Abstract
Background
The nematode Caenhorhabditis elegans offers great power for the identification and characterization of genes that regulate behavior. In support of this effort, analytical methods are required that provide dimensional analyses of subcomponents of behavior. Previously, we demonstrated that loss of the presynaptic dopamine (DA) transporter, dat-1, evokes DA-dependent Swimming Induced Paralysis (Swip) (Mcdonald et al. 2007), a behavior compatible with forward genetic screens (Hardaway et al. 2012).
New Method
Here, we detail the development and implementation of SwimR, a set of tools that provide for an automated, kinetic analysis of C. elegans Swip. SwimR relies on open source programs that can be freely implemented and modified.
Results
We show that SwimR can display time-dependent alterations of swimming behavior induced by drug-treatment, illustrating this capacity with the dat-1 blocker and tricyclic antidepressant imipramine (IMI). We demonstrate the capacity of SwimR to extract multiple kinetic parameters that are impractical to obtain in manual assays.
Comparison with Existing Methods
Standard measurements of C. elegans swimming utilizes manual assessments of the number of animals exhibiting swimming versus paralysis. Our approach deconstructs the time course and rates of movement in an automated fashion, offering a significant increase in the information that can be obtained from swimming behavior.
Conclusions
The SwimR platform is a powerful tool for the deconstruction of worm thrashing behavior in the context of both genetic and pharmacological manipulations that can be used to segregate pathways that underlie nematode swimming mechanics.
Keywords: C. elegans, swimming, dopamine, paralysis, imipramine, transporter
1. Introduction
Since the completion of the human genome project, and the growing annotation of the genomes of several model organisms, neuroscientists have exploited these systems to identify novel conserved genes and determine their impact on behavior. The nematode Caenhorhabditis elegans, possesses a repertoire of simple, reproducible behaviors that facilitate a link between genotype to phenotype and that can be manipulated by drugs that act to modulate behavior in more complex organisms, including humans. Importantly, the nematode utilizes a set highly conserved signaling molecules that include fast-acting neurotransmitters such as acetylcholine, γ-aminobutyric acid (GABA), and glutamate, as well as slow-acting neuromodulators such as dopamine (DA), serotonin (5-HT), and neuropeptides (Jorgensen 2005; Brockie and Maricq 2006; Chase 2007). DA was among the first neurotransmitters demonstrated to modulate locomotion in the worm (Schafer and Kenyon 1995) and since that time, the genes necessary for DA synthesis, packaging, release, reuptake, and signaling at pre- and postsynaptic receptors have been identified and their impact on worm behavior studied (Mcdonald et al. 2006). Although the categorical methods associated with worm behavioral analyses (e.g. movement vs paralysis) have proved efficient for genetic screens, these approaches provide only a limited window into components of behavior that respond to DA and the drugs and/or genetic manipulations that perturb DA signaling.
The need for novel methods to decompose worm swimming arose concomitantly with our discovery that worms with loss of function mutations of the DA transporter (DAT), a presynaptic protein necessary for synaptic DA clearance, demonstrate DA and DA receptor (dop-3) dependent, Swimming-Induced Paralysis (Swip) (Mcdonald et al. 2007). Our prior in vitro studies (Jayanthi et al. 1998) demonstrated that DAT-1 is antagonized by cocaine, as well as by several drugs that block mammalian norepinephrine and serotonin transporters, incuding nisoxetine and the tricyclic antidepressant imipramine (IMI). Recently (Hardaway et al. 2012), we implemented an initial version of an open-source toolkit (SwimR) to capture the kinetic features of Swip in mutant lines identified from a forward genetic screen. Here, we describe the generation and implementation of an optimized version of SwimR and illustrate the enhanced capabilities of the tool for the decomposition of Swip induced by pharmacological DAT-1 blockade in vivo, as assessed with worms treated with the tricyclic antidepressant imipramine (IMI).
2. Materials and Methods
2.1 C. elegans Strains and Husbandry
C. elegans strains were cultured on bacterial lawns of OP50 and maintained at 12 to 20°C using standard methods (Brenner 1974) unless otherwise noted. The wild type strain is N2 Bristol. The dat-1(ok157)III strain was obtained from J. Duerr and J. Rand (Oklahoma Medical Research Foundation, Oklahoma City), and is a complete loss of function mutation that eliminates the majority of the DAT-1 coding sequence. A strain producing a loss of function disruption of DOP-3 (dop-3(vs106)X) was obtained from the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We obtained cat-2(tm2261) from Shohei Mitani at the National Bioresource Project at Tokyo Women’s Medical University. See Supplementary Methods for more information on these strains.
2.2 Assessment of Swip Behavior
Specific details of swimming assays of wild type, mutant and drug-treated animals are provided in Supplementary Methods. Imipramine HCl was obtained from Sigma (St. Louis, MO). Swip assays were performed as described in Hardaway and Hardie et al. Concentration response curves were fit by nonlinear regression methods as implemented in Prism 6.0 (GraphPad, Inc, La Jolla, CA). Average frequency values were analyzed by 2-way ANOVA with Dunnet’s post tests of each time point in the drug condition to the corresponding basal time point.
2.3 SwimR Tools
Individual worm videos were tracked using an updated version of our open source morphometric tracking program (Tracker 2.0 (Mcdonald et al. 2007). Tracker output files were imported into SwimR and analyzed based on user settings, as described in a manual provided in the Supplementary Materials. Software tools are deposited at Biconductor: http://www.bioconductor.org/packages/devel/bioc/html/SwimR.html
3. Results and Discussion
3.1 Development of a Novel, R-based Analytical Platform (SwimR) to Extract Components of Swimming Behavior
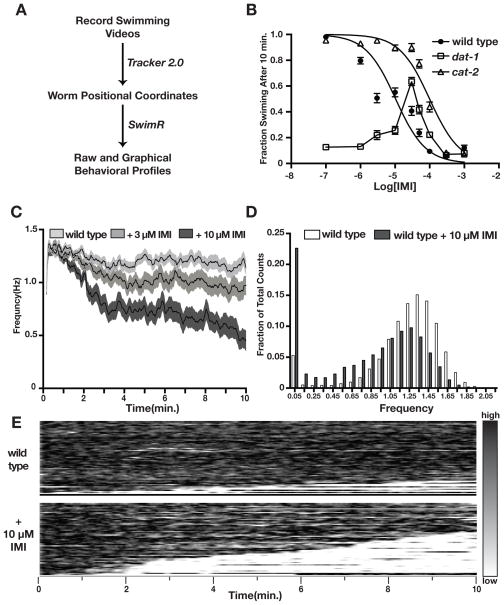
To quantify worm thrashing behavior in liquid medium, we implement Tracker 2.0 to plot the swimming frequency of individual nematodes (Matthies et al. 2006; Mcdonald et al. 2007). This software provides an output of the worm positional coordinates during swimming, which is then imported into SwimR for further data analysis and graphical rendering (Fig. 1A). Since C. elegans swim by the propagation of C-shaped bends on their dorsal and ventral sides, Tracker 2.0 fits a 3-segment spine to a worm in each frame of a video. The software uses a particle filter algorithm to perform this tracking. The particle filter is composed of many spine particles, each of which represents an estimate of the true location and orientation of the worm in each frame. For each frame, the particles are weighted by comparing the spine with a difference image between the current and previous frame. The particles are then resampled and perturbed according to their weighted values and perturbed by small random motion and a representative particle is selected to be the location of the spine for each frame. Particle filters were chosen for their robustness and flexible computation time (Thrun et al. 2005). Within this program, parameters related to spine length, the number of particles, the maximum angle between two points along the spine, and the allowable displacement of the spine between frames can be altered to maximize tracking fidelity (See Supplementary Materials).
Figure 1.
Paralytic effects of IMI on C. elegans wild type (N2) animals as determined by manual and automated assays. A) Diagram of the workflow for C. elegans swimming analysis. B) Response of wild type (filled circles), cat-2 (open triangles), and dat-1 (open squares) to increasing concentrations of IMI. Dose-response data points for dat-1 were connected with a line between data points. C) Effect of IMI on swimming frequency of wild type animals. Shaded areas represent SEM. 3 μM IMI triggered a significant (P<.05) decrease in the average frequency beginning at the 4th minute (P<.05). 10 μM IMI induced a significant decrease in average frequency (P<.05) with individual time points reaching significance in the 2nd minute. D) Distribution of wild type swimming frequency values in the presence or absence of IMI (10 μM). IMI treatment induces a shift to lower frequency values. E) Heat map representations of wild type swimming frequency wjth or without 10 μM IMI.
To analyze subcomponents of worm swimming, we developed SwimR, an open source analytical suite generated in R (http://www.r-project.org/). SwimR analyzes Tracker 2.0 files and implements multiple approaches (Fast Fourier Transform, Extrema Count, Peak Delta and Racetrack Filter plus Get Peaks) to determine spine movement frequency. After the analysis of each worm track, SwimR constructs a matrix of frequency versus time for each worm in the data set that is used for subsequent calculations and graphical output. SwimR analyzes the frequency matrix to extract kinetic parameters related to swimming frequency, paralysis probability and duration, and the probability and duration of movement reversion.
3.2 Manual Analysis of IMI-Triggered Paralysis and Dose Selection
Previously we implemented a preliminary version of SwimR to capture and analyze the swimming behavior of worm mutants identified in a forward genetic screen (Hardaway et al. 2012). Here, we fully operationalize SwimR and provide a user manual to permit the tailoring of analyses based on user preference. Additionally, we demonstrate the ability of SwimR to define the kinetic components of animals subjected to a pharmacological challenge. Specifically, we treated N2 and dat-1 mutant animals with the dat-1 antagonist IMI and determine characteristics and DAT-specificity of IMI-induced Swip behavior.
We first validated the ability of IMI to produce Swip using conventional, manual assays, counting the number of animals that paralyze 10 minutes after incubation in water containing varying concentrations of IMI (Fig. 1B). These studies revealed an IC50 of IMI on N2 animals of 10 +/−1.5 μM. When these assays were conducted with cat-2 animals that lack expression of tyrosine hydroxylase, the rate limiting enzyme in the DA biosynthetic pathway, we observed a ~10-fold shift in the IMI IC50 (99 +/−12 μM). We also observed a reduction in IMI potency with animals deficient in the D2-like DA receptor (dop-3) (Sup. Fig. 1A) that we have shown to be required for Swip behavior in DAT-1 deficient animals, though the shift was more modest (~5 fold). As most dat-1 animals paralyze within 10 minutes, manual assays are not able to evaluate the role of DAT-1 in IMI-induced Swip (Fig 1B). Further complicating this evaluation is the presence of an IMI-induced reversal of Swip at some concentrations (see Fig 1B, 33 and 50 μM IMI). Although we have not explored further the mechanism(s) leading to Swip reversal, we note that IMI has been reported to inhibit the 5-HT transporter mod-5 (Ranganathan et al. 2001; Mcdonald et al. 2006), and may also act on ion channels like egl-2 (Weinshenker et al. 1999; Mcdonald et al. 2007) where the drug has been reported to modulate egg-laying through this pathway.
3.3 Automated Analysis of IMI-induced Swip Behavior
To evaluate IMI-induced Swip as a function of time, we generated single worm tracking files from videos using Tracker 2.0 and extracted kinetic features of these tracks by genotype and drug using SwimR. As shown in the average frequency plots in Fig. 1C, IMI produced a dose-dependent decrease in mean thrashing frequency with the wild type strain. At both 3μM and 10μM, SwimR analyses revealed that IMI reduced the average maximal swimming frequency across the entire recording (Fig. 1D, Table 1, Sup. Fig. 3A). We hypothesize that a reduction in maximal swimming frequency in dat-1 animals, as well as the DA-dependent Swip lines we isolated in our forward genetic screen (Hardaway et al. 2012) derives from the presence of paralyzers in this population that reduce the overall time frame at which high swimming frequencies can be reached. To determine whether the decrease in average thrashing frequency of IMI-treated animals was due to an increase in Swip latency or in penetrance, we used a feature of SwimR that aligns the swimming profiles of individual animals, yielding a heat map of population swimming frequency over time (Fig. 1E and Sup. Fig. 2A). In these plots, a running average of the frequency of movement is calculated for each animal and plotted as a fraction of time in single rows, one above the other. Thrashing frequency is scaled according by color where black (Fig 1E)/yellow (Sup Fig. 2) represents highest swimming frequency and white/blue represents paralysis. SwimR orders the maps so that the strongest paralyzers are placed at the bottom.
Table 1.
Swimming and Paralytic Measurements
| Maximal Frequency (Hz) | Latency to Paralyze (sec) | Reversion IncidenceA | Reversion Frequency (events/animal) | Reversion ProbabilityB | Time to 1st Reversion (sec) | Average Reversion Event Length (sec) | |
|---|---|---|---|---|---|---|---|
| N2(n=63) | 1.82+/−0.141 | 305+/−165 | 0.455 | 3.80 +/−1.93 | 1070+/−693 | 93.9+/−132 | 17.5+/−24.0 |
| N2 + 3 μM IMI(46) | 1.60+/−0.262*** | 274+/−141 | 0.25 | 3.33+/−3.21 | 432+/−584 | 147+/−89.8 | 20.0+/−31.5 |
| N2 + 10 μM IMI(38) | 1.65+/−0.137*** | 297+/−169 | 0.2 | 1.25+/−0.50 | 42.7+/−52.3* | 175+/−97.0 | 7.62+/−8.79 |
| dat-1(ok157)(117) | 1.46+/−0.436 | 206+/−124 | 0.366 | 0.336+/−2.63 | 394+/−500 | 140+/−127 | 13.4+/−26.6 |
| dat-1 + 3 μM IMI(31) | 1.61+/−0.193 | 261+/−109 | 0.308 | 1.38+/−0.744 | 237+/−417 | 171+/−121 | 5.36+/−6.20 |
| dat-1 + 10 μM IMI(27) | 1.56+/−0.260 | 260+/−141 | 0.273 | 0.273+/−2.33 | 162+/−264 | 235+/−84.2 | 6.40+/−3.19 |
| cat-2(tm2261)(38) | 1.75+/−0.091 | N/A | N/A | N/A | N/A | N/A | N/A |
| cat-2 + 3 μM IMI(23) | 1.82+/−0.097 | 438+/−138 | 0.333 | 0.333+/−3.5 | 0.333+/−3.50 | 177+/−44.3 | 1.15+/−0.54 |
| cat-2 + 10 μM IMI(27) | 1.53+/−0.183*** | 140+/68.3**C | 0.25 | 9 | 715 | 98.2 | 5.87 |
| dop-3(vs106)(22) | 1.88+/−0.034 | 223+/−206 | 1 | 2.67+/−1.15 | 1500+/−1550 | 157+/−139 | 15.89+/−10.0 |
| dop-3 + 3 μM IMI(27) | 1.81+/−0.050 | 379+/−165 | 0.333 | 0.333+/−2.00 | 986+/−1380 | 50.1+/−3.15 | 9.08+/−12.8 |
| dop-3 + 10 μM IMI(34) | 1.50+/−0.278*** | 188+/−144 | 0D* | N/A | N/A | N/A | N/A |
Reversion Probability = total time spent in reversion/total time after paralysis onset
Reversion Incidence = fraction of animals that reverse from paralysis
Asterisks(*) indicate a significant difference to control genotype as determined by one-way ANOVA with Dunnet’s multiple comparision test where
p < 0.05,
p < 0.01, and
p < 0.001.
unpaired student’s t test to cat-2 + 3 μM IMI
Chi-square test
Inspection of the heat map plots and the accompanying quantitative analyses of IMI-treated animals revealed a higher incidence of Swip with 10 μM IMI relative to N2 animals without drug treatment, with no change in Swip latency (Table 1). The increased penetrance of Swip can also be detected in frequency histograms produced by SwimR that reveal an increased in a shift of swimming profiles to low frequency values with 10 μM IMI treatment (Fig. 1D and Sup. Fig. 3). In our experience, a small number of paralyzers arise during Swip assays in N2 animals. Whereas N2 paralyzers in the absence of drug treatment displayed a high level of reversion from paralysis, IMI (10 μM)-treated animals displayed a lower incidence of reversion, though this parameter did not reach statistical significance (Table 1). Nonetheless, these findings resulted in a statistically significant reduction in reversion probability. In contrast, no change in the time to 1st reversion or reversion event length was induced by IMI. Thus, IMI-induced paralysis increases the proportion of animals becoming paralyzed as a function of time by progressively reducing thrashing frequency and decreasing the likelihood that paralyzed animals return to a mobile state.
Whereas our manual assays indicated no DA-independent effects of IMI at 3 μM, Swim R analyses of cat-2 and dop-3 animals detected IMI responses (Sup. Fig. 2 and Table 1). 3 μM IMI increased the average thrashing frequency of cat-2 animals during the first two minutes of assay and decreased thrashing frequency during the final minute. For dop-3 animals, 3 μM IMI triggered a significant reduction in thrashing frequency in the final minute of recording whereas reductions were detected by the third minute of recordings for 10 μM IMI. Unlike wild type animals, only 10 μM had a significant effect on the maximal frequency reached by cat-2 and dop-3 worms. Inspection of the population plots for cat-2 and dop-3 revealed a higher incidence of paralyzed worms at both doses, and an increase in Swip latency for cat-2 paralyzers treated with 10 μM IMI relative to 3 μM IMI. Consistent with the significant reduction in average thrashing frequency evident in the final minute, 3 μM IMI triggered paralysis at the end of several worm tracks (asterisks in Sup. Fig. 2). Inspection of the frequency distribution for cat-2 and dop-3 treated with IMI revealed that the increase in average thrashing frequency during the first two minutes was reflected in a shift in the density of higher frequency value bins (Sup. Fig. 3). As we only detected a few paralyzers in either dose, we were underpowered to make any significant observations regarding IMI’s effects on cat-2 or dop-3 reversion from paralysis. Nonetheless, the evaluations with SwimR noted above provide clear evidence of DA-independent actions of IMI even at low drug concentrations.
As noted above, manual assays preclude assessment of the DAT-1 specificity of IMI action, possibly due to a requirement for the transporter in the action of the antidepressant. However, time course analysis of frequency changes clearly demonstrates that IMI produces, at both 3 and 10 μM, an increase in average thrashing frequency across all time points on a dat-1 mutant background (Sup. Fig. 1D). Unlike cat-2 and dop-3, IMI did not alter the maximal frequency reached at either dose. Consistent with our manual assays, inspection of heat map frequency plots (Sup. Fig. 2D), frequency distributions (Sup. Fig. 3B), and tabulated values of Swip latency and reversion (Table 1) in dat-1 animals treated with 3 μM and 10 μM IMI revealed that this increase in average thrashing frequency is due to a decrease in Swip penetrance. These observations represent the first demonstration of a pharmacological agent that can acutely rescue dat-1 Swip. The apparent affinities of IMI for dat-1 and mod-5 suggest that IMI is more likely to act on dat-1 (Jayanthi et al. 1998; Ranganathan et al. 2001; Hardaway et al. 2012). In the absence of DAT-1, however, we hypothesize that IMI acts at mod-5 or egl-2 (Weinshenker et al. 1995). Considering the reported antagonistic roles for DA and serotonin in multiple worm behaviors including egg-laying and movement on plates, it seems plausible that swimming behavior engages both dat-1 and mod-5 function, limiting the extrasynaptic availability of DA and serotonin respectively to effect worm swimming patterns.
Together, these studies demonstrate the utility and sensitivity of Tracker 2.0 and SwimR for the presentation and analysis of both genetic- and pharmacologically-induced patterns of C. elegans swimming. In manual assays, we demonstrated that IMI induces paralysis consistent with DA and dat-1 dependence. Analysis of IMI effects using SwimR revealed that whereas the behavioral effects of IMI clearly require endogenous DA signaling, additional pathways are likely to regulate responses even at low concentrations of the drug. SwimR enabled us to depict and quantify novel subcomponents of IMI effects. Time-dependent changes in gene- and drug-modulated swimming behavior can be missed in end-point, “yes-no” manual paralysis assays. We conclude that SwimR is a useful tool for exploring the differential impact of mutations and drugs on distinct signaling networks, one that, due to its freely available, open-platform nature, can be tailored to the specific interests of C. elegans researchers.
Supplementary Material
IMI effects on swimming of dat-1, cat-2 and dop-3 animals A) Dose response effects of IMI on dop-3(vs106) as measured manually. Assays and statistical analyses were performed as described in Fig. 1 and in the methods. r2 of fitted curves for wild type and dop-3 were 0.91 and 0.83 and the IC50 values for wild type and dop-3 were 10 +/−1.5 μM and 56 +/− 7 μM respectively. B) Automated recordings and analysis using Tracker 2.0. Average frequency of cat-2(tm2261) without drug, 3 μM IMI, and 10 μM IMI. Shaded area
ETHICAL STANDARDS.
The authors declare that all experiments on human subjects were conducted in accordance with the Declaration of Helsinki http://www.wma.net and that all procedures were carried out with the adequate understanding and written consent of the subjects.
The authors also certify that formal approval to conduct the experiments described has been obtained from the human subjects review board of their institution and could be provided upon request.
if the studies deal with animal experiments, the authors certify that they were carried out in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80–23) revised 1996 or the UK Animals (Scientific Procedures) Act 1986 and associated guidelines, or the European Communities Council Directive of 24 November 1986 (86/609/EEC).
The authors also certify that formal approval to conduct the experiments described has been obtained from the animal subjects review board of their institution and could be provided upon request.
The authors further attest that all efforts were made to minimize the number of animals used and their suffering.
If the ethical standard governing the reported research is different from those guidelines indicated above, the authors must provide information in the submission cover letter about which guidelines and oversight procedures were followed.
The Editors reserve the right to return manuscripts in which there is any question as to the appropriate and ethical use of human or animal subjects.
Acknowledgments
The authors thank Sarah Baas, Daniel Bermingham, Hussain Jinnah, Kevin Erreger, David Airey, and Heinrich Matthies for helpful feedback during the performance of this work. This study was supported by MH064913 and MH093102 (J.L.H.), DA027739 (S.L.H), MH078028 and MH095044 (R.D.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974 May 1;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Maricq AV. Ionotropic glutamate receptors: genetics, behavior and electrophysiology. WormBook: the online review of C elegans biology. 2006:1–16. doi: 10.1895/wormbook.1.61.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase DL. Biogenic amine neurotransmitters in C. elegans. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.132.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardaway JA, Hardie SL, Whitaker SM, Baas S, Zhang B, Bermingham DP, et al. Forward Genetic Analysis to Identify Determinants of Dopamine Signaling in Caenorhabditis elegans Using Swimming-Induced Paralysis. G3: Genes| Genomes|. 2012 doi: 10.1534/g3.112.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanthi LD, Apparsundaram S, Malone MD, Ward E, Miller DM, Eppler M, et al. The Caenorhabditis elegans gene T23G5.5 encodes an antidepressant- and cocaine-sensitive dopamine transporter. Mol Pharmacol. 1998 Oct 1;54(4):601–9. [PubMed] [Google Scholar]
- Jorgensen EM. GABA. WormBook. WormBook; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies DS, Fleming PA, Wilkes DM, Blakely RD. The Caenorhabditis elegans choline transporter CHO-1 sustains acetylcholine synthesis and motor function in an activity-dependent manner. Journal of Neuroscience. 2006 Jun 7;26(23):6200–12. doi: 10.1523/JNEUROSCI.5036-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonald PW, Hardie SL, Jessen TN, Carvelli L, Matthies DS, Blakely RD. Vigorous motor activity in Caenorhabditis elegans requires efficient clearance of dopamine mediated by synaptic localization of the dopamine transporter DAT-1. Journal of Neuroscience. 2007 Dec 19;27(51):14216–27. doi: 10.1523/JNEUROSCI.2992-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonald PW, Jessen T, Field JR, Blakely RD. Dopamine Signaling Architecture in Caenorhabditis elegans. Cell Mol Neurobiol. 2006 Nov 20;26(4–6):591–616. doi: 10.1007/s10571-006-9003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R, Sawin ER, Trent C, Horvitz HR. Mutations in the Caenorhabditis elegans serotonin reuptake transporter MOD-5 reveal serotonin-dependent and -independent activities of fluoxetine. J Neurosci. 2001 Aug 15;21(16):5871–84. doi: 10.1523/JNEUROSCI.21-16-05871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer WR, Kenyon CJ. A calcium-channel homologue required for adaptation to dopamine and serotonin in Caenorhabditis elegans. Nature. 1995 May 4;375(6526):73–8. doi: 10.1038/375073a0. [DOI] [PubMed] [Google Scholar]
- Thrun S, Burgard W, Fox D. Robotics. MIT Press (MA); 2005. [Google Scholar]
- Weinshenker D, Garriga G, Thomas JH. Genetic and pharmacological analysis of neurotransmitters controlling egg laying in C. elegans. J Neurosci. 1995 Oct 1;15(10):6975–85. doi: 10.1523/JNEUROSCI.15-10-06975.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshenker D, Wei A, Salkoff L, Thomas JH. Block of an ether-a-go-go-like K(+) channel by imipramine rescues egl-2 excitation defects in Caenorhabditis elegans. J Neurosci. 1999 Nov 15;19(22):9831–40. doi: 10.1523/JNEUROSCI.19-22-09831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IMI effects on swimming of dat-1, cat-2 and dop-3 animals A) Dose response effects of IMI on dop-3(vs106) as measured manually. Assays and statistical analyses were performed as described in Fig. 1 and in the methods. r2 of fitted curves for wild type and dop-3 were 0.91 and 0.83 and the IC50 values for wild type and dop-3 were 10 +/−1.5 μM and 56 +/− 7 μM respectively. B) Automated recordings and analysis using Tracker 2.0. Average frequency of cat-2(tm2261) without drug, 3 μM IMI, and 10 μM IMI. Shaded area