The RNA-binding protein Arrest occupies a novel intranuclear domain and directs flight muscle–specific patterns of alternative splicing in flies.
Abstract
Drosophila melanogaster flight muscles are distinct from other skeletal muscles, such as jump muscles, and express several uniquely spliced muscle-associated transcripts. We sought to identify factors mediating splicing differences between the flight and jump muscle fiber types. We found that the ribonucleic acid–binding protein Arrest (Aret) is expressed in flight muscles: in founder cells, Aret accumulates in a novel intranuclear compartment that we termed the Bruno body, and after the onset of muscle differentiation, Aret disperses in the nucleus. Down-regulation of the aret gene led to ultrastructural changes and functional impairment of flight muscles, and transcripts of structural genes expressed in the flight muscles became spliced in a manner characteristic of jump muscles. Aret also potently promoted flight muscle splicing patterns when ectopically expressed in jump muscles or tissue culture cells. Genetically, aret is located downstream of exd (extradenticle), hth (homothorax), and salm (spalt major), transcription factors that control fiber identity. Our observations provide insight into a transcriptional and splicing regulatory network for muscle fiber specification.
Introduction
Skeletal muscle is a highly heterogeneous tissue. Muscle fibers, the basic cellular units of muscles, demonstrate distinctive features at the physiological, structural, and molecular levels (Punkt, 2002; Zierath and Hawley, 2004; Schiaffino and Reggiani, 2011). In vertebrates, different fiber types coexist in every large muscle, and their relative ratio determines the functional properties of the muscle (Schiaffino et al., 1970; Zierath and Hawley, 2004). In Drosophila melanogaster, skeletal muscles are tuned to perform specialized functions, such as flying, jumping, and walking. Although individual muscles are made of the same fiber type, fibers from different muscles can differ substantially (O’Donnell et al., 1989; Bernstein et al., 1993; Bryantsev et al., 2012b). The most dramatic distinction in fiber-type characteristics can be found between the two largest Drosophila muscles, the indirect flight muscle (or flight muscle) and the tergal depressor of the trochanter (or jump muscle). The fibers of the flight and jump muscles belong to the fibrillar and tubular types, respectively. Therefore, these muscles are useful models to dissect fiber-type specification. The differences between fibrillar and tubular fibers are based on transcriptome diversity arising from differential gene expression and from alternative transcript splicing. Although insight into the transcriptional network controlling expression of fiber-specific muscle genes was obtained in recent studies (Schönbauer et al., 2011; Bryantsev et al., 2012b), the regulation of fiber-specific splicing remains largely unknown.
Alternative mRNA spicing provides the means to achieve greater genetic variety without the necessity for additional genes. Transcripts of many muscle-specific genes in vertebrates and Drosophila undergo alternative splicing depending on the muscle lineage or fiber type (Bernstein et al., 1993; Venables et al., 2012; Spletter and Schnorrer, 2014). The potential of alternative splicing in fine-tuning of muscle properties is thought to be of a significant value. For instance, rescue studies for the Mhc (Myosin heavy chain) gene showed that expression of stage-inappropriate Mhc isoforms was sufficient to restore normal muscle morphology but, at the same time, could not rescue proper function in adult muscles (Wells et al., 1996; Swank et al., 2000). Nevertheless, the ultimate role of alternative splicing in muscle morphology has not been extensively studied as a result of the inability to switch fiber-specific splicing en masse.
Drosophila has been a valuable research model for studying alternative splicing regulation (Venables et al., 2012). Mechanistically, the choice of splicing sites on pre-mRNA transcripts is determined through interactions with RNA-binding proteins acting as splicing factors (SFs; Black, 2003). Systemic analyses reveal that knockdown (KD) of a single SF may affect hundreds of splicing events (Blanchette et al., 2005, 2009). Furthermore, a single SF can act as a master regulator that directly and indirectly controls an entire chain of specific alternative splicing events, as implemented in sex-specific splicing regulation (Black, 2003; Förch and Valcárcel, 2003). Whether the principles and organization of sex-specific splicing regulation are also applicable to other instances of alternative splicing control remains to be determined.
Regulation of alternative splicing in muscles has drawn steadily increasing interest, in the light of several human muscular pathological conditions that impact alternative splicing, including DM1 (myotonic dystrophy 1) and congenital heart diseases (Dhaenens et al., 2011). Prominent among muscle-specific SFs are members of the CELF (CUG-binding protein and ETR-3–like factor) family of RNA-binding proteins (Barreau et al., 2006). In muscles of DM1 patients, changes in the expression of the CELF founding member CUG-binding protein have been implicated in aberrant splicing of several muscle transcripts, which correlates with impaired fiber-type differentiation (Farkas-Bargeton et al., 1988; Philips et al., 1998; Savkur et al., 2001; Charlet-B et al., 2002). In the mouse model, modulation of the function of CELF proteins affects alternative splicing and also causes changes to relative distribution of fiber types within several muscles studied (Timchenko et al., 2004; Berger et al., 2011). Detailed analysis of mammalian CELF proteins is complicated by the overlapping expression of multiple members with potentially redundant functions (Barreau et al., 2006). A Drosophila orthologue of CELF, named Arrest (Aret; also known as Bruno), has long been studied as a transcriptional repressor in ovaries (Kim-Ha et al., 1995; Webster et al., 1997); however, a role for Aret in muscle biology has not been investigated.
In this study, we identified the Drosophila CELF protein Aret as a central regulator of fiber-specific splicing in flight muscles. We demonstrate that it is expressed in the flight muscles and is both required and sufficient for flight muscle–specific patterns of mRNA splicing. Experimental down-regulation of aret expression leads to dramatic changes in myofibrillar architecture of flight muscles. In addition, we demonstrate that aret is genetically downstream of the transcription factor Spalt major, which, together with the evolutionarily conserved transcription factors Extradenticle and Homothorax, specifies flight muscle fate.
Results
Identification of Aret as a regulator of alternative splicing in adult flight muscles
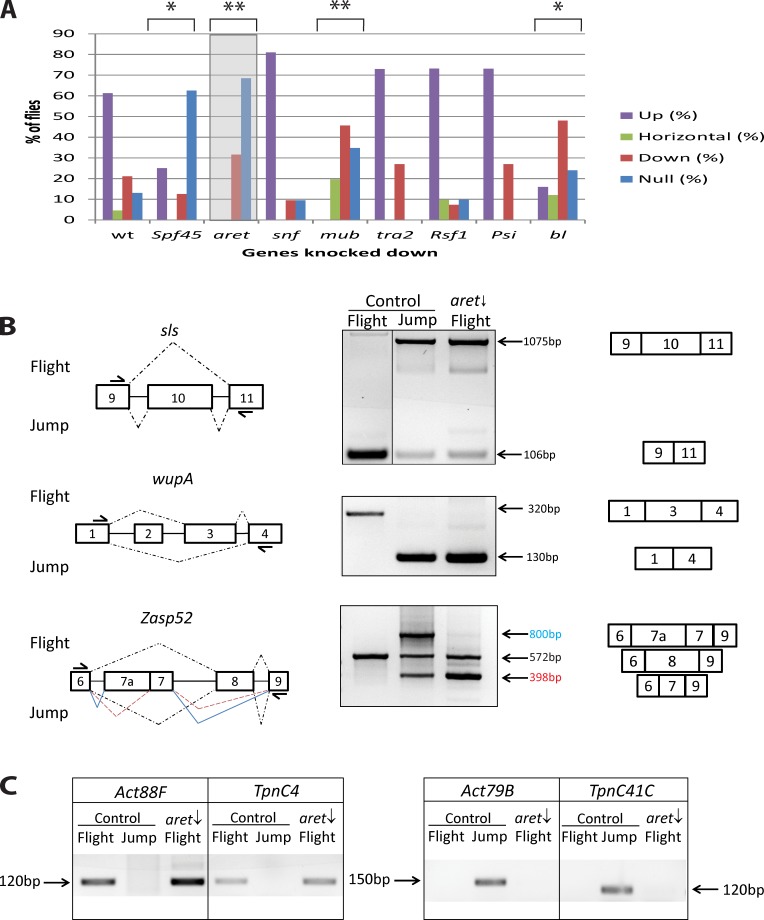
To identify regulators of fiber-specific alternative splicing, we first identified genes encoding proteins with predicted RNA binding ability through their annotations at FlyBase and whose expression was enriched in the flight muscles that were identified in the dataset published by Schönbauer et al. (2011). Transgenic flies harboring a flight muscle–specific driver, Actin88F-Gal4 (Bryantsev et al., 2012a), were crossed to upstream activating sequence (UAS)–RNAi lines targeting the potential splicing regulators. Each KD was assessed for flight ability (Fig. 1 A).
Figure 1.
Identification of Aret as a regulator of alternative splicing in adult flight muscles. (A) Flight test results of KDs in the flight muscles of putative splicing regulators. Flies with RNAi-mediated KD of the indicated genes in flight muscles were assessed for four types of flight behavior: up for normal flight, horizontal and down for weak flight, and not at all for flightless. Control flies are heterozygous Act88F-Gal4 driver flies. The shaded box highlights the results of aret KD, which is the focus of this work. Asterisks indicate the degree of statistically significant deviation from flight of control flies: *, P < 10−5; **, P < 10−15. (B) Analysis of fiber-specific splicing. (left) Gene regions that are differentially spliced between flight and jump muscles. Broken lines indicate splice junctions; junctions of multispliced variants are color coded. Arrows indicate primers used for amplification. (middle) RT-PCR of splicing products analyzed on gels identifies splicing preferences in normal (control) flight and jump muscles and detects splice switching in aret KD (aret ↓) flight muscles. Black line indicates that intervening lanes have been spliced out. (right) Schematics of spliced transcripts corresponding to the amplified products detected on gels. (C) Expression of fiber-specific genes identified by RT-PCR in normal and experimental muscle samples. (left) Expression of flight muscle markers. (right) Expression of jump muscle markers. Note that aret KD (aret ↓) does not alter the expression of fiber-specific genes.
In parallel, we designed an assay to identify flight muscle–specific splicing events among commonly expressed muscle transcripts. In brief, we dissected flight and jump muscles from young adults, isolated RNA from those tissues, and performed RT-PCR for selected genes, to identify patterns of alternative exon usage. This molecular assay allowed rapid, unambiguous identification of a variety of flight muscle–specific splicing events, including selective exon skipping (Fig. 1 B, sls) and exon inclusion (Fig. 1 B, wupA) as well as more complex splicing patterns (Fig. 1 B, Zasp52), as compared with the splicing patterns of the same genes expressed in the jump muscle. The high abundance of transcripts of the selected markers in muscles made this assay amenable to microsampling.
The KD flies exhibiting the most statistically significant loss of flight when compared with controls were mub, aret, bl, and spf45 (Fig. 1 A), and these flies were tested for defects in alternative splicing. Of the genes tested, only aret uniquely satisfied our criteria for a potential splicing regulator because KD animals were flightless, and there was a switch in patterns of splicing within the flight muscles. For mub, bl, and spf45 KDs, there were no detectable changes in the alternative splicing pattern of flight muscle transcripts (unpublished data).
When samples of aret KD flight muscles were analyzed for their patterns of splicing, we observed a dramatic effect on flight muscle–specific splicing, in which all (seven out of seven) of the markers tested reverted to splicing patterns characteristic of the jump muscle (Figs. 1 B and S1 A). Thus, aret affects fiber-specific splicing control in flight muscles.
Next, we wanted to determine whether the observed changes in flight muscle splicing were caused by an overall change in muscle identity. We analyzed expression of flight and jump muscle–specific genes that served as molecular hallmarks of correct muscle fiber identity. Specifically, we tested expression of Act88F and TpnC4, which are almost exclusively expressed in flight muscles (Karlik et al., 1984; Herranz et al., 2004), and Act79B and TpnC41C, which are expressed in the jump muscle but not flight muscles (Fyrberg et al., 1983; Herranz et al., 2004). Our analysis determined that fiber-specific genes did not change their normal expression patterns (Fig. 1 C), indicating that aret down-regulation does not lead to identity transformation of flight muscles. Altogether, our data demonstrate that aret is important for proper functioning of flight muscles, where it is involved in the regulation of flight muscle–specific splicing events.
Developmental dynamics of Aret during flight muscle formation
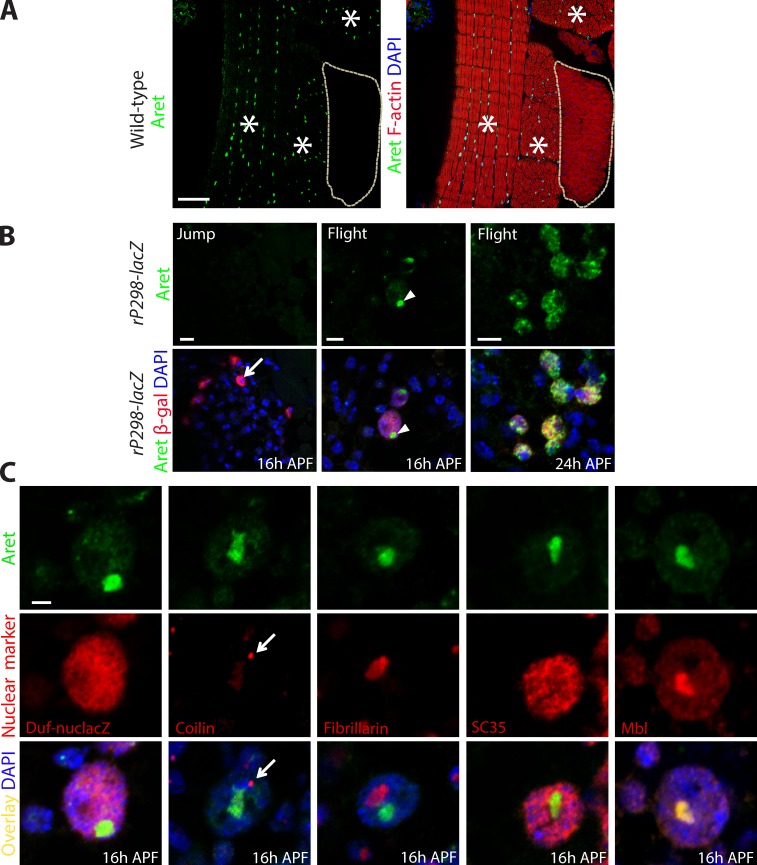
We applied immunofluorescence to analyze Aret protein distribution in mature adult muscles as well as during the course of adult myogenesis in pupa. We observed Aret expression exclusively in flight muscles, where the protein was localized to the nuclei (Fig. 2 A). As seen before, the only other location of Aret in adult females was in the ovaries, where the protein was localized in the cytoplasm of developing oocytes (Webster et al., 1997). No other adult muscle group, including the jump muscle, expressed Aret at immunologically detectable levels. We also confirmed that aret KD affected Aret accumulation in the adult muscles, in which KD animals showed no detectable Aret accumulation (Fig. S1 B). We further characterized aret transcript expression in samples of dissected flight muscles, by carrying out RT-PCR analysis using exon-specific primers. We found that the most likely flight muscle–specific transcript of aret is the RA isoform annotated at FlyBase (Fig. S2).
Figure 2.
Aret shows a dynamic localization in the nuclei of flight muscles and flight muscle precursors. (A) Immunofluorescent analysis of Aret expression in the thorax of pharate adults (96 h APF). Aret accumulates in nuclei of flight muscles (red; asterisks) but not jump muscles (red; outlined). High magnification views of these images are shown again in Fig. 6 A. (B) Early in development (16 h APF), jump muscle FCs (arrow) are devoid of Aret (left), whereas flight muscle FCs have Aret accumulated in a large nuclear granule (center, arrowheads). In fusing nascent flight muscle myofibers (24 h APF; right), Aret is dispersed throughout the nuclei and forms small, brighter foci. (C) Aret accumulates in a novel nuclear domain within nuclei of flight muscle FCs. Nuclei (blue) of FCs are positive for β-galactosidase (β-gal) in rP298-LacZ flies and contain distinct nuclear domains: Cajal body (detected with antibody to Coilin, arrows), nucleolus (Fibrillarin), and nuclear speckles (SC35). Aret does not colocalize with any of these domains (merged images are in the bottom row). The SF Muscleblind (Mbl) colocalizes with Aret in this novel nuclear domain. Bars: (A) 50 µm; (B) 5 µm; (C) 2 µm.
To better understand the origin of Aret expression in flight muscles, we analyzed early stages of adult myogenesis. At 16 h after puparium formation (APF), myotubes are not yet formed, and the myogenic cells are represented by two types: predominant fusion-competent myoblasts and rare founder cells (FCs; Jaramillo et al., 2009). The latter have larger nuclei and express the molecular marker Duf, which can be detected with the lacZ enhancer trap transgene rP298 (Nose et al., 1998; Ruiz-Gómez et al., 2000). At 16 h APF, we observed Aret expression only in the rP298-positive flight muscle FCs, where it was concentrated in large intranuclear foci (Fig. 2 B, flight, 16 h APF). In contrast, Aret was not detectable in FCs for the jump muscle (Fig. 2 B, jump, 16 h APF). At 24 h APF, during fusion of FCs with myoblasts, Aret dispersed from its large nuclear focus, to a more broad nuclear distribution with sporadic small foci (Fig. 2 B, flight, 24 h APF).
To map the large Aret-accumulating foci to a known subnuclear domain, we performed a series of colocalization experiments, costaining FC nuclei with antibodies to markers of the Cajal body (positive for Coilin), nucleolus (Fibrillarin), and nuclear speckles (SC35). None of the tested nuclear domains demonstrated colocalization with the Aret-containing nuclear structures (Fig. 2 C). These observations suggest that Aret accumulates in a separate or previously uncharacterized nuclear domain. We named this location in the nucleus as the Bruno body (B body). To further characterize B-body composition, we probed for localization of another SF, Muscleblind (Mbl), which has been previously implicated in the regulation of alternative splicing of muscle transcripts (Vicente et al., 2007; Vicente-Crespo et al., 2008). Here, we observed a precise overlap in localization between Mbl and Aret (Fig. 2 C, Mbl). Thus, we tentatively assign the B body a role in accumulation or storage of SFs involved in muscle alternative splicing regulation, with two known members, Aret and Mbl.
Overall, our findings of Aret’s exclusive localization in the flight muscles are consistent with a role for Aret in controlling flight muscle–specific alternative splicing. This also suggests that the physical presence of Aret might by itself be a sufficient determinant for flight muscle–specific splicing.
Flight muscles with down-regulated aret exhibit alterations in myofibrillar architecture
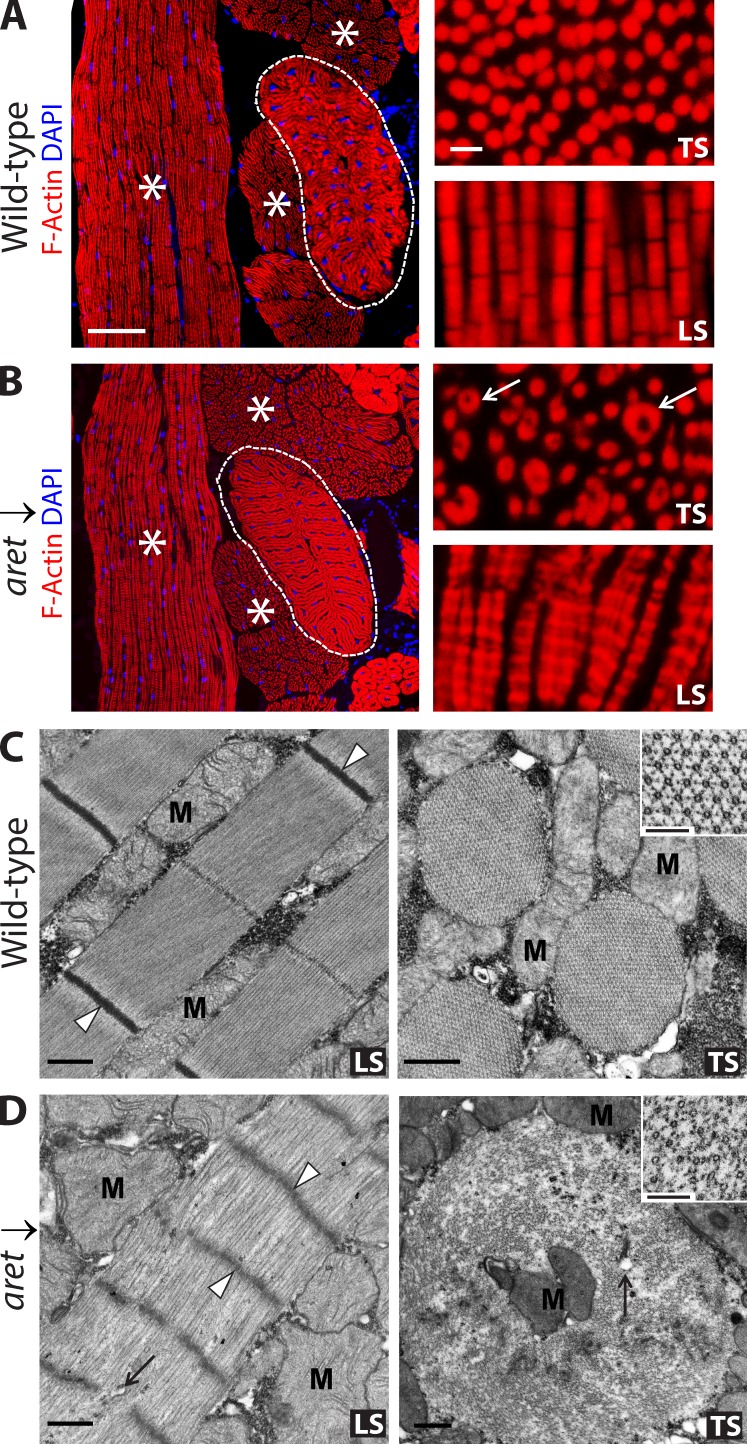
To investigate the cause of the flight impairment in aret KD flies, we examined control and aret KD flight muscles. We found that aret KD flight muscles were located normally within the thorax, demonstrated correct attachments to the cuticle, contained evenly dispersed nuclei, and overall, were readily recognizable as flight muscles (Fig. 3 A, left). At a higher magnification, myofibrils of control flight muscles that had been stained with phalloidin demonstrated uniform, round shapes in transverse section (TS; Fig. 3 A, TS) and a characteristic striation pattern in longitudinal section (LS; Fig. 3 A, LS). In contrast, in aret KD flight muscles, the myofibrils demonstrated a wide range of shapes and sizes and had altered and striation patterns in which adjacent myofibrils were held in register to one another (Fig. 3 B, LS). Notably, on TSs of aret KD flight muscles, we often observed myofibrils with a hollow core (Fig. 3 B, TS). This prompted us to use electron microscopy to obtain a better understanding of myofibrillar alterations upon aret KD.
Figure 3.
Effect of aret KD on the morphology of the flight muscles. (A and B) Muscle morphology in control (WT; A) and aret KD (aret ↓; B) flies, assessed using fluorescent microscopy. Flight muscles (asterisks) do not change in size and position upon aret KD. The jump muscle (outlined) is also unaffected. At higher magnification (right), WT myofibrils are cylindrical, appearing circular in transverse section (TS) and showing regular striations in longitudinal section (LS). In aret KDs, the myofibrils are irregular in shape, often enlarged and with hollow centers in TS, and showing increased width and changes in shape and striation pattern in LS (arrows). Bars: (left) 50 µm; (right) 5 µm. (C) Electron microscopy of control sarcomeres shows electron-dense Z discs (arrowheads) and sharp myofibril boundaries, interspersed with mitochondria (M) in LS. In TS, there are hollow thick filaments and hexagonal packing of thin filaments around the thick filaments (inset). (D) In aret KDs, there was a loss of normal sarcomere structure, fuzzy and penetrated Z lines (arrowheads), loose filament packing, diffuse myofibril boundaries, and incorporated sarcoplasmic elements (arrow) in LS. In TS, myofibril alterations included increased cross-sectional area, loose packing of filaments, incorporation of mitochondria (M) and other membranous elements in the myofibril vicinity (arrow), and loss of regular hexagonal organization of the thin filaments around the thick filaments (inset). Bars: (C and D, main images) 0.5 µm; (C and D, insets) 100 nm.
At the electron microscope level, it was evident that myofibrils from aret KD flight muscles were made of significantly less densely packed myofilaments, which resulted in severe structural perturbations. Remarkably, aret KD myofibrils lost normal sarcomeres that are defined by dark, sharp Z lines in control myofibrils. Instead, aret KD myofibrils had fuzzy, often penetrated, broad Z lines (Fig. 3, compare C [LS] with D [LS]). Moreover, the sarcomere length was significantly shorter in aret KD flight muscle myofibrils: WT sarcomere length was 3.53 ± 0.05 µm, whereas sarcomeres in aret KD flight muscle myofibrils were 1.08 ± 0.06 µm (P = 3.3 × 10−9). On cross sections, thin and thick filaments were recognizable in aret KD fibrils, but the typical hexagonal lattice organization of normal fibrils in which six thin filaments surround each thick filament was lost (Fig. 3, C and D, TS, compare insets). The loose packing of filaments within aret KD myofibrils allowed incorporation of cytoplasmic elements, such as mitochondria and membranous structures, corresponding to the hollow-centered fibrils detected at the light microscopy level (Fig. 3, C and D, compare TS images). The large diameters of aret KD myofibrils might result from fusion of several adjacent myofibrils. The coincidence of structural alterations and fiber-specific alternative splicing deregulation that we observed upon aret KD suggests a substantial role that alternative splicing plays in the organization of Drosophila flight muscle ultrastructure, although it is unknown to what extent other aspects of Aret function might contribute to this phenotype.
Aret is a potent regulator of flight muscle–specific splicing
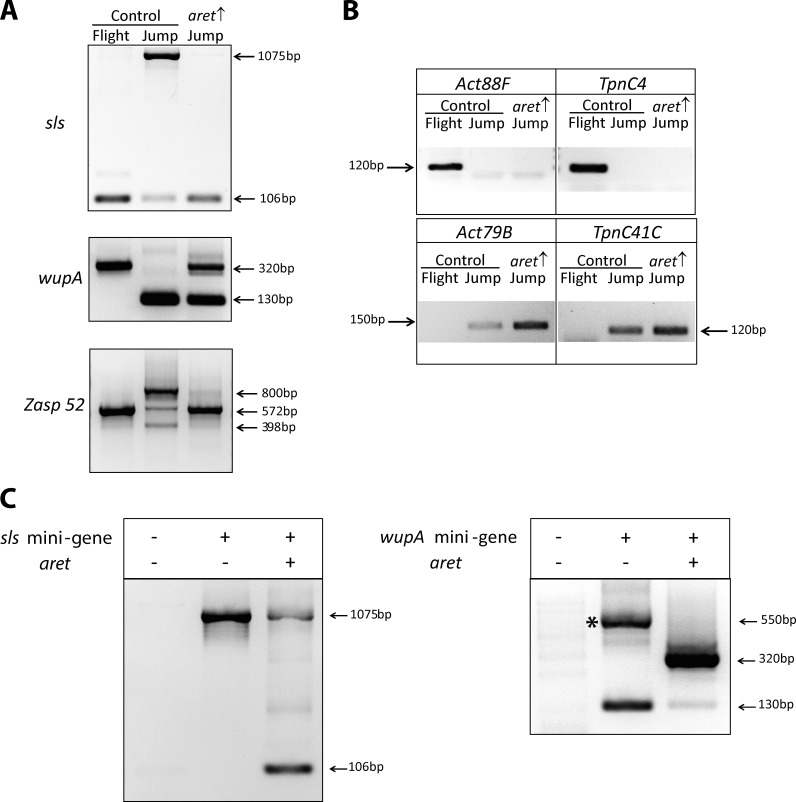
To investigate whether Aret has the capacity to promote flight muscle–specific alternative splicing outside the flight muscle environment, we ectopically expressed the gene in the jump muscle and used the same panel of spliced structural genes to monitor changes in fiber-specific splicing. Ectopic expression of aret in early jump muscles, immediately after fusion, was deleterious to muscle morphology and resulted in loss of jump muscles by the end of pupal development (unpublished data). Therefore, Aret expression was initiated later in jump muscle development, during the hypertrophic growth phase (48 h APF) and expression was continued until the end of pupal development (96 h APF). Note that the driver system that we used for this experiment (tubulin-Gal80TS; Mef2-Gal4) is active in all muscles when animals are switched to elevated temperatures. Under these conditions, jump muscles developed and appeared morphologically normal, but their functional analysis was precluded by the inability of the adults to eclose, presumably because of general muscle weakness caused by ectopic aret expression. Molecular analysis showed that the presence of Aret in the jump muscle significantly promoted flight muscle–specific splicing of the selected muscle transcripts (Fig. 4 A). Some residual presence of jump muscle isoforms could be a result of the late initiation of Aret expression. Importantly, Aret introduction to the jump muscle did not alter normal jump muscle identity, as seen by unaltered expressing of fiber-specific markers (Fig. 4 B). This analysis ruled out the possibility that the changes in alternative splicing were caused by transformation of the muscle fiber type.
Figure 4.
Aret is sufficient to enforce flight muscle–specific splicing. (A) Ectopic expression of aret in the jump muscle (aret ↑), changes the splicing of fiber-specific spliced genes to the state of the flight muscle. Refer to Fig. 1 for key to splice patterns and products. (B) Ectopic expression of aret in the jump muscle does not alter muscle identity. RT-PCR detection of fiber-specific gene expression in samples of control (WT) flight muscles and jump muscles and jump muscles ectopically expressing aret (aret ↑). (C) aret promotes flight muscle splicing choices in the naive environment of cultured S2 cells. Nontransfected cells do not express muscle transcripts (left lanes), but transcripts of transfected sls and wupA minigenes readily assume jump muscle splicing patterns (middle lanes). Note that the 550-bp wupA product (asterisk) is a result of erroneous splicing. Cotransfection of aret along with the minigenes changes the splicing of minigene transcripts to the flight muscle state (right lanes). Refer to Fig. 1 for key to splice patterns and products.
Next, we wanted to determine whether Aret could promote flight muscle–specific splicing choices in the molecular environment of cultured cells. We generated minigenes for sls and wupA, in which a constitutive promoter drove transcription of alternatively spliced exons as well as the introns and exons on either side. When transfected into Drosophila S2 cells, transcripts from the minigenes assumed the splicing patterns seen in the jump muscle (Fig. 4 C, center lanes), suggesting that for these exons, the jump muscle pattern is a default splicing choice. Note that the wupA minigene, in addition to the correct jump muscle isoform, also produced an unexpected 550-bp band that we identified as a splicing artifact resulting from intron retention (Fig. 4 C, right, asterisk). Remarkably, when we cotransfected an Aret expression plasmid with the minigenes, both minigenes produced flight muscle spliced transcripts (Fig. 4 C, right lanes). Collectively, the Aret ectopic expression experiments confirmed that Aret is a potent regulator, promoting flight muscle–specific splicing even in the molecular contexts of the jump muscle and cultured cells.
Aret regulates inclusion of an alternatively spliced exon through conserved intron sequences
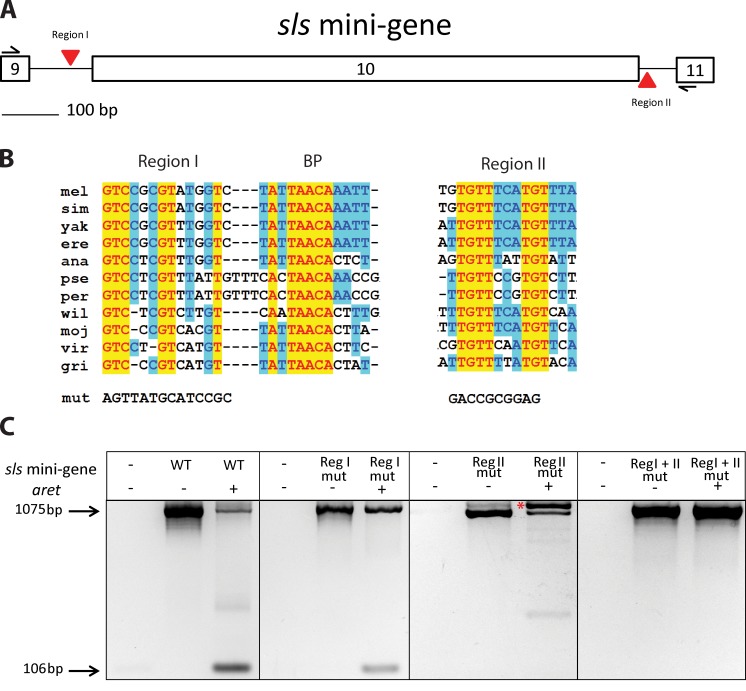
To identify cis-acting elements that could mediate Aret-directed splicing selection, we searched for evolutionarily conserved intron sequences. We focused upon the introns flanking the alternatively spliced exon 10 of sls because exon 10 was excluded from mature transcripts in the presence of Aret in our cell culture experiments. After aligning the corresponding intron sequences from 12 Drosophila species, we located two conserved regions in addition to the expected conservation at the splice junctions. Conserved region I and region II were located within the upstream and downstream introns, respectively (Fig. 5, A and B). Upon closer examination, part of the sequence in region I was identified as a possible splicing branchpoint (Fig. 5 B, labeled BP), and its subsequent mutation caused permanent intron retention (not depicted). Thus, for functional analysis in the sls minigene, we mutated individually and in combination the 5′ half of region I and the highly conserved sequences in region II (Fig. 5 C). The resulting mutated minigenes were used in transfection experiments.
Figure 5.
RNA cis-regulatory sequences participating in Aret-dependent splicing. (A) Schematic of the sls minigene, indicating locations of the evolutionarily conserved areas termed region I and region II. (B) Sequence and conservation of region I (reg I) and region II (reg II). Yellow and blue highlights indicate absolute and significant nucleotide conservation across 12 Drosophila species (mel, Drosophila melanogaster; sim, Drosophila simulans; yak, Drosophila yakuba; ere, Drosophila erecta; ana, Drosophila annanassae; pse, Drosophila pseudoobscura; per, Drosophila persimilis; wil, Drosophila willistoni; moj, Drosophila mojavensis; vir, Drosophila virilis; gri, Drosophila grimshawi). BP indicates a consensus branch point. Mut refers to the sequence of the mutated region I and region II tested in C. (C) Effects of mutated region I and region II on splicing of the sls minigene in S2 cells. Nontransfected cells do not express sls transcripts (left lanes in each section). Expression of minigenes of native sequence (WT) or with mutated region I (reg I mut) or region II (reg II mut) or with a combination of the mutations (reg I + II mut) invariably results in exon 10 inclusion in spliced transcripts, which corresponds to jump muscle splicing (middle lanes). With aret coexpression (right lanes), region I mutated reduces the relative amount of transcripts; with exon 10 exclusion, region II mutated abolishes exon 10 exclusion but leads to abnormally spliced transcripts with retained upstream intron (asterisk); and region I + II mutated effectively prevents exon 10 exclusion and results in jump muscle splicing despite the presence of Aret.
In the absence of cotransfected Aret, all mutated constructs produced unaltered jump muscle–specific splice products, similar to the wild-type (WT) minigene in the absence of Aret, indicating that exon 10 was recognized normally, and the adjacent introns were removed (Fig. 5 C). However, in the presence of Aret, the region I mutant produced noticeably less flight muscle–specific product than the WT minigene, and the region II mutant encountered significant problems in correct splicing, generating incorrect transcripts retaining the upstream intron. When both region I and region II were mutated, sls minigene transcripts displayed jump muscle–specific splicing even in the presence of Aret (Fig. 5 C). These results indicate that individually, the mutation of region I and region II only attenuate correct sls gene splicing in the presence of Aret, whereas disruption of both regions eliminated Aret-dependent flight muscle splicing. The results of our mutation observations support the idea that Aret-dependent action is mediated by multiple conserved sequences within introns of muscle genes.
aret is a component of the genetic network regulating flight muscle identity
Previous studies showed that the homeodomain transcription factor genes exd and hth and the zinc-finger transcription factor gene salm are involved in the regulation of flight muscle identity. exd and hth work in concert to promote flight muscle fate and are genetically upstream of salm (Schönbauer et al., 2011; Bryantsev et al., 2012b). Although many flight muscle–specific genes appear to be under salm-dependent control, some genes (such as Act88F) receive direct regulation from exd/hth (Bryantsev et al., 2012b).
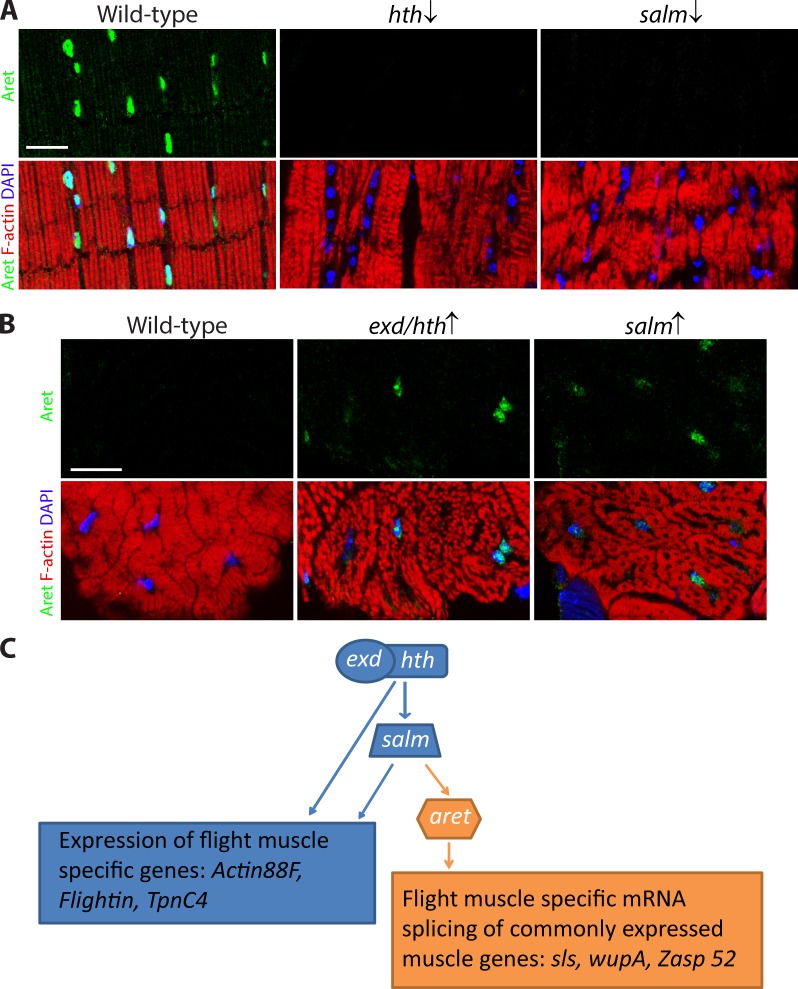
We determined the location of aret in this transcriptional framework by genetically manipulating the transcription factors that impact fiber fate. KD of either hth or salm in flight muscles invariably suppressed aret expression, as detected by immunofluorescence (Fig. 6 A). Along with aret inactivation, the KD flight muscles acquired the morphology of the jump muscle as a result of identity transformation (Fig. 6 A). In converse experiments, when either salm or exd plus hth were forcefully overexpressed in the jump muscle, aret expression was ectopically activated (Fig. 6 B). These results place aret within the salm-dependent branch of the genetic network controlling flight muscle identity (Fig. 6 C).
Figure 6.
aret location in the transcriptional pathway specifying flight muscle identity. (A) Immunological detection of Aret expression in control (WT), hth KD (hth ↓), and salm KD (salm ↓) flight muscles, counterstained for F-actin and nuclei (blue). Note that the KD muscles do not accumulate Aret and have transformed myofibrils resulting from identity transformation. Low-magnification views of the images for WT were shown previously in Fig. 2 A. (B) Localization of Aret in jump muscles of control (left), jump muscles ectopically expressing exd plus hth (exd/hth ↑), and jump muscles expressing salm (salm ↑). Note the transformed fibrillar-type jump muscles accumulate Aret in the nuclei. Muscle morphology was assessed using phalloidin to highlight actin filaments and DAPI to stain nuclei. (C) Scheme of genetic interactions in the flight muscle identity pathway. The identity genes exd/hth and salm encode proteins that regulate expression of flight muscle structural genes; salm is genetically upstream of aret. Aret promotes flight muscle–specific alternative splicing of muscle transcripts. Bars, 20 µm.
Discussion
In this paper, we demonstrate that the Drosophila CELF orthologue Aret promotes flight muscle–specific patterns of alternative splicing during adult muscle development. aret expression within the adult musculature occurs early during flight muscle specification and persists until the end of pupal development. aret KDs show switches in splicing, such that transcripts in the flight muscles are spliced in a pattern characteristic of the jump muscles, and there is a concomitant disruption of flight muscle ultrastructure. Collectively, the loss of flight muscle splicing in aret KDs, the ability of Aret to promote flight muscle splicing in jump muscles and in S2 cells, and the identification of sequences surrounding an alternatively spliced exon that are essential for Aret function indicate that Aret is a regulator of alternative splicing in the flight muscles.
Is Aret a master regulator of flight muscle alternative splicing?
According to FlyBase, there are ∼250 muscle-related genes having more than one annotated transcript isoform. This number implies that transcript splicing in muscles probably involves hundreds of events that have to be properly controlled in different fiber types. Although the pathways of fiber-specific splicing are far from being understood, our study suggests that Aret could be a master regulator of such splicing in the flight muscles. This statement is supported by the significant impact imposed by aret KD on the structure of the flight muscles and by the fact that all of the randomly selected and analyzed flight muscle–specific splicing events were Aret dependent (Figs. 1 B and S1). In the much better discerned sex-specific splicing pathway, the master regulator Sxl (Bell et al., 1988) controls a cascade of splicing events that ultimately lead to sex determination. In the flight muscles, it remains to be determined whether Aret controls all fiber-specific splicing events directly or influences some events through alternatively spliced SF proxies, similar to the action of Sxl (Salz, 2011).
Importance of alternative splicing to myofibril structure
One parameter that is used to differentiate between muscle fiber types in a wide spectrum of species, including humans, is the morphological appearance of myofibrils (Payne et al., 1975; Gauthier, 1979). In Drosophila, myofibrils of the flight muscles bear unique morphological features distinguishing them from myofibrils of other adult muscles (O’Donnell et al., 1989). In aret KD flight muscles, abnormal myofibrillar organization was the most visible effect. Because fiber-specific genes were expressed in aret KD flight muscles properly, we tentatively attribute the myofibrillar damage to the expression of fiber-inappropriate isoforms of structural proteins, hence highlighting the importance of alternative splicing in the formation of myofibrils. Meanwhile, it should be noted that other documented Aret functions include translational regulation and control of mRNA oligomerization. Whether these activities are part of the role of Aret in flight muscles remains to be determined, thus it is conceivable that a component of the Aret KD phenotype might arise from derangement of these other activities. The aret KD results also suggest that myofibrillar morphology and muscle function can serve as a sensitive readout of misregulation in muscle splicing. Indeed, myofibrillar abnormalities have been observed in human myotonic dystrophy (Silver et al., 1984) and experimental animal models (Machuca-Tzili et al., 2011; Majczenko et al., 2012), reportedly affecting normal splicing of muscle transcripts.
Reconciling the dual functions of Aret
We also show that Aret can function as a SF, although previously this protein (also known as Bruno) was extensively studied as a potent translational repressor (Webster et al., 1997; Kim-Ha et al., 1995; Filardo and Ephrussi, 2003; Moore et al., 2009). How can these two Aret functions be reconciled? We note from the immunofluorescent analysis that Aret is present in two distinct locations in adult flies: the gonads and the flight muscles. Moreover, the intracellular localization of Aret in the cytoplasm in ovaries (Snee et al., 2008) and in the nucleus in muscles (this study) is also distinct. However, the ovarian and muscle isoforms of Aret do not differ in their amino acid sequences (Fig. S2; Webster et al., 1997), thus ruling out the possibility that the functional divergence of Aret can lie in sequence differences of its protein isoforms. We therefore speculate that the molecular environment and, especially, interacting partners could have a determining impact on Aret localization and function. For example, in the ovary Aret physically interacts with the germline-specific protein Vasa, which is not expressed in muscles (Webster et al., 1997). A flight muscle partner for Aret has yet to be identified, although the colocalization of Aret and Mbl in the B bodies and observation of their genetic interaction (Vicente-Crespo et al., 2008) suggest that Mbl would be a candidate protein for such a role.
Localization of Aret to a nuclear domain in FCs
The earliest accumulation of Aret during flight muscle development was in flight muscle FCs, in which Aret localized to a large body within the nucleus. Numerous intranuclear domains have been characterized in the last 20 yr, and many have been shown to have critical functions in particle storage or segregation of functional domains within the nucleus (Spector, 2001). However, the localization of Aret does not correspond to any known nuclear domain in Drosophila, and we termed this accumulation the B body.
What is the nature of the B body? Its most likely role is as a storage granule within the nucleus. We reason that large amounts of Aret are segregated into the B bodies of FCs, such that upon fusion with non-Aret–positive fusion competent myoblasts, this reservoir of Aret protein can be dispersed into the other nuclei, where it can facilitate correct fiber-specific splicing of muscle genes.
The fact that Aret colocalizes with Mbl in the B body argues that the B body has a role in storage of at least some alternative SFs because both Aret (this study) and Mbl impact alternative splicing in muscle (Ho et al., 2004; Vicente-Crespo et al., 2008). Moreover, the presence of at least two factors in this structure implies that it represents an organized nuclear storage location.
Our data also demonstrate that Aret is released from the B body around the time of myoblast fusion, at which time Aret becomes more broadly dispersed in the founder nuclei, and in nuclei arising from myoblasts that fused with the FC. This is presumably the time at which Aret becomes functional in regulating flight muscle alternative splicing. How this dynamic control of Aret location is regulated has yet to be defined. One possibility is that a trigger for its release may be the same as that which promotes myoblast fusion and activation of muscle structural gene expression. An alternative model is that Aret localizes passively in the B body in the absence of splicing targets, but upon transcription of muscle genes, the muscle transcripts serve as a sink for Aret distribution. Similar observations have been made for nuclear speckles, in which the sizes of the speckles increase when splicing targets are removed through inhibition of transcription (O’Keefe et al., 1994).
Nuclear foci of CELF proteins have not been observed in other systems. However, given the sensitivity of certain fiber types to manipulations with CELF proteins (see following paragraph), the appropriate time and place to investigate such accumulations would be in subsets of skeletal myoblasts at the onset of myoblast fusion.
A regulatory network for muscle fiber identity
A role for CELF proteins in vertebrate alternative splicing has been demonstrated during cardiac development, in which developmental increase in CELF protein accumulation in the heart correlates with adult patterns of splicing predominating over fetal patterns. Moreover, CELF factors directly regulate this process, through interacting with several pre-mRNA molecules to influence their splicing (Ladd et al., 2001, 2005; Kalsotra et al., 2008). Inappropriate expression of CELF, as well as resulting missplicing of muscle transcripts, is also a major contributor to the pathology of adult onset–type muscular dystrophy (Philips et al., 1998; Savkur et al., 2001; Charlet-B et al., 2002). Recently, expression of a dominant-negative CELF isoform in mouse skeletal muscles resulted in some defects of alternative splicing and an increase in the relative presence of slow fibers in the muscle (Berger et al., 2011). This may be a result of selective elimination of fast fiber types as a result of their sensitivity to altered CELF function. Interestingly, fast fiber fate also depends on vertebrate Pbx and Meis proteins in zebrafish and probably mice (Heidt et al., 2007; Maves et al., 2007). Our experiments for the first time place these observations into an integrative context, in which we demonstrate that the Drosophila orthologues of Pbx, Meis, and CELF (Exd, Hth, and Aret, respectively) are part of a regulatory pathway, which links transcriptional control with alternative splicing regulation to promote flight muscle fiber fate.
Materials and methods
Flies and crosses
Fly stocks were obtained from the Bloomington Drosophila Stock Center (BDSC) or Vienna Drosophila RNAi Center (VDRC) and maintained on Jazz Mix medium (Thermo Fisher Scientific). Screening crosses were performed at 29°C using the Act88F-Gal4 driver line (Bryantsev et al., 2012a). Act88F-Gal4 is a transgenic line containing enhancer sequences from the flight muscle actin gene Act88F fused to a minimal heat shock promoter and a Gal4 cDNA.
The following RNAi-inducible fly lines were used in this study: 32948 (targeting Spf45; obtained from VDRC), 48237 and 41567 (aret; VDRC), 104334 (snf; VDRC), 105495 (mub; VDRC), 100805 (tra2; VDRC), 22186 (Rsf1; VDRC), 105135 (psi; VDRC), 2912 (bl; VDRC), 34637 (hth; BDSC), 100687 (exd; VDRC), and 101052 (salm; VDRC). In flight muscle transformation experiments, hth, exd, or salm RNAi lines were crossed with the 1151-Gal4 enhancer trap driver line that is active in adult myoblasts (Anant et al., 1998; Bryantsev et al., 2012b). In jump muscle transformation experiments, UAS-salm flies, comprising the salm coding sequence under the control of a promoter containing UAS (obtained from F. Schnorrer, Max Planck Institute of Biochemistry, Martinsried, Germany), were crossed with Mef2-Gal4 (Ranganayakulu et al., 1998), or UAS-exd; UAS-hth flies were crossed with Act79B-Gal4 flies (Bryantsev et al., 2012a). UAS-exd and UAS-hth comprise the coding sequences of the respective genes under the control of a promoter containing UAS, and the Act79B-Gal4 line comprises enhancer sequences from the tubular muscle actin gene Act79B fused to a minimal heat shock promoter and a Gal4 cDNA. The UAS-aret transgenic flies were generated via P element–mediated transgenesis by Rainbow Transgenic Flies, Inc., using the molecular construct described under Molecular cloning. As a result of the severe damage of developing jump muscles, in aret ectopic expression experiments, the Act79B-Gal4 driver was substituted for the temperature-sensitive inducible driver system, consisting of Mef2-Gal4 (comprising 9 kb of Mef2 regulatory sequence upstream of a minimal promoter and Gal4 cDNA and expressed in all muscles) and tubulin-Gal80TS (BDSC stock 7016; McGuire et al., 2003). Activation of aret expression was then initiated in the jump muscles, as well as other adult muscles, at 48 h APF by raising the incubation temperature from 18 to 29°C and maintaining animals at the elevated temperature until the end of pupal development.
Flight testing was performed in a flight chamber as previously described (Drummond et al., 1991), In brief, flies released inside the chamber were scored for whether they flew upwards, horizontally, downward, or not at all. At least 20 flies per genotype were assayed.
Molecular cloning
To create an inducible Aret-expressing construct, clone LD29068, encoding the Aret-PA protein isoform, was obtained from the Drosophila Genomics Resource Center. The coding region with attached full-size 5′ and 3′ UTRs was amplified by PCR and recombined into pUASTattB (Bischof et al., 2007) using a cloning kit (GeneArt Seamless; Invitrogen). The pPacPl-Gal4 construct was created by conventional ligation-based subcloning of the Gal4 coding sequence from pAct79B-Gal4 (Bryantsev et al., 2012a) into pPacPl (FBmc0001179; FlyBase) at the SpeI sites. To create sls and wupA minigenes, appropriate genomic fragments were PCR amplified using the primers listed in this paragraph and then recombined into pUASTattB precut with NotI and KnpI using the GeneArt kit: sls forward, 5′-TCGTTAACAGATCTGCCGCGCAGTATGTGCAAAAT-3′; sls reverse, 5′-GATCCTCTAGAGGTACAAACCGTTCCACGAAAAGTG-3′; wupA forward, 5′-TCGTTAACAGATCTGCCCAGTTCCTTGAGTTCACCTCTC-3′; and wupA reverse, 5′-GATCCTCTAGAGGTACGTCTACGTTCAGCCGCTTTG-3′. Bold font denotes introduced sequences required for product recombination with the vector.
The sls mutant constructs were PCR amplified, with the introduced mutations engineered to generate restriction enzyme sites: NsiI for sls region I and SacII for sls region II. The PCR fragments were recombined into pUASTattB using the GeneArt kit. The following primers were used to produce the mutations: sls region I mutant forward, 5′-TAGTTATGCATCCGCTATTAACAAATTATTTCGTGTTTTGTTG-3′; sls region I mutant reverse, 5′-AGCGGATGCATAACTAGTGACAAATAATTTTAATGGTTGACA-3′; sls region II mutant forward, 5′-TTCTGGACCGCGGAGTTAGGTCTACTATAAATGTTGT-3′; and sls region II mutant reverse, 5′-CCTAACTCCGCGGTCCAGAACTTACCTTCGATTGTTA-3′. The bold sequences are sequences required to recombine the PCR fragments with the rest of the sls sequence. For each mutant construct, the mutant primers were used in combination with the corresponding cloning sls primers described in the previous paragraph, to obtain two PCR products that were next recombined in a single reaction between themselves and into the KpnI−NotI precut pUASTattB vector. The double mutant was made similarly, by introducing a mutation in region II in a construct already bearing a region I mutation.
Cell culture
Drosophila S2 cells were maintained at 28°C in standard Schneider’s medium (Gibco) supplemented with 10% of fetal bovine serum (HyClone). Transfections were performed with TransIT-2020 (Mirus Bio LLC) according to the manufacturer’s instructions, with 3 µl of reagent per 1 µg of total plasmid DNA. Typically, plasmid DNA mixes contained pUAS-aret, a minigene construct, and pPacPl-Gal4 premixed at a 9:9:2 ratio. For negative controls, pUAS-aret was substituted with empty parental vector pUASTattB. Transfected cells were used for RNA extraction at ∼24 h after transfection. Details of RNA extraction, cDNA synthesis, and RT-PCR analysis were as described below (Expression analysis).
Expression analysis
Either flight or jump muscles were dissected out from thoraces and used for RNA extraction with the RNeasy Mini kit (QIAGEN). cDNA was synthesized from 50–100 ng of collected RNA using SuperScript II Reverse Transcriptase (Invitrogen) and random hexamer primers (Roche). Diluted cDNA was used as template for subsequent PCR analysis with Pfx Polymerase (Invitrogen) and the following pairs of gene-specific primers: sls, 5′-CGCGCAGTATGTGCAAAAT-3′ and 5′-AAACCGTTCCACGAAAAGTG-3′; wupA, 5′-ACACAAATCAAAATGGCTGATG-3′ and 5′-GGGGTCATGAAACCCTTCTT-3′; Zasp52, 5′-ATCGCTTCCGACGTTCTGAAG-3′ and 5′-GTCGCAGTAGAGCTTGTTGTTG-3′; Zasp66, 5′-TCCACAAGCAATTCAACTCG-3′ and 5′-GATACTGGCGCTGATACTGG-3′; Act88F, 5′-AGCTCTTCAAAGGCAGCAAC-3′ and 5′-ATTGTTGTGCGATGGGTTC-3′; TpnC4, 5′-TGGCAGCTCGCTTTATTG-3′ and 5′-GTGTTGCAACTGTCAGGTATCC-3′; Act79B, 5′-TGTCTCCAGCGTAAGACATCC-3′ and 5′-TTCCGGTCTTTTCTCGTCTC-3′; and TpnC41C, 5′-CGCCTTTACGACAAAGAAGG-3′ and 5′-CATGTCCAGGTCGTCATTTG-3′.
The PCRs were run for 30–40 cycles. The appropriate dilution of cDNA was determined in separate, quantitative PCR amplifications, to yield equal amplification of the reference gene Mhc across samples (Bryantsev et al., 2012b). Final amplification products were resolved in 2% agarose gels.
Immunofluorescence and electron microscopy
Cryosections were made and analyzed as described previously (Morriss et al., 2012). In brief, pupae of the desired stages were collected, and the pupal cases were removed. Samples were frozen in Tissue-Tek optimal cutting temperature compound and frozen in liquid nitrogen. Sections were cut at a thickness of 10 µm at −18°C using a cryomicrotome (Minotome Plus; Triangle Biomedical Sciences) and collected on slides. Sections were fixed in 3.7% formaldehyde in PBS for 8 min at room temperature and then washed in PBS containing 0.1% (vol/vol) Triton X-100 before incubation with primary antibodies. We used the following primary antibodies and sera: rabbit anti-Aret (obtained from P. MacDonald, University of Texas at Austin, Austin, TX), guinea pig anti-Coilin (J. Gall, Carnegie Institution, Baltimore, MD), mouse anti-SC35 (Sigma-Aldrich), mouse anti-Fibrillarin (Abcam; provided by O. Pontes, University of New Mexico, Albuquerque, NM), mouse anti–β-galactosidase (Promega), and sheep anti-Mbl (provided by D. Monckton, University of Glasgow, Glasgow, Scotland, UK). Phalloidin and secondary antibodies, labeled with Alexa Fluor dyes 488, 568, and 633, were obtained from Molecular Probes. Stained samples were mounted in mounting medium containing 10% (vol/vol) polyvinyl alcohol and 10% (vol/vol) glycerol. A confocal microscope (LSM 780; Carl Zeiss) was used for confocal microscopy at room temperature using the Plan Apochromat 40×, 1.40 NA oil differential interference contrast M27 objective or the Plan Apochromat 100×, 1.40 NA oil differential interference contrast objective. All images were acquired through Zen 2011 (black edition; Carl Zeiss) and edited using Photoshop (Adobe) and Illustrator (Adobe).
Electron microscopy followed an established protocol (O’Donnell et al., 1989). In brief, bisected thoraces were fixed in 1% (vol/vol) paraformaldehyde/5% (vol/vol) glutaraldehyde, in 100 mM sodium phosphate buffer, pH 7.2, and 100 mM sucrose, overnight on ice. Washed samples were then postfixed in 1% (wt/vol) osmium tetroxide in 100 mM phosphate buffer before dehydration and embedding in Spurr’s resin for sectioning. Sections were cut at 70-nm thickness using a diamond knife. Sections were stained with 2% uranyl acetate and Reynolds’ lead citrate, before viewing on a transmission electron microscope (H-7500; Hitachi). Images were captured using an imaging camera (XR60; Advanced Microscopy Techniques).
Bioinformatics
Exon 10 of the D. melanogaster sls gene was used as a query sequence to identify homologous exons in other Drosophila species in a BLAST (Basic Local Alignment Search Tool) search. Sequences adjacent to the identified BLAST hits were aligned against sequences of D. melanogaster introns flanking sls exon 10 using AlignX (Vector NTI software suit; Invitrogen).
Statistical analysis
All statistical analysis was performed using R Studio. For the flight data, analysis was performed by segregating flight performance into successes (up behavior) versus failure (horizontal, down, and not at all) using a proportion test. The flies were counted in total, and the proportions of up behavior in KDs were compared with the proportion of up behavior in WT flies. Bonferroni correction was used for eight different comparisons, and as a consequence, the p-value was adjusted to 0.05/8. The analysis resulted in p-values of 10−25, 10−16, 10−9, and 10−5 for mub, aret, bl, and Spf45, respectively. The sarcomere length was measured using ImageJ software (National Institutes of Health) and analyzed using a Wilcoxon rank-sum test.
Online supplemental material
Fig. S1 shows splicing alterations upon aret KD for additional muscle structural genes and shows that aret KD results in the inability to detect Aret by immunofluorescence. Fig. S2 shows the organization of the aret locus and data demonstrating how we identified the flight muscle isoform of Aret. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201405058/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201405058.dv.
Supplementary Material
Acknowledgments
We thank Drs. Paul Macdonald, Darren Monckton, Joseph Gall, and Olga Pontes for antibodies and Dr. Frank Schnorrer for flies. Electron microscopy was performed by Dr. Stephen Jett at the University of New Mexico Health Sciences Center Electron Microscopy Facility.
This work was supported by GM061738 awarded by the National Institutes of Health to R.M. Cripps. We acknowledge technical support from the Molecular Biology Facility at the Department of Biology, University of New Mexico, supported by National Institutes of Health grant P20 GM103452 from the Institute Development Award Program of the National Institute of General Medical Sciences.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- APF
- after puparium formation
- Aret
- Arrest
- B body
- Bruno body
- FC
- founder cell
- KD
- knockdown
- LS
- longitudinal section
- Mbl
- Muscleblind
- SF
- splicing factor
- TS
- transverse section
- UAS
- upstream activating sequence
- WT
- wild type
References
- Anant, S., Roy S., and VijayRaghavan K.. 1998. Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Development. 125:1361–1369 [DOI] [PubMed] [Google Scholar]
- Barreau, C., Paillard L., Méreau A., and Osborne H.B.. 2006. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 88:515–525 10.1016/j.biochi.2005.10.011 [DOI] [PubMed] [Google Scholar]
- Bell, L.R., Maine E.M., Schedl P., and Cline T.W.. 1988. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell. 55:1037–1046 10.1016/0092-8674(88)90248-6 [DOI] [PubMed] [Google Scholar]
- Berger, D.S., Moyer M., Kliment G.M., van Lunteren E., and Ladd A.N.. 2011. Expression of a dominant negative CELF protein in vivo leads to altered muscle organization, fiber size, and subtype. PLoS ONE. 6:e19274 10.1371/journal.pone.0019274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, S.I., O’Donnell P.T., and Cripps R.M.. 1993. Molecular genetic analysis of muscle development, structure, and function in Drosophila. Int. Rev. Cytol. 143:63–152 10.1016/S0074-7696(08)61874-4 [DOI] [PubMed] [Google Scholar]
- Bischof, J., Maeda R.K., Hediger M., Karch F., and Basler K.. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 104:3312–3317 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L.2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291–336 10.1146/annurev.biochem.72.121801.161720 [DOI] [PubMed] [Google Scholar]
- Blanchette, M., Green R.E., Brenner S.E., and Rio D.C.. 2005. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 19:1306–1314 10.1101/gad.1314205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M., Green R.E., MacArthur S., Brooks A.N., Brenner S.E., Eisen M.B., and Rio D.C.. 2009. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Mol. Cell. 33:438–449 10.1016/j.molcel.2009.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryantsev, A.L., Baker P.W., Lovato T.L., Jaramillo M.S., and Cripps R.M.. 2012a. Differential requirements for Myocyte Enhancer Factor-2 during adult myogenesis in Drosophila. Dev. Biol. 361:191–207 10.1016/j.ydbio.2011.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryantsev, A.L., Duong S., Brunetti T.M., Chechenova M.B., Lovato T.L., Nelson C., Shaw E., Uhl J.D., Gebelein B., and Cripps R.M.. 2012b. Extradenticle and homothorax control adult muscle fiber identity in Drosophila. Dev. Cell. 23:664–673 10.1016/j.devcel.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet-B, N., Savkur R.S., Singh G., Philips A.V., Grice E.A., and Cooper T.A.. 2002. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 10:45–53 10.1016/S1097-2765(02)00572-5 [DOI] [PubMed] [Google Scholar]
- Dhaenens, C.M., Tran H., Frandemiche M.L., Carpentier C., Schraen-Maschke S., Sistiaga A., Goicoechea M., Eddarkaoui S., Van Brussels E., Obriot H., et al. 2011. Mis-splicing of Tau exon 10 in myotonic dystrophy type 1 is reproduced by overexpression of CELF2 but not by MBNL1 silencing. Biochim. Biophys. Acta. 1812:732–742 10.1016/j.bbadis.2011.03.010 [DOI] [PubMed] [Google Scholar]
- Drummond, D.R., Hennessey E.S., and Sparrow J.C.. 1991. Characterisation of missense mutations in the Act88F gene of Drosophila melanogaster. Mol. Gen. Genet. 226:70–80 10.1007/BF00273589 [DOI] [PubMed] [Google Scholar]
- Farkas-Bargeton, E., Barbet J.P., Dancea S., Wehrle R., Checouri A., and Dulac O.. 1988. Immaturity of muscle fibers in the congenital form of myotonic dystrophy: its consequences and its origin. J. Neurol. Sci. 83:145–159 10.1016/0022-510X(88)90064-0 [DOI] [PubMed] [Google Scholar]
- Filardo, P., and Ephrussi A.. 2003. Bruno regulates gurken during Drosophila oogenesis. Mech. Dev. 120:289–297 10.1016/S0925-4773(02)00454-9 [DOI] [PubMed] [Google Scholar]
- Förch, P., and Valcárcel J.. 2003. Splicing regulation in Drosophila sex determination. Prog. Mol. Subcell. Biol. 31:127–151 10.1007/978-3-662-09728-1_5 [DOI] [PubMed] [Google Scholar]
- Fyrberg, E.A., Mahaffey J.W., Bond B.J., and Davidson N.. 1983. Transcripts of the six Drosophila actin genes accumulate in a stage- and tissue-specific manner. Cell. 33:115–123 10.1016/0092-8674(83)90340-9 [DOI] [PubMed] [Google Scholar]
- Gauthier, G.F.1979. Ultrastructural identification of muscle fiber types by immunocytochemistry. J. Cell Biol. 82:391–400 10.1083/jcb.82.2.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidt, A.B., Rojas A., Harris I.S., and Black B.L.. 2007. Determinants of myogenic specificity within MyoD are required for noncanonical E box binding. Mol. Cell. Biol. 27:5910–5920 10.1128/MCB.01700-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz, R., Díaz-Castillo C., Nguyen T.P., Lovato T.L., Cripps R.M., and Marco R.. 2004. Expression patterns of the whole troponin C gene repertoire during Drosophila development. Gene Expr. Patterns. 4:183–190 10.1016/j.modgep.2003.09.008 [DOI] [PubMed] [Google Scholar]
- Ho, T.H., Charlet-B N., Poulos M.G., Singh G., Swanson M.S., and Cooper T.A.. 2004. Muscleblind proteins regulate alternative splicing. EMBO J. 23:3103–3112 10.1038/sj.emboj.7600300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo, M.S., Lovato C.V., Baca E.M., and Cripps R.M.. 2009. Crossveinless and the TGFbeta pathway regulate fiber number in the Drosophila adult jump muscle. Development. 136:1105–1113 10.1242/dev.031567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra, A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., and Cooper T.A.. 2008. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. USA. 105:20333–20338 10.1073/pnas.0809045105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlik, C.C., Coutu M.D., and Fyrberg E.A.. 1984. A nonsense mutation within the act88F actin gene disrupts myofibril formation in Drosophila indirect flight muscles. Cell. 38:711–719 10.1016/0092-8674(84)90266-6 [DOI] [PubMed] [Google Scholar]
- Kim-Ha, J., Kerr K., and Macdonald P.M.. 1995. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 81:403–412 10.1016/0092-8674(95)90393-3 [DOI] [PubMed] [Google Scholar]
- Ladd, A.N., Charlet N., and Cooper T.A.. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21:1285–1296 10.1128/MCB.21.4.1285-1296.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd, A.N., Taffet G., Hartley C., Kearney D.L., and Cooper T.A.. 2005. Cardiac tissue-specific repression of CELF activity disrupts alternative splicing and causes cardiomyopathy. Mol. Cell. Biol. 25:6267–6278 10.1128/MCB.25.14.6267-6278.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machuca-Tzili, L.E., Buxton S., Thorpe A., Timson C.M., Wigmore P., Luther P.K., and Brook J.D.. 2011. Zebrafish deficient for Muscleblind-like 2 exhibit features of myotonic dystrophy. Dis. Model. Mech. 4:381–392 10.1242/dmm.004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majczenko, K., Davidson A.E., Camelo-Piragua S., Agrawal P.B., Manfready R.A., Li X., Joshi S., Xu J., Peng W., Beggs A.H., et al. 2012. Dominant mutation of CCDC78 in a unique congenital myopathy with prominent internal nuclei and atypical cores. Am. J. Hum. Genet. 91:365–371 10.1016/j.ajhg.2012.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves, L., Waskiewicz A.J., Paul B., Cao Y., Tyler A., Moens C.B., and Tapscott S.J.. 2007. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development. 134:3371–3382 10.1242/dev.003905 [DOI] [PubMed] [Google Scholar]
- McGuire, S.E., Le P.T., Osborn A.J., Matsumoto K., and Davis R.L.. 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 302:1765–1768 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Moore, J., Han H., and Lasko P.. 2009. Bruno negatively regulates germ cell-less expression in a BRE-independent manner. Mech. Dev. 126:503–516 10.1016/j.mod.2009.04.002 [DOI] [PubMed] [Google Scholar]
- Morriss, G.R., Bryantsev A.L., Chechenova M., LaBeau E.M., Lovato T.L., Ryan K.M., and Cripps R.M.. 2012. Analysis of skeletal muscle development in Drosophila. Methods Mol. Biol. 798:127–152 10.1007/978-1-61779-343-1_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose, A., Isshiki T., and Takeichi M.. 1998. Regional specification of muscle progenitors in Drosophila: the role of the msh homeobox gene. Development. 125:215–223 [DOI] [PubMed] [Google Scholar]
- O’Donnell, P.T., Collier V.L., Mogami K., and Bernstein S.I.. 1989. Ultrastructural and molecular analyses of homozygous-viable Drosophila melanogaster muscle mutants indicate there is a complex pattern of myosin heavy-chain isoform distribution. Genes Dev. 3:1233–1246 10.1101/gad.3.8.1233 [DOI] [PubMed] [Google Scholar]
- O’Keefe, R.T., Mayeda A., Sadowski C.L., Krainer A.R., and Spector D.L.. 1994. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124:249–260 10.1083/jcb.124.3.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, C.M., Stern L.Z., Curless R.G., and Hannapel L.K.. 1975. Ultrastructural fiber typing in normal and diseased human muscle. J. Neurol. Sci. 25:99–108 10.1016/0022-510X(75)90190-2 [DOI] [PubMed] [Google Scholar]
- Philips, A.V., Timchenko L.T., and Cooper T.A.. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 280:737–741 10.1126/science.280.5364.737 [DOI] [PubMed] [Google Scholar]
- Punkt, K.2002. Fibre types in skeletal muscles. Adv. Anat. Embryol. Cell Biol. 162:1–109 [DOI] [PubMed] [Google Scholar]
- Ranganayakulu, G., Elliott D.A., Harvey R.P., and Olson E.N.. 1998. Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development. 125:3037–3048 [DOI] [PubMed] [Google Scholar]
- Ruiz-Gómez, M., Coutts N., Price A., Taylor M.V., and Bate M.. 2000. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell. 102:189–198 10.1016/S0092-8674(00)00024-6 [DOI] [PubMed] [Google Scholar]
- Salz, H.K.2011. Sex determination in insects: a binary decision based on alternative splicing. Curr. Opin. Genet. Dev. 21:395–400 10.1016/j.gde.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur, R.S., Philips A.V., and Cooper T.A.. 2001. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 29:40–47 10.1038/ng704 [DOI] [PubMed] [Google Scholar]
- Schiaffino, S., and Reggiani C.. 2011. Fiber types in mammalian skeletal muscles. Physiol. Rev. 91:1447–1531 10.1152/physrev.00031.2010 [DOI] [PubMed] [Google Scholar]
- Schiaffino, S., Hanzlíková V., and Pierobon S.. 1970. Relations between structure and function in rat skeletal muscle fibers. J. Cell Biol. 47:107–119 10.1083/jcb.47.1.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönbauer, C., Distler J., Jährling N., Radolf M., Dodt H.U., Frasch M., and Schnorrer F.. 2011. Spalt mediates an evolutionarily conserved switch to fibrillar muscle fate in insects. Nature. 479:406–409 10.1038/nature10559 [DOI] [PubMed] [Google Scholar]
- Silver, M.M., Vilos G.A., Silver M.D., Shaheed W.S., and Turner K.L.. 1984. Morphologic and morphometric analyses of muscle in the neonatal myotonic dystrophy syndrome. Hum. Pathol. 15:1171–1182 10.1016/S0046-8177(84)80312-3 [DOI] [PubMed] [Google Scholar]
- Snee, M., Benz D., Jen J., and Macdonald P.M.. 2008. Two distinct domains of Bruno bind specifically to the oskar mRNA. RNA Biol. 5:49–57 10.4161/rna.5.1.5735 [DOI] [PubMed] [Google Scholar]
- Spector, D.L.2001. Nuclear domains. J. Cell Sci. 114:2891–2893 [DOI] [PubMed] [Google Scholar]
- Spletter, M.L., and Schnorrer F.. 2014. Transcriptional regulation and alternative splicing cooperate in muscle fiber-type specification in flies and mammals. Exp. Cell Res. 321:90–98 10.1016/j.yexcr.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swank, D.M., Wells L., Kronert W.A., Morrill G.E., and Bernstein S.I.. 2000. Determining structure/function relationships for sarcomeric myosin heavy chain by genetic and transgenic manipulation of Drosophila. Microsc. Res. Tech. 50:430–442 [DOI] [PubMed] [Google Scholar]
- Timchenko, N.A., Patel R., Iakova P., Cai Z.J., Quan L., and Timchenko L.T.. 2004. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 279:13129–13139 10.1074/jbc.M312923200 [DOI] [PubMed] [Google Scholar]
- Venables, J.P., Tazi J., and Juge F.. 2012. Regulated functional alternative splicing in Drosophila. Nucleic Acids Res. 40:1–10 10.1093/nar/gkr648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente, M., Monferrer L., Poulos M.G., Houseley J., Monckton D.G., O’dell K.M., Swanson M.S., and Artero R.D.. 2007. Muscleblind isoforms are functionally distinct and regulate alpha-actinin splicing. Differentiation. 75:427–440 10.1111/j.1432-0436.2006.00156.x [DOI] [PubMed] [Google Scholar]
- Vicente-Crespo, M., Pascual M., Fernandez-Costa J.M., Garcia-Lopez A., Monferrer L., Miranda M.E., Zhou L., and Artero R.D.. 2008. Drosophila muscleblind is involved in troponin T alternative splicing and apoptosis. PLoS ONE. 3:e1613 10.1371/journal.pone.0001613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, P.J., Liang L., Berg C.A., Lasko P., and Macdonald P.M.. 1997. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 11:2510–2521 10.1101/gad.11.19.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, L., Edwards K.A., and Bernstein S.I.. 1996. Myosin heavy chain isoforms regulate muscle function but not myofibril assembly. EMBO J. 15:4454–4459 [PMC free article] [PubMed] [Google Scholar]
- Zierath, J.R., and Hawley J.A.. 2004. Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS Biol. 2:e348 10.1371/journal.pbio.0020348 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.