Abstract
Identifying the species on which hematophagous arthropods feed is crucial for studying the factors that affect pathogen distributions and that can aid public health. Here we describe a protocol to identify the species a parasitic arthropod has previously fed upon by identifying the source of the remnants of a previous blood meal via shotgun proteomics and spectral matching. The protocol is a nontargeted approach that uses the entire detected blood proteome for source identification; it does not require a priori knowledge of genome or protein sequences. Instead, reference spectral libraries are compiled from the blood of multiple host species by using SpectraST, which takes ~4 d; the identification of the species from which a previous blood meal of a hematophagous arthropod was taken is achieved with spectral matching against the reference spectral libraries, which takes approximately another 4 d. This method is robust against random degradation of the blood meal and can identify unknown blood remnants months after the feeding event.
INTRODUCTION
Many hematophagous arthropods transmit microbial pathogens that are the primary cause of infectious diseases1. The blood-feeding behavior of the parasitic arthropod has a crucial role in the transmission and maintenance of vector-borne pathogens in natural systems2–4. For zoonotic diseases, which involve non-human vertebrate reservoirs, little is known about the extent to which different vertebrate species affect the dynamics of most parasite species. Traditionally, labor-intensive quantification of the parasite burden on all potential wildlife species was necessary to assess the relevance of each host species to parasite population dynamics5, 6, although these data were often heavily biased by the variation in trapping success among animal species4, 7. Analysis of the vertebrate blood remaining in hematophagous arthropods is an effective alternative method for determining the importance of each species to the parasite population8–13.
Early blood meal identification methods relied on traditional serological techniques, including precipitin tests and ELISA9, 10, 12, 14. However, these techniques are time consuming and have limited sensitivity and specificity. Methods that were subsequently developed to elevate sensitivity and accuracy in the identification of the origins of blood meals used DNA-based analyses8, 11. However, such DNA-based PCR techniques are still limited by the quality and quantity of the host’s DNA inside the disease vector. For example, host DNA is quickly degraded during blood digestion after feeding, resulting in a rapid decrease in the success of the identification of blood meal sources over time. Therefore, the success of this technique is limited by accessibility to recently fed free-living vectors. Advances in proteomics technology offer new tools to approach this problem at the protein level13, 15, 16. Mass spectrometry (MS)-based proteomics has the ability to detect small remnants of blood meals even months after feeding15. This is an advantage, as disease vector organisms that have not fed for some time are easier to capture in nature as they are actively seeking further sources of blood. Moreover, as modern proteomics technologies, in particular bottom-up methods, effectively sample many short peptides in the proteome, this approach is more robust against random degradation of the blood meal compared with DNA-based methods.
Whether DNA- or protein-based, most current methods rely heavily on the known sequences of each parasitized organism13, 16–22. Unfortunately, limited sequence information exists for most of the wildlife species that are potential hosts of important disease vectors, thus limiting the use of such methods to well-characterized model organisms. A number of improving bioinformatic tools, such as de novo sequencing and homology searching in proteomics, can assist in overcoming these difficulties16, but owing to the difficulty of applying such methods on a large scale, only one or several a priori–selected target molecule(s) among a set of a priori–selected host species can be used as markers13, 16. Nevertheless, such target molecules may be identical among related host species that are not included in a study, thus reducing confidence in these procedures. Furthermore, random degradation or digestion of the target molecules over time further limits the success of these methodologies, even though the non-target molecules that can distinguish among host species may still be present13, 16, 18. To overcome these deficiencies, we developed a robust, universal proteomics-based method15 that can eliminate the need for genome sequences and that uses the entire detected proteome, rather than particular target molecules, as a marker for blood meal identification.
Experimental design
The methodology presented here involves the use of a non-targeted, genome-free, proteomics platform based on spectral matching technology23–28. The described methodology is designed to identify the source of blood meal remnants in hematophagous arthropods, and it has so far been used to identify the source of blood remnants in black-legged ticks (Ixodes scapularis) that had fed on different vertebrate species15. The described protocol will use I. scapularis as a model for its application. This protocol can also be applied to study other hematophagous arthropods to determine the ecological importance of each vertebrate species in the community to the maintenance of the parasite population. The experimental workflow can be divided into two main sections: building unidentified spectral reference libraries from whole blood, and blood meal proteome analysis by spectral matching against the reference spectral libraries.
Building unidentified spectral reference libraries (Steps 1–18)
Outlined in this protocol are the steps involved in constructing a reference spectral library from multiple potential host species by using SpectraST (version 4.0), which we adapted to build spectral libraries from unidentified spectra15. Species-specific spectral reference libraries are built from liquid chromatography-tandem MS (LC-MS/MS) analyses of trypsin-digested whole blood from potential host vertebrates. In theory, once reference spectral libraries are built for the blood proteomes of all potential hosts, the same libraries can also be used to identify the blood meal source of any other hematophagous arthropods.
We use a Thermo LCQ Deca XP Plus mass spectrometer coupled to an LC Packings/Thermo Ultimate Nano liquid chromatography system, controlled by Thermo Xcalibur 2.0 software for shotgun proteomics (procedures described in detail below)29, 30. However, any LC-MS system equipped with the requisite data acquisition and analysis capabilities to perform tandem MS analyses can be used. As in any shotgun proteomics analysis, the use of faster and more sensitive MS systems will improve the peptide and protein coverage, which can yield more comprehensive reference libraries.
Blood meal proteome analysis (Steps 19–26)
The second part of the experimental workflow is the identification of the blood meal source(s) in a hematophagous arthropod via spectral matching methodology and reference spectral libraries. The method presented compares the entire tandem mass spectral data set derived from individual vectors with the spectral reference libraries derived from the whole blood of individual species, to determine the source of the blood remaining in the disease vector. The Spectral Data set Similarity Scoring (SDSS) algorithm provides a quantitative similarity measure between two sets of spectra15. The reference spectral library to which the data from hematophagous arthropod is most similar (greatest SDSS score) can be identified, using statistical confidence calculations, as the source of the remnants of blood in that parasite.
MATERIALS
REAGENTS
Blood samples of vertebrate animals of interest ! CAUTION Adhere to all relevant ethical regulations and guidelines for the collection and use of animal blood. ! CAUTION To avoid potential contact with blood-borne pathogens, perform all work with appropriate personal protective equipment.
Ammonium bicarbonate (NH4HCO3; Sigma-Aldrich, cat. no. A6141) ! CAUTION NH4HCO3 is toxic. Wear eye protection, gloves and a dust mask while working with this reagent, and wash skin thoroughly after handling it.
Milli-Q ultrahigh-purity water (Millipore)
Acetonitrile (C2H3N; Thermo-Fisher, cat. no. A996-4) ! CAUTION Acetonitrile is highly flammable and toxic if swallowed.
NaH2PO4·2H2O (Fisher Scientific, cat. no. S381)
Na2HPO4·2H2O (Fisher Scientific, cat. no. S472)
Sodium chloride (NaCl; Sigma, cat. no. S9888)
DTT (Sigma-Aldrich, cat. no. D0632) ! CAUTION DTT causes acute toxicity. Wear gloves and safety glasses when handling it.
Iodoacetamide (IAA; Sigma-Aldrich, cat. no. I1149) ! CAUTION IAA causes acute toxicity and damage to the organs. Wear gloves and safety glasses when handling it, and use a respirator when handling the powder.
Trypsin (Promega, cat. no. V5111)
ZipTip C18 (Millipore, cat. no. ZTC18S960; alternatively, StageTips can be used)
Trifluoroacetic acid (TFA; Pierce, cat. no. 28904) ! CAUTION Trifluoroacetic acid is highly corrosive. It is harmful if swallowed, inhaled or absorbed through skin. The reagent is extremely destructive to the upper respiratory tract, eyes and skin.
Formic acid (Fluka, cat. no. 94318) ! CAUTION Formic acid is corrosive and volatile. Handle it in a fume hood, and wear gloves and safety glasses.
Bradford reagent (Sigma-Aldrich, cat. no. B6916) ! CAUTION This reagent contains phosphoric acid, which is a corrosive liquid.
Standard cytochrome c digest (Dionex/Thermo, cat. no. 161089)
EQUIPMENT
▲ CRITICAL Do not use autoclaved glassware, plasticware, buffers or solutions, as they can introduce unwanted chemical contamination into the samples, which compromises the LC-MS/MS analyses. If absolutely necessary, sterilize the stock solutions by filtration. Minimize contamination by keratin by keeping consumables in a dust-free environment and by always wearing nitrile gloves.
Temperature-controlled incubator, set to 37 °C
Milli-Q ultrapure water system (Millipore)
Microcentrifuge (Eppendorf, cat. no. 5424 or 5430 or similar)
SpeedVac concentrator (Eppendorf Vacufuge Concentrator, cat. no. 022820109, or similar)
UV-vis spectrophotometer (Hitachi or similar)
HPLC instrument configured for nanoflow delivery of solvent (UltiMate nanoscale LC system with FAMOS microautosampler (LC Packings/Thermo))
Tandem mass spectrometer (Thermo, LCQ-DECA XP Plus quadrupole ion trap (QIT) mass spectrometer)
Analytical column, Acclaim PepMap100, C18, 3 µm, 100 Å, 75 µm inner diameter (i.d.) × 150 mm (Dionex/Thermo, cat. no. 160321)
μ-Precolumn, Acclaim PepMap100, C18, 5 µm, 100 Å, 300 µm i.d. × 5 mm (Dionex/Thermo, cat. no. 160454)
SilicaTip emitters (New Objective, cat. no. FS360-75-8-N)
Blood collection tubes with anticoagulant (BD Microtainer green tubes, lithium heparin, cat. no. 365971 or similar)
Tubes, 1.5 ml (Eppendorf, cat. no. 22363204 or similar high-quality tubes) ▲ CRITICAL Tubes made of high-quality polypropylene must be used to avoid plasticizer contamination.
ProteoWizard (Download the ProteoWizard31 toolkit from http://proteowizard.sourceforge.net/downloads.shtml.)
Trans-Proteomic Pipeline (Download the Trans-Proteomic Pipeline suite from http://sourceforge.net/projects/sashimi/files/Trans-Proteomic%20Pipeline%20%28TPP%29/.)
REAGENT SETUP
Ammonium bicarbonate, 100 mM
Dissolve 0.4 g ammonium bicarbonate in 50 ml of deionized water. ▲ CRITICAL This must be freshly prepared; discard the unused solution.
Acetic acid, 50 mM
Add 71.4 µl of glacial acetic acid to 25 ml of deionized water. ▲ CRITICAL Use MS-grade acetic acid (100%). The molecular weight of acetic acid is 60.05 g/mol, and the density is 1.051 g/ml. Acetic acid solution can be stored at −20 °C (or below) for several months.
Reduction solution (200 mM DTT in 100 mM ammonium bicarbonate)
Dissolve 30.86 mg of DTT in 1 ml of 100 mM ammonium bicarbonate buffer immediately before use. ▲ CRITICAL The solution must be freshly prepared; discard the unused solution.
Alkylation solution (1 M iodoacetamide in 100 mM ammonium bicarbonate)
Dissolve 37 mg of iodoacetamide in 200 µl of 100 mM ammonium bicarbonate buffer immediately before use. ▲ CRITICAL Iodoacetamide is an unstable and light-sensitive reagent. Protect it from light at all times. It must be freshly prepared; discard the unused portion.
Trypsin digestion solution (6 ng/µl trypsin in 50 mM ammonium bicarbonate)
To prepare 100 ng/µl trypsin stock solution, add 200 µl of 50 mM acetic acid to 20 µg trypsin powder. Immediately before use, prepare 6 ng/µl trypsin digestion solution by diluting 5 µl of 100 ng/µl trypsin stock solution with 80 µl of 50 mM ammonium bicarbonate. ▲ CRITICAL Trypsin stock solution can be stored at −20 °C (or below) for several weeks. Use freshly thawed aliquots of trypsin. Discard the unused portions of both the thawed trypsin stock and the freshly prepared digestion solution after use.
HPLC mobile phases
To prepare fresh LC buffer A, mix 5% (vol/vol) acetonitrile in high-grade water with 0.1% (vol/vol) formic acid. To prepare fresh LC buffer B, mix 80% (vol/vol) acetonitrile with 0.1% (vol/vol) formic acid.
PBS buffer (1.9 mM NaH2PO4·2H2O, 8.1 mM Na2HPO4·2H2O, 154 mM NaCl)
Dissolve 0.296 g of NaH2PO4·2H2O, 1.442 g of Na2HPO4·2H2O and 9 g of NaCl in 900 ml deionized water. After adjusting the pH to 7.2, add deionized water to a final volume of 1 liter and filter-sterilize the solution. This solution can be stored for several months.
EQUIPMENT SETUP
nanoLC setup
The nanoLC system needs to be optimized and tested to provide the maximum resolution, sensitivity and selectivity with minimal sample sizes. We recommend using a simple protein digest (such as a standard cytochrome c digest from Dionex/Thermo) to verify the integrity of the HPLC system. This sample can be used for quality control of the HPLC system by injecting a defined amount (such as 300 fmol, or less for more-sensitive systems) of the peptide mix and monitoring the number of peptides identified with a specific false discovery rate (FDR). We recommend running a standard sample before and after each set of analyses to ensure that the system is running in optimal conditions throughout the analysis. The system can also be tested between samples to evaluate quality if large numbers of samples are running without breaks.
Gradients should be optimized for the samples at hand and the post-sample column washing and re-equilibration should be optimized to prevent sample carryover. In the setup described in this protocol, tryptic peptide mixtures were first loaded onto a precolumn (Acclaim PepMap100 C18-µ-precolumn; Dionex/Thermo) by using a FAMOS autosampler (LC Packings/Thermo) in batch mode with a user-defined program to withdraw 1 µl from each sample, and they were then washed for 4 min at a flow rate of 250 nl min−1 with LC buffer A (5% (vol/vol) acetonitrile, 0.1% (vol/vol) formic acid). The peptide mixtures were then transferred onto an analytical C18 nanocapillary HPLC column (Acclaim PepMap100 C18 peptide column; Dionex/Thermo) by using a nanocolumn-switching device (Switchos, LC Packings/Thermo) to direct the flow either to waste or to the analytical column. The peptide mixture was fractionated using a linear gradient slope of increasing LC buffer B (80% (vol/vol) acetonitrile, 0.1% (vol/vol) formic acid) up to 50% over 90 min, and the LC buffers were delivered by an Ultimate (LC Packings/Thermo) micropump. A fused silica-tip emitter with an 8-µm aperture (New Objectives) was used for ionization of peptides29, 30.
Mass Spectrometer setup
It is important to tune and calibrate the mass spectrometer and optimize the spray before each set of samples. In general, a wide m/z range (m/z 200–2,000) is recommended for mass detection.
ProteoWizard
Download the ProteoWizard31 toolkit from http://proteowizard.sourceforge.net/downloads.shtml. Among the many MS data analysis tools in ProteoWizard, msconvertGUI.exe allows one to convert the vendor-specific data format to an open XML format (mzXML or mzML). Supported vendor formats include Agilent (MassHunter .d), Bruker (Compass .d, FID, YEP, BAF), Thermo Fisher (RAW) and Waters (MassLynx .raw).
Trans-proteomic pipeline
Download the Trans-Proteomic Pipeline suite from http://sourceforge.net/projects/sashimi/files/Trans-Proteomic%20Pipeline%20%28TPP%29/. Linux users can obtain the source code by downloading the zipped .tgz archive or through subversion (SVN).
Windows users can download a one-click installer. Instructions and documentations are available at http://tools.proteomecenter.org/wiki/index.php?title=Software:SpectraST. After compilation or installation, type the command ‘spectrast’ on the command line and hit the Enter key. If you see the usage statement, then SpectraST is properly installed. On Linux, you must locate the executable spectrast and precede your SpectraST commands in Steps 16, 17 and 24, outlined below, with the full path.
PROCEDURE
Blood sample collection ● TIMING 5–10 min per animal
! CAUTION For all blood and blood-derived samples, take appropriate precautions. Handle all blood samples as a potential source of pathogens, use the appropriate protective attire (lab coats, safety glasses, gloves and other measures) and dispose of all biohazardous materials properly.
▲ CRITICAL The procedure comprises two main sections: building unidentified spectral reference libraries (Steps 1–18) and blood meal proteome analysis (Steps 19–26).
-
1|
Collect the blood samples of vertebrate animals of interest into blood collection tubes containing anticoagulants (preferably sodium citrate, or EDTA or heparin). Mix each tube as soon as it is collected by gently inverting the tube several times to ensure even distribution of the additive.
▲ CRITICAL STEP Use the blood samples as collected with anticoagulant in analyses; do not separate the blood cells.
-
2|
After collection and before further processing, immediately freeze all blood samples at −20 °C (or below) overnight to break the blood cells. Multiple freeze-thaw cycles may increase the amount of protein released, but excessive freeze-thaw cycles should be avoided (no more than four). Transport blood samples to the MS laboratory on dry ice.
! CAUTION Dry ice is an extremely cold solid (−79 °C) that can cause frostbite on contact with skin; breathing its vapors can cause suffocation.
▲ CRITICAL STEP Samples are more stable at a lower temperature. Therefore, we recommend using the lowest storage temperature available (−80 °C, if possible).
■ PAUSE POINT Blood sample vials can be kept frozen for years at −80 °C.
Protein digestion (in-solution) ● TIMING 18 h–2 d
▲ CRITICAL All solutions should be freshly prepared just before use. Work in a clean environment (e.g., sterile hood) while wearing gloves to reduce contamination of the digest with common contaminant proteins.
-
3|
Thaw the frozen blood samples.
-
4|
Dilute a blood sample in a 1.5-ml tube with 100 µl of 50 mM NH4HCO3 buffer (~0.5–1 µl of blood sample into 100–200 µl of 50 mM NH4HCO3 buffer would be a good starting concentration), and then determine the protein concentration of diluted blood sample by using the Bradford reagent kit according to the manufacturer’s instructions. Typical protein quantity should not exceed ~5 µg or ~50 pmol.
-
5|
Add 5 µl of reduction solution to the sample. Mix and incubate the sample at room temperature (20–25 °C) for 45–60 min with gentle shaking to reduce protein disulfide bonds.
-
6|
Add 5 µl of alkylation solution to the sample. Mix and incubate the sample at room temperature for 45–60 min with gentle shaking in the dark to alkylate the free thiol groups.
-
7|
Add another 20 µl of reduction solution. Mix and incubate the sample at room temperature for 45–60 min with gentle shaking to stop the reaction.
-
8|
Add 30 µl of trypsin digestion solution, mix and quickly spin to collect all droplets at the bottom of tubes. Incubate at 37 °C for 12–15 h (overnight) to digest the proteins into peptides.
-
9|
Dry the sample completely by using a SpeedVac concentrator. Store the dried sample at −80 °C until further analyses.
■ PAUSE POINT Digested samples can be stored at −80 °C for several days.
Peptide desalting (ZipTip C18) ● TIMING 30 min per sample
-
10|
Reconstitute each sample in 100 µl of reconstitution solution and clear the solution with a brief centrifugation.
-
11|
Perform a desalting step with the ZipTip C18 pipette tips according to the manufacturer’s instructions.
-
12|
Dry the eluted peptide sample completely on a SpeedVac concentrator. Store the sample at −80 °C until further analyses.
■ PAUSE POINT Dried peptides can be stored at −20 °C or below for several weeks to months.
Nano-LC-MS/MS analysis of peptides ● TIMING ~2 h per sample
▲ CRITICAL Because of the complexity of the sample, use a nano-LC setup with a 15-cm (or more) column filled with 3-µm (or smaller) C18 particles to increase the number of peptides measurable and to enlarge the overall dynamic range, and thus improve the coverage.
-
13|
Reconstitute the dried peptides in ~10 µl of LC running buffer A (see ‘HPLC mobile phases’ in Reagent Setup) for nano-LC-ESI-MS/MS analysis.
▲ CRITICAL STEP Several LC-MS systems can be used to analyze the samples, e.g., an UltiMate nanoscale LC system equipped with a FAMOS microautosampler and a C18 PepMap 100 column coupled to an LC-QIT-MS/MS instrument for mass-spectrometric analysis.
-
14|
Load sample onto a nanocapillary column, perform online RP-HPLC separation of peptides by using nanoflow LC (see suggested gradient in Equipment Setup) and directly elute it into an electrospray ionization tandem mass spectrometer operated in data-dependent mode with dynamic exclusion enabled, in which the top three peaks in every full MS scan are subjected to MS/MS analysis. Collected data will be used in the next step to build the spectral library (Fig. 1).
▲ CRITICAL STEP Sample carryover can cause serious problems in the quality of spectral libraries and thus can compromise the spectral library search. It is very important to completely flush the system and re-equilibrate the column before analyzing subsequent samples. Choosing an appropriate washing condition can minimize carryover for a particular analysis. Method conditions also affect carryover and need to be properly adjusted.
? TROUBLESHOOTING
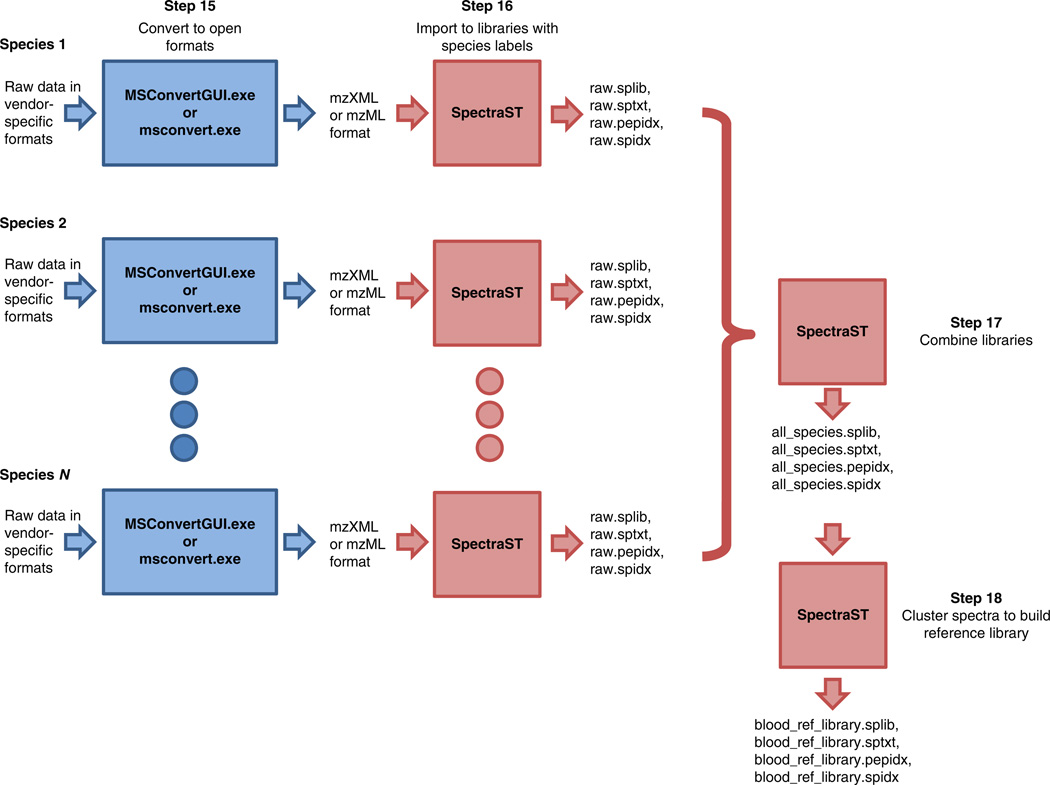
Figure 1.
Data analysis workflow as described in Steps 15–18.
Unidentified spectral library building ● TIMING 30 min–2 h
-
15|
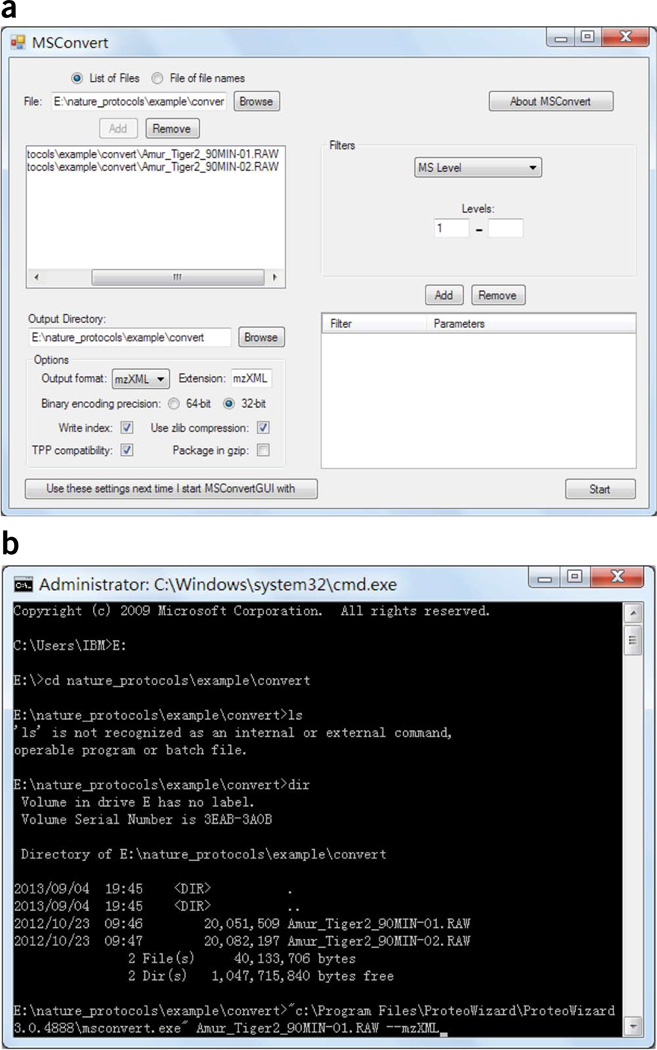
Convert raw data in vendor-specific formats into mzXML or mzML format (Fig. 1, ‘Step 15’). Run the ProteoWizard application MSConvertGUI.exe, select the list of files, choose the output directory, choose the output format and then click ‘start’ to begin the conversion (Fig. 2a). Alternatively, execute msconvert.exe at the Windows command line (Fig. 2b).
-
16|Import the data into an unidentified spectral library by using SpectraST (Fig. 1, ‘Step 16’). For each species, create a raw spectral library specific to the blood source (species) by using the command:
spectrast –cN<LIBRARY_NAME> –cn<ANIMAL_NAME> <LIST_OF_ALL_DATA_FILES>
in which ‘<LIBRARY_NAME>’ is the name of the spectral library to be written. Use meaningful names for organizational purposes; these names will be the input values in the following step. ‘<ANIMAL_NAME>’ is a word to indicate the animal source of the blood. This identifies the source of all MS/MS spectra from the data files, and will show up in the final report. For both ‘<LIBRARY NAME>’ and ‘<ANIMAL NAME>’, use no space or special characters; if needed, underscores can be used as space. ‘<LIST_OF_ALL_DATA_FILES>’ is a list of space-delimited names of the data files (.mzXML or .mzML files converted from the vendor-specific formats). Directory paths to the files can also be specified. If files from the same species are organized in the same directory, it is convenient to use a wildcard, e.g., ‘*.mzXML’ to denote all .mzXML files in that directory. This step should create four files: ‘<LIBRARY_NAME>.splib’, ‘<LIBRARY_NAME>.sptxt’, ‘<LIBRARY_NAME>.pepidx’ and ‘<LIBRARY_NAME>.spidx’. Do not move the files or rename them once they are created.▲ CRITICAL STEP For better organization of data sets, it is highly recommended that data be separated into separate folders on the basis of the corresponding vertebrate source of blood. To avoid confusion, use meaningful file or directory names.
-
17|Create a consensus spectral library of all species by using SpectraST. First, combine all the raw spectral libraries created in the previous step (Fig. 1, ‘Step 17’) by the command:
spectrast –cNall_species <LIST_OF_ALL_RAW_LIBRARIES>
This creates a spectral library named ‘all_species.splib’, which is just a union of all the raw libraries.‘<LIST_OF_ALL_RAW_LIBRARIES>’ is a space-delimited list of all .splib files created in the previous step. Directory paths can be included. As an example, if the raw libraries, named ‘raw.splib’, are located in two sub-directories called ‘lion’ and ‘tiger’, the command would be:spectrast –cNall_species lion/raw.splib tiger/raw.splib
-
18|Perform spectrum clustering and merge replicates to produce a consensus spectral library by the following command (Fig. 1, ‘Step 18’):
spectrast –cNblood_ref_library –cAS –c_RRS all_species.splib
This step will create a spectral library named ‘blood_ref_library.splib’, which is the reference library to be searched for identifying unknown samples. In this step, spectra that are found to be similar (replicates), regardless of their species sources, will be merged to form a consensus by the algorithm described in ref. 25 to improve the spectral quality. The number of replicates contributed by each species to the consensus will be recorded. Spectra without replicates will be left alone with some minimal de-noising.
Figure 2.
Screenshots of running msconvert (ProteoWizard) to convert vendor-specific formats to open XML data formats. (a) Using msconvertGUI.exe with a graphical user interface. (b) Running msconvert.exe at the Windows command line.
Extraction of total proteins from hematophagous arthropods ● TIMING 15 min per sample + 1 h
▲ CRITICAL Although blood meal proteome analysis is described for a tick parasite, it can be adapted to analyze other hematophagous arthropods with similar success.
-
19|
Transport the collected tick samples to the MS laboratory either alive or frozen (on dry ice) in a 1.5-ml tube. Immediately store all ticks (whether they are blood engorged or flat) at −20 °C or below. This will keep the proteins more stable and prevent further degradation of the blood meal.
■ PAUSE POINT Ticks can be kept frozen for several months to years at −80 °C.
-
20|
Thaw a single tick into a 1.5-ml tube and then homogenize it in 20 µl of PBS by manual crushing with a P1000 pipette tip until everything but the hard shell and the legs gets incorporated into the solution.
-
21|
Remove the insoluble materials by centrifugation for 5 min at 25,000g at room temperature.
-
22|
Remove supernatant to a new tube and determine its protein concentration by using the Bradford reagent kit according to the manufacturer’s instructions.
In-solution digestion and desalting ● TIMING 18 h–2 d
-
23|
Perform in-solution trypsin digestion without previous gel separation before performing the LC-MS/MS analysis as described in Steps 4–12.
Nano-LC-MS/MS analysis of peptides ● TIMING 2 h per sample
-
24|
Collect spectra from the trypsin-digested peptide sample as described in Steps 13 and 14.
Identification of blood meal by spectral library search ● TIMING 30 min–2 h
-
25|
Convert raw data in vendor-specific formats into mzXML or mzML format, following the same procedure as in Step 15.
-
26|Search the files in question against the reference spectral library by using the command:
spectrast -sLblood_ref_library.splib –sExls -s_FIN<REPORT_FILE_NAME> <LIST_OF_QUERY_FILES>
in which ‘-sLblood_ref_library.splib’ specifies that the reference library to search is named ‘blood_ref_library.splib’; replace this with the name of the reference library if another name was specified in Step 17. ‘<LIST_OF_QUERY_FILES>’ is a space-delimited list of data files (in .mzXML or .mzML format) from the unknown sample. The search will produce a tab-delimited file for each query file, listing the search results of all query spectra, which can be viewed in spreadsheet software if desired. The option ‘-s_FIN<REPORT_FILE_NAME>’ turns on the fingerprinting mode, in which SpectraST will produce a text file called ‘<REPORT_FILE_NAME>’ summarizing the results.▲ CRITICAL STEP For better results, more than one MS run can be acquired on the same tick sample, whether as repeated technical replicates or as multiple fractions. All files acquired from the same tick sample can be listed in ‘<LIST_OF_QUERY_FILES>’ to be analyzed in aggregate.
▲ CRITICAL STEP We recommend acquiring data from a sample(s) of known origin with available genome sequences and then searching the data to check for the accuracy of the method. Ideally, these test data should be from a species included in the library, but the exact same file would not be used to build the library.
? TROUBLESHOOTING
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
TABLE 1.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 14 | Low number of spectra | Sample of high complexity (samples’ co-elution) | Further fractionate or simplify the sample (i.e., by adjusting the HPLC gradient to make it more gradual to better separate peptides) and thus increase the number of spectra observed |
| Insufficient amount of injection or sample | Determine the optimal amount of the sample for the type of MS instrument and LC gradient used. Adjust the amount of the sample loaded. Check the autosampler for possible failure | ||
| 26 | Low-quality tandem mass spectral data sets | Problems with the chromatography column or system | Use a simple protein digest (such as cytochrome c digest) to verify the integrity of the HPLC system |
| Insufficient amount of injection or sample | Determine the optimal amount of the sample for the type of MS instrument and LC gradient used. Adjust the amount of the sample loaded. Check the autosampler for possible failure | ||
| Heavy contamination, such as polymers leaching from laboratory plasticware | Use MS-compatible grades of plastic tubes, tips and powder-free gloves as indicated throughout the protocol | ||
| Low ‘spectrum-spectrum match ratio’ | Low-quality tandem mass spectral data sets | Open the raw data file and examine the total ion chromatogram by using the mass spectrometer’s software to check the quality of the data set. A good-quality data set should have numerous peaks that rise above the noise in the total ion chromatogram. If the quality is not good, see the troubleshooting section for low-quality tandem mass spectral data sets | |
| The species of the query sample is not represented in the library | Extend the library data set by adding more species |
● TIMING
Building unidentified spectral reference libraries
Steps 1 and 2, blood sample collection: 5–10 min per animal
Days 1 and 2 of experiment
Steps 3–9, protein digestion (in-solution): 18 h–2 d (4 h + overnight incubation + 2–3-h SpeedVac concentration)
Steps 10–12, peptide desalting (ZipTip C18): 30 min per sample
Day 3 of experiment
Steps 13 and 14, nano-LC-MS/MS analysis of peptides: variable; once the instrument is set up, a typical run takes 2 h per sample
Day 4 of experiment
Steps 15–18, unidentified spectral library building: 30 min–2 h
Blood meal proteome analysis
Days 1 and 2 of experiment
Steps 19–22, extraction of total proteins from hematophagous arthropods: 15 min per sample + 1-h protein assay
Step 23, in-solution protein digestion and peptide desalting: 18 h–2d (4 h + overnight incubation + 2–3-h SpeedVac concentration + 30 min of desalting per sample).
Day 3 of experiment
Step 24, nano-LC-MS/MS analysis of peptides: variable; once the instrument is set up, a typical run takes 2 h per sample
Day 4 of experiment
Steps 25 and 26, identification of blood meal by spectral library search: 30 min–2 h
ANTICIPATED RESULTS
The anticipated fingerprinting summary file, whose name is specified in ‘<REPORT_FILE_NAME>’ in Step 26, will have the following format (Fig. 3). The first line lists the animal sources (as specified in ‘<ANIMAL_NAME>’ in Step 16) and their column indices in square brackets. The second line is the fractions of bootstrap samples that identify each of the animals as the top-scoring match, in the order of animals specified in the first line. The third line is the match ratio (the fraction of good matches among all MS/MS spectra in the query files) as well as the actual number of spectral matches and the number of query spectra. The fourth line further breaks down the number of spectral matches by species, in the order listed on the first line, in the form of: (number of spectral matches to library spectra unique to that species)/(total number of spectral matches to library spectra that have contributing replicates from that species).
Figure 3.
A snapshot illustrating the anticipated fingerprinting summary file. The first line is the column legend that lists the animal sources (as specified in <ANIMAL_NAME> in Step 16) and their column indices in square brackets. The second line is the fractions of the top-scoring match that are in the order of animals specified in the first line. The third line is the match ratio, showing the fraction of good matches among all MS/MS spectra in the query files and the actual number of spectral matches versus the number of query spectra. The fourth line is the ratio of the number of spectral matches to library spectra unique to a species versus the total number of spectral matches to library spectra that have contributing replicates from that species, and it is in the same order as listed on the first line. The next five lines are the different SDSS values. After the SDSS scores, the actual spectral fingerprint is shown as a table. See the text for details.
The next five lines are the different SDSS scores that have been previously described15. The score ‘dot_product_unique_ sqrt’ was found to be the best and is used for calculating the bootstrap support, although the other scores are also reported and may be optimal in different applications (e.g., when there are more similar species). After the SDSS scores, the actual spectral fingerprint is shown as a table formatted as: * i * mq(i) * r1(i) r2(i) r3(i) …rs(i), where i is the library spectrum index, mq(i) is the number of spectral matches to library spectrum i and rs(i) is the number of replicates from animal s contributing to library spectrum i. The correspondence of the index s and the animal is specified in the first line. This information can also be read into a spreadsheet software and visualized, similar to what is shown in ref. 15.
The ‘spectrum-spectrum match ratio’ on the second line is usually in the range 0.05–0.40. If this ratio is too low, the results may not be reliable. Very low values indicate that few spectral matches were found, probably reflecting low-quality tandem mass spectral data sets, which could be because of insufficient amount of injection, problems with the chromatography column, heavy contamination such as polymers leaching from various laboratory plastic-ware or incorrect settings. This could also be because the species of the query sample are not represented in the library, nor are any close relatives. To eliminate the possibility of experimental mishandling, it is useful to open the raw data file and examine the total ion chromatogram in the mass spectrometer’s software. A good-quality data set should have numerous peaks that rise above the noise in the total ion chromatogram.
Although the methodology presented here was developed by using Ixodes ticks that feed only once per life stage, the spectral matching methodology’s ability to correctly identify the source of blood from vector organisms that had fed multiple times has been determined experimentally by mixing whole blood from different species at specified ratios15. This methodology can successfully identify at least one of the true sources of the blood in a vector that has taken two blood meals in rapid succession with high confidence. When one source of blood is at much greater concentrations than the other (e.g., 1:5 and 1:10 ratios), the more concentrated source is to be identified as the top-scoring hit, whereas the SDSS for the less-abundant source will vary15.
ACKNOWLEDGMENTS
This work was supported by a grant from the US National Institutes of Health (AI076342); the US Center for Disease Control and Prevention (U01CK000170); and the University Grant Council, Hong Kong Special Administrative Region Government (HKUST RPC10EG08, DAG12EG01S).
Footnotes
AUTHOR CONTRIBUTIONS D.B., H.L. and Ö.Ö. conceived the study and formulated the experimental and computational strategy. Ö.Ö. optimized all the experimental protocols. H.L. and W.S. adapted SpectraST to build spectral libraries from unidentified spectra. Ö.Ö., H.L. and D.B. co-wrote the manuscript, with input from all authors.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Philos. Trans. Royal Soc. Lond. B Biol. Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leibold MA. A graphical model of keystone predators in food webs: trophic regulation of abundance, incidence, and diversity patterns in communities. Am. Naturalist. 1996;147:784–812. [Google Scholar]
- 3.Rose MD, Polis GA. The distribution and abundance of coyotes: the effects of allochthonous food subsidies from the sea. Ecology. 1998;79:998–1007. [Google Scholar]
- 4.LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc. Natl. Acad. Sci. USA. 2003;100:567–571. doi: 10.1073/pnas.0233733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lord RD, Lord VR, Humphreys JG, McLean RG. Distribution of Borrelia burgdorferi in host mice in Pennsylvania. J. Clin. Microbiol. 1994;32:2501–2504. doi: 10.1128/jcm.32.10.2501-2504.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matuschka FR, Fischer P, Musgrave K, Richter D, Spielman A. Hosts on which nymphal Ixodes ricinus most abundantly feed. Am. J. Trop. Med. Hyg. 1991;44:100–107. doi: 10.4269/ajtmh.1991.44.100. [DOI] [PubMed] [Google Scholar]
- 7.Brisson D, Dykhuizen DE, Ostfeld RS. Conspicuous impacts of inconspicuous hosts on the Lyme disease epidemic. Proc. Roy. Soc. Biol. Sci. 2008;275:227–235. doi: 10.1098/rspb.2007.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allan BF, Goessling LS, Storch GA, Thach RE. Blood meal analysis to identify reservoir hosts for Amblyomma americanum ticks. Emerg. Infect. Dis. 2010;16:433–440. doi: 10.3201/eid1603.090911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold EH, Simmons SW, Fawcett DG. Precipitin technique for determining mosquito blood meals. Public Health Rep. 1946;61:1244–1249. [PubMed] [Google Scholar]
- 10.Burkot TR, Goodman WG, DeFoliart GR. Identification of mosquito blood meals by enzyme-linked immunosorbent assay. Am. J. Trop. Med. Hyg. 1981;30:1336–1341. doi: 10.4269/ajtmh.1981.30.1336. [DOI] [PubMed] [Google Scholar]
- 11.Kent RJ. Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol. Ecol. Resour. 2009;9:4–18. doi: 10.1111/j.1755-0998.2008.02469.x. [DOI] [PubMed] [Google Scholar]
- 12.Tempelis CH, Rodrick ML. Passive hemagglutination inhibition technique for the identification of arthropod blood meals. Am. J. Trop. Med. Hyg. 1972;21:238–245. doi: 10.4269/ajtmh.1972.21.238. [DOI] [PubMed] [Google Scholar]
- 13.Wickramasekara S, Bunikis J, Wysocki V, Barbour AG. Identification of residual blood proteins in ticks by mass spectrometry proteomics. Emerg. Infect. Dis. 2008;14:1273–1275. doi: 10.3201/eid1408.080227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schubert JH, Holdeman LV. A modified precipitin technique for determining the source of mosquito blood-meals. Am. J. Trop. Med. Hyg. 1956;5:272–273. doi: 10.4269/ajtmh.1956.5.272. [DOI] [PubMed] [Google Scholar]
- 15.Onder O, Shao W, Kemps B, Lam H, Brisson D. Blood meal source tracking through genome-free proteomics technology using unidentified tandem mass spectral libraries. Nat. Commun. 2013;4:1746. doi: 10.1038/ncomms2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskay UA, et al. Development of a host blood meal database: de novo sequencing of hemoglobin from nine small mammals using mass spectrometry. Biol. Chem. 2012;393:195–201. doi: 10.1515/hsz-2011-0196. [DOI] [PubMed] [Google Scholar]
- 17.Gariepy TD, Lindsay R, Ogden N, Gregory TR. Identifying the last supper: utility of the DNA barcode library for bloodmeal identification in ticks. Mol. Ecol. Resour. 2012;12:646–652. doi: 10.1111/j.1755-0998.2012.03140.x. [DOI] [PubMed] [Google Scholar]
- 18.Mukabana WR, Takken W, Knols BG. Analysis of arthropod bloodmeals using molecular genetic markers. Trends Parasitol. 2002;18:505–509. doi: 10.1016/s1471-4922(02)02364-4. [DOI] [PubMed] [Google Scholar]
- 19.Humair PF, et al. Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J. Med. Entomol. 2007;44:869–880. doi: 10.1603/0022-2585(2007)44[869:miobsi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Pizarro JC, Stevens L. A new method for forensic DNA analysis of the blood meal in Chagas disease vectors demonstrated using Triatoma infestans from Chuquisaca, Bolivia. PLoS ONE. 2008;3:e3585. doi: 10.1371/journal.pone.0003585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mota J, et al. Identification of blood meal source and infection with Trypanosoma cruzi of Chagas disease vectors using a multiplex cytochrome b polymerase chain reaction assay. Vector Borne Zoonotic Dis. 2007;7:617–627. doi: 10.1089/vbz.2007.0106. [DOI] [PubMed] [Google Scholar]
- 22.Moran Cadenas F, et al. Identification of host bloodmeal source and Borrelia burgdorferi sensu lato in field-collected Ixodes ricinus ticks in Chaumont (Switzerland) J. Med. Entomol. 2007;44:1109–1117. doi: 10.1603/0022-2585(2007)44[1109:iohbsa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 23.Dasari S, et al. Pepitome: evaluating improved spectral library search for identification complementarity and quality assessment. J. Proteome Res. 2012;11:1686–1695. doi: 10.1021/pr200874e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam H, Aebersold R. Building and searching tandem mass (MS/MS) spectral libraries for peptide identification in proteomics. Methods. 2011;54:424–431. doi: 10.1016/j.ymeth.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Lam H, et al. Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods. 2008;5:873–875. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 2006;5:1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
- 27.Frewen BE, Merrihew GE, Wu CC, Noble WS, MacCoss MJ. Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 2006;78:5678–5684. doi: 10.1021/ac060279n. [DOI] [PubMed] [Google Scholar]
- 28.Stein SE, Scott DR. Optimization and testing of mass-spectral library search algorithms for compound identification. J. Am. Soc. Mass Spectr. 1994;5:859–866. doi: 10.1016/1044-0305(94)87009-8. [DOI] [PubMed] [Google Scholar]
- 29.Onder O, Turkarslan S, Sun D, Daldal F. Overproduction or absence of the periplasmic protease DegP severely compromises bacterial growth in the absence of the dithiol: disulfide oxidoreductase DsbA. Mol. Cell Proteomics. 2008;7:875–890. doi: 10.1074/mcp.M700433-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onder O, et al. Modifications of the lipoamide-containing mitochondrial subproteome in a yeast mutant defective in cysteine desulfurase. Mol. Cell Proteomics. 2006;5:1426–1436. doi: 10.1074/mcp.M600099-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]