Abstract
The ability to isolate and analyze rare circulating tumor cells (CTCs) has the potential to further our understanding of cancer metastasis and enhance the care of cancer patients. In this protocol, we describe the procedure for isolating rare CTCs from blood samples by using tumor antigen–independent microfluidic CTC-iChip technology. The CTC-iChip uses deterministic lateral displacement, inertial focusing and magnetophoresis to sort up to 107 cells/s. By using two-stage magnetophoresis and depletion antibodies against leukocytes, we achieve 3.8-log depletion of white blood cells and a 97% yield of rare cells with a sample processing rate of 8 ml of whole blood/h. The CTC-iChip is compatible with standard cytopathological and RNA-based characterization methods. This protocol describes device production, assembly, blood sample preparation, system setup and the CTC isolation process. Sorting 8 ml of blood sample requires 2 h including setup time, and chip production requires 2–5 d.
INTRODUCTION
Systematic studies of metastasis require numerous unbiased observations of patient-derived CTCs1. Efforts aimed at analyzing CTCs have spurred the development of several technologies for isolating these rare cells from the blood of patients2,3; this has, in turn, enabled studies of metastasis in human cancer4–14. A better understanding of CTC biology and the development of more advanced technologies could enable real-time analysis of CTCs probing for noninvasive screening of tumor evolution and for predictive biomarkers to guide therapy15.
Methods for isolating and analyzing CTCs
A variety of specialized methods to isolate and analyze CTCs have been developed (reviewed in Yu et al.3). Direct analysis of unpurified nucleated cells from blood16–18 or diluted blood with tumor-specific staining19 excels in its simplicity, but it is limited in its applications. Bulk blood-processing methods, such as flow cytometry20,21 and magnetophoresis 22–25, despite their tendency to lose rare cells26, have been popular for CTC enrichment, because they require simple and readily available tools. Enrichment of CTCs on the basis of physical properties such as size27–30, deformability 31,32 and density 33 have also been applied. CellSearch, the only US Food and Drug Administration (FDA)-cleared CTC isolation method for prognostic use to date, uses an epithelial cell adhesion molecule (EpCAM)34,35–dependent immunomagnetic enrichment method commercialized by Veridex 36,37.
Microfluidic methods were first introduced by our group to enable gentle and sensitive capture of CTCs using epithelial cell–specific markers 38. Since their introduction, our laboratory has reported a variety of methods for isolation from patient blood 3,5,38,39, paving the way for systematic study of these cells in the cancers of the pancreas, breast, lung and prostate 4,6,7,40. The microfluidic CTC isolation methods developed to date use antibodies against a surface antigen, such as EpCAM, to select for cells of epithelial origin, thus potentially missing tumor cells that lose EpCAM expression because of the dynamic epithelial-mesenchymal transition 6,41,42. Similarly, CTCs from cancers of non-epithelial origin are not detectable with EpCAM-based enrichment. In addition, the binding of antibodies to EpCAM may induce cytotoxicity 43, thus altering the original state of CTCs and reducing the reliability of further biological research. It is possible to widen the cell capture abilities of CTC-antigen–dependent capture methods by using antibody cocktails against cancer type–specific antigens6,44. These limitations can be overcome by using methods that enrich aberrant cells from blood by depleting cells that normally exist in blood13,26,45–52. This strategy provides tumor antigen–independent enrichment performance, and thus it is applicable to cells disseminated from virtually any tumor type. This strategy is commonly referred to as negative depletion due to removal of the leukocytes with antibodies directed against them, as opposed to isolating cancerous (positive) cells in clinical blood samples of advanced cancer patients.
Introduction of magnetophoretic cell separation via antibody-covered superparamagnetic beads53 enabled the depletion of white blood cells (WBCs) and the analysis of CTCs in studies of head and neck squamous cell carcinoma48, colorectal cancer45 and renal cell carcinoma46. These studies used density-gradient centrifugation and subsequent immunomagnetic separation by using anti-CD45–coated superparamagnetic beads and a magnetic column to remove CD45+ cells and enrich for CTCs. These cell-sorting methods could achieve up to a 1.4-log depletion of nucleated cells with a 40–90% cancer cell yield. Lara et al.26 developed a flow-through quadrupole magnetic cell sorter that relies both on magnetism-driven deflection and attachment rather than solely on attachment of labeled cells. By using RBC lysis pre-processing, this system resulted in 2.5-log WBC depletion and a 46% yield of spiked MCF-7 cells. Zhang et al.54 found a reduction in antibody affinity upon its immobilization on a magnetic particle. Incorporation of tetrameric antibody complexes55,56 to this negative enrichment method improved the depletion to 3.1 log with a cancer cell recovery of 84% (ref. 57). These reported methods have partial recovery of CTCs owing to manual sample pre-processing steps such as red blood cell lysis (loss quantified as ~11% (ref. 26)) and density-gradient centrifugation (loss quantified as ~27% (ref. 26)). Although the advantages of tumor antigen–independent detection have been put forward with the previous studies, a robust and automated platform that exhibits high efficiency while performing negative depletion is still lacking.
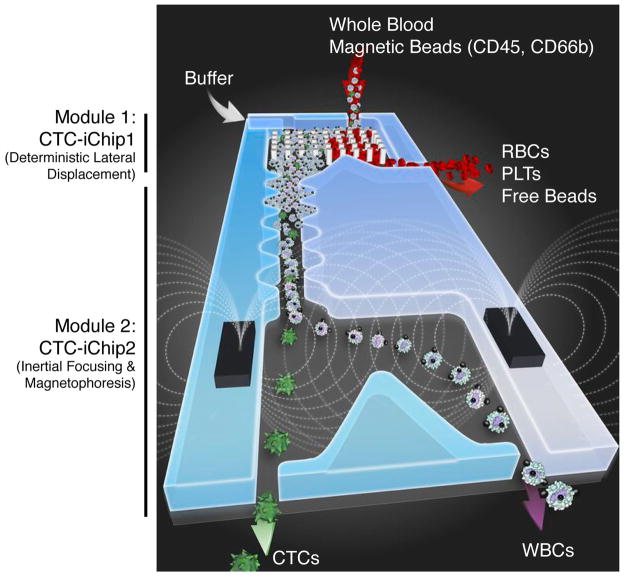
The current protocol describes the isolation of CTCs by using a modified CTC-iChip architecture58 to improve hematopoietic cell depletion. The CTC-iChip uses continuous deterministic lateral displacement (DLD)59 for size-based separation of WBCs and tumor cells from whole blood, inertial focusing for precise positioning of these cells in a micro-channel60, and then microfluidic magnetophoresis for immunomagnetic isolation of circulating tumor cells (Fig. 1). It is currently composed of two separate chips (CTC-iChip1 and CTC-iChip2), used serially (in-line); each chip is produced with different manufacturing procedures.
Figure 1.
CTC-iChip schematic. The CTC-iChip is composed of two separate microfluidic devices that house three different microfluidic components engineered for inline operation: DLD to remove nucleated cells from whole blood by size-based deflection by using a specially designed array of posts performed in CTC-iChip1, inertial focusing to line up cells to prepare for precise magnetic separation and magnetophoresis for sensitive separation of bead-labeled WBCs and unlabeled CTCs, which are performed in CTC-iChip2. PLTs, platelets.
Development of the protocol
Throughout the process of developing the current protocol, we used our experience from the development of two prior CTC isolation technologies. In our previous devices, antibody-functionalized microposts or herringbone structures were used to manipulate fluid flow, enhance cell-surface interactions in microfluidic channels coated with anti-EpCAM and efficiently capture CTCs on the walls of the device. The CTC-Chip enabled microfluidic capture of CTCs with high yield and purity 38, but difficulties in production limited its use to small-scale studies. The herringbone-Chip used an innovative microvortex-generating device geometry, which could isolate EpCAM-positive CTCs with practically no loss and a 2.5 ml/h throughput 39. The herringbone-Chip could be mass-produced and translated into the clinic; its ease of use, with no blood sample preparation steps, enabled a variety of large-scale studies, and was also developed for use with cocktail capture antibodies6. However, systematic analyses of CTCs require running larger volumes of blood and an unbiased approach for truly capturing their heterogeneity. Thus, we aimed at developing an antigen-independent technique that can process larger volumes. In addition, considering the variety of applications developed to date, we aimed to isolate CTCs in a state that was compatible for both clinical and sophisticated molecular analyses: suspended and not bound to a device. Hence we developed CTC-iChip; it has all the features that we aimed for, and work is underway to translate it to a form usable in clinics.
In our previous study with the CTC-iChip system, we demonstrated successful isolation of CTCs from non-epithelial cancers (e.g., melanoma), as well as mesenchymal tumors58. To establish a baseline comparison with the CellSearch system, we used the positive-selection mode of the CTC-iChip in which CTCs were captured with EpCAM-labeled magnetic beads, and we stained the isolated cells with the same reagent cocktail before the enumeration process. This reagent cocktail consisted of antibodies against cytokeratins as epithelial markers, CD45 as a leukocyte marker and DAPI as a nuclear marker. Sensitivity of this modified CTC-iChip was higher for samples from patients with lower CTC numbers (<30 CTCs/7.5ml) of captured CTCs58. The positive-selection modality achieved >3.5-log purification (a background of 1,500 WBCs/ml of blood) compared with the negative-depletion strategy, which yielded an order of magnitude less purity (32,000 WBCs/ml of blood, 2.5-log depletion). This prompted us to re-design the CTC-iChip to improve leukocyte depletion. To date, WBC depletion using these standard technologies has been insufficient to allow detailed analysis of CTCs.
We performed systematic studies to re-design CTC-iChip, which led to substantial performance enhancements. We performed identification and quantification of unlabeled WBC populations that contaminate the product by using various bead-labeling protocols. Although CD45 is the most widely expressed leukocyte marker, the degree of its expression is variable in the leukocyte population. We found that immunomagnetic enrichment targeting solely CD45 provided insufficient purity, as a large granulocyte population was consistently found in the product. To target this population, we chose to supplement CD45 with CD66b, as it is highly expressed in granulocytes and not observed in tumor cells. We markedly improved magnetic labeling of the WBC population by first introducing magnetic bead–free biotinylated CD45 and CD66b and subsequent attachment of the magnetic beads. To overcome the problem of large WBC and magnetic bead aggregates forming plaques on the side walls of the CTC-iChip, leading to contamination in the product, we re-designed the magnetophoresis geometry such that labeled WBCs were directed to the middle of the deflection channel. The design change included splitting magnetophoresis into two stages with low and high magnetic field strength to increase the dynamic range. By using the re-designed CTC-iChip system, we achieved an average of 3.8-log depletion of WBCs at a rate of 8 ml of whole blood/h and a cancer cell yield of 97 ± 2.7%.
It is a challenging task to rapidly separate a rare cell population without loss from whole blood, a complex fluid, which makes most of the CTC isolation methods laborious. The CTC-iChip successfully brings together sophisticated fluidic and magnetic components. The protocol is relatively straightforward after the production and assembly of all the chips and manifolds. It involves manipulating pumps and tubing for sequentially running the buffer through the fluidic system for priming; pipetting antibodies and magnetic beads to the clinical blood sample; and manipulating tubing and pumps again for running and stopping the system. However, preparation of the system (Steps 1–44; Fig. 2) is lengthy and manual because the many different pieces are produced and assembled separately. We envision the CTC-iChip to eventually be a monolithic device that does not require the user to perform Steps 1–44. However, this information is useful if the reader is modifying the device or using a single module of the device. We discuss the details of the design of separation methods below in the ‘Principles of the present protocol’ section.
Figure 2.
Protocol flowchart. Flowchart of the protocol including references to the steps in the text and related figures.
During the development of the CTC-iChip, we aimed for a high-throughput, high-recovery and gentle method to immobilize the cells on glass slides for applications such as clinical cytopathology. For this purpose, we designed a closed centrifugation system, the Spintrap, that concentrates cells while preserving cellular integrity and quantity (Supplementary Figs. 1 and 2, Supplementary Video 1). In our previous study, the Spintrap was used to process CTCs isolated by the CTC-iChip system58.
Applications of protocol
The most common application of CTC analysis is quantification of CTCs, which is done by using antibody stains. Fluorescence in situ hybridization (FISH) and RNA in situ hybridization 6,14 (RNA-ISH) techniques can also be used to interrogate CTCs. In addition to fluorescence-based methods, the cytopathology of CTCs can also be evaluated with traditional stains such as Papanicolaou or H&E and characterized further by immunocytochemistry using antibodies against tumor markers. CTCs can also be analyzed by RNA analytical methods, even at the single-cell level.
Two distinct features of the CTC-iChip enable a variety of applications for research and diagnostics of CTCs and other rare cells: the cells of interest are in suspension rather than immobilized on a chip, and the mode of CTC isolation is tumor antigen–independent. The combination of these factors enables high-quality cytopathological evaluation of cells, single-cell RNA and genotyping analysis, and culture of CTCs58.
Principles of the present protocol
In microfluidic magnetophoresis, micrometer-sized paramagnetic beads are functionalized with antibodies to target cells of interest and then added to a suspension containing cells expressing the antigen of interest. Upon injection of the cell suspension into the microfluidic chip, a magnetic field is applied to direct the flow of cells within the microfluidic channel. Previous adaptations of magnetophoretic separation into microfluidic systems61–63 resulted in devices with low throughput and/or yield, making them unsuitable for biomedical applications. To address the challenges of isolating CTCs from whole blood, we used two microfluidic principles to prepare nucleated cells for magnetophoretic sorting.
Design of CTC-iChip1: blood debulking
On the basis of the work published by Austin and co-workers59, we developed a continuous-flow system using DLD that separates nucleated cells from whole blood (Fig. 3). DLD uses an array of posts with a pillar size and array offset designed to deflect particles above a certain size, thereby separating them from the main suspension64. The key parameter for DLD arrays is the critical deflection diameter (Dc), which is the minimum particle hydrodynamic diameter deflected by the DLD array. More specifically, particles whose hydrodynamic diameter is smaller than the array’s Dc are not deflected by the presence of the pillar array, and they follow the primary fluid streamlines around the posts (Supplementary Fig. 3). Conversely, particles whose hydrodynamic diameter is larger than Dc are deflected by the array (Supplementary Video 2).
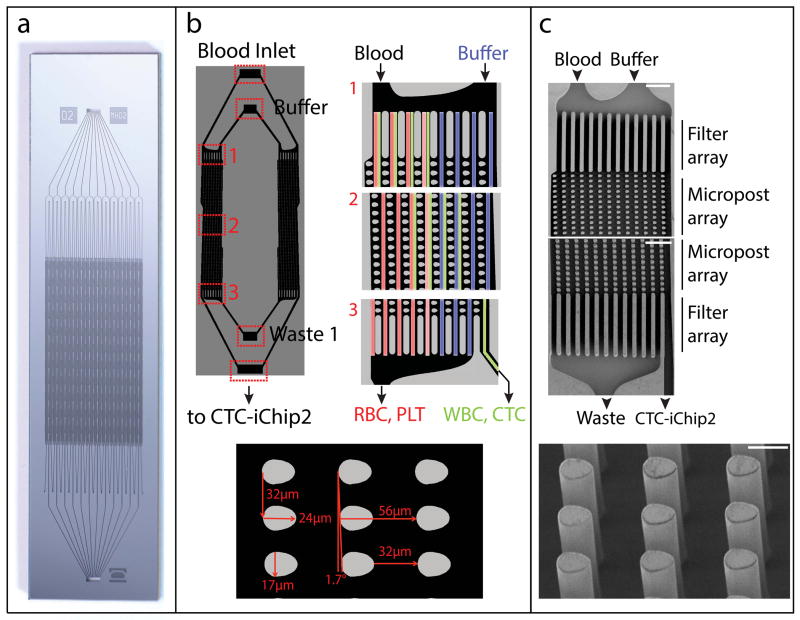
Figure 3.
Structure of the CTC-iChip1. DLD is designed to separate nucleated cells from blood, and it is performed in CTC-iChip1. (a) High-resolution photograph of the fabricated chip. (b) Schematic of CTC-iChip1 (left image shows only two lanes, whereas the device is composed of many). Whole blood and buffer inlets enter from opposite top corners of the post array (right, 1). The post geometry (bottom, egg-shaped with a width • length • height of 24 • 17 • 150 μm) is engineered to deflect particles with a diameter >4 μm. As such, posts deflect nucleated cells (green-labeled lanes) away from smaller RBCs, platelets and plasma (red-labeled lanes) and toward the buffer (blue-labeled lanes). As the device operates in a laminar flow regime (Supplementary Fig. 3), RBCs, platelets (PLTs) and free beads remain in their lateral positions. (c) Electron micrographs of CTC-iChip1. Start and end of post array (top; scale bars, 100 μm) and posts (bottom; scale bar, 20 μm).
The critical deflection diameter depends on three array parameters65: row shift fraction (ε), horizontal gap between adjacent pillars (gH) and the array geometrical factor (η). A mathematical expression for Dc can then be written as follows:
The array geometrical factor η accounts for nonuniform flow through the gap, and it depends on array arrangement, pillar shape, as well as material and surface properties. Determination of η requires one to resolve the flow profile within the gap (u(x)), from which the array geometrical factor can be computed as follows:
where β= ηgHε. Numerical tools (e.g., COMSOL, Ansys) can be used to model different array configurations and determine their associated Dc, although analytical solutions are available for certain array configurations where the flow profile is known a priori (e.g., array of cylindrical micropillars in square arrangement).
The CTC-iChip2 contains 24 debulking microarrays, with inlets for blood and running buffer, and outlets for deflected product and undeflected waste (Fig. 3). The inlets and outlets of arrays get combined into single inlets and outlets (Fig. 3a). The blood and running buffer enter from opposite sides of a microarray, so that they do not mix in the laminar flow conditions, and the debulking microarray deflects cells above Dc toward the buffer lanes, which are collected by the product outlet (Fig. 3b). The debulking microarrays are characterized by a 32-μm gap, a 1.7° slope and ‘egg-shaped’ micropillars (Fig. 3b) to provide a smaller DC compared with cylindrical pillars66. These array properties yield a 4-μm critical deflection diameter for the microarray. The relative injection of blood sample to running buffer is set to 20%. The percentage of volume collected as product is set to 20% as well. This design has been shown to provide complete removal of red blood cells and platelets while yielding a large operational range for particles between 4 μm and 32 μm (F.F., unpublished data). Unlabeled WBC retention for the microarray was quantified as 60%, whereas magnetic bead labeling increases the retention to ~85–90% owing to increased hydrodynamic diameter. It is possible to design a debulking array to remove most of the leukocytes along with red blood cells and platelets for size-based sorting, but CTCs that have similar sizes as leukocytes58 would be lost.
Design of CTC-iChip2: Magnetic separation
The second stage of the CTC-iChip, CTC-iChip2, uses inertial focusing and magnetophoresis to sort nucleated cells according to their magnetic bead load (Fig. 4). Inertial focusing governs cell sorting precision by positioning all the cells in a single line, whereas magnetophoresis governs cell-sorting efficiency by removing nontarget cells as they are moving with the fluid flow.
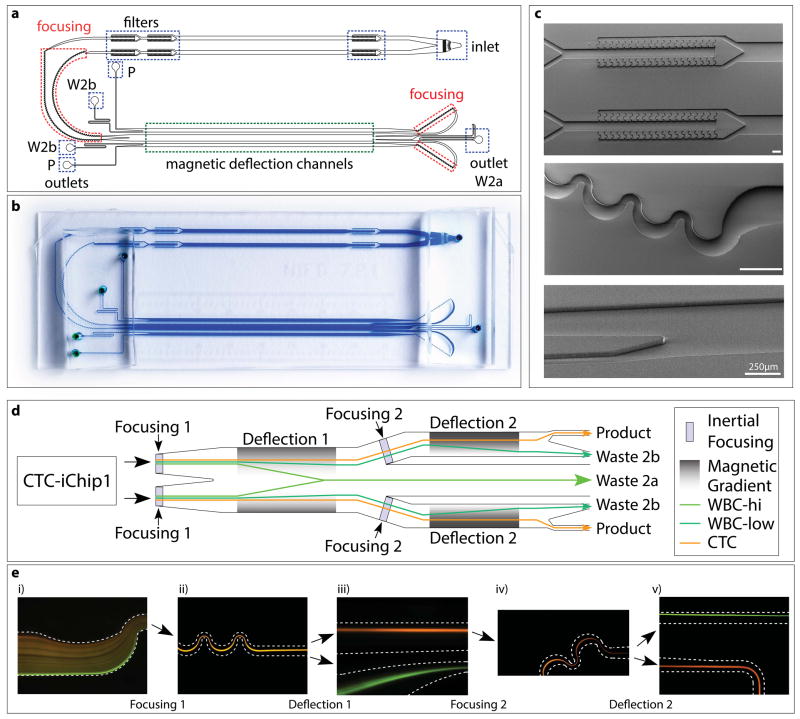
Figure 4.
Structure of CTC-iChip2. (a–c) CAD drawing (a), high-resolution photograph (b) and electron micrographs (c) of CTC-iChip2. Highlighted are inlet and outlets, filters, focusing regions and magnetic deflection channels (width • length of magnetic deflection stage 1 = 1 mm • 37 mm; width • length of stage 2 = 0.5 mm • 37 mm). Scale bars, 250 μm. W2, waste 2a; W2b, waste 2b; P, product. (d) Schematic of CTC-iChip2. Note that, for simplification, the schematic was linearized; in the actual device, before entering the second stage, fluid flow turns around 180° and thus moves in the opposite direction of the first stage. (e) Fluorescence images of cell streaks formed during CTC-iChip2 operation showing both stages of inertial focusing and magnetophoresis of WBCs with (green) or without (orange) magnetic load. CTC-iChip2 initially separates the flow equally into two parallel and equivalent channels for higher throughput. Cells get tightly focused in the inertial focusing region, and subsequently separated in the magnetophoresis region (dashed lines symbolize channel wall). In the first separation stage, the magnetic force is directed toward the middle of the channel, and thus labeled cells deflect toward the middle and exit through waste 2a (d, Fig. 5). The first magnetophoresis stage is designed to have a relatively lower magnetic gradient, as a result only the portion of the WBC population with more than seven beads is successfully deflected. Channels on each side of waste 2a lead to the second stage, where cells are refocused and deflected with higher sensitivity (cells with more than two beads are deflected). Separated populations exit through their respective outlets (WBC → waste 2b, CTC → product).
It is important to align the nucleated cell population such that the cells’ lateral positions within the channel before magnetophoretic separation are restricted and overlapping. Conventional flow cytometry and magnetophoresis use hydrodynamic focusing with sheath flows for critically higher accuracy in sorting and analysis of cells. Although this achieves well-controlled positioning of cells, the use of sheath flow drastically reduces the sample flow rate (it effectively dilutes the sample) and the overall throughput of such systems. Instead, we developed and integrated another microfluidic design that is capable of aligning cells with high precision by using inertial forces in microchannels, a technique that was pioneered in our laboratory60. Inertial focusing does not require dilution, which enables high-throughput magnetophoresis of millions of cells.
Inertial focusing enables the precise lateral positioning of particles within a microfluidic channel. Positioning all cells, both labeled and unlabeled, to the same position allows highly sensitive separation of the two populations by using magnetophoresis. The cells that are not labeled do not deflect and follow the streamlines on which they are located after inertial focusing, which are directed into a narrow microchannel at the physical end of the magnetophoresis stage (Fig. 4d and Supplementary Video 3). A key design requirement is consistent focusing of cells over a large size range, as a CTC population has a heterogeneous size distribution 39,58. Particle ordering through inertial focusing has been shown in microchannels of various sizes and geometries, including straight, curved, spiral, symmetric and nonsymmetric serpentine channels60,67. We have observed that nonsymmetric serpentine channels (Fig. 4c) satisfy the functional requirements better than the other channel geometries. Therefore, this geometry was selected as the primary focusing channel design.
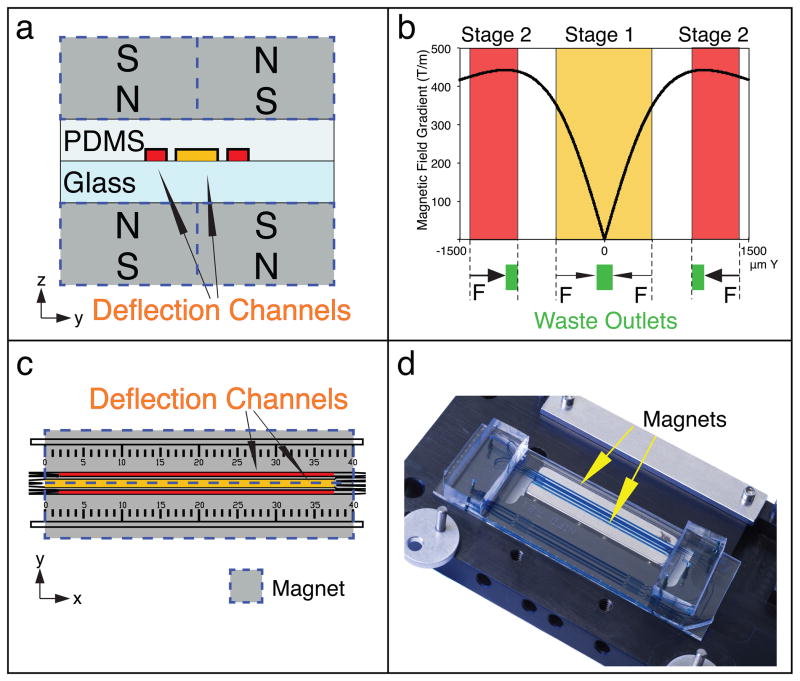
In magnetophoresis, the magnetic field gradient and the residence time of bead-labeled cells within the magnetic field (flow rate) determine the separation performance and thus the final purity of the output. As the magnetic field gradient increases, so does separation performance. A greater residence time within the magnetic gradient (lower flow rate) will also increase magnetic sensitivity, but at the expense of throughput.
The CTC-iChip performs magnetic separation in two serial stages (see Fig. 4d for overall schematic and Fig. 5 for magnetophoretic design). In the first stage, cells travel at ~50 mm/s, and they are sorted at a rate of ~10,000 nucleated cells/s with a magnetic sensitivity of ~7 magnetic beads/cell. This stage was designed to be extremely robust and resistant to aggregations of magnetic beads and WBCs while still having a sufficiently large magnetic gradient to remove 99.9% of WBCs under typical conditions. This is accomplished by deflecting bead-labeled cells toward the center of the flow channel rather than against a side wall where they may accumulate. Cells that are not deflected in the first stage (CTCs and WBCs with few beads) continue to the second stage, which comprises two parallel microfluidic channels in which cells travel at ~30 mm/s and are sorted at a rate of ~10–20 nucleated cells/s. The second stage has greater residence time and a larger magnetic field gradient than the first stage (Fig. 5b), which gives the second stage (and the device as a whole) a magnetic sensitivity of ~2 magnetic beads/cell. An important design consideration was avoidance of magnetic aggregate generation. Deflecting bead-labeled cells toward the center of the flow channel rather than the side walls prevented the generation of aggregates, which upon break-off can be collected in the product and reduce purity.
Figure 5.
Magnetophoresis setup. Deflection channel positioning in the CTC-iChip2 magnetic manifold is crucial in the success of the protocol. (a) zy plane schematic of the magnetophoretic setup for both stage 1 (yellow) and stage 2 (red). (b) The plot on top shows the calculated magnetic field gradient on points across the y axis, where the indicated stages of the deflection channels are. Stage 2 positioning is designed to enable the highest sensitivity separation possible, whereas stage 1 positioning is designed to push the cells toward the middle with a softer magnetic field gradient. (c) CTC-iChip2 design includes a ruler to help align CTC-iChip2 with its magnetic manifold. (d) When aligned, the stage 1 deflection channel is positioned in the middle of two magnets.
Production and operation
The current CTC-iChip technology consists of two separate microfluidic devices, specifically using two different materials for the fluidic layers: silicon and polydimethylsiloxane (PDMS). For both devices, glass is used as the cover layer, providing both a tight seal (>200 kPa (30 p.s.i.) pressure range) and optical transparency.
CTC-iChip1 production
Silicon was selected as the material for the first device of the CTC-iChip, which consists of micropost arrays arranged in patterns that enable deterministic displacement of nucleated cells. Current silicon-processing technologies enable very-high-fidelity reproduction of large-height (up to hundreds of micrometers), high-aspect-ratio features (1:100 ratios are easily achievable). Silicon is therefore particularly suitable for our micropillar technology, where the device depth is 150 μm and where aspect ratios up to 1:50 are required. In this device, manufacturing of the pillar array devices is outsourced to the Swedish company Silex, a microelectromechanical systems (MEMS) company that uses silicon deep reactive ion etching68 to produce the necessary structures. Translation from silicon to lower-cost plastic manufacturing is currently under investigation.
CTC-iChip2 production
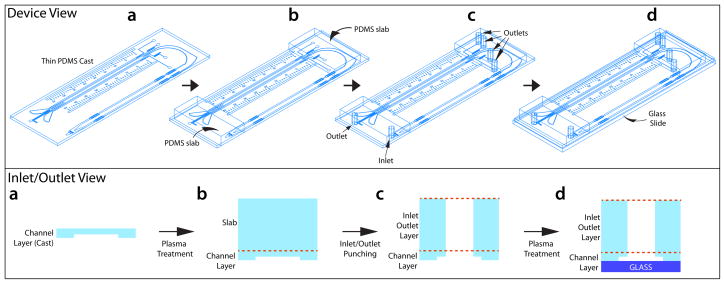
CTC-iChip2 is produced via initial microfabrication of silicon masters via soft lithography with SU-8 and a mask, and a subsequent mold made of PDMS. This procedure is relatively inexpensive, and suitable especially when the design is still under development. One important design criterion we had in CTC-iChip2 is the ability for the chip to fit into its magnetic manifold that needs to apply a strong magnetic field. To enable a high-magnetic-field gradient, the magnetic manifold should be in as close proximity to the microfluidic channel as possible, but it also should not deform the channel. Thus, we produced CTC-iChip2 as a relatively thin device (~1-mm channel layer, Fig. 6). In contrast to this thickness requirement of magnetophoresis, fluidic coupling of the microfluidic channels to tubing requires relatively thick inlet and outlet for the holding tubing against pressure. To facilitate this, during production of the chip, we added a layer of thick PDMS slab via plasma bonding and subsequently punched the inlet and outlets (Fig. 6). To avoid failures due to the CTC-iChip operation, we perform quality control of CTC-iChip2 under the microscope by inspecting channels for delamination or blockage. If they are missed during the inspection, such problems can also be found during the initial priming steps of the protocol and avoided before blood sorting begins. Problematic chips are discarded at these early steps.
Figure 6.
Schematic of the production of CTC-iChip2. (a–d) Schematic showing the production steps of CTC-iChip2 in the clean room in device view (top) or inlet/outlet view (bottom panel). Thin PDMS is produced in Steps 1–13 and bonded to a second layer of thick PDMS pieces in Steps 14–16; inlet and outlets are punched in Step 17 and bonded to glass slides in Steps 18–22.
Custom-made CTC-iChip1 manifold
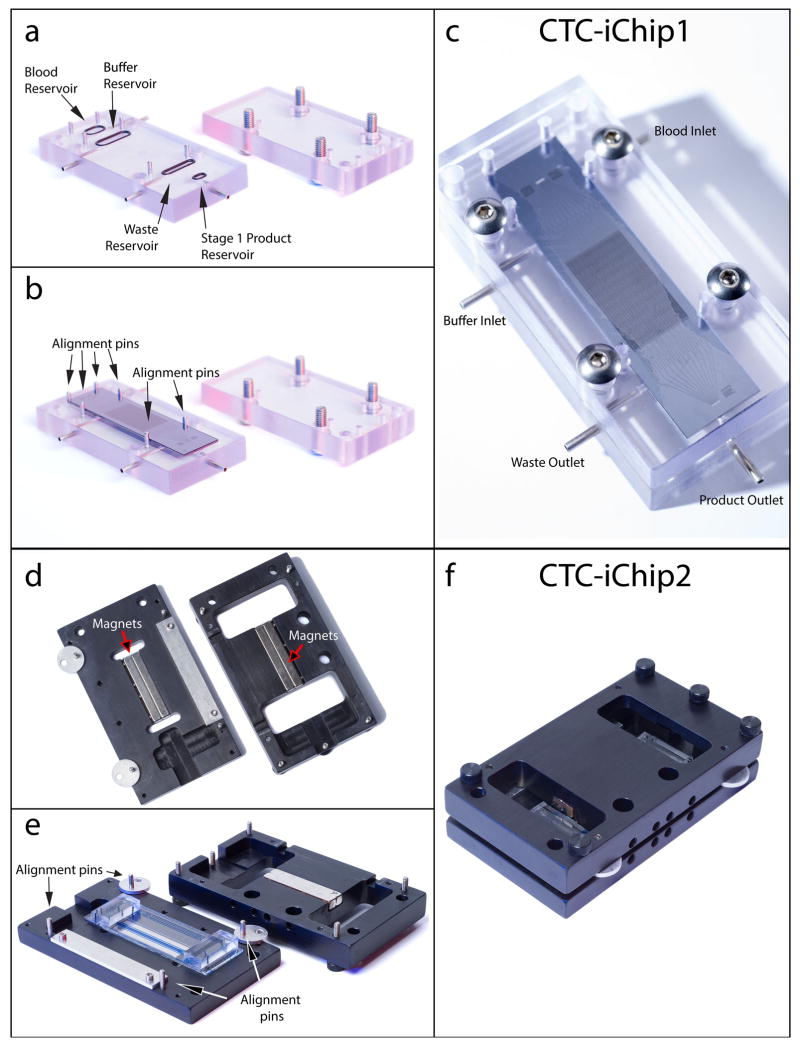
The CTC-iChip1 manifold (Fig. 7, top) is designed to hold the CTC-iChip1 chip during the experiment and provide a leak-free interface between the tubing and the microfluidic channels. The main body is composed of a top and a bottom piece, milled from polycarbonate, that can be screwed together. The bottom piece has (i) chambers for incoming and outgoing fluids, (ii) O-ring grooves around the chambers and (iii) fluidic channels that connect the chambers to (iv) short stainless steel tube fittings for connecting the silicone tubing. Also see Supplementary Figure 4 for CTC-iChip1 assembly.
Figure 7.
CTC-iChip assembly. (a) CTC-iChip1 manifold bottom piece with inlet and outlet O-rings attached (left) and top piece with screws in place (right). (b) CTC-iChip1 is placed on top of O-rings and fits between the metal guide posts. Fluid reservoirs, O-rings and CTC-iChip1 inlets are automatically aligned in this step. (c) The top piece is screwed into the bottom piece with the chip. (d) CTC-iChip2 manifold bottom piece (left) and top piece (right). CTC-iChip2 manifold houses the permanent magnets oriented to form a quadrupole (Fig. 5). (e,f) CTC-iChip2 is placed on the manifold bottom piece (e), is aligned (as detailed in Fig. 5 and Supplementary Video 2) and the top piece is pushed and screwed in f. When connected with tubing, these assembled devices form the CTC-iChip (Fig. 8).
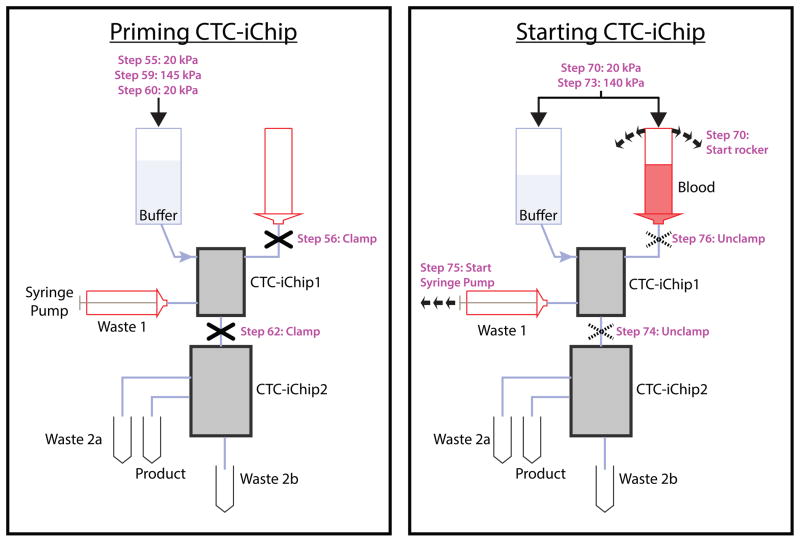
Custom-made CTC-iChip2 manifold
The CTC-iChip2 manifold is designed to hold the CTC-iChip2 and the four permanent magnets for magnetophoresis (Fig. 7, bottom). It is composed of a top and a bottom piece with permanent magnet grooves and screw holes. See Supplementary Video 3 for CTC-iChip2 assembly.
CTC-iChip system design for lossless operation
The current CTC-iChip technology consists of a multidevice platform, including manifolds (Fig. 7) and macroscale fluidic components in addition to microfluidic devices that are described above. The system and instrumentation components were designed to guarantee lossless interfaces, especially past the debulking device where the sample is highly enriched for target particles, and accurate process control to ensure that each device performs within pre-established operational ranges (e.g., flow rate, pressures, sample condition). In our experience, macro-micro interfacing and operational coupling between CTC-iChip1 and CTC-iChip2 are two key areas of system design that need particular focus.
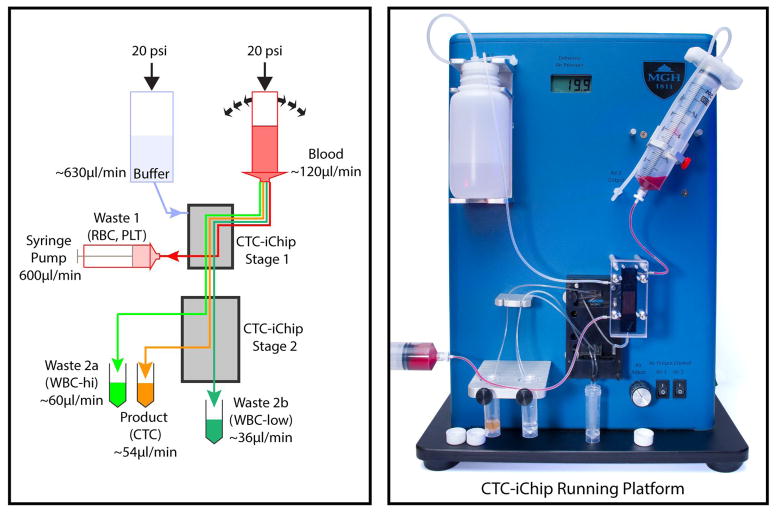
As the sample is processed through the CTC-iChip2 with the inertial focusing and magnetophoresis stages, the relative number (purity) of possible target cells (i.e., CTCs) to background particles (mostly WBCs) increases significantly. As such, the system must be carefully designed to avoid parasitic losses (e.g., cell settling in tubing and dead zones in manifolds). For the CTC-iChip1, the micro-macro interfacing exploits a manifold approach where the sample is driven from a blood hopper into a smaller blood reservoir within the manifold (Fig. 7a), and from there into the chip via through-holes on the chip. The manifold reservoir is small enough to ensure minimal sample settling, but it also possesses some additional volume to accommodate possible micro-bubbles and prevent them from entering the devices. Next to the blood reservoir, the CTC-iChip1 manifold also includes a buffer reservoir, a waste reservoir and a product reservoir (Fig. 7a). Whereas the design of the buffer and waste reservoirs is not crucial, the product reservoir must be designed to be of minimal volume, as at this stage the sample is already significantly enriched for target cells. In our platform, the volume for this product reservoir is 47 μl. The first and second devices are connected by a 10-cm, 0.5-mm diameter tubing that feeds from the first debulking device product reservoir and that is inserted directly into the CTC-iChip2 (Fig. 8).
Figure 8.
Running setup. Schematic (left) and picture (right) of CTC-iChip operation. A constant pressure of 140 kPa (20 p.s.i.) is applied to the buffer and blood, which are both connected to CTC-iChip1 inlets via tubing. Blood is rocked with a period of 5 s during the operation. CTC-iChip1 separates WBCs and CTCs from whole blood. Purified nucleated cell suspension exit from the bottom outlet and is transferred to CTC-iChip2. The remaining blood, which gets diluted with buffer, is collected in a 60-ml syringe at a rate of 600 μl/min controlled by using a syringe pump running in withdrawal mode. CTC-iChip2 separates CTCs from WBCs in two stages. The output containing highly bead-labeled WBCs exit from waste 2a. Sparsely bead-labeled cells exit from waste 2b collected at the bottom of the manifold. The product containing CTCs exit from the outermost two outlets and are collected together.
Operational coupling between CTC-iChip1 and CTC-iChip2
CTC-iChip1 and CTC-iChip2 are characterized by different operational parameters. For example, the inertial focusing stages in CTC-iChip2 require a flow rate approximately between 130 and 180 μl/min to guarantee proper particle focusing, whereas the DLD in CTC-iChip1 requires that the blood sample be mixed during the run. Used as stand-alone devices, both devices would intrinsically perform within their operational windows, without the need for any system control to guarantee their proper operation. Yet when the devices are coupled, system controls are required to ensure that adequate process windows are maintained for each device. The following four parameters uniquely describe the CTC-iChip operation: blood throughput; relative injection between blood and buffer in the CTC-iChip1; relative volume collection between the product and waste in the CTC-iChip1; and CTC-iChip2 flow rate. Only three of these four parameters are independent variables. In our platform, we selected blood throughput, blood injection ration and first-device waste collection ration to define the process parameters. These parameters were, respectively, dialed in through the pressure setting control, by hard-coded on-chip blood injection resistors (blood injection was set to 20%) and by externally controlling the first-device waste flow rate through a syringe pump (Fig. 8). In addition, mixing of the blood sample is achieved through gentle rocking of the blood hopper with a 5-s period (Fig. 8).
Limitations
The CTC-iChip is an immunoaffinity-based selection system. Thus, it is limited by the antibody used; if there are double-positive cells, such as CD45+/cytokeratin+, these are pushed to waste. Although we do not use size-based selection of CTCs from other nucleated cells, we use DLD to separate all nucleated cells from whole blood. Thus, owing to the critical hydrodynamic diameter during DLD process (4.0 μm) and the small size of inertial focusing channels, larger cells are filtered before inertial focusing and smaller cells are deflected to waste 1. The gap size of the DLD array is 32 μm, but we have experimentally observed pass-through of particles and aggregates up to 60 μm in size. This can be attributed to particle deformability, which enables these large cells to squeeze between the DLD array posts and reach the second stage of CTC-iChip. Although the system yields extraordinary purity compared with other systems, it is still limited for direct use with methods such as DNA sequencing, in which highly pure samples are required. Current setup and device fabrication require a clean room and manual labor, and expensive deep reactive ion etching procedures (that can be outsourced). For rigorous clinical studies, the CTC-iChip system needs to be manufactured in plastic within a single monolithic device.
Work is underway to minimize, if not eliminate, some of these limitations. We are currently working on designing a plastic monolithic device for cheaper production and easier use. This plastic device would markedly reduce the price and difficulties in use and production. Improvement of the product purity could be possible by altering the antibody cocktail with the addition of leukocyte-specific antigens. In addition, moving to a larger-band-pass DLD stage, where the lower cutoff is 2.8 μm, would allow efficient passage of possible smaller CTCs. As a result of these possible improvements, the CTC-iChip could be cheaper, more sensitive and yield purer product.
MATERIALS
REAGENTS
Dynabeads MyOne streptavidin T1 1-μm-diameter magnetic beads (Invitrogen, 656-02)
PBS, 1• (Mediatech, 21-040-CV)
Anti-human CD45 antibody, biotinylated (R&D Biosystems, clone 2d1, cat. no. BAF960)
Anti-human CD66b antibody (AbD Serotec, clone 80H3, cat. no. MCA216)
EZ-Link NHS-PEG-biotin kit (Thermo Scientific, cat. no. 21455)
Kolliphor P188 (Sigma-Aldrich, cat. no. K4894)
Vybrant DyeCycle Green stain (Life Technologies, cat. no. V35004)
DAPI (Life Technologies, cat. no. D1306)
CellTracker Green, Red and Orange (Life Technologies, cat. nos. C2925, C34552, C34551)
Fresh blood sample drawn into EDTA Vacutainer tube (BD, cat. no. 366643) and analyzed within 6 h of the draw
! CAUTION Treat all blood as potentially infectious. Wear protective gloves, a lab coat and a face shield while handling blood. Dispose of contaminated waste according to regulations. All experiments with human blood should follow guidelines and regulations.
! CAUTION Informed consent must be obtained for blood collection from human subjects.
EQUIPMENT
Silicon wafer, 4 inches (Silicon Sense)
Blue tape (Ultron Systems, cat. no. 1007R-4.0)
Ultraclean glass slides (Thermo Scientific, cat. no. C22-5128-M20)
PDMS - Sylgard 184 silicone elastomer kit (Dow Corning, Ellsworth Adhesives, cat. no. 184 Sil Elast kit, 0.5 kg)
Plasma etcher
Hot plate, 85 °C
Oven
Harris Uni-Core 1.2-mm hole puncher (Ted Pella, cat. no. 15074)
O-ring, 3 mm (McMaster-Carr, cat. no. 9262K441)
O-ring, 6 mm (McMaster-Carr, cat. no. 9262K103)
O-ring, 9 mm (McMaster-Carr, cat. no. 9262K443)
O-ring, 10 mm (McMaster-Carr, cat. no. 9262K104)
Pipe-to-tube adapter, • 4 (McMaster-Carr, cat. no. 5047K11)
Female Lluer fitting (Cole Parmer, cat. no. EW06539-25)
Stainless steel button-head screws, • 4, 1 inch (McMaster-Carr, 92949A116)
Screws with caps, • 4, 7/8 inch (McMaster-Carr)
Pieces (2-cm-, 13.5-cm-, 28-cm- and 31-cm-long) of silicone tubing, inner diameter (i.d.): 0.031 inch, outer diameter (o.d.): 0.094 inch, wall: 0.0315 inch (Cole Parmer 06411-60)
Pieces (10-cm- and 20-cm-long) of Tygon Microbore tubing, i.d.: 0.02 inch, o.d.: 0.06 inch (Cole-Parmer, EW-06418-02)
Swing-rotor centrifuge
Custom-made CTC-iChip1 manifold (Fig. 7a)
Custom-made CTC-iChip2 manifold (Fig. 7d)
Custom-made Spintrap (Supplementary Fig. 1)
Conical tubes, 15 ml (Corning, cat. no. 430766)
Conical tubes, 50 ml (Corning, cat. no. 430897)
Syringes, 60 ml (BD Falcon, 309653)
Nalgene bottle, 250 ml (Fisher, cat. no. 09-740-25E)
Nalgene low-particulate bottle, 250 ml (Fisher, cat. no. 03-313-87B)
Stir bar (Fisher, cat. no. 14-511-60B)
Size 122 O-ring (McMaster, cat. no. 9452K86)
Nalgene filter system, 250 ml (Fisher, cat. no. 09-740-2A)
Serological pipette tips, 10 ml and 25 ml (Fisher, cat. no. 13-675-31)
Timer (Fisher, cat. no. S407992)
Removable sample rack (Invitrogen, cat. no. 123-22D)
HulaMixer sample mixer (Invitrogen, cat. no. 15920D)
Sysmex blood analyzer (Sysmex, cat. no. KX-21N)
Bolt driver, 3/32 inch (Bondhus, cat. no. 10632)
Syringe pump (New Era pump system, NE-1000)
Surgical clamp
Binder clips
Vortex mixer (Scientific Industries, cat. no. SI-0236)
Microscope slide (Fisher Scientific, cat. no. 12-550B)
Cover glass (Fisher Scientific, cat. no. 12-545-100)
Vacuum degassing system (Fisher, cat. no. 13-880-30)
Vacuum chamber (Nalgene, cat. no. 5300-0507)
Custom-made equipment
Snorkel buffer cap (Supplementary Fig. 5)
Syringe cap (Supplementary Fig. 5)
CTC-iChip1 manifold (Fig. 7a)
CTC-iChip2 manifold (Fig. 7d)
CTC-iChip running platform (Fig. 8)
REAGENT SETUP
Poloxamer running buffer
We use Kolliphor or Pluronic nonionic copolymer-based buffers for their effect in reducing nonspecific cell attachment to walls (of channels, tubing, etc.)69. Weigh out 2.5 g of Kolliphor-P188 and place it in an empty Nalgene bottle (250 ml). Transfer 250 ml of 1 • PBS into the Nalgene bottle. Place a stir bar in the Nalgene bottle, close the cap and place the bottle on a magnetic stirrer for 20 min. Filter the solution through a Nalgene filtration system with a 250-ml receiver. Degas with an ultrasonic bath or a vacuum system. The Poloxamer running buffer can be stored at room temperature (20–24 °C) for up to 1 week.
Biotinylated anti-CD66b
Use the EZ-Link NHS-PEG-biotin kit to functionalize anti-CD66b with biotin according to the kit’s instructions with a 30:1 biotin:antibody molar ratio. Biotinylated antibody can be stored at 4 °C for up to 2 weeks.
EQUIPMENT SETUP
Custom-made CTC-iChip running platform
The running platform is custom-made and contains pumps, a syringe rocker and posts to hold chips (Fig. 8). It is designed to hold the buffer container, blood sample syringe and CTC-iChip manifolds; to apply specified pressures to the buffer and blood containers; and to position all components at the same heights and distances for each CTC-iChip operation to ensure consistency in air pressure.
Snorkel buffer cap
A snorkel buffer cap is needed to interface between the air-pressure system and the buffer container. This cap must be airtight; it should have an inlet for air pressure and an outlet that is directly connected to the buffer (by dipping). We use a plastic body with an O-ring to seal the buffer container, an inlet for receiving air pressure via plastic tubing and a stainless steel tube that is dipped into the buffer for transferring buffer to a plastic tube, and we have this custom-made (Supplementary Fig. 5).
Syringe cap
A syringe cap is needed to interface between the air-pressure system and the blood sample in the syringe. This cap must be airtight, and it should have an inlet for air pressure. We use a plastic body with an O-ring to seal the syringe and an inlet for receiving air pressure via plastic tubing, and we have this custom-made (Supplementary Fig. 5). Place an O-ring to the syringe cap’s groove, remove the plunger of a 60-ml syringe and push the syringe cap to the bottom of the syringe to seal the cap.
PROCEDURE
Design and mold preparation of CTC-iChip2 TIMING 2–5 d
-
1|
Draw the microfluidic device design in AutoCAD software (the specific design used for CTC-iChip2 is available as Supplementary Data).
-
2|
Submit the design to a vendor for mask printing (we work with Advance Reproductions Corporation, Andover, Massachusetts). We usually receive the mask in 2–3 business days.
-
3|
Use negative photoresist (SU-8 50) and standard photolithography methods68 (see also http://www.biomemsrc.org/resources/protocols-procedures) to prepare a silicon wafer to serve as the master mold for the CTC-iChip2. We find that for our flow rates and typical cell sizes, 50 μm ± 2 μm is an optimal channel height. Place the silicon wafer into a Petri dish and secure the edges with vinyl clean-room tape.
PAUSE POINT This wafer can be reused, and it can be kept in the clean room for extended periods.
CTC-iChip2 production TIMING 2–3 h
CRITICAL Steps 4–23 must be performed in a clean room.
-
4|
Mix PDMS base with PDMS curing agent homogeneously at a weight ratio of 10:1. With the silicon wafer secured in the center of a 15-cm-diameter Petri dish, pour 70 g of the PDMS mixture into the dish. This is termed the ‘thick pour.’
-
5|
Degas the PDMS in a vacuum chamber for a minimum of 45 min. When the PDMS is bubble-free, bake the dish at 70–75 °C in a convection oven for at least 2 h to cure the PDMS.
-
6|
Cut and peel away the center of the thick pour so that the devices are exposed but the edges of the wafer are still secured by the thick PDMS layer. This creates a thick PDMS wall that prevents subsequent deposition of fresh PDMS from seeping underneath the wafer, preventing the master from lying level in the Petri dish.
-
7|
Pour 4.2 g of mixed PDMS into the now-vacated center area of the silicon wafer. Ensure that the PDMS is evenly distributed across the surface of the wafer.
CRITICAL STEP The thickness of the PDMS should not exceed 1 mm in height from the surface of the silicon wafer.
-
8|
In an empty Petri dish, pour 70 g of mixed PDMS. This thick PDMS will be used as the inlet/outlet part of the device.
-
9|
Repeat Step 5 for both Petri dishes to cure the PDMS.
CRITICAL STEP Ensure that the wafer is left on a level surface for the entire duration of the curing process. The variation in thickness may cause problems later when interfacing the device to the magnetic manifold.
PAUSE POINT It is possible to keep cured uncut PDMS devices in a clean room for extended periods.
-
10|
With the Petri dish containing the blank PDMS layer, peel and cut the PDMS into 10 • 25 mm blocks. These will serve as the inlet/outlet interfaces.
-
11|
With the Petri dish containing the device features, cut out the thin layer of PDMS. As with Step 6, ensure that your cut diameter is just slightly less than the diameter of the silicon wafer. Peel this layer off with flat-head tweezers.
-
12|
Cut and trim the devices from the thin PDMS layer; ensure that all uneven edges have been removed (Fig. 6a).
CRITICAL STEP Uneven thickness of the device may prevent the device from fitting into the magnetic manifold if the device thickness exceeds 1.8 mm
-
13|
For each device, clean the flat side of the PDMS (without features) and two 10 • 25 mm blocks by using clean-room adhesive tape by leaving them on the tape for no longer than 10 s. Press gently on the PDMS to ensure that the entire area is in contact with the blue tape. Repeat this step twice.
CRITICAL STEP Leaving PDMS on tape for extended periods inhibits permanent bonding to PDMS and glass.
-
14|
Plasma-etch all the devices and the blank PDMS with surfaces to be bonded facing up. We use the plasma-etching parameters described in the table below.
Parameter Value Etcher model March PX-250 Chamber dimensions 9.0 × 9.0 × 12.0 inches RF frequency 13.56 MHz RF power 50 W DC bias 0 W Operating pressure 70 mTorr End point 100% Time 35 s BP/RP 70 mTorr Oxygen flow rate 2 s.c.c.m. Ambient temperature 22 °C Ambient humidity 25–45% -
15|
Bond the blank PDMS to the device inlet and outlet regions (Fig. 6b). If there is an air bubble, press on the area to remove it.
CRITICAL STEP Misaligning in this step may cause the device to leak or not to fit into the CTC-iChip2 manifold properly, which in turn causes misalignment with the magnetic field. If there is any delamination across the device features, check that every part of the bonding surface is even. In addition, this may be a result of leaving the PDMS on the tape for too long, or it may be due to extremely low humidity (<25%).
-
16|
Place PDMS on an 85 °C hot plate for 10 min, with the device features facing up. Allow cooling for 3–4 min.
-
17|
Use a 1.2-mm hole puncher to punch holes for the inlet and outlet (Fig. 6c).
-
18|
Clean the side with device features by leaving them on the blue tape for no longer than 10 s. Press gently along the entire channel to ensure that the entire area is in contact with the blue tape. Repeat this step twice.
-
19|
Plasma-etch all the devices with the ultraclean glass slides. The settings should be the same as in Step 14.
-
20|
Bond the PDMS to the glass slides (Fig. 6d). Gently press on the features to make sure that every part is well bonded.
CRITICAL STEP Misaligning in this step may cause the device to leak or not to fit into CTC-iChip2 manifold properly, which in turn causes misalignment with the magnetic field. If there is any delamination across the device features, check that every part of the bonding surface is even. In addition, this may be a result of leaving the PDMS on the tape for too long, or it may be due to extremely low humidity (<25%).
-
21|
Bake devices on an 85 °C hot plate for 10 min, with the glass side facing the hot plate.
-
22|
Allow the devices to cool on an insulating surface for 3–4 min.
-
23|
Inspect the devices under the microscope for delamination, and check the channels for blockage.
PAUSE POINT Store CTC-iChip2 devices in Petri dishes at room temperature until use for up to 1 year.
CTC-iChip1 assembly TIMING 10 min
CRITICAL Steps 24–36 and Steps 37–44 can be performed in parallel if desired.
-
24|
Position the 6-mm O-ring in the top oval groove around the blood reservoir on the manifold (Fig. 7a and Supplementary Fig. 4a).© 2014 Nature America, Inc. All rights reserved. protocol 704 | VOL.9 NO.3 | 2014 | nature protocols
-
25|
Position the 10-mm O-ring in the second oval groove from the top of the manifold (around the buffer reservoir).
-
26|
Position the 9-mm O-ring in the third oval groove from the top of the manifold (around the waste reservoir).
-
27|
Position the 3-mm O-ring on the bottom oval groove on the manifold (around the product reservoir).
-
28|
Connect one female Luer to the stage 1 waste outlet tubing (Supplementary Fig. 4b).
-
29|
Connect one female Luer to the blood-to–stage 1 inlet tubing.
-
30|
Connect the 2-cm silicone tubing to the stage 1–to–stage 2 inlet tubing.
-
31|
Place the microfluidic chip into the manifold, between the alignment pins (Fig. 7b and Supplementary Fig. 4b).
-
32|
Place the manifold cover carefully over the manifold base without displacing the microfluidic chip.
-
33|
Place the four screws in the holes and use a bolt driver to tighten them gently in a criss-cross method (Fig. 7c and Supplementary Fig. 4c).
CRITICAL STEP Check that the O-rings are still in place. If they are not properly seated, unscrew the manifold and reposition the O-rings. Repeat Steps 24–32.
-
34|
Connect the 31-cm buffer-to–stage 1 inlet tubing into the top left steel barb on the manifold (Supplementary Fig. 4d).
-
35|
Connect the 28-cm stage 1 waste outlet tubing into the bottom left steel barb on the manifold.
-
36|
Connect the open end of the 13.5-cm blood-to–stage 1 inlet tubing into the top right steel tubing on the manifold. Place the assembled stage 1 device to posts on the running platform and connect it to the buffer cap, blood syringe cap and waste syringe.
CTC-iChip2 assembly TIMING 10 min
-
37|
Prepare five lengths of tubing: two 5.5-cm pieces of Tygon (i.d. 0.5 mm, o.d. 1.5 mm; for waste 2b) and three 22-cm pieces of Tygon (i.d. 0.5 mm, o.d. 1.5 mm; for waste 2a, product). Also prepare one length of 7.5-cm Tygon tubing (i.d. 0.5 mm) attached to a 2-cm piece of silicone tubing (i.d. 0.031 inch, o.d. 0.094 inch).
-
38|
Install the tubing into the inlets and outlets.
-
39|
Carefully place the microfluidic chip to the bottom piece of the manifold (see Supplementary Video 4 for Steps 39–43).
-
40|
Align the microfluidic chip to the manifold (Fig. 5a and Supplementary Video 4). In the alignment process, the main aim is to align the center of the first deflection channel to the line where two magnets meet (and where the magnetic field is 0) (Fig. 5b). To aid this, the device has a ruler that marks where the magnets should be positioned (Fig. 5c,d). First, ensure that the edges of these magnets sit between numbers 0 and 40 on the ruler.
-
41|
For finer alignment, place the bottom piece with the chip onto a microscope; use ~4• magnification. Align the first-stage deflection channel according to the line where the two magnets meet. The center of the channel should fall on this line. After this fine alignment, tape the chip to the bottom piece. The top piece can now be screwed in.
-
42|
Screw in the top piece. Be aware that the top piece will repel the bottom piece because of the same magnetic orientation. Be sure not to move the chip to perturb the alignment.
-
43|
Carefully tighten the CTC-iChip2 manifold.
-
44|
Make sure that the alignment of the microfluidic chip with the magnets is preserved. If the chip has shifted, correct the alignment.
CTC-iChip priming TIMING 30–45 min
CRITICAL Steps 45–52 and Steps 63–68 can be performed in parallel if desired.
-
45|
Check the syringe pump settings: syringe diameter = 26.7 mm, rate = 600 μl/min, mode = withdraw.
-
46|
Push the plunger of one of the 60-ml syringes all the way into the empty air space, and then lock it into the syringe pump. This collects the waste 1.
-
47|
Remove the plunger from the other 60-ml syringe and lock it on the rocker. This syringe holds the blood sample.
-
48|
Add ~100 ml of Pluronic running buffer to the buffer bottle. Cap the buffer container with the snorkel buffer cap.
-
49|
Connect the air inlets to the snorkel buffer cap and syringe cap.
-
50|
Weigh all product and waste containers, and be sure to record these values.
-
51|
Place the CTC-iChip1 device on the post (Fig. 8).
-
52|
Connect the buffer-to-CTC-iChip1 inlet tubing to the buffer.
-
53|
Connect the female Luer of the blood-to–CTC-iChip1 inlet tubing to the 60-ml blood sample syringe on the rocker (Fig. 8). Please note that the air supply is not connected to the blood syringe.
-
54|
Connect the CTC-iChip1–to–CTC-iChip2 tubing of the assembled CTC-iChip2 device to the CTC-iChip1 product outlet. Carefully place the CTC-iChip2 device on the posts. This is the first step of the clamping sequence, schematized in Figure 9.
CRITICAL STEP The order of the clamping sequence that starts at this step is crucial to priming the system properly.
-
55|
Set the air pressure to 20 kPa (3.0 p.s.i.). Turn on air pressure #1 to pressurize the buffer.
! CAUTIONWear a face shield and a lab coat while handling pressurized systems.
-
56|
Watch as the buffer fills the Blood-to–CTC-iChip1 inlet tubing, and clamp it once the buffer has formed a droplet in the bottom of the 60-ml syringe. Check for bubbles at the back of the CTC-iChip1 manifold and try to remove the bubbles by tilting the manifold.
? TROUBLESHOOTING
-
57|
Watch the waste 1 outlet fill with buffer. Once it has filled, connect the female Luer of the waste 1 outlet tubing to the 60-ml waste 1 syringe in the syringe pump.
-
58|
To prime CTC-iChip2, increase the pressure gradually from 20 kPa to 140 kPa (3.0 p.s.i. to 20.0 p.s.i.). Watch carefully as the buffer fills the CTC-iChip2 channels. Check for bubbles; if bubbles are seen, try to push them out by gently pressing on the PDMS and increasing the pressure.
CRITICAL STEP Pressing too forcefully on the PDMS may move the CTC-iChip2 device out of alignment with the magnets. If CTC-iChip2 becomes misaligned, carefully realign the device before proceeding.
CRITICAL STEP If bubbles cannot be extracted, a new CTC-iChip2 device should be used.
? TROUBLESHOOTING
-
59|
Once all CTC-iChip2 output tubings are filled with buffer, proceed with pressure testing: raise the pressure to 145 kPa (21.0 p.s.i.). Check for any leaks or faulty tubing connections.
CRITICAL STEP Identification of leaks at this step is critical, as any leaks will affect flow rates and may cause a dirty run or loss of sample.
-
60|
Reduce the pressure back to 20 kPa (3.0 p.s.i.).
-
61|
Run the buffer through the system for >15 min.
Ĩ PAUSEPOINT The CTC-iChip system can be left at this position for up to 1 d before proceeding to step 62; however, sufficient buffer must be provided to ensure the buffer does not run out before proceeding to the next step.
-
62|
Clamp CTC-iChip1 to the CTC-iChip2 tubing. The CTC-iChip is now ready to use.
Figure 9.
Sequence of steps that are crucial to priming and starting the CTC-iChip. Schematic showing critical clamping sequence during priming (left) and starting (right) the CTC-iChip.
Blood preparation TIMING 40 min
-
63|
Take 200 μl of blood sample into an Eppendorf tube and take a complete blood count (CBC), which quantifies the numbers of WBCs and other cells in whole blood (the WBC reading is accurate to ± 0.2 • 106 cells/ml).
! CAUTION Treat all blood as potentially infectious. Wear protective gloves, a lab coat and a face shield while handling blood.
-
64|
Calculate the amount of antibody and magnetic beads required on the basis of the donor’s WBC count (reported in the CBC in Step 19) and the volume of blood to be run. For CD45 antibody, use 50 fg/WBC. For CD66b antibody, use 37.5 fg/WBC. Use 125 beads/WBC. Dilute the antibodies to 0.1 mg/ml and then pipette an appropriate aliquot out on the basis of the WBC count.
CRITICAL STEP The accuracy of this step substantially affects the purity of the product.
-
65|
Take the desired blood volume in a PBS-rinsed Vacutainer tube. The optimal volume range is 1–20 ml.
-
66|
Add the calculated volumes of CD45 and CD66b antibodies to the whole blood. Pipette up and down >10 times with a 1-ml pipette to fully mix the antibody and blood. Incubate the mixture for 20 min at room temperature (20–24 °C) on the rocker.
-
67|
Pipette the full volume of Dynabeads to thoroughly resuspend the beads in solution.
CRITICAL STEP Make sure that the Dynabeads are not clumped at this step. Check by pipetting a small amount on a glass slide and inspecting the beads under the microscope at 10• magnification. If clumps are observed, gently vortex for 1 min.
-
68|
Add the calculated amount of Dynabeads. To mix, pipette the mixture up and down 10 times and incubate the tube for 20 min on the rocker.
Running the CTC-iChip TIMING 30–90 min
-
69|
Use a serological pipette to transfer the blood sample to the blood syringe.
-
70|
Cap the blood sample syringe. Switch on air pressure #2. Turn on the blood sample rocker.
-
71|
Place the waste 2 tubing inside a 15-ml conical tube.
-
72|
Place the product tubing inside a 5-ml tube at the CTC-iChip2 manifold to collect the product.
-
73|
Raise the pressures to 140 kPa (20 p.s.i.).
CRITICALSTEP Upon completion of this step, the CTC-iChip is ready for operation. Visually verify all connections (the running setup is shown in Fig. 8). The subsequent five steps are time-sensitive, and they must be performed promptly.
-
74|
Unclamp the CTC-iChip1–to–CTC-iChip2 inlet tubing.
-
75|
Wait for 2 s, and then start the syringe pump.
-
76|
Wait for 2 s, and then unclamp the blood-to–CTC-iChip1 inlet tubing.
-
77|
Start the timer to record the length of the experiment.
-
78|
Watch the blood run into the CTC-iChip1.
? TROUBLESHOOTING
-
79|
Perform visual checks to confirm the expected running conditions: Ensure that the blood lanes in the CTC-iChip1 device are straight, that waste 1 is pulling RBCs (tubing and syringe are red), that the product is clear, and that waste 2a is brown.
? TROUBLESHOOTING
Stopping the CTC-iChip run TIMING 15 min
-
80|
Turn off the rocker when the remaining blood is just enough to cover the tip of the syringe. CRITICALSTEP It is important that no air enters the blood-to–CTC-iChip1 tubing.
-
81|
As soon as the end of the blood reaches the entrance of the CTC-iChip1, clamp the blood-to–CTC-iChip1 inlet tubing.
? TROUBLESHOOTING
-
82|
Wait 5 min for the buffer to wash the remaining cells from the system.
-
83|
Stop the timer. Clamp both inlet tubings. Stop the syringe pump. Record the run time of the experiment.
-
84|
Decrease the pressure back to 7 kPa (1.0 p.s.i.).
-
85|
Turn off the air pressures.
-
86|
Slowly remove the syringe from the waste 1 outlet tubing while gently pulling the syringe.
-
87|
Weigh all product and waste containers and record these values for evaluation.
Immobilizing the CTC-iChip product using Spintrap • TIMING 15 min
-
88|
Install a poly-lysine–coated glass slide to Spintrap bottom piece (Supplementary Fig. 1a,b). Place the top piece (with the O-ring in place) on top. Screw the nuts to seal the chamber.
-
89|
Pipette the cell suspension through the Spintrap inlet. If the suspension is less than the volume of the chamber, you can fill the chamber completely by adding Pluronic buffer.
CRITICAL STEP While doing this step, avoid generating bubbles. If bubbles travel to the glass surface, they can damage or remove the cells.
-
90|
Place the Spintrap into a swing-bucket centrifuge with a 96-well-plate adapter installed. Balance the centrifuge.
-
91|
Centrifuge the Spintrap for 5 min at 50g at room temperature. After the centrifugation, incubate the Spintrap for 10 min to complete cell attachment.
-
92|
Remove the Spintrap from the centrifuge.
PAUSE POINT The slide containing CTCs is ready for cell fixation and for histological and immunological staining.
TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 12 | Air bubble trapped in CTC-iChip1 lane | Bubbles in buffer | Clamp CTC-iChip1-to-CTC-iChip2 tubing, increase pressure to 140 kPa (20 psi) and tap on CTC-iChip1 to flush out bubbles via waste outlet |
| 14 | Air bubble trapped in CTC-iChip2 channel | Bubbles in buffer | Unclamp CTC-iChip1-to-CTC-iChip2 tubing, press gently to guide the bubbles out from the outlets |
| 34 | Blood level rises or blood not flowing through the system | Pressure problem | Make sure air pressure # 2 is on |
| 35 | RBC carryover to CTC-iChip2 | Pressure or syringe pump problem | Check pressure and syringe pump settings, confirm that there is no leak |
| 35 | Majority of CTC-iChip1 lanes blocked | Blood has large aggregates | Terminate the run |
| 37 | Bubbles get into CTC-iChip1 | Blood sample is finished, blood inlet not clamped on time | Stop the run |
TIMING
Steps 1–3, design and mold preparation of CTC-iChip2: 2–5 d
Steps 4–23, CTC-iChip2 production (in a clean room): 2–3 h
Steps 24–36, CTC-iChip1 assembly: 10 min
Steps 37–44, CTC-iChip2 assembly: 10 min (can be performed in parallel with Steps 24–36)
Steps 45–62, CTC-iChip priming: 30–45 min
Steps 63–68, blood preparation: 40 min
Steps 69–79, running the CTC-iChip: 30–90 min (8 ml of blood/h)
Steps 80–87, stopping the CTC-iChip run: 15 min
Steps 88–92, immobilizing the CTC-iChip product using Spintrap: 15 min
ANTICIPATED RESULTS
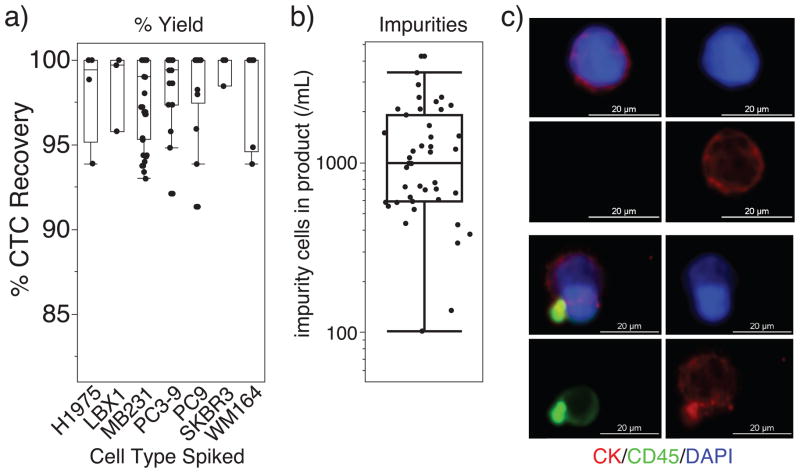
The described protocol allows for CTC isolation from blood. We characterized the system using healthy donor blood spiked with known number of cancer cells grown in vitro. We used WM164, MB231, PC9, PC3-9, SKBR3 and LBX1 (ref. 70) cell lines to represent the different types of tumor cells that occur in patients. Percent yield of cancer cells was determined by (i) staining cell lines with CellTracker (Invitrogen), (ii) counting cells to be added with a hemocytometer and an epifluorescence microscope, (iii) spiking a healthy donor blood sample (~1,000 cancer cells/ml blood), (iv) performing the CTC-iChip protocol and (v) counting the product with a Nageotte chamber, which is designed to count cells in dilute suspensions. The average yield of our experimental data set is 97 ± 2.7% (±s.d.), showing almost complete target cell recovery, regardless of the cell type (Fig. 10a). We analyzed the number of cellular impurities in the CTC-iChip product from healthy or patient donor experiments. We used DyeCycle Green live-cell DNA stain to count all nucleated cells in the CTC-iChip product and subtracted observed tumor cells. Work is underway to identify and further deplete this population of cells, which are observed to be not labeled with any magnetic beads.
Figure 10.
Expected results. (a) Characterization of the CTC-iChip protocol shows high yield of spiked cancer cells isolated from whole blood that is independent of spiked cell type with low number of nucleated cell impurities. Mean yield: 97 ± 2.7% (± s.d.). (b) Logarithmic plot of the number of nucleated cell impurities after application of the CTC-iChip to healthy donor samples and clinical samples from patients of breast and pancreatic cancers. Mean carryover: 1,200 ± 900 (± s.d.) nucleated cells/ml of blood. Box plots were generated using JMP (SAS Institute) with the following settings: boxes represent first and third quartiles, lines represent upper and lower data points and exclude outliers. (c) Fluorescence micrographs of CTCs and impurities from the CTC-iChip product (20× objective, Nikon). Rare cell suspension is immobilized using Spintrap (Supplementary Fig. 1), fixed using 4% (wt/vol) paraformaldehyde and stained for immunofluorescence microscopy using antibodies against cytokeratins (CK; red, CTC marker), CD45 (green, leukocyte marker) and DAPI (nuclear stain, blue). Bright green dots indicate anti-CD45–coated magnetic beads.
Once the clinical blood sample is sorted, a variety of pathological, immunological or molecular analyses are possible (see the INTRODUCTION). To enumerate CTCs from clinical blood samples, we typically perform immunofluorescence microscopy by using antibodies against CTC markers (i.e., cytokeratins) in one color, WBC markers (i.e., CD45) in another and DAPI as the nuclear marker (Fig. 10c, experimental details of immunofluorescence imaging reported in ref. 58). Automated immunofluorescence microscopy yields the number of CTCs in the enriched clinical sample, which can be converted to the number of CTCs/ml of blood analyzed. When this procedure is repeated for a patient, the number of CTCs/ml of blood can be tracked6.
Single-cell RNA-profiling methods can be applied to isolated CTCs for exploring gene expression6. We applied single-cell micromanipulation approaches to interrogate individual CTCs isolated from patients with prostate cancer by using the CTC-iChip58. Application of this technology to limited number of cells showed a high degree of heterogeneity in RNA expression of CTCs from a single patient.
Once chips and manifolds are produced, the CTC-iChip is a system is relatively easy to learn and apply. All of the results reported in Figure 10 were from CTC-iChip operated by laboratory technicians, most of who lacked prior experience in microfluidics. After two or three iterations of the protocol, they achieved consistently successful results. Rare system failures occur, usually because of production errors or sample problems. There are rare blood samples that cause blockage of CTC-iChip1; this causes RBC carryover to the product, and it can result in discarding the product if the number of RBC impurities is too high.
Supplementary Material
Acknowledgments
We express our gratitude to all patients and healthy volunteers who participated in this study and contributed blood samples. We thank L. Libby, O. Hurtado and A.J. Aranyosi for coordination of the research laboratories; D.M. Lewis and B. Hamza for expertise in device fabrication; and L. Nieman and J. Walsh for expertise in microscopy. This work was supported by the US National Institutes of Health (NIH) P41 Resource Center (M.T.); a NIH National Institute of Biomedical Imaging and Bioengineering Quantum Grant (M.T. and D.A.H.); Stand Up to Cancer (D.A.H., M.T. and S.M.); the Howard Hughes Medical Institute (D.A.H.); the Prostate Cancer Foundation and the Charles Evans Foundation (D.A.H. and M.T.); and Johnson and Johnson (M.T. and S.M.).
Footnotes
AUTHOR CONTRIBUTIONS: All authors contributed to design of the CTC-iChip. N.M.K., P.S.S., F.F., V.P., E.O., N.K., K.S. and M.T. prepared the manuscript. E.J.L., J.M.M., P.C., J.Y., H.H., B.M., J.T., T.A.B., S.L.S., S.M., R.K. and D.A.H. commented on the manuscript.
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
References
- 1.Lustberg M, Jatana KR, Zborowski M, Chalmers JJ. Emerging technologies for CTC detection based on depletion of normal cells. Recent Results Cancer Res. 2012;195:97–110. doi: 10.1007/978-3-642-28160-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Med. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 3.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stott SL, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu M, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Science Transl Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baccelli I, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 10.Ramsköld D, et al. Full-length mRNA-seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heitzer E, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 12.Sakaizawa K, et al. Mutation analysis of BRAF and KIT in circulating melanoma cells at the single-cell level. Br J Cancer. 2012;106:939–946. doi: 10.1038/bjc.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balasubramanian P, et al. Multiparameter analysis, including EMT markers, on negatively enriched blood samples from patients with squamous cell carcinoma of the head and neck. PLoS ONE. 2012;7:e42048. doi: 10.1371/journal.pone.0042048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payne RE, et al. Viable circulating tumour cell detection using multiplex RNA in situ hybridisation predicts progression-free survival in metastatic breast cancer patients. Br J Cancer. 2012;106:1790–1797. doi: 10.1038/bjc.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell Res. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 16.Issadore D, et al. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci Transl Med. 2012;4:141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krivacic RT, et al. A rare-cell detector for cancer. Proc Natl Acad Sci USA. 2004;101:10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh HB, et al. High-speed detection of circulating tumor cells. Biosens Bioelectron. 2006;21:1893–1899. doi: 10.1016/j.bios.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 19.Zhao M, et al. An automated high-throughput counting method for screening circulating tumor cells in peripheral blood. Anal Chem. 2013;85:2465–2471. doi: 10.1021/ac400193b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghossein RA, et al. Detection of circulating tumor cells in patients with localized and metastatic prostatic carcinoma: clinical implications. J Clin Oncol. 1995;13:1195–1200. doi: 10.1200/JCO.1995.13.5.1195. [DOI] [PubMed] [Google Scholar]
- 21.Simpson SJ, et al. Detection of tumor cells in the bone marrow, peripheral blood, and apheresis products of breast cancer patients using flow cytometry. Exp Hematol. 1995;23:1062–1068. [PubMed] [Google Scholar]
- 22.Witzig TE, et al. Detection of circulating cytokeratin-positive cells in the blood of breast cancer patients using immunomagnetic enrichment and digital microscopy. Clin Cancer Res. 2002;8:1085–1091. [PubMed] [Google Scholar]
- 23.Deng G, et al. Enrichment with anti-cytokeratin alone or combined with anti-EpCAM antibodies significantly increases the sensitivity for circulating tumor cell detection in metastatic breast cancer patients. Breast Cancer Res. 2008;10:R69. doi: 10.1186/bcr2131. © 2014 Nature America, Inc. All rights reserved. protocol 710 | VOL.9 NO.3 | 2014 | nature protocols. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talasaz AH, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci USA. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riethdorf S, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 26.Lara O, Tong X, Zborowski M, Chalmers JJ. Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Exp Hematol. 2004;32:891–904. doi: 10.1016/j.exphem.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Zheng S, et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162:154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 28.Vona G, et al. Isolation by size of epithelial tumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seal SH. A sieve for the isolation of cancer cells and other large cells from the blood. Cancer. 1964;17:637–642. doi: 10.1002/1097-0142(196405)17:5<637::aid-cncr2820170512>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 30.Hou HW, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohamed H, Murray M, Turner JN, Caggana M. Isolation of tumor cells using size and deformation. J Chromatogr A. 2009;1216:8289–8295. doi: 10.1016/j.chroma.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 32.Hur SC, Henderson-MacLennan NK, McCabe ERB, Di Carlo D. Deformability-based cell classification and enrichment using inertial microfluidics. Lab Chip. 2011;11:912–920. doi: 10.1039/c0lc00595a. [DOI] [PubMed] [Google Scholar]
- 33.Gertler R, et al. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res. 2003;162:149–155. doi: 10.1007/978-3-642-59349-9_13. [DOI] [PubMed] [Google Scholar]
- 34.Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, Warnaar SO. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J Cell Biol. 1994;125:437–446. doi: 10.1083/jcb.125.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herlyn M, Steplewski Z, Herlyn D, Koprowski H. Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies. Proc Natl Acad Sci USA. 1979;76:1438–1442. doi: 10.1073/pnas.76.3.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allard WJ. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 37.de Bono JS, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 38.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyamoto DT, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 42.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Münz M, et al. Side-by-side analysis of five clinically tested anti-EpCAM monoclonal antibodies. Cancer Cell Int. 2010;10:44. doi: 10.1186/1475-2867-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pecot CV, et al. A novel platform for detection of CK+ and CK– CTCs. Cancer Discov. 2011;1:580–586. doi: 10.1158/2159-8290.CD-11-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iinuma H, et al. Detection of tumor cells in blood using CD45 magnetic cell separation followed by nested mutant allele-specific amplification of p53 and K-ras genes in patients with colorectal cancer. Int J Cancer. 2000;89:337–344. doi: 10.1002/1097-0215(20000720)89:4<337::aid-ijc4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 46.Bilkenroth U, et al. Detection and enrichment of disseminated renal carcinoma cells from peripheral blood by immunomagnetic cell separation. Int J Cancer. 2001;92:577–582. doi: 10.1002/ijc.1217. [DOI] [PubMed] [Google Scholar]
- 47.Zigeuner RE, Riesenberg R, Pohla H, Hofstetter A, Oberneder R. Immunomagnetic cell enrichment detects more disseminated cancer cells than immunocytochemistry in vitro. J Urol. 2000;164:1834–1837. [PubMed] [Google Scholar]
- 48.Partridge M, Phillips E, Francis R, Li SR. Immunomagnetic separation for enrichment and sensitive detection of disseminated tumour cells in patients with head and neck SCC. J Pathol. 1999;189:368–377. doi: 10.1002/(SICI)1096-9896(199911)189:3<368::AID-PATH441>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 49.Tong X, Yang L, Lang JC, Zborowski M, Chalmers JJ. Application of immunomagnetic cell enrichment in combination with RT-PCR for the detection of rare circulating head and neck tumor cells in human peripheral blood. Cytometry. 2007;72B:310–323. doi: 10.1002/cyto.b.20177. [DOI] [PubMed] [Google Scholar]
- 50.Tkaczuk KHR, et al. The significance of circulating epithelial cells in breast cancer patients by a novel negative selection method. Breast Cancer Res Treat. 2007;111:355–364. doi: 10.1007/s10549-007-9771-9. [DOI] [PubMed] [Google Scholar]
- 51.Liu Z, et al. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J Transl Med. 2011;9:70. doi: 10.1186/1479-5876-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin VM, et al. Immunomagnetic enrichment of disseminated epithelial tumor cells from peripheral blood by MACS. Exp Hematol. 1998;26:252–264. [PubMed] [Google Scholar]
- 53.Miltenyi S, Müller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, Williams PS, Zborowski M, Chalmers JJ. Binding affinities/avidities of antibody–antigen interactions: Quantification and scale-up implications. Biotechnol Bioeng. 2006;95:812–829. doi: 10.1002/bit.21024. [DOI] [PubMed] [Google Scholar]
- 55.Lansdorp PM, Aalberse RC, Bos R, Schutter WG, Van Bruggen EFJ. Cyclic tetramolecular complexes of monoclonal antibodies: a new type of cross-linking reagent. Eur J Immunol. 1986;16:679–683. doi: 10.1002/eji.1830160615. [DOI] [PubMed] [Google Scholar]
- 56.Thomas TE, Sutherland HJ, Lansdorp PM. Specific binding and release of cells from beads using cleavable tetrameric antibody complexes. J Immunol Methods. 1989;120:221–231. doi: 10.1016/0022-1759(89)90246-9. [DOI] [PubMed] [Google Scholar]
- 57.Yang L, et al. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol Bioeng. 2009;102:521–534. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ozkumur E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 60.Di Carlo D, Irimia D, Tompkins RG, Toner M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc Natl Acad Sci USA. 2007;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pamme N, Wilhelm C. Continuous sorting of magnetic cells via on-chip free-flow magnetophoresis. Lab Chip. 2006;6:974–980. doi: 10.1039/b604542a. [DOI] [PubMed] [Google Scholar]
- 62.Berger M, Castelino J, Huang R, Shah M, Austin RH. Design of a microfabricated magnetic cell separator. Electrophoresis. 2001;22:3883–3892. doi: 10.1002/1522-2683(200110)22:18<3883::AID-ELPS3883>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 63.Inglis DW, Riehn R, Austin RH, Sturm JC. Continuous microfluidic immunomagnetic cell separation. Appl Phys Lett. 2004;85:5093–5095. [Google Scholar]
- 64.Davis JA, et al. Deterministic hydrodynamics: taking blood apart. Proc Natl Acad Sci USA. 2006;103:14779–14784. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inglis DW, Davis JA, Austin RH, Sturm JC. Critical particle size for fractionation by deterministic lateral displacement. Lab Chip. 2006;6:655–658. doi: 10.1039/b515371a. [DOI] [PubMed] [Google Scholar]
- 66.Loutherback K, et al. Improved performance of deterministic lateral displacement arrays with triangular posts. Microfluid Nanofluid. 2010;9:1143–1149. [Google Scholar]
- 67.Martel JM, Toner M. Inertial focusing dynamics in spiral microchannels. Phys Fluids. 2012;24:032001–032013. doi: 10.1063/1.3681228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Madou MJ. Fundamentals of Microfabrication. CRC Press; 2002. [Google Scholar]
- 69.Boxshall K, et al. Simple surface treatments to modify protein adsorption and cell attachment properties within a poly(dimethylsiloxane) micro-bioreactor. Surf Interface Anal. 2006;38:198–201. [Google Scholar]
- 70.Yu M, et al. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev. 2009;23:1737–1742. doi: 10.1101/gad.1809309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.