using the In Vitro Motility Assay
The development of the in vitro motility assay led to many important fundamental observations about the mechanism of actin and myosin interaction. The discovery that S1 moved actin filaments [7] proved fatal to several models for muscle contraction that required an organized sarcomere or the collapse of the coiled coil of myosin to power sarcomere shortening [9, 10]. Subsequent studies revealed important features of the assay. Single-headed myosins moved actin filaments demonstrating that a two-headed myosin was not necessary [7, 11]. The rate of movement of actin was largely independent of the concentration of myosin on the surface or on the length of the actin filament [7, 12]. Once the threshold of myosin density for movement was achieved, the movement occurred at the peak velocity. This meant that assays could be performed easily and quickly without having to control for actin filament length or myosin head density over a specific threshold.
The assay showed that there was amazing functional diversity among myosins and that the rate of actin filament sliding, even within the myosin class II family, varied from 0.02 µm/s for nonmuscle myosin 2B to 4 µm/s for fast skeletal muscle myosin [13]. Unconventional myosins, and not just class II myosins, were shown to be competent motors using this assay [14–17].
The best kinetic correlation for the rate at which the actin is moved in the motility assay is the rate at which ADP is released from actomyosin [18]. This means that the motility assay correlates well with the measurement of unloaded shortening velocity in muscle contraction, as they are both constrained by the rate of detachment of myosin from actin. The unloaded shortening velocity is a standard physiological parameter measured when a muscle mounted in a force transducer is allowed to shorten with no load applied. It is considered to be a measure of the maximal rate of myosin function in the muscle.
Structural data had proposed that myosin moved actin by swinging its neck region (also known as light-chain binding region or lever arm) [19]. This hypothesis was tested by showing a proportionality between the rate of movement and the neck length of mutant myosins created by inserting or deleting light-chain binding sites [20, 21].
9.3 Sliding Actin In Vitro Motility Assay
While the sliding actin in vitro motility assay has many variations in protocol, broadly, the assay observes fluorescently labeled actin filaments gliding over a cover slip that has been coated with myosin in the presence of ATP [22] (Fig. 9.1). This movement is captured with a sensitive video camera, often an ICCD (charged-coupled device) camera or other low-light imaging system. The movement of the labeled actin is tracked using software to estimate velocity.
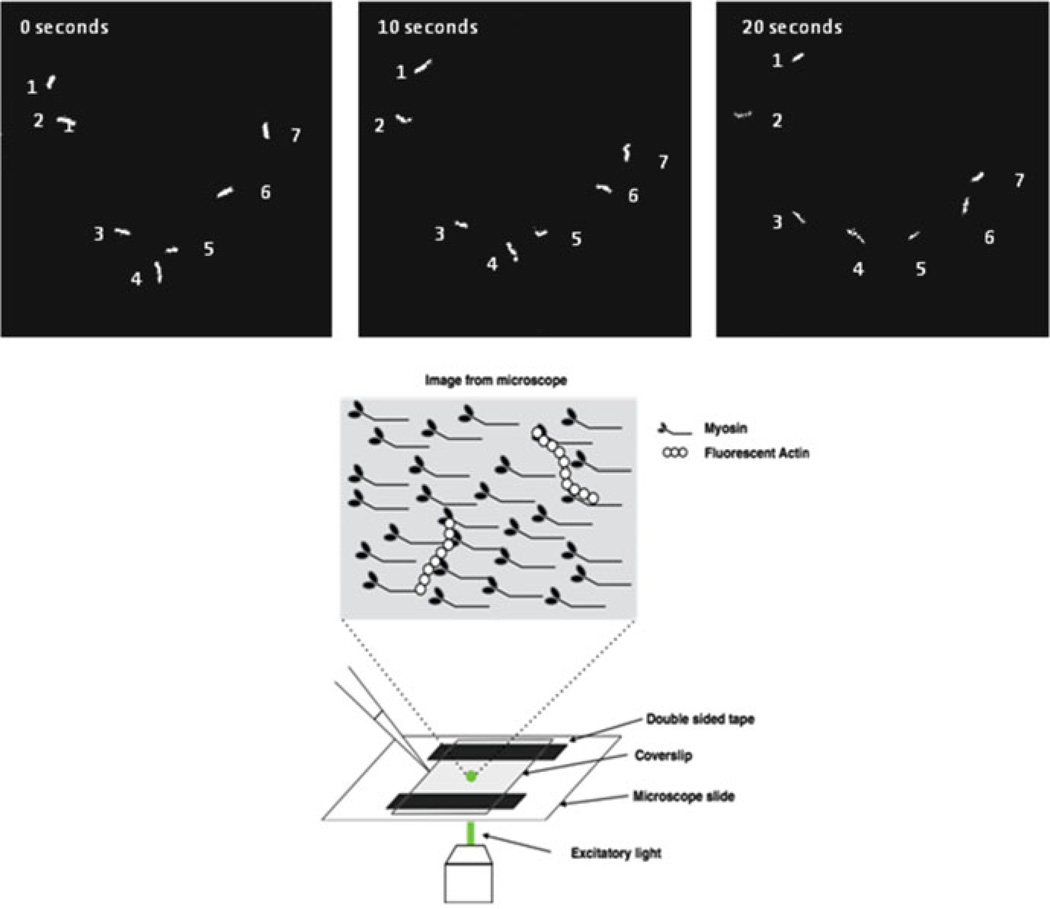
Fig. 9.1.
Sliding actin in vitro motility assay. Upper panel, shown are the position of 7 actin filaments at 0, 10, and 20 s while moving over a surface coated with myosin 5. Lower panel, schematic of the assay flow chamber
9.3.1 General Design of the Motility Assay
9.3.1.1 Myosin Preparation
Ideally in vitro motility assays are performed with freshly prepared myosin. However, myosin that has been flash frozen and stored in liquid nitrogen is often used. Damaged myosin that no longer hydrolyzes ATP, but binds actin strongly (dead heads) can often interfere with motility and reduce the quality of actin movement. There are two techniques to minimize this problem. The myosin-coated coverslip can be washed with unlabeled actin (termed “black actin”) in the presence of ATP that binds and blocks the damaged myosins before introducing the fluorescently labeled actin. Another technique, known as a “dead head spin,” mixes stoichiometric amount of F-actin with myosin in the presence of ATP at high ionic strength, followed by sedimentation in an ultracentrifuge. Under these conditions, actively cycling myosins are dissociated from actin, and the dead heads remain tightly bound to actin and are removed from the supernatant. Finally, during the measurement, a viscosity-enhancing reagent, methylcellulose (at 0.7 % concentration), can be added to the motility buffer to increase the likelihood of continuous movement over surfaces sparsely coated with myosins or at higher ionic strength, where the affinity of myosin for actin is reduced. Methylcellulose restricts the Brownian diffusion of actin to its long axis and inhibits “escape” of the actin filament, if it is transiently free of any strong myosin interactions.
9.3.1.2 Actin Preparation
The actin is most often labeled with stoichiometric concentrations of rhodamine phalloidin. Phalloidin is a fungal toxin that stabilizes filamentous actin and can be conjugated to various organic dyes to fluorescently label the actin and reduce the critical concentration for polymerization, so that actin filaments remain polymerized even at nanomolar concentrations. Actin is most commonly purified from rabbit skeletal muscle, but actin from other species or different isoforms of actin can also be used.
9.3.1.3 Assay Setup
Several articles describing the methodologies used in this assay have been published [22–24]. An assay chamber is easily constructed with a microscope slide, coverslip, and two strips of double-sided scotch tape to create a flow track. The flow cell is constructed to be open ended so that experimental solutions can be flushed through the chamber. Solution is added at one end and drawn through the chamber by wicking the solution away from the other end with filter paper. Coverslips are typically pre-coated with nitrocellulose or silicon which act as a nonspecific binding surface for the myosin molecules that are added. The assay needs only small amounts of protein due to the small chamber volumes (typically 10–100 µl). Myosin can be added in many different forms to the coverslip. It can be bound in its monomeric form, filamentous form (in case of myosin 2 molecules), or can be in the form of proteolytic or truncation fragments such as HMM or S1. After myosin is bound, the flow cell is washed with blocking buffer containing a moderate concentration of bovine serum albumin to prevent nonspecific binding of actin filaments. Fluorescently labeled F-actin is added to the flow chamber. The actin is left for a short period to bind to myosin in the absence of ATP. Excess actin is washed from the flow chamber by the addition of a few volumes of buffer to the flow cell. Finally, a solution with ATP and an oxygen-scavenging cocktail containing glucose, glucose oxidase, and catalase is added to initiate motility. The oxygen-scavenging cocktail helps to reduce photobleaching of the fluorophore that occurs when free radicals are generated by the illumination of the sample. Video data can then be analyzed using tracking software to track individual filament movement and then extract sliding velocities and percentage of motile filaments.
9.3.2 Variations of the In Vitro Motility Assay
9.3.2.1 Myosin Filaments
Variations on the classical in vitro motility assay have been developed creating tailored techniques to investigate different forms and classes of myosin. Studies have shown that myosin 2 molecules bound to the coverslip surface in monomeric form move actin at the same speed as does this myosin when bound as filaments [7]. Several studies observed the sliding of actin over intact native thick filaments imaged by dark field microscopy [25, 26], demonstrating that actin moves most efficiently when moving toward the center of the thick filament.
9.3.2.2 Measurements Under Load
The movement of actin filaments in the standard sliding actin in vitro motility assay is under unloaded conditions. Several methods have been developed to measure the effect of load on myosin in this assay. Actin-cross-linking proteins, such as α-actinin, bound to the coverslip surface along with myosin create a frictional load by transiently interacting with actin filaments. This results in a slowing of the actin filament sliding speed [27, 28]. This technique allows the measurement of isotonic kinetics, force-velocity relationships, and power curves in the motility assay. Motility assays have been performed with mixtures of fast and slow myosins to produce relationships that describe the ability of the slower myosin to reduce the movement velocity of the actin over the surface of the mixture [29, 30].
9.3.2.3 Calcium Regulation
Calcium-dependent actin filament regulation was studied by the addition of the skeletal muscle regulatory proteins troponin and tropomyosin [31, 32] or the smooth muscle regulatory proteins caldesmon and calmodulin [33, 34]. Smooth muscle and nonmuscle myosin 2 are purified in an autoinhibited off state, which require phosphorylation by myosin light-chain kinase for activity. Molluscan muscle myosins require calcium for activity [25, 35, 36].
9.3.2.4 Nonrecombinant/Myosin from Cell Extracts
The motility assay can be tailored to select a specific myosin from an impure mixture by first binding an antibody that binds an epitope of a particular myosin to the coverslip. The surface is then coated with bovine serum albumin to block nonspecific binding of other proteins. The antibody selects for the desired myosin and unwanted myosins or other proteins are washed out [37, 38]. The selecting antibody should be one that does not interfere with the function of the desired myosin. Antibodies directed toward the tails (C-terminal regions) of myosin often are appropriate for this use. Such antibodies can also be used to attach the myosin to the surface rather than relying on nonspecific attachment of the myosin to the surface [39].
9.3.2.5 Polarity/Directionality
The F-actin filament is polar with a “+” end and a “−” end. Actin can be fluorescently labeled to demonstrate its polarity by using small actin “seeds” labeled with one fluorophore to nucleate the assembly of longer actin filaments which will then be labeled with another fluorophore [16, 40]. These types of actin filaments can be used to determine the directionality of the movement of F-actin by myosin. Thus far, all myosins move actin filaments with the + end leading except for myosin 6 [16].
9.4 Single-Molecule Motility Assays
The development of TIRF (total internal reflection fluorescent) microscopy allowed for the design of single-molecule motility assays where the movement of processive myosins along a surface-attached actin filament can be observed [41]. In TIRF microscopy, a laser beam is introduced to the coverslip at a critical angle, which results in complete internal reflection of the laser light at the coverslip-buffer boundary. This sets up an evanescent wave that penetrates the assay chamber to a depth of only 150–200 nm leaving all fluorophores above this in a dark, unexcited state. The result is a dramatic improvement in the signal-to-noise ratio to allow visualization of a single fluorophore.
9.4.1 Assay Setup
9.4.1.1 Myosin Labeling
There are several such ways of labeling myosins for this purpose. A fluorescently labeled light chain can be exchanged into myosin. The light chain is usually labeled with organic dyes [41] although light chains fused with fluorescent fusion proteins such as GFP can also be used [42]. Myosins with GFP fused to the heavy chain at either the amino (N-) or the carboxy (C-) terminus can also be used [43], or the myosin can be tagged with a quantum dot [44]. There are advantages and disadvantages to each of these approaches. Fusion of GFP molecules to the N- or C-termini of myosins does not seem to have a major effect on their activity, and these fluorescent fusion proteins are available in a variety of colors by expression of recombinant DNA in the baculovirus/Sf9 system [45]. However, the brightness and photostability of the fluorescent fusion proteins varies with the type of fusion protein selected, and these proteins, in general, are not as bright or as photostable as organic fluorophore dyes which can be introduced by chemical modification of surface lysines or cysteines residues. The use of quantum dots also has benefits and problems. Quantum dots come in different colors, are very bright, and maintain fluorescence over an extended period of time. However, they suffer from the blinking phenomena, which means that they periodically go dark and then emit light again. Their size is variable, but they are usually at least the size of a GFP molecule. The most common way to attach quantum dots to myosin is by introducing a biotin tag onto either the N-terminus or the C-terminus of a myosin and using streptavidin-functionalized quantum dots. If the quantum dot is attached to the N-terminus of a single head of myosin, the movement of that head is visualized, whereas if the quantum dot is attached to the C-terminus, the movement of the center of mass of the molecule is followed. Here, one must be very careful to only have one myosin attached per quantum dot, if single-molecule mechanics are to be explored. The way to accomplish this is to add only 1–4 myosin molecules per quantum dot in the coupling reaction. One can create an artificial “cargo”-bearing multiple myosins per quantum dot by increasing this labeling ratio.
Performing the Assay
In the single-molecule in vitro motility assay, actin filaments are bound to the coverslip surface via NEM-inactivated skeletal muscle HMM or via the use of lightly biotinylated actin filaments which can be tethered to a biotin-streptavidin-coated coverslip. Fluorescently labeled myosin is added and is visualized in a TIRF microscope. The images of the diffraction limited moving myosin spots are typically captured by a high-sensitivity camera (Fig. 9.2). Several parameters can be gleaned from this assay including speed, run length, and run frequency. Run length is usually determined by compiling a histogram of many events and fitting the curve with an exponential decay. This version of the single-molecule motility assay is now in routine use in many laboratories to study the processive movement of several classes of myosin, including myosin 5, myosin 6, myosin 7, and myosin 10 [17, 21, 41, 46, 47]. There is good agreement between rates that single processive molecules move along actin filaments and the rates at which these myosins slide actin filaments in the gliding assay.
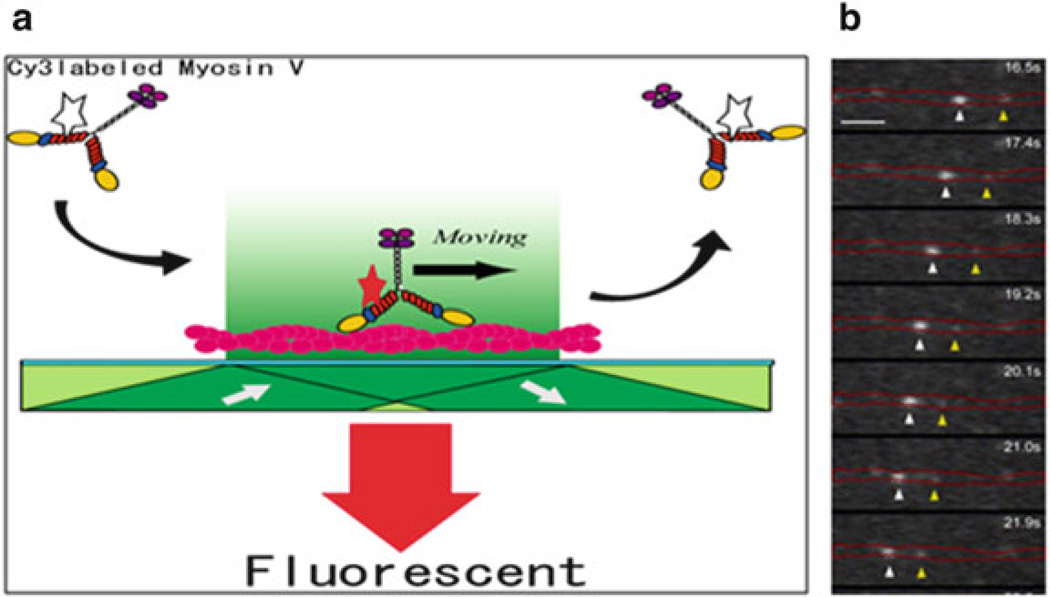
Fig. 9.2.
Single-molecule in vitro motility assay. (a) Diagram of the geometry of the assay using myosin 5a. The star on myosin 5a represents a fluorophore bound to a calmodulin light chain. When it is in the evanescent field, it is depicted in red showing that the fluorophore is excited. The dark green shading shows the direction of the laser excitation. The actin filament bound to the surface is colored red. (b) The position of a moving fluorophore along actin as a function of time (top to bottom). In this case, the fluorophore is GFP-Rab27a which is bound to the motor, myosin 5a, via melanophilin. The image is taken from Wu et al. [63]. The position of the stationary actin filament is marked by red lines, and the positions of two moving spots are marked with arrow-heads. The scale bar is 3.2 µm
9.5 Assay Variations
9.5.1 Attachment Lifetimes
There is at least one case, where this assay geometry was used to measure the lifetime of attachment of a nonprocessive myosin. Nagy et al. [48] used the singlemolecule motility assay with nonmuscle myosin 2B HMM which was shown to be nonprocessive as a single molecule in optical trapping experiments, but to have a long attachment lifetime due to its very slow kinetics. GFP-tagged nonmuscle myosin 2B HMM molecules could be observed to bind to and then detach from surface-bound actin filaments, and the kinetics of these events matched those observed in the optical trapping experiments. Full-length nonmuscle myosin 2B molecules form short bipolar filaments in vitro at low ionic strengths and in cells. When GFP-labeled filaments of nonmuscle myosin 2B are formed, they move along the surface-attached actin filament for several microns at a slow rate consistent with the rate of actin filament translocation in the sliding actin assay.
9.5.2 Types of Actin Filaments
Actin filaments exist in pleiotropic forms in cells including single actin filaments, branched arbor-like networks, meshworks, loose bundles, and very tightly packed bundles [49]. These various states are formed and maintained by a slew of actin-binding proteins. Myosins are capable of interacting with different forms of actin, and different myosin isoforms are specifically localized to different areas of the cell or are found in actin-containing areas of specialized cells such as in the sarcomere of muscle fibers or the stereocilia of inner ear hair cells. The single-molecule motility assay can be performed on bundles of actin [50, 51] or with actin-containing regulatory proteins such as tropomyosin [52]. Actin filament meshworks can be created in vitro, and it is seen that myosin 5a can readily make abrupt turns and switch from one filament to another or can “step over” an intersecting filament [53]. Myosin 6, in contrast, prefers to stay on the initial filament [51]. Several studies have “deroofed” tissue culture cells by washing with a detergent which exposed the underlying actin cytoskeleton and then have added fluorescently tagged myosins to these cells to examine their motility in this more cellular-like in vitro setting [54, 55]. Interestingly, different classes of myosins prefer to move on different actin structures in these assays.
9.5.3 Calcium Regulation
Regulation can also be studied in the single-molecule motility assay. Purified full-length myosin 5a exists in an autoinhibited state where the C-terminal cargo-binding domain folds back and contacts the motor domain to adopt a state with very low actin-activated MgATPase and inability to move on actin [56–58]. This is clearly seen by examining the frequency of movement of full-length myosin 5a compared to that of the constitutively active myosin 5a HMM [57]: the latter moves much more frequently than the former. The ATPase activity of full-length myosin 5a in the presence of actin is activated by calcium ions, but paradoxically, this impairs the ability of the myosin to move processively on actin [59]. The reason for this apparent contradiction is that some calmodulin molecules dissociate from myosin 5a upon addition of calcium. This leaves the alpha-helical region of the IQ motif exposed and unstable, so the myosin is unable to convert its enzymatic output into a mechanical output [59, 60].
9.5.4 Cargo Regulation
Myosin 5a is involved in the actin-dependent movement of melanosomes which are membrane-bound pigment granules found in melanocytes. The linkage of myosin 5a to melanocytes is via two other proteins (see [3] for review). The cargo-binding domain of myosin 5a interacts with the C-terminus of a protein called melanophilin which in turn interacts through its N-terminus with Rab27a, a resident melanosome membrane-binding protein. Two studies have examined aspects of this complex. Wu et al. [61] mixed GFP-tagged Rab27a with melanophilin and myosin 5a and observed that fluorescent spots moved processively along actin filaments. No movement was seen if melanophilin or myosin 5a was omitted, and thus, it was concluded that the tripartite complex could be reconstituted in vitro at low concentrations. Melanophilin also has an actin-binding site which is important for enhancing the frequency of processive runs. A more recent study used fluorescently labeled myosin 5a and melanophilin to examine this in more detail [62]. It was shown that melanophilin significantly enhanced the run frequency and slowed the rate of movement of myosin 5a by acting as a tether.
Several classes of myosin including myosins 6, 7, and 10 do not appear to form stable dimers as purified from expression systems, despite the presence of a predicted coiled-coil motif in their tail sequence [46, 63–66]. Some of these myosins appear to be able to dimerize and move processively in single-molecule TIRF motility assays when “clustered” by first binding to actin filaments at high density in the absence of ATP or by interaction with certain binding partners that induce dimerization [46, 67, 68].
9.6 Super-Resolution Tracking of Moving Myosins
The single-molecule in vitro motility assay can be used to measure the step size of myosins through the use of super-resolution microscopy. The resolution of the conventional epifluorescence microscope is limited by the wavelength of light and is typically 200–300 nm. However, if sufficient photons can be collected from a single fluorophore, its point spread function can be fitted to a two-dimensional Gaussian whose peak can be localized with 2–3 nm resolution (Fig. 9.3). This technique was first used for myosins by Yildiz et al. [69] who coined the term FIONA (Fluorescence Imaging at One Nanometer Accuracy) for the technique. It allows the user to measure the size of the steps taken by myosin and to get information on the stepping kinetics. By lowering the ATP concentration to decrease the rate of stepping, which allows for the collection of more photons at each time point, Yildiz et al. [69] showed that a Cy3-labeled myosin 5a took 36 nm steps moving along actin, consistent with values obtained by optical trapping and EM imaging; see [70] for review. Examining the stepping distance and kinetics of molecules in which only one of the two heads was labeled provided evidence that the molecule stepped in a hand-over-hand mechanism where the trailing head dissociated, moved forward to a new actin-binding site 72 nm away. Interestingly, this stepping pattern precisely matches the crossover repeat of the helical actin filament and allows the myosin 5a to walk straight along an actin filament. The ability to observe single steps of myosin 5a was subsequently shown to be applicable to GFP-labeled myosins [43] and to myosins with bound quantum dots [71]. The latter were used in an elegant study in which each head of a myosin 5a molecule was tagged with a quantum dot of a different color [44]. The two colors could be shown to alternate positions (separated by 36 nm) as the molecule moved processively along actin. The assay was used to confirm that the neck region acted as a rigid lever arm by examining the step size of the same set of myosin 5a lever mutants as was used for the in vitro motility studies described earlier [72]. Another study used simultaneous imaging of a labeled myosin 5a molecule and a fluorescent ATP analog, deac-aminoATP, whose fluorescence was enhanced 15-fold upon binding to myosin, to demonstrate that one ATP molecule was hydrolyzed for each step, demonstrating tight coupling between the enzymology and mechanics of this myosin [73].
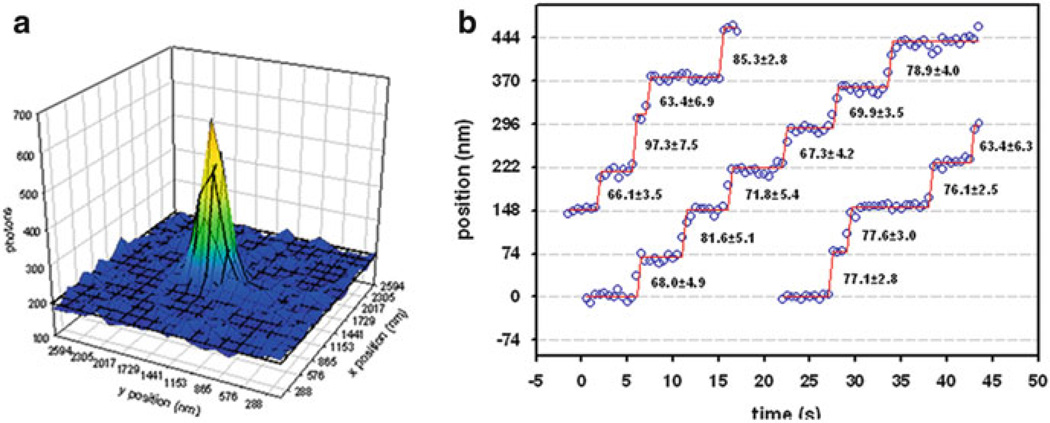
Fig. 9.3.
Nanometer tracking of a myosin’s movement by FIONA. (a) Cartoon demonstrating the fitting of a point spread function of a single fluorophore to a two-dimensional Gaussian. (b) Three traces of GFP-myosin 5a moving along actin with the position determined at each time point by the FIONA technique. Only a single head of each myosin is labeling with GFP. The numbers in between steps show the distance between successive steps along with the standard deviation. The data is taken from Snyder et al. [43]
The FIONA assay has also been applied to artificially dimerized myosin 6 [74], myosin 7a [46], and myosin 10 [75] molecules. The results show that myosin 6 moves along actin with a much more variable step size and that myosin 7a takes somewhat shorter steps than myosin 5a. FIONA measurements of myosin 10 are somewhat controversial with a range of values reported [75–77]. The longer step size of 36 nm is surprisingly similar in magnitude to that of myosin 5a as the lever arm of myosin 10 binds only three calmodulins. However, this myosin has a stable single alpha-helical (or SAH) domain which serves to extend the lever arm as determined by electron microscopy [65]. A chimeric molecule in which the motor domain plus first three IQ motifs of myosin 5a was fused to a SAH domain with similar length and dimerized by a portion of the coiled-coil domain took 36 nm steps consistent with the SAH domain being part of a rigid lever arm [78].
Another variation of the single-molecule motility assay allows one to gain insight into the detailed mechanisms of myosin movement. polTIRF is a technique which measures the polarization of a fluorescent probe rigidly bound to one of the calmodulin molecules via a bifunctional linkage. This technique allows for the angular position of the probe, and hence the lever arm, to be measured, while the myosin is walking along actin. The data showed that lever arm of both myosin 5a and myosin 6 alternates between pre- and post-powerstroke angles giving strong support for the hand-over-hand model for walking [79, 80].
9.7 Conclusions
As can be seen from the above, in vitro motility assays have dramatically increased our understanding of molecular mechanism of myosin-actin interactions and have given insight into the regulation of this interaction and to the functional diversity found in the myosin superfamily. The assay has been adapted to study myosins from 13 of the known classes. The fundamental principles involved in the assay, the ability to observe fluorescently labeled actin filaments and to bind myosin to a surface, led to the development of optical trapping techniques that allows one to measure lifetimes of interaction, power stroke size, and force production of singlemyosin molecules [81]. Variants of the assay are being continually developed. These include using zero-mode waveguides to further increase the sensitivity of the assay at higher fluorophore concentrations [82] and the use of DNA origami to create scaffolds for binding more than one motor [83].
Acknowledgments
We thank Yasuharu Takagi for the images shown in Fig. 9.1.
Abbreviations
- TIRF
Total internal refection fluorescence
- CCD
Charged-coupled device
- HMM
Heavy meromyosin
- S1
Subfragment-1
- FIONA
Fluorescence Imaging at One Nanometer with Accuracy
- GFP
Green fluorescent protein
References
- 1.Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 2.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammer JA, III, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol. 2012;13:13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 4.Sheetz MP, Spudich JA. Movement of myosin-coated fluorescent beads on actin cables in vitro. Nature. 1983;303:31–35. doi: 10.1038/303031a0. [DOI] [PubMed] [Google Scholar]
- 5.Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanagida T, Nakase M, Nishiyama K, Oosawa F. Direct observation of motion of single F-actin filaments in the presence of myosin. Nature. 1984;307:58–60. doi: 10.1038/307058a0. [DOI] [PubMed] [Google Scholar]
- 7.Toyoshima YY, Kron SJ, McNally EM, Niebling KR, Toyoshima C, Spudich J. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature. 1987;328:536–539. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- 8.Homsher E, Wang F, Sellers JR. Factors affecting movement of F-actin filaments propelled by skeletal muscle heavy meromyosin. Am J Physiol Cell Physiol. 1992;262:C714–C723. doi: 10.1152/ajpcell.1992.262.3.C714. [DOI] [PubMed] [Google Scholar]
- 9.Harrington WF. A mechanochemical mechanism for muscle contraction. Proc Natl Acad Sci USA. 1971;68:685–689. doi: 10.1073/pnas.68.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oplatka A. The role of water in the mechanism of muscular contraction. FEBS Lett. 1994;355:1–3. doi: 10.1016/0014-5793(94)01158-3. [DOI] [PubMed] [Google Scholar]
- 11.Harada Y, Noguchi A, Kishino A, Yanagida T. Sliding movement of single actin filaments on one-headed myosin filaments. Nature. 1987;326:8005–8808. doi: 10.1038/326805a0. [DOI] [PubMed] [Google Scholar]
- 12.Harada Y, Yanagida T. Direct observation of molecular motility by light microscopy. Cell Motil Cytoskeleton. 1988;10:71–76. doi: 10.1002/cm.970100112. [DOI] [PubMed] [Google Scholar]
- 13.Sellers JR. Myosins. 2nd edn. Oxford: Oxford University Press; 1999. [Google Scholar]
- 14.Collins K, Sellers JR, Matsudaira P. Calmodulin dissociation regulates brush border myosin I 110-kD-calmodulin) mechanochemical activity in vitro. J Cell Biol. 1990;110:1137–1147. doi: 10.1083/jcb.110.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nascimento AAC, Cheney RE, Tauhata SBF, Larson RE, Mooseker MS. Enzymatic characterization and functional domain mapping of brain myosin-V. J Biol Chem. 1996;271:17561–17569. doi: 10.1074/jbc.271.29.17561. [DOI] [PubMed] [Google Scholar]
- 16.Wells A, Lin AW, Chen L-Q, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 17.Nagy S, Ricca BL, Norstrom MF, Courson DS, Brawley CM, Smithback PA, Rock RS. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci USA. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yengo CM, Takagi Y, Sellers JR. Temperature dependent measurements reveal similarities between muscle and non-muscle myosin motility. J Muscle Res Cell Motil. 2012;33:385–394. doi: 10.1007/s10974-012-9316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 20.Warshaw DM, Guilford WH, Freyzon Y, Krementsova E, Palmiter KA, Tyska MJ, Baker JE, Trybus KM. The light chain binding domain of expressed smooth muscle heavy meromyosin acts as a mechanical lever. J Biol Chem. 2000;275:37167–37172. doi: 10.1074/jbc.M006438200. [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto T, Wang F, Schmitz S, Xu YH, Xu Q, Molloy JE, Veigel C, Sellers JR. Neck length and processivity of myosin V. J Biol Chem. 2003;278:29201–29207. doi: 10.1074/jbc.M303662200. [DOI] [PubMed] [Google Scholar]
- 22.Sellers JR, Cuda G, Wang F, Homsher E. Myosin-Specific Adaptations of Motility Assays. In: Scholey JM, editor. Motility Assays for Motor Proteins. San Diego: CA, Academic Press, Inc.; 1993. pp. 23–49. [DOI] [PubMed] [Google Scholar]
- 23.Sellers JR. In vitro motility assays with actin. In: Celis JE, editor. Cell biology. A laboratory handbook. Vol. 2. Amsterdam: Academic; 2006. pp. 387–392. [Google Scholar]
- 24.Kron SJ, Toyoshima YY, Uyeda TQP, Spudich JA. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 1991;196:399–416. doi: 10.1016/0076-6879(91)96035-p. [DOI] [PubMed] [Google Scholar]
- 25.Sellers JR, Kachar B. Polarity and velocity of sliding filaments. Control of direction by actin and of speed by myosin. Science. 1990;249:406–408. doi: 10.1126/science.2377894. [DOI] [PubMed] [Google Scholar]
- 26.Previs MJ, Beck PS, Gulick J, Robbins J, Warshaw DM. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337:1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janson LW, Sellers JR, Taylor DL. Actin-binding proteins regulate the work performed by myosin II motors on single actin filaments. Cell Motil Cytoskeleton. 1992;22:274–280. doi: 10.1002/cm.970220407. [DOI] [PubMed] [Google Scholar]
- 28.Greenberg MJ, Moore JR. The molecular basis of frictional loads in the in vitro motility assay with applications to the study of the loaded mechanochemistry of molecular motors. Cytoskeleton (Hoboken) 2010;67:273–285. doi: 10.1002/cm.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris DE, Work SS, Wright RK, Alpert NR, Warshaw DM. Smooth, cardiac and skeletal muscle myosin force and motion generation assessed by cross-bridge mechanical interactions in vitro. J Muscle Res Cell Motil. 1994;15:11–19. doi: 10.1007/BF00123828. [DOI] [PubMed] [Google Scholar]
- 30.Cuda G, Pate E, Cooke R, Sellers JR. In vitro actin filament sliding velocities produced by mixtures of different types of myosin. Biophys J. 1997;72:1767–1779. doi: 10.1016/S0006-3495(97)78823-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin D, Bobkova A, Homsher E, Tobacman LS. Altered cardiac troponin T in vitro function in the presence of a mutation implicated in familial hypertrophic cardiomyopathy. J Clin Invest. 1996;97:2842–2848. doi: 10.1172/JCI118740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser IDC, Marston SB. In vitro motility analysis of actin-tropomyosin regulation by troponin and calcium. The thin filament is switched as a single cooperative unit. J Biol Chem. 1995;270:7836–7841. doi: 10.1074/jbc.270.14.7836. [DOI] [PubMed] [Google Scholar]
- 33.Shirinsky VP, Biryukov KG, Hettasch JM, Sellers JR. Inhibition of the relative movement of actin and myosin by caldesmon and calponin. J Biol Chem. 1992;267:15886–15892. [PubMed] [Google Scholar]
- 34.Fraser IDC, Marston SB. In vitro motility analysis of smooth muscle caldesmon control of actin-tropomyosin filament movement. J Biol Chem. 1995;270:19688–19693. doi: 10.1074/jbc.270.34.19688. [DOI] [PubMed] [Google Scholar]
- 35.Umemoto S, Sellers JR. Characterization of in vitro motility assays using smooth muscle and cytoplasmic myosins. J Biol Chem. 1990;265:14864–14869. [PubMed] [Google Scholar]
- 36.Kurzawa-Goertz SE, Perreault-Micale CL, Trybus KM, Szent-Gyorgyi AG, Geeves MA. Loop I can modulate ADP affinity, ATPase activity, and motility of different scallop myosins. Transient kinetic analysis of S1 isoforms. Biochemistry. 1998;37:7517–7525. doi: 10.1021/bi972844+. [DOI] [PubMed] [Google Scholar]
- 37.Cuda G, Fananapazir L, Zhu W-S, Sellers JR, Epstein ND. Skeletal muscle expression and abnormal function of b-myosin in hypertrophic cardiomyopathy. J Clin Invest. 1993;91:2861–2865. doi: 10.1172/JCI116530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reck-Peterson SL, Tyska MJ, Novick PJ, Mooseker MS. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin- based motors. J Cell Biol. 2001;153:1121–1126. doi: 10.1083/jcb.153.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweeney HL, Rosenfeld SS, Brown F, Faust L, Smith J, Xing J, Stein LA, Sellers JR. Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket. J Biol Chem. 1998;273:6262–6270. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- 40.Liao JC, Elting MW, Delp SL, Spudich JA, Bryant Z. Engineered myosin VI motors reveal minimal structural determinants of directionality and processivity. J Mol Biol. 2009;392:862–867. doi: 10.1016/j.jmb.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakamoto T, Amitani I, Yokota E, Ando T. Direct observation of processive movement by individual myosin V molecules. Biochem Biophys Res Commun. 2000;272:586–590. doi: 10.1006/bbrc.2000.2819. [DOI] [PubMed] [Google Scholar]
- 42.Kengyel A, Wolf WA, Chisholm RL, Sellers JR. Nonmuscle myosin IIA with a GFP fused to the N-terminus of the regulatory light chain is regulated normally. J Muscle Res Cell Motil. 2010;31:163–170. doi: 10.1007/s10974-010-9220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snyder GE, Sakamoto T, Hammer JA, III, Sellers JR, Selvin PR. Nanometer localization of single green fluorescent proteins: evidence that myosin V walks hand-over-hand via telemark configuration. Biophys J. 2004;87:1776–1783. doi: 10.1529/biophysj.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warshaw DM, Kennedy GG, Work SS, Krementsova EB, Beck S, Trybus KM. Differential labeling of myosin V heads with quantum dots allows direct visualisation of hand-over-hand processivity. Biophys J. 2005;88:L30–L32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, Kovacs M, Sakamoto T, Zhang F, Kiehart DP, Sellers JR. Dimerized Drosophila myosin VIIa: a processive motor. Proc Natl Acad Sci USA. 2006;103:5746–5751. doi: 10.1073/pnas.0509935103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagy A, Takagi Y, Billington N, Sun SA, Hong DK, Homsher E, Wang A, Sellers JR. Kinetic Characterization of Nonmuscle Myosin IIB at the Single Molecule Level. J Biol Chem. 2012;288:709–722. doi: 10.1074/jbc.M112.424671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagy S, Rock RS. Structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J Biol Chem. 2010;285:26608–26617. doi: 10.1074/jbc.M110.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ali MY, Previs SB, Trybus KM, Sweeney HL, Warshaw DM. Myosin VI has a one track mind versus myosin Va when moving on actin bundles or at an intersection. Traffic. 2013;14:70–81. doi: 10.1111/tra.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hodges AR, Krementsova EB, Bookwalter CS, Fagnant PM, Sladewski TE, Trybus KM. Tropomyosin is essential for processive movement of a class v Myosin from budding yeast. Curr Biol. 2012;22:1410–1416. doi: 10.1016/j.cub.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ali MY, Krementsova EB, Kennedy GG, Mahaffy R, Pollard TD, Trybus KM, Warshaw DM. From the Cover: Myosin Va maneuvers through actin intersections and diffuses along microtubules. Proc Natl Acad Sci USA. 2007;104:4332–4336. doi: 10.1073/pnas.0611471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brawley CM, Rock RS. Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc Natl Acad Sci USA. 2009;106:9685–9690. doi: 10.1073/pnas.0810451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sivaramakrishnan S, Spudich JA. Coupled myosin VI motors facilitate unidirectional movement on an F-actin network. J Cell Biol. 2009;187:53–60. doi: 10.1083/jcb.200906133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. J Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thirumurugan K, Sakamoto T, Hammer JA, III, Sellers JR, Knight PJ. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li XD, Jung HS, Mabuchi K, Craig R, Ikebe M. The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. J Biol Chem. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- 59.Lu H, Krementsova EB, Trybus KM. Regulation of myosin V processivity by calcium at the single molecule level. J Biol Chem. 2006;281:31987–31994. doi: 10.1074/jbc.M605181200. [DOI] [PubMed] [Google Scholar]
- 60.Koide H, Kinoshita T, Tanaka Y, Tanaka S, Nagura N, Meyer zu HG, Miyagi A, Ando T. Identification of the single specific IQ motif of myosin V from which calmodulin dissociates in the presence of Ca2+ Biochemistry. 2006;45:11598–11604. doi: 10.1021/bi0613877. [DOI] [PubMed] [Google Scholar]
- 61.Wu X, Sakamoto T, Zhang F, Sellers JR, Hammer JA., III In vitro reconstitution of a transport complex containing Rab27a, melanophilin and myosin Va. FEBS Lett. 2006;580:5863–5868. doi: 10.1016/j.febslet.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 62.Sckolnick M, Krementsova EB, Warshaw DM, Trybus KM. More Than Just a Cargo Adapter: Melanophilin Prolongs and Slows Processive Runs of Myosin Va. J Biol Chem. 2013;288:29313–291322. doi: 10.1074/jbc.M113.476929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lister I, Schmitz S, Walker M, Trinick J, Buss F, Veigel C, Kendrick-Jones J. A monomeric myosin VI with a large working stroke. EMBO J. 2004;23:1729–1738. doi: 10.1038/sj.emboj.7600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Umeki N, Jung HS, Watanabe S, Sakai T, Li XD, Ikebe R, Craig R, Ikebe M. The tail binds to the head-neck domain, inhibiting ATPase activity of myosin VIIA. Proc Natl Acad Sci USA. 2009;106:8483–8488. doi: 10.1073/pnas.0812930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knight PJ, Thirumurugan K, Yu Y, Wang F, Kalverda AP, Stafford WF, III, Sellers JR, Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 66.Umeki N, Jung HS, Sakai T, Sato O, Ikebe R, Ikebe M. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011;18:783–788. doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
- 67.Park H, Ramamurthy B, Travaglia M, Safer D, Li-Qiong C, Franzini-Armstrong C, Selvin PR, Sweeney HL. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 68.Phichith D, Travaglia M, Yang Z, Liu X, Zong AB, Safer D, Sweeney HL. Cargo binding induces dimerization of myosin VI. Proc Natl Acad Sci USA. 2009;106:17320–17324. doi: 10.1073/pnas.0909748106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 70.Sellers JR, Veigel C. Walking with myosin V. Curr Opin Cell Biol. 2006;18:68–73. doi: 10.1016/j.ceb.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 71.Ali MY, Lu H, Bookwalter CS, Warshaw DM, Trybus KM. Myosin V and Kinesin act as tethers to enhance each others’ processivity. Proc Natl Acad Sci USA. 2008;105:4691–4696. doi: 10.1073/pnas.0711531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakamoto T, Yildiz A, Selvin PR, Sellers JR. Step-size is determined by neck length in myosin V. Biochemistry. 2005;44:16203–16210. doi: 10.1021/bi0512086. [DOI] [PubMed] [Google Scholar]
- 73.Sakamoto T, Webb MR, Forgacs E, White HD, Sellers JR. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature. 2008;455:128–132. doi: 10.1038/nature07188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yildiz A, Park H, Safer D, Yang Z, Chen LQ, Selvin PR, Sweeney HL. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J Biol Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 75.Ricca BL, Rock RS. The stepping pattern of myosin X is adapted for processive motility on bundled actin. Biophys J. 2010;99:1818–1826. doi: 10.1016/j.bpj.2010.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun Y, Sato O, Ruhnow F, Arsenault ME, Ikebe M, Goldman YE. Single-molecule stepping and structural dynamics of myosin X. Nat Struct Mol Biol. 2010;17:485–491. doi: 10.1038/nsmb.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bao J, Huck D, Gunther LK, Sellers JR, Sakamoto T. Actin Structure-Dependent Stepping of Myosin 5a and 10 during Processive Movement. PLoS One. 2013;8:e74936. doi: 10.1371/journal.pone.0074936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baboolal TG, Sakamoto T, Forgacs E, White HD, Jackson SM, Takagi Y, Farrow RE, Molloy JE, Knight PJ, Sellers JR, Peckham M. The SAH domain extends the functional length of the myosin lever. Proc Natl Acad Sci USA. 2009;106:22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Syed S, Snyder GE, Franzini-Armstrong C, Selvin PR, Goldman YE. Adaptability of myosin V studied by simultaneous detection of position and orientation. EMBO J. 2006;25:1795–1803. doi: 10.1038/sj.emboj.7601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun Y, Schroeder HW, III, Beausang JF, Homma K, Ikebe M, Goldman YE. Myosin VI walks “wiggly” on actin with large and variable tilting. Mol Cell. 2007;28:954–964. doi: 10.1016/j.molcel.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 82.Elting MW, Leslie SR, Churchman LS, Korlach J, McFaul CM, Leith JS, Levene MJ, Cohen AE, Spudich JA. Single-molecule fluorescence imaging of processive myosin with enhanced background suppression using linear zero-mode waveguides (ZMWs) and convex lens induced confinement (CLIC) Opt Express. 2013;21:1189–1202. doi: 10.1364/OE.21.001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu H, Efremov AK, Bookwalter CS, Krementsova EB, Driver JW, Trybus KM, Diehl MR. Collective dynamics of elastically coupled myosin V motors. J Biol Chem. 2012;287:27753–27761. doi: 10.1074/jbc.M112.371393. [DOI] [PMC free article] [PubMed] [Google Scholar]