Abstract
Pharmacogenomics is research to study the drug treatment responses in subgroups of patients according to their genetic variants or genetic expression information. Methadone maintenance treatment, which is usually prescribed for patients with heroin dependence, was launched in Taiwan by the government in 2006. In this study, 366 patients who had taken methadone continually in the previous 7 days were examined. Data from administration of the Treatment Outcomes Profile (TOP), Severity of Dependence Scale (SDS), Clinical Opioid Withdrawal Scale (COWS), and Treatment Emergent Symptoms Scale (TESS) were obtained from patients' report records. Genes encoding the liver cytochrome P-450 (CYP) enzymes that are involved with the metabolism of methadone (CYP2B6, 3A4 and 2C19) were selected and genotyped in this cohort. We found that the SNPs on CYP2B6 were associated with plasma S-methadone concentration; SNPs on CYP3A4 were associated with withdrawal symptoms and side effects; and SNPs on CYP2C19 were associated with methadone dose. SNPs in the genes encoding the morphine phase II metabolic enzyme, UGT2B7, were associated with withdrawal symptom scores. In pharmacodynamic genes, the SNPs on OPRM1 were associated with insomnia and change in libido side effects. We conclude that SNP markers may be useful for future methadone dosage adjustment and to reduce adverse reactions.
Keywords: COWS, CYP2B6, methadone, opioid receptors, pharmacogenomics, TESS
1. Introduction
Individual steady state plasma drug concentrations may vary from person to person following the drug's administration. Research reports suggest that the steady state plasma drug concentrations may change from 5 to 10 times in a male population with the same body weight of around 70 kg [1]. If gender, age, and pregnancy status are taken into consideration, the steady state plasma drug concentrations may vary from 50 to100 times. Based on this result, using therapeutic drug monitoring to place patients in subgroups in order to examine treatment responses is impossible. Pharmacogenomics is trying to use information about individual's genomic make-up to subgroup patients and match them with a more optimal medication choice. This is a new field of pharmacology, and single nucleotide polymorphism (SNP) is often used as a genomic marker in subgrouping patients for their treatment responses.
A successful pharmacogenomics clinical trial requires a few set criteria. The most important is to ensure the participating patients' compliance through both observation by research nurses and the measurement of the steady state drug concentrations of each patient. A well-designed clinical trial is another key to achieving a successful pharmacogenomics study.
The Taiwanese government started a methadone maintenance treatment (MMT) program in 2006. Since then, more than 90 hospitals have provided MMT, and over 11,000 heroin-dependent patients have participated in this program. The purpose of this program is to reduce heroin abuse, the spread of infectious diseases, such as HIV and HCV infection, and the drug-related crime rate. As individual methadone doses range from 5 to 180 mg/day, the drug-drug interaction may cause overdose and might induce lethal complications. The National Health Research Institutes (NHRI), therefore, launched the study of pharmacogenomics in a MMT cohort.
2. Methods
2.1 Subjects
This study was designed as a cross-sectional methadone clinical trial and was approved by the Institutional Review Boards of the National Health Research Institutes (Miaoli County, Taiwan) and all seven participating hospitals (Tao-Yuan Mental Hospital, En-Chu-Kong Hospital, Far-Eastern Memorial Hospital, Taipei City Hospital Song-De and Yang-Ming Branches, China Medical University Hospital, and Wei Gong Memorial Hospital). A written informed consent was obtained from each participant. The project also has been registered with the U.S. National Institutes of Health Clinical Trial database (http://www.clinicaltrial.gov/ct/show/NCT01059747). We recruited 366 subjects with heroin dependence who were undergoing MMT as outpatients. The inclusion criteria included an age of 18 years or above, being under MMT for at least 3 months with regular attendance for the past 7 days, and a methadone dosage adjustment of no more than 10 mg in the past 7 days. Exclusion criteria included co-morbidity with physical or other mental disorders requiring immediate treatment as well as pregnancy.
2.2 Clinical assessments
Demographics, clinical characteristics, and methadone treatment courses, including the dose and treatment duration, and treatment adherence over the previous week were obtained from patients' report records (case report form; CRF). Information regarding the possible co-administration of other medications in the previous week was obtained either from medical records or the subjects' reports [2]. Several interviewer-administered assessments, including the Treatment Outcomes Profile (TOP) [3], which measures the amount and frequency of alcohol and other illicit substance used in the past 28 days, the Severity of Dependence Scale (SDS), and the Clinical Opioid Withdrawal Scale (COWS), which measures the severity of 11 opioid withdrawal symptoms [4], were conducted before methadone was administered.
Methadone-related adverse events were assessed by research nurses using the Treatment Emergent Symptoms Scale (TESS) [5]. The TESS consists of 43 treatment emergent symptoms, and only 31 were complained from the patients in this study. Only symptoms that occurred after the initiation of MMT were counted as adverse events related to methadone. The severity of each symptom was rated on a 3-point Likert scale ranging from mild, moderate, to severe. In the present study, only adverse events with a frequency of 15% or above were included in the analyses.
2.3.1 Serum and urine drug test
Levels of glutamate oxaloacetate transaminase (GOT, reference range: <38 U/L), glutamic pyruvic transaminase (GPT, reference range: <41 U/L) and gamma-glutamyl transpeptidase (γ-GT, reference range: 8-61 U/L) from serum samples of patients were measured at the Taipei Institute of Pathology (Taipei, Taiwan).
Urine specimens were collected prior to the administration of methadone on the study day. The morphine screen test was performed via a kinetic interaction of microparticles (KIMS) on an Integra 800 device (Roche Diagnostics, Basel, Switzerland). In our present analyses and previous reports [2,6,7], the urine morphine test was used as a surrogate measurement for the methadone treatment outcome.
2.4 Analyses of methadone and its metabolites in the plasma
Whole blood samples of 12 ml were taken at around 24 ± 2 hrs after the last methadone dose, when the plasma concentration of methadone is likely to be at its lowest. Plasma concentrations of racemic methadone and its metabolite, 2-ethylidene-1,5-dimethyl-3,3-diphenylpyrrolidine (EDDP) were measured using high-performance liquid chromatography (HPLC) using the settings described in our previous study [8].
2.5 SNP selection and genotyping
Selection of SNPs was based on literature reports [2,6,9-13], a minor allele frequency of greater than 0.1 on the HapMap in Chinese ethnic groups (http://hapmap.ncbi.nlm.nih.gov/index.html.en), and possible functionalities predicted by the bioinformatics tool FastSNP [14].
Genomic DNA was extracted from the buffy coat of 6 ml whole blood lymphocyte pellets using the Puregene® Blood kit C (QIAGEN Sciences; Maryland, USA). Genotypes of the selected 17 SNPs were identified using matrix-assisted laser desorption/ ionization-time of flight mass spectrometry (MALDI-TOF MS) [15]. This genotyping method has been utilized in a broad variety of clinical applications due to the accuracy of SNP detection, sufficient sensitivity to score SNPs from small amounts of template, flexibility of the procedure, and cost-effectiveness [16].
2.6 Statistics
All statistical analyses were conducted using SAS software, Version 9.1 (SAS Institute, Inc., Cary, NC). The Wilcoxon rank-sum test for continuous data and chi-square test or Fisher's exact test for categorical data were applied in clinical character, TESS, and COWS comparisons between patients testing positive for urine morphine and patients testing negative. Association analyses between the selected SNPs and treatment outcomes, withdrawal symptoms, and adverse events were calculated using a generalized linear model (GLM) procedure. False discovery rate (FDR) test of the MULTTEST procedure was employed to correct for multiple hypotheses tests of p-values [17]. The Hardy-Weinberg equilibrium test and haplotype block association analysis were performed using HAPLOVIEW version 4.1 [18]. A statistical significance was designed with p-value less than 0.05.
3. Results
3.1 General clinical characteristics
Half of these 366 MMT patients tested positive for urine morphine (Table 1). The average age of these patients was 38.2 ± 7.7 years and almost all of the participants smoked cigarettes (99.5%). Males were predominant (n = 297, 81.1%) in this cohort. The average starting dose of methadone was 32 ± 11 mg/day and the dosage at the time of testing was 54.7 ± 28 mg/day. The ratios of plasma R- and S-methadone concentration/methadone dose were higher in the patients who tested negative for urine morphine than those who tested positive. The HCV incidence in these patients was 95%. There were no significant differences between those who tested positive for urine morphine and those who tested negative in regard to BMI, HIV, and HCV tests.
Table 1. General demograph of 366 methadone maintenance treatment patients.
| Overall N=366 | Urine Morphine Positive n=185 | Urine Morphine Negative n=178 | P-value | |
|---|---|---|---|---|
|
|
|
|
||
| Mean ± SD | Mean ± SD | Mean ± SD | ||
| Starting dose of Methadone (mg/day) | 32.04 ± 11.15 | 31.64 ± 10.07 | 32.44 ± 12.22 | 0.77a |
| Current dose of Methadone (mg/day) | 54.67 ± 28.12 | 54.53 ± 26.07 | 55.32 ± 30.13 | 0.88a |
| Age (years) | 38.17 ± 7.72 | 38.37 ± 7.96 | 37.87 ± 7.46 | 0.57a |
| Male | 297 (81.2%) | 152 (82.2%) | 142 (79.8%) | 0.56b |
| BMI (kg/m-1) | 23.58 ± 3.52 | 23.62 ± 3.57 | 23.59 ± 3.49 | 0.89a |
| R-Methadone/methadone dose ratio | 3.86 ± 2.32 | 3.7 ± 2.71 | 4.03 ± 1.82 | 0.001a |
| S-Methadone/methadone dose ratio | 2.77 ± 1.57 | 2.58 ± 1.45 | 2.98 ± 1.66 | 0.012a |
| R-EDDP/methadone dose ratio | 0.31 ± 0.5 | 0.26 ± 0.33 | 0.33 ± 0.54 | 0.26a |
| S-EDDP/methadone dose ratio | 0.33 ± 0.49 | 0.31 ± 0.38 | 0.33 ± 0.58 | 0.95a |
| Human immunodeficiency virus (HIV) | 86 (24.0%) | 50 (27.3%) | 36 (20.8%) | 0.15b |
| Hepatitis C virus (HCV) | 334 (94.9%) | 173 (96.1%) | 158 (93.5%) | 0.27b |
Values are shown as mean ± SD or N (%).
Bold P-value: P<0.05.
Wilcoxon rank-sum test.
Chi-Square test.
The average withdrawal symptom score rated by COWS was 1.49 ± 1.86 (Table 2). There was a statistically significant higher incidence of bone or joint aches symptom scores in patients testing positive for urine morphine (13% and 0.14 average score) than for patients who tested negative (6.2% and 0.08 average score). The top 15 (by percentage) side effects rated by TESS are listed in Table 3. The side effect of impaired mentation showed a significantly higher percentage of incidence in the patients who tested positive for urine morphine than those who tested negative (22.2% vs. 20.8%).
Table 2. Withdrawal Symptoms of Methadone Maintenance Subjects (N=366).
| Overall N=366 |
Urine Morphine Positive N=185 |
Urine Morphine Negative N=178 |
P-valuea | P-valueb,c | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| n | % | Mean ± SD | n | % | Mean ± SD | n | % | Mean ± SD | |||
| Sum of COWS | 366 | 1.49 ± 1.86 | 185 | 1.51 ± 1.93 | 178 | 1.44 ± 1.78 | 0.85 | ||||
| Heart Rate | 365 | 77.55 ± 11.86 | 184 | 76.83 ± 11.29 | 178 | 78.20 ± 12.24 | 0.37 | ||||
| Sweating | 41 | 11.20% | 0.13 ± 0.40 | 26 | 14.10% | 0.17 ± 0.48 | 14 | 7.90% | 0.08 ± 0.30 | 0.06 | 0.06b |
| Restlessness | 24 | 6.60% | 0.07 ± 0.25 | 15 | 8.10% | 0.08 ± 0.27 | 8 | 4.50% | 0.04 ± 0.21 | 0.16 | 0.16b |
| Pupil size | 69 | 18.90% | 0.21 ± 0.46 | 34 | 18.50% | 0.20 ± 0.44 | 35 | 19.70% | 0.22 ± 0.48 | 0.74 | 0.77b |
| Bone or Joint aches | 36 | 9.80% | 0.11 ± 0.36 | 24 | 13.00% | 0.14 ± 0.38 | 11 | 6.20% | 0.08 ± 0.33 | 0.033 | 0.028b |
| Runny nose or tearing | 31 | 8.50% | 0.11 ± 0.39 | 14 | 7.60% | 0.10 ± 0.36 | 17 | 9.60% | 0.13 ± 0.43 | 0.49 | 0.50b |
| GI Upset | 16 | 4.40% | 0.07 ± 0.33 | 9 | 4.90% | 0.08 ± 0.39 | 7 | 3.90% | 0.05 ± 0.27 | 0.65 | 0.67b |
| Tremor | 39 | 10.70% | 0.15 ± 0.46 | 20 | 10.80% | 0.13 ± 0.40 | 19 | 10.70% | 0.17 ± 0.53 | 0.91 | 0.97b |
| Yawning | 10 | 2.70% | 0.04 ± 0.23 | 6 | 3.20% | 0.04 ± 0.25 | 3 | 1.70% | 0.02 ± 0.18 | 0.34 | 0.50c |
| Anxiety or Irritability | 39 | 10.70% | 0.12 ± 0.35 | 19 | 10.30% | 0.12 ± 0.37 | 19 | 10.70% | 0.11 ± 0.31 | 0.94 | 0.90b |
| Gooseflesh skin | 8 | 2.20% | 0.07 ± 0.44 | 4 | 2.20% | 0.06 ± 0.44 | 4 | 2.30% | 0.07 ± 0.45 | 0.96 | 1.00c |
Values are shown as mean ± SD or n, %.
Bold P-value: P<0.05.
Wilcoxon rank-sum test.
Chi-Square test.
Fisher Exact test.
Table 3. The top 15 Side Effects of Methadone Maintenance Subjects (N=366).
| Overall N=366 |
Urine Morphine Positive N=185 |
Urine Morphine Negative N=178 |
P-valuea | P-valueb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| n | % | Mean ± SD | n | % | Mean ± SD | n | % | Mean ± SD | |||
| Constipation | 248 | 67.80% | 1.96 ± 0.84 | 125 | 67.60% | 2.01 ± 0.85 | 121 | 68.00% | 1.92 ± 0.83 | 0.4 | 0.93 |
| Sedation | 172 | 47.00% | 1.51 ± 0.70 | 94 | 50.80% | 1.51 ± 0.68 | 77 | 43.30% | 1.49 ± 0.72 | 0.73 | 0.15 |
| Change in Libido | 111 | 30.30% | 1.72 ± 0.79 | 63 | 34.10% | 1.70 ± 0.75 | 47 | 26.40% | 1.77 ± 0.84 | 0.76 | 0.11 |
| Dry Mouth | 101 | 27.60% | 1.55 ± 0.70 | 59 | 31.90% | 1.61 ± 0.74 | 41 | 23.00% | 1.49 ± 0.64 | 0.49 | 0.06 |
| Excessive Sweating | 71 | 19.40% | 1.86 ± 0.85 | 38 | 20.50% | 1.95 ± 0.87 | 32 | 18.00% | 1.75 ± 0.84 | 0.34 | 0.54 |
| Insomnia | 67 | 18.30% | 1.93 ± 0.78 | 32 | 17.30% | 2.03 ± 0.78 | 34 | 19.10% | 1.85 ± 0.78 | 0.36 | 0.66 |
| Impaired Mentation | 79 | 21.60% | 1.61 ± 0.72 | 41 | 22.20% | 1.78 ± 0.76 | 37 | 20.80% | 1.43 ± 0.65 | 0.032 | 0.75 |
| Fatigue | 65 | 17.80% | 1.62 ± 0.78 | 32 | 17.30% | 1.78 ± 0.79 | 32 | 18.00% | 1.47 ± 0.76 | 0.08 | 0.87 |
| Difficulty with Urination | 52 | 14.20% | 1.38 ± 0.60 | 28 | 15.10% | 1.50 ± 0.69 | 23 | 12.90% | 1.26 ± 0.45 | 0.24 | 0.54 |
| Increase in Appetite | 46 | 12.60% | 1.48 ± 0.59 | 25 | 13.50% | 1.52 ± 0.51 | 21 | 11.80% | 1.43 ± 0.68 | 0.37 | 0.62 |
| Decrease in Appetite | 44 | 12.00% | 1.55 ± 0.70 | 26 | 14.10% | 1.50 ± 0.71 | 18 | 10.10% | 1.61 ± 0.70 | 0.54 | 0.25 |
| Weight Gain | 38 | 10.40% | 1.68 ± 0.74 | 18 | 9.70% | 1.67 ± 0.59 | 20 | 11.20% | 1.70 ± 0.86 | 0.86 | 0.64 |
| Weakness | 33 | 9.00% | 1.36 ± 0.70 | 18 | 9.70% | 1.56 ± 0.86 | 14 | 7.90% | 1.14 ± 0.36 | 0.17 | 0.53 |
| Malaise | 27 | 7.40% | 1.48 ± 0.64 | 13 | 7.00% | 1.62 ± 0.65 | 13 | 7.30% | 1.38 ± 0.65 | 0.31 | 0.92 |
| Tachycardia/Palpitations | 25 | 6.80% | 1.44 ± 0.71 | 14 | 7.60% | 1.43 ± 0.65 | 10 | 5.60% | 1.50 ± 0.85 | 1.00 | 0.45 |
Values are shown as mean ± SD or n, %.
Bold P-value: P<0.05.
Wilcoxon rank-sum test.
Chi-Square test.
3.2 Association analyses of Pharmacokinetic genes and methadone dose
Methadone is mainly metabolized in the liver through specific isoforms of the cytochrome P-450 (CYP) enzyme system [19]. These isoforms include CYP2B6, CYP2C19, and CYP3A4, which have metabolic preferences toward the two different methadone enantiomers; the R-form is preferably metabolized by CYP2C19 and the S-form is preferably metabolized by CYP2B6 [19]. In our previous studies, the SNPs on CYP2B6 were significantly associated with plasma S-methadone concentration, the ratio of S-methadone/methadone dose, and the S-methadone clearance [6]. We also found that the CYP2B6 has a higher level of activity in the HCV-positive patients [20]. SNPs in CYP3A4 were significantly associated with COWS, TESS, and betel nut use [12]. The higher the withdrawal symptoms scores, the higher the side effect symptom score, but the lower the betel nut use. The gene dose of CYP2C19 showed significant associations with methadone dose, R-methadone/methadone dose, and R-EDDP/methadone dose [13]. Gene dose is associated with TESS scores in patients testing positive for urine morphine. The extensive metabolizers had a higher side effect score than did poor metabolizers.
In gene-gene interaction association analyses, the CYP2C19 gene may not interact with either CYP2B6 or CYP2C19. However, the allele types in either CYP2B6 or CYP3A4 may subgroup the methadone dose into six different dosage ranges and increase the level of significant associations between the gene dose of CYP2C19 and methadone dose. In three-gene interaction association analyses, the CYP2C19 did not interact with either CYP2B6 or CYP3A4. The allelic combination on CYP2B6 and CYP3A4 further subgrouped the methadone dose into 12 different dose ranges and increased the level of significant associations between CYP2C19 and methadone dose from a p-value of 0.018 to less than 10-4. This CYP2C19 interaction was also significantly associated with methadone tolerance when it was defined as the dose difference between the current dose and the starting dose of methadone.
In phase II morphine metabolic enzyme UDP glucuronosyltransferase 2 family, polypeptide B7 (UGT2B7), the SNPs in this coding region showed significant associations with the severity of withdrawal symptom scores rated by COWS [7].
3.3 Association analyses of a Pharmacodynamic gene OPRM1
The pharmacodynamic gene was reported first in the mu-opioid receptors (OPRM1). The non-synonymous OPRM1 SNP rs1799971 (exon 1; A118G polymorphism; Asn40Asp) showed a marginally significant association with methadone dose (adjusted GLM, p-value=0.027). The intron 1 region of SNPs demonstrated significant associations with the side effects of change in libido and insomnia [2]. It was also significantly associated with plasma nicotine metabolite cotinine [21]. In the recessive model of association analyses, the patients who had a higher plasma cotinine level had a lower insomnia symptom score. The pharmacokinetic genes may be correlated to physical dependence, whereas the pharmacodynamic genes are correlated to psychological dependence.
4. Discussion
Using this methadone maintenance cohort, we found that pharmacogenomics studies in heroin dependence are impossible to explain using a single gene association analysis. The metabolism of methadone involves more than one single cytochrome P-450 (CYP) enzyme; e.g., 2B6, 2C19 and 3A4. Each CYP isozyme may play different roles in their genetic variants. The patients in this cohort were exposed to multiple environmental influences; 95% patients tested positive for the HCV antibody. All patients reported smoking cigarettes. It is impossible to report these important environmental influences with a single genetic association analyses.
In the current study, the pharmacokinetic genes especially, the CYP2C19 x CYP2B6 x CYP3A4 interaction, may subgroup the patients into different methadone dosage groups with a p-value lower than that from the p-value of a pharmacodynamic OPRM1 single gene. This result indicated that the required methadone dosage is regulated by genes in combination with pharmacokinetic and pharmacodynamic influences. SNPs in the pharmacokinetic genes, such as CYP2B6, CYP2C19, and CYP3A4, may be indicators for the plasma methadone concentrations, methadone dose, and severity of withdrawal symptoms.
SNPs in the pharmacodynamic genes may influence the dosage prediction and the treatment side effects. SNPs in the pharmacodynamic genes, especially the OPRM1, were associated with methadone dose, and may be predictors for methadone side effects, such as change in libido and insomnia.
Pharmacogenomics studies promise the advent of personalized medicine. In association analyses of SNPs in single genes, we found that each single gene covers different numbers of exons and introns. Using single genetic variants, studies may provide further information for genetic regulation and its functional correlation with the responses to direct further biological study or to be replicated in the study of other ethnic groups.
The limitation of this cohort include gender imbalance (81% are male), 95% HCV positive, around 50% of patients tested positive for urine morphine , and it was a relatively small sample. Replication studies are needed in order to establish future treatment guidelines.
Additional methods of optimizing drug therapy, with respect to patients' genotype, warrant further investigation to ensure maximum efficacy with minimal adverse effects. This study considered the potential in a single gene of both pharmacokinetics genes (e.g., CYP2B6, 3A4, 2C19, UGT2B7 and PXR) and pharmacodynamic genes (e.g., OPRM1). Studying random combinations of these genes and then making association analyses without having the results of each single gene would not reveal the individual contributions from each single gene in the systemic methadone treatment response. Gene-gene interaction should be based upon single gene discoveries, not nebulous theory. As the pharmacogenomics studies are still in their early stage, studies of a number of SNP markers in each single gene, which may potentially represent the function of a gene, are essential. We therefore showed the SNP IDs, so that it would be easier for researchers who are interested in replicating the study with other ethnic groups.
Methadone pharmacogenomics study may provide a key to deciphering the mechanisms of opioid dependence. There is great potential for pharmacogenomics to be a clinical practice guideline for future individualized medicine. Our studies provide essential information in terms of bridging the basic biological functional study, through understanding the exon and intron roles in each gene, and the potential drug-targeting of novel treatment processes, through the association analyses of treatment responses.
Figure 1.
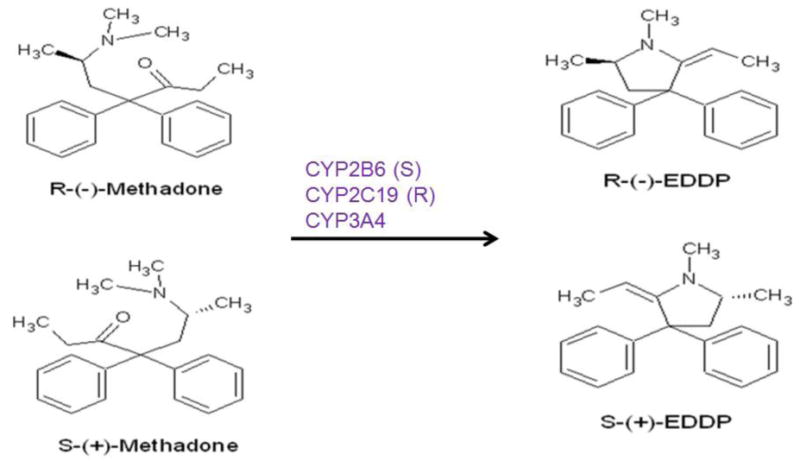
Methadone metabolic pathway.
Figure 2.
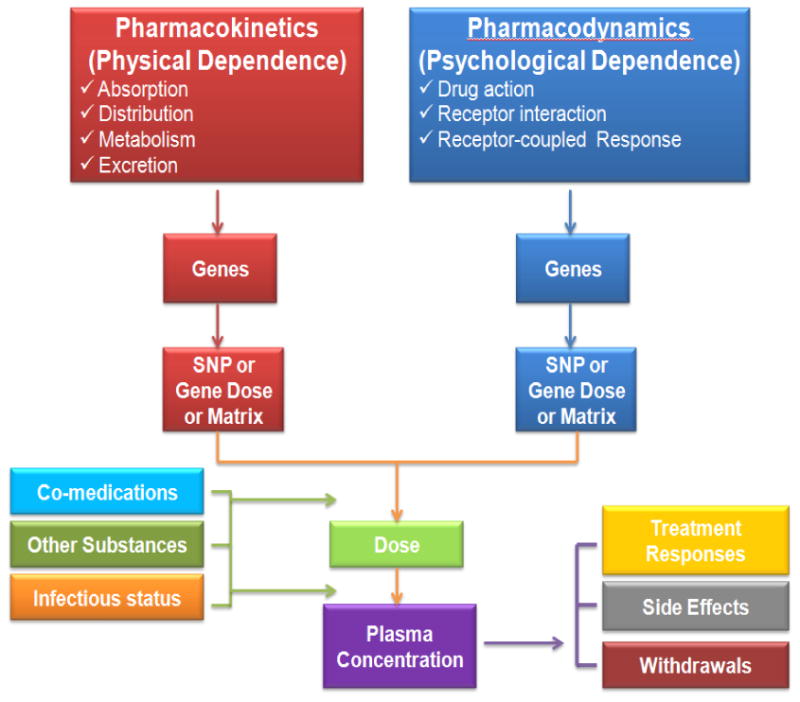
Pharmacogenomics study diagram for methadone maintenance treatment patients.
Acknowledgments
I thank the collaborators Dr. Shang-Chang Wang who recruited the patients, Dr. Hsiao-Hui Tsou who performed the statistical analyses, Prof. Ing-Kang Ho who led this study, and Prof. Keh-Ming Lin who designed the pharmacogenomics study. I thank Ming-Chu Tseng, Pei-Fang Li, Shu-Chuan Ting, Yu-Ching Lin, Miao-Fang Lee, Chi-Yun Huang, and Yu-Hun Tsai of the nursing staff from the seven participating hospitals for interviewing the patients. I also thank the Clinical Trial Information Management System (CTIMeS) at NHRI for the data collection. I thank the National Center for Genome Medicine at Academia Sinica, Taiwan, for genotyping/technical support. This Center was supported by grants from the National Core Facility Program for Biotechnology of National Science Council, Taiwan.
I acknowledge the significant contributions of the Tao-Yuan Mental Hospital, En-Chu-Kong Hospital, Far-Eastern Memorial Hospital, Taipei City Hospital Song-De and Yang-Ming Branches, China Medical University Hospital, and Wei-Gong Memorial Hospital.
These studies were supported by grants from the National Research Program for Genomic Medicine (NSC 98-3112-B-400-011, NSC 99-3112-B-400-003 and NSC 100-3112-B-400-015), the National Science Council (NSC 97-2314-B-400-001-MY3 and NSC 100-2314-B-400-002-MY3) and National Health Research Institutes (PH-098-PP-41, PH-098-PP-46, PH-098-PP-38, PH-098, 99-PP-42, PH-100-PP-37, PH-101-PP-32 and PH-98, 99, 100-SP-11, NHRI-101A1-PDCO-1312141), Taiwan.
Footnotes
Conflicts of Interests: The author declared no conflicts of interests.
References
- 1.Bengtsson F. therapeutic drug monitoring of psychotropic drugs: TDM “Nouveau. Ther Drug Monit. 2004;26:145–151. doi: 10.1097/00007691-200404000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Wang SC, Tsou HH, Chen CH, et al. Genetic polymorphisms in the opioid receptor mu1 gene are associated with changes in libido and insomnia in methadone maintenance patients. Eur Neuropsychopharmacol. 2012;22:695–703. doi: 10.1016/j.euroneuro.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Marsden J, Farrell M, Bradbury C, et al. Development of the Treatment Outcomes Profile. Addiction. 2008;103:1450–1460. doi: 10.1111/j.1360-0443.2008.02284.x. [DOI] [PubMed] [Google Scholar]
- 4.Wesson DR, Ling W. The Clinical Opiate Withdrawal Scale (COWS) J Psychoactive Drugs. 2003;35:253–259. doi: 10.1080/02791072.2003.10400007. [DOI] [PubMed] [Google Scholar]
- 5.Guy W. ECDEU assessment manual for psychopharmacology. Revised edition. Rockville, MD: U.S. Dept. of Health, Education, and Welfare, Public Health Service, Alcohol, Drug Abuse, and Mental Health Administration, National Institute of Mental Health, Psychopharmacology Research Branch, Division of Extramural Research Programs; 1976. [Google Scholar]
- 6.Wang SC, Ho IK, Tsou HH, et al. CYP2B6 polymorphisms influence the plasma concentration and clearance of the methadone S-enantiomer. J Clin Psychopharmacol. 2011;31:463–469. doi: 10.1097/JCP.0b013e318222b5dd. [DOI] [PubMed] [Google Scholar]
- 7.Tian JN, Ho IK, Tsou HH, et al. UGT2B7 genetic polymorphisms are associated with the withdrawal symptoms in methadone maintenance patients. Pharmacogenomics. 2012;13:879–888. doi: 10.2217/pgs.12.69. [DOI] [PubMed] [Google Scholar]
- 8.Wang SC, Ho IK, Wu SL, et al. Development of a method to measure methadone enantiomers and its metabolites without enantiomer standard compounds for the plasma of methadone maintenance patients. Biomed Chromatogr. 2010;24:782–788. doi: 10.1002/bmc.1363. [DOI] [PubMed] [Google Scholar]
- 9.Gerra G, Leonardi C, Cortese E, et al. Human kappa opioid receptor gene (OPRK1) polymorphism is associated with opiate addiction. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:771–775. doi: 10.1002/ajmg.b.30510. [DOI] [PubMed] [Google Scholar]
- 10.Luo X, Zuo L, Kranzler H, et al. Multiple OPR genes influence personality traits in substance dependent and healthy subjects in two American populations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1028–1039. doi: 10.1002/ajmg.b.30701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar D, Chakraborty J, Das S. Epistatic effects between variants of kappa-opioid receptor gene and A118G of mu-opioid receptor gene increase susceptibility to addiction in Indian population. Prog Neuro-Psychopharmacol Biol Psychiatry. 2012;36:225–230. doi: 10.1016/j.pnpbp.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 12.Chen CH, Wang SC, Tsou HH, et al. Genetic polymorphisms in CYP3A4 are associated with withdrawal symptoms and adverse reactions in methadone maintenance patients. Pharmacogenomics. 2011;12:1397–1406. doi: 10.2217/pgs.11.103. [DOI] [PubMed] [Google Scholar]
- 13.Wang SC, Ho IK, Tsou HH, et al. Functional genetic polymorphisms in CYP2C19 gene in relation to cardiac side effects and treatment dose in a methadone maintenance cohort. OMICS. 2013 doi: 10.1089/omi.2012.0068. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan HY, Chiou JJ, Tseng WH, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodi CP, Darnhofer-Patel B, Stanssens P, et al. A strategy for the rapid discovery of disease markers using the MassARRAY (TM) system. BioTechniques. 2002;32:S62–S69. [PubMed] [Google Scholar]
- 16.Tost J, Gut IG. Genotyping single nucleotide polymorphisms by MALDI mass spectrometry in clinical applications. Clin Biochem. 2005;38:335–350. doi: 10.1016/j.clinbiochem.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 18.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 19.Gerber JG, Rhodes RJ, Gal J. Stereoselective metabolism of methadone N-demethylation by cytochrome P450 2B6 and 2C19. Chirality. 2004;16:36–44. doi: 10.1002/chir.10303. [DOI] [PubMed] [Google Scholar]
- 20.Wu SL, Wang SC, Tsou HH, et al. Hepatitis C virus infection influences the S-methadone metabolite plasma concentration. PLoS ONE. 2013 doi: 10.1371/journal.pone.0069310. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen YT, Tsou HH, Kuo HW, et al. OPRM1 genetic polymorphisms are associated with the plasma nicotine metabolite cotinine concentration in methadone maintenance patients: a cross sectional study. J Hum Genet. 2013;58:84–90. doi: 10.1038/jhg.2012.139. [DOI] [PubMed] [Google Scholar]


