Abstract
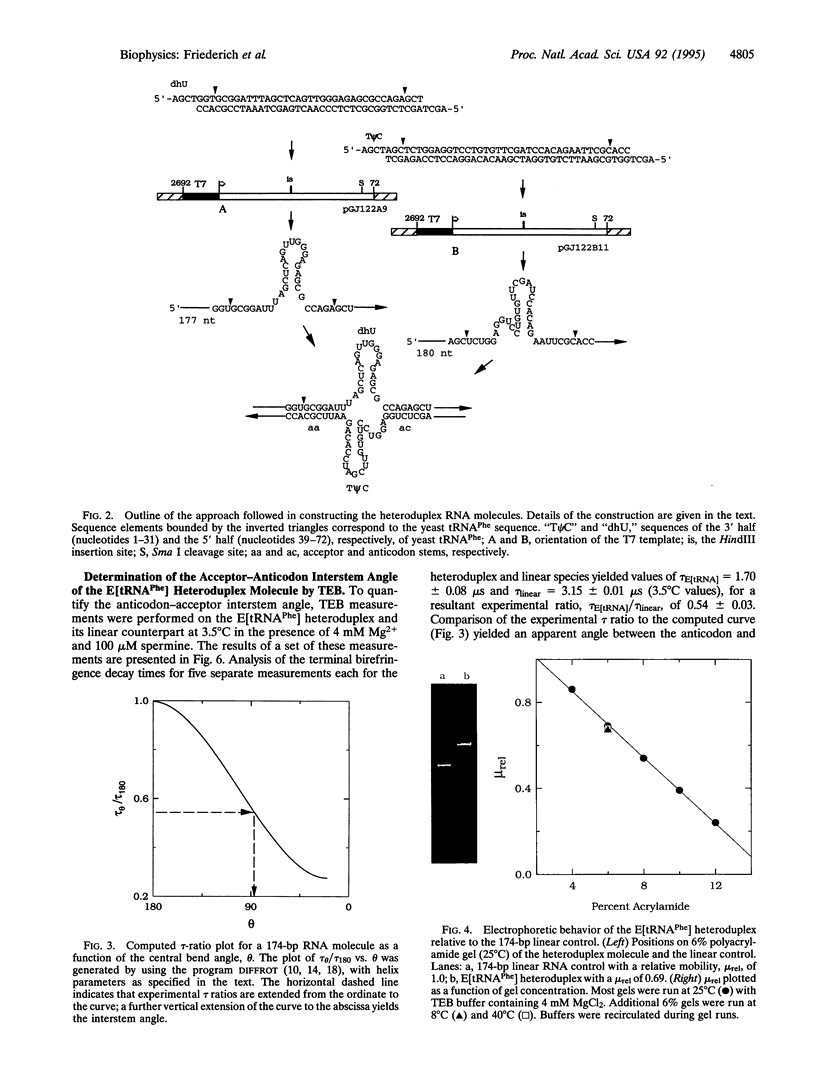
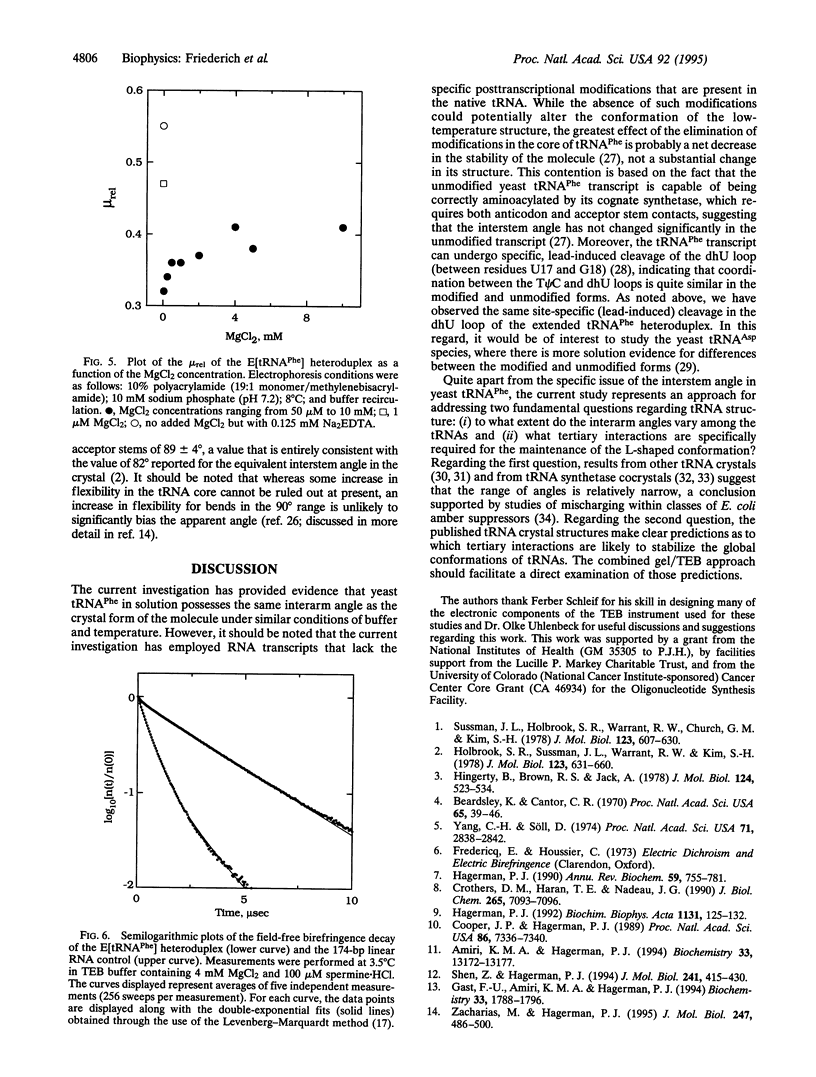
A principal feature of the crystal structures of tRNAs is an L-shaped tertiary conformation in which the aminoacyl acceptor stem and the anticodon stem are approximately perpendicular. However, the anticodon-acceptor interstem angle has not been precisely quantified in solution for any tRNA. Such a determination would represent an important test of the predicted global conformation of tRNAs in solution. To this end, we have constructed a yeast tRNA(Phe) heteroduplex RNA molecule in which the anticodon and acceptor stems of the tRNA have each been extended by approximately 70 base pairs. A comparison of the rotational decay times of the heteroduplex molecule and a linear control yields an interstem angle of 89 +/- 4 degrees in 4 mM magnesium chloride/100 microM spermine hydrochloride, essentially identical to the corresponding angle observed in the crystal under similar buffer and temperature conditions. The current approach is applicable to the study of a wide variety of RNA molecules that possess elements of nonhelical structure.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amiri K. M., Hagerman P. J. Global conformation of a self-cleaving hammerhead RNA. Biochemistry. 1994 Nov 15;33(45):13172–13177. doi: 10.1021/bi00249a003. [DOI] [PubMed] [Google Scholar]
- Basavappa R., Sigler P. B. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991 Oct;10(10):3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardsley K., Cantor C. R. Studies of transfer RNA tertiary structure by singlet-singlet energy transfer. Proc Natl Acad Sci U S A. 1970 Jan;65(1):39–46. doi: 10.1073/pnas.65.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlen L. S., Sampson J. R., DiRenzo A. B., Uhlenbeck O. C. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry. 1990 Mar 13;29(10):2515–2523. doi: 10.1021/bi00462a013. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., Lilley D. M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990 Feb 1;343(6257):484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- Brown R. S., Dewan J. C., Klug A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry. 1985 Aug 27;24(18):4785–4801. doi: 10.1021/bi00339a012. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Geometry of a branched DNA structure in solution. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7336–7340. doi: 10.1073/pnas.86.19.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- Gast F. U., Amiri K. M., Hagerman P. J. Interhelix geometry of stems I and II of a self-cleaving hammerhead RNA. Biochemistry. 1994 Feb 22;33(7):1788–1796. doi: 10.1021/bi00173a023. [DOI] [PubMed] [Google Scholar]
- Gast F. U., Hagerman P. J. Electrophoretic and hydrodynamic properties of duplex ribonucleic acid molecules transcribed in vitro: evidence that A-tracts do not generate curvature in RNA. Biochemistry. 1991 Apr 30;30(17):4268–4277. doi: 10.1021/bi00231a024. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Straightening out the bends in curved DNA. Biochim Biophys Acta. 1992 Jun 15;1131(2):125–132. doi: 10.1016/0167-4781(92)90066-9. [DOI] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Krzyzosiak W. J., Marciniec T., Wiewiorowski M., Romby P., Ebel J. P., Giegé R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988 Jul 26;27(15):5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J. P., Florentz C., Giegé R. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie. 1990 Oct;72(10):735–743. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Hagerman P. J. Conformation of the central, three-helix junction of the 5 S ribosomal RNA of Sulfolobus acidocaldarius. J Mol Biol. 1994 Aug 19;241(3):415–430. doi: 10.1006/jmbi.1994.1517. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- Tang R. S., Draper D. E. Bulge loops used to measure the helical twist of RNA in solution. Biochemistry. 1990 Jun 5;29(22):5232–5237. doi: 10.1021/bi00474a003. [DOI] [PubMed] [Google Scholar]
- Tang R. S., Draper D. E. On the use of phasing experiments to measure helical repeat and bulge loop-associated twist in RNA. Nucleic Acids Res. 1994 Mar 11;22(5):835–841. doi: 10.1093/nar/22.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner C., Krebs B., Keith G., Dirheimer G. Specific cleavages of pure tRNAs by plumbous ions. Biochim Biophys Acta. 1976 May 3;432(2):161–175. doi: 10.1016/0005-2787(76)90158-1. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Yang C. H., Söll D. Studies of transfer RNA tertiary structure of singlet-singlet energy transfer. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2838–2842. doi: 10.1073/pnas.71.7.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M., Hagerman P. J. Bulge-induced bends in RNA: quantification by transient electric birefringence. J Mol Biol. 1995 Mar 31;247(3):486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]