Abstract
Most cancer-associated genetic variants identified from genome-wide association studies (GWAS) do not obviously change protein structure, leading to the hypothesis that the associations are attributable to regulatory polymorphisms. Translating genetic associations into mechanistic insights can be facilitated by knowledge of the causal regulatory variant (or variants) responsible for the statistical signal. Experimental validation of candidate functional variants is onerous, making bioinformatic approaches necessary to prioritize candidates for laboratory analysis. Thus, a systematic approach for recognizing functional (and, therefore, likely causal) variants in noncoding regions is an important step toward interpreting cancer risk loci. This review provides a detailed introduction to current regulatory variant annotations, followed by an overview of how to leverage these resources to prioritize candidate functional polymorphisms in regulatory regions.
Keywords: bioinformatics, variant annotation, regulatory prediction, functional follow-up
Introduction
Genome-wide association studies (GWAS) have successfully identified hundreds of cancer susceptibility loci, and have been particularly powerful in the discovery of distal regulatory elements such as enhancers. Understanding the mechanisms underlying GWAS findings requires the functional characterization of causal alleles and is therefore an important next step for gaining deeper insight into cancer susceptibility. If the single-nucleotide polymorphism (SNP) with the strongest statistical association (ie, the “index SNP”) was guaranteed to be causal, then functional validation would likely occur more often. However, SNPs that are represented on genotyping platforms are explicitly chosen as markers for “bins” of SNPs having highly correlated genotypes, with the consequence that SNPs within the same bin will have similar levels of statistical association, even when the association is driven by a single functional variant. The present generation of genotyping platforms contain roughly one million markers that tag a total of nearly six million SNPs, for a mean bin size of approximately six SNPs. The median number of SNPs within a bin is somewhat smaller, but still greater than three. Under the assumption that most associated bins contain only one functional variant, a large proportion of index SNPs with reported associations are not functional (ie, they are not causally linked to the phenotype). Thus, winnowing the list of associated variants to those with hypothesized function is an important step prior to laboratory-based validation.
The large number of GWAS loci in noncoding regions1,2 has illuminated two important facets of genetic influences on sporadic cancer: (1) many of the loci contributing to cancer susceptibility are likely to impact gene regulatory elements, and (2) the individual effects of common risk variants are associated with small quantitative differences in expression rather than qualitative changes in protein structure.3 Given these findings, it is not surprising that the rapid discovery of genetic associations with cancer outcomes has not been easily translated into improved clinical practice. In part, the identification of causal variants within known loci will likely improve genetic risk prediction and thereby enable more tailored prevention strategies. Furthermore, improved understanding of the mechanisms underlying genetic associations might enable development of interventions that mimic the effects of the protective allele. As noted, testing variants for functional follow-up in the laboratory can be labor- and resource-intensive, making the selection of candidate variants a critical next step.
When studying familial cancer, the vast majority of findings were germline mutations that disrupt the coding sequence of tumor suppressor genes, DNA repair genes, or oncogenes. In protein-coding regions, there are many annotation resources for assigning functional information. For example, PMD,4 PhenCode,5 and HumDiv/HumVar/Polyphen26 are catalogs constructed from one or more variant databases or from the literature with the intent of annotating specific variants for functional effect. Similarly, nonsynonymous SNPs (nsSNPs) can be assigned function based on their impact on structural stability through databases like ProTherm7 and ProNit.8 In contrast, noncoding SNP annotations are constrained by a dearth of knowledge pertaining to the phenotypic impact of regulatory variation. However, the number of genome annotations relevant to regulation is increasing rapidly, and is finally maturing to the point that appropriate annotations can be leveraged by researchers to enrich lists of variation for candidates to test in the laboratory.
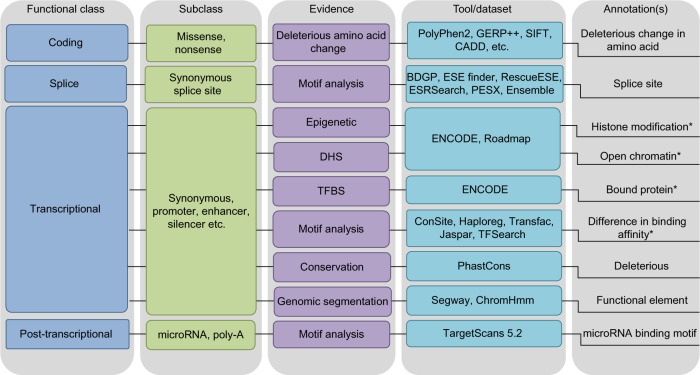
Most regulatory annotations attempt to define, or segment, noncoding regions into short stretches of the genome that associate with biochemical properties exhibited by a small number of experimentally validated regulatory elements. These segments are defined by: (1) biofeatures such as epigenetic remodeling of chromatin, (2) particular sequence motifs like conserved microRNA binding sites, (3) gene expression data, and (4) evolutionary constraint (see Fig. 1). This review will cover the existing tools that are available to annotate cancer loci with regulatory features (see Table 1).
Figure 1.
Tools for bioinformatic annotation.
Note: *Combined evidence across histone modification, open chromatin, ChIP-seq protein and motif annotations provides finer demarcation of functional elements and stronger evidence for the regulatory potential.
Table 1.
Description of bioinformatics tools used for functional follow-up of noncoding regions.
| DATASET | GENOMIC CLASS | DESCRIPTION | DATA SOURCE/PROGRAM |
|---|---|---|---|
| 1 | Non-synonymous coding | Exonic positions leading to amino acid replacement | Nonsense-Ensembl,61 Missense-Polypen2,13 GERP++,12 SIFT,74 SnpEff,75 LS-SNP,20 FoldX76 |
| 2 | Promoter | 1kb regions upstream of annotated transcription start sites | RefSeq, UCSC Genome Browser50: ENCODE Histone Modifications9 and Uniformly processed signal from Roadmap Project tracks |
| 3 | TFBS | Transcription factor binding sites (TFBS) predicted in promoter & non-promoter regulatory elements | UCSC Genome Browser50: ChIP-seq Transcription factor,77 PWM-scana JASPAR,17 CONSITE,18 HaploReg19 |
| 4 | Non-coding RNA | All types of experimentally supported non-coding RNA, including microRNAs | RNAdb 2.078 & miRBase 17.079 |
| 5 | MicroRNA target site | Computationally predicted microRNA target sites within 3′ UTRs | TargetScanS 5.280 |
| 6 | Enhancer element | Experimentally supported enhancer elements in any tissue | VISTA Enhancer Browser UCSC Table Browser81: ENCODE Histone Modifications9 |
| 7 | Candidate non-specific regulatory element | Open chromatin loci in at least one human cell type, as assessed by DNase I hypersensitivity (DHS) mapping | UCSC Table Browser81: Duke and UW DNase I HS data from > 50 cell types77 |
| 8 | Insulator elements | CTCF binding sites assessed by ChIP-seq technology | UCSC Table Browser81: ChIP-seq TFBS77 |
| 9 | eQTL | Allele-specific differences in expression levels | GTEx eQTL Browser,82 TCGA51 |
| 10 | Conserved Element | UCSC Table Browser81: PhastCons 46-way conservation83 | |
| 11 | Splice Site | BDGP |
Note:
PWM-scan was applied using positional weight matrices (PWMs) from the Transfac database.
Defining Regulatory Elements
There are now many biological datasets that have been made available through the extensive efforts of the Encyclopedia of DNA Elements (ENCODE)9 and the NIH Roadmap Epigenomics10 projects. These datasets provide information that enables functional annotation of noncoding regions harboring association signals.11 The ENCODE and Roadmap projects were created with the intention of cataloging the genome-wide regulatory landscape across many cell types9 and tissues.10 These experimental datasets can be summarized into three general biofeatures: (1) chromatin structure as detected through DNaseI hypersensitivity (DHS) assays, (2) histone modifications detected through chromatin immunoprecipitation sequencing (ChIP-seq), and (3) protein-binding detected through ChIP-seq. In contrast to tissue-specific features, we can use known binding motifs to predict variants that disrupt poly-A signals or binding of post-transcriptional regulators like miRNAs. Comparison of sequence alignment across species can also be used to identify conserved sequences reflective of regions that are important to organismal fitness.12 PhastCons13 is a particularly useful conservation measurement for detection of regulatory elements as it factors in the constraint of neighboring nucleotides. Taken together, these datasets can identify specific remodeling and general accessibility of chromatin for the binding of regulatory machinery (eg, transcription factors [TFs]) or for other post-transcriptional mechanisms.
Since regulatory elements such as enhancers often facilitate cell-type specific expression,14 it is helpful to look for evidence in a variety of tissues or cell lines in addition to those specifically related to the cancer of interest. Examination of multiple cell types allows differentiation between tissue-specific regulatory mechanisms likely to contribute to cancer susceptibility and regulatory elements shared by many cell types. Epigenetic chromatin modifications are useful for identifying cell-specific regulatory elements. The methylation and acetylation of histone proteins changes chromatin accessibility for transcription, and such marks can serve as a powerful tool for identifying enhancers, promoters, and other regulatory regions like insulators. There are three histone modification marks that are particularly informative for the identification of most active enhancer and promoter regulatory regions, namely H3k4me1, H3k27ac, and H3k4me3. The H3k4me1 histone mark is associated with enhancers downstream of transcription start sites, and the H3k27ac signal is similarly thought to enhance transcription. Alternatively, the H3k4me3 mark is associated with active promoters. In addition to these three epigenetic signals, H3k27me3 is useful to distinguish between active and poised promoters/enhancers.
There are just over 350 cell lines assayed through ENCODE and 264 phenotypically normal tissue samples assayed through Roadmap. The “Histone Modification” tracks within the ENCODE Integrative Analysis Data Hub to the Genome Browser public hubs page (http://genome.ucsc.edu/cgi-bin/hgHubConnect) and “Uniformly Signal” within the Roadmap Epigenomics Data Complete Collection at Wash U VizHub (http://vizhub.wustl.edu/VizHub/RoadmapReleaseAll.txt) can be used to query data from multiple cell lines/tissues that have been processed through a uniform quality pipeline. Importantly, these datasets have been taken through a uniform processing pipeline developed by the ENCODE and Roadmap Analysis Working Groups (AWG) to reduce cross-lab differences and ensure comparability between datasets (http://genome.ucsc.edu/ENCODE/qualityMetrics.html).
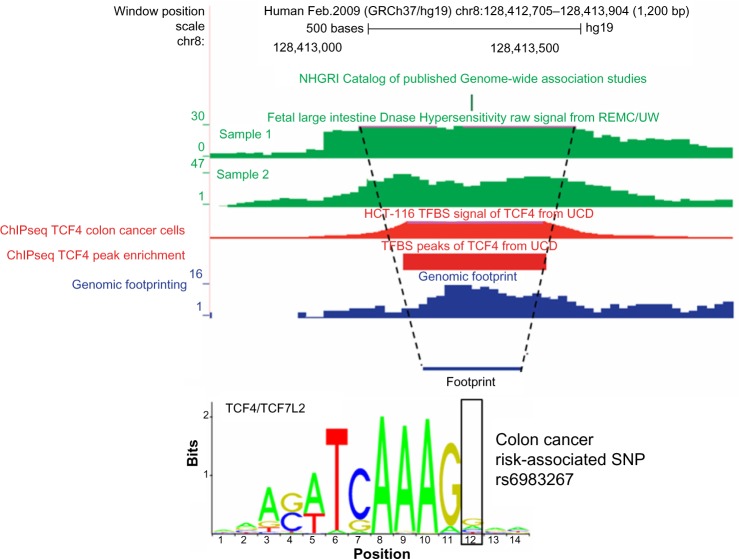
DHS demarks regions of open chromatin by partial digestion of intact chromatin.9 Chromatin bound by TFs tends to be more accessible than chromatin bound to histones. As such, DHS can be used to identify nonspecific protein binding at finer resolution than histone modifications. ChIP-seq methods provide an independent mapping of TF occupancy, which can reveal the identity of the TF contributing to a DHS, and also can narrow peaks of histone modification to a relatively small region (a few hundred base pairs).15 Although the binding of TFs creates a DHS peak, protein occupancy blocks nuclease cutting at the nucleotides specifically bound by protein. Therefore, at nucleotide resolution a TF-bound region would be expected to have low DHS signal flanked by two strong DHS peaks (see Fig. 3). This pattern, referred to as a DHS footprint, has been used to provide nucleotide resolution of evidence for TF binding. Algorithms have been designed to detect this signature for genome-wide detection of TF occupancy in the absence of appropriate antibodies for ChIP-seq experiments.16 However, since DHS signals are an aggregate of many cells, for which protein occupancy is inconsistent, the boundaries of these two flanking peaks are not always clearly defined and can diminish the DHS valley in protein-binding regions. Thus, although this can be a powerful alternative for detecting novel protein-binding regions, the sensitivity of DHS may be imperfect.
Figure 3.
DHS footprinting annotation.
Notes: The rs6983267 variant is a laboratory confirmed functional variant in a distal MYC enhancer associated with colorectal cancer risk. Within the DHS peaks (shown in green) and broad ChIP-seq signal for TCF4 (shown in red), we find a slight valley between two higher DHS peaks. This valley corresponds to a region that actually binds TCF4 and can be detected through DHS footprinting (shown in blue). Motif analysis reveals that rs6983267 falls within the conserved binding motif of TCF4. It is important to note that because there are a number of TFs that bind to this region, as is the case with many enhancers, the valley is less pronounced than would be expected of a strong regulatory element binding a single TF.
ChIP-seq TF binding site (TFBS) data provide evidence for regions of the genome that bind a specific TF in a given cell line or tissue sample. Using ChIP-seq datasets, sequences of several hundred base pairs are identified, each nominally containing one or more motifs that bind to a specific TF. After ChIP-seq identification of motifs that bind to a particular region, position weight matrix (PWM) motifs can be used to screen for candidate regions that might bind a given TF, or to identify allelic motifs that might perturb TF binding. Motif recognition databases trained from ChIP-seq and similar methods include JASPAR,17 ConSite,18 and HaploReg19 PWM. Although the identification of altered affinities for allelic binding sequences positioned within a specific protein’s ChIP-seq signal provides strong supporting evidence for a variant having function, many ChIP-seq peaks lack a clear copy of the consensus motif. It is possible that the lack of known motifs reflects TF binding to weak motifs influenced by cooperative binding of multi-TF complexes or the existence of unknown binding motifs. It is important to note that many consensus TF motifs throughout the genome are not necessarily ever accessible for binding to the relevant TF. As such, the co-occurrence of both epigenetic remodeling and protein binding from independent assays is considered stronger evidence of a regulatory element than the presence of either signal alone. Each of these annotation features (histone marks, DHS, and ChIP-seq) captures a different snapshot of regulatory potential of a region, making the combination of annotations stronger evidence of function than any single annotation alone.
Segmentation through machine learning techniques
For nsSNPs there exist a number of annotation tools that exploit existing annotation data for their predictions, such as LS-SNP,20 SNPs&GO,21 PMUT,22 and SNAP.23 In noncoding regions, similar techniques can be used to aggregate biofeatures and predict regulatory elements. Several groups have independently applied machine learning methodologies to combine epigenetic datasets with chromatin structure, such as those available through ENCODE and the RoadMap project, into discrete annotations of regulatory regions.24,25 Segway implements a dynamic Bayesian network method using the aforementioned biofeatures (ChIP-seq and DHS signals) to identify patterns associated with transcription start sites, gene boundaries, enhancers, and other transcription regulators in an unsupervised approach.24 Software and genome browser tracks using ENCODE data are available at http://noble.gs.washington.edu/proj/segway/. Alternatively, ChromHMM first labels each biochemical assay as high or low signal in 200-bp bins across the genome and then runs a 25-state multivariate Hidden Markov Model that can similarly use the presence or absence of chromatin signals to map chromatin states.25 ChromHMM and Segway data for ENCODE cell lines can be downloaded from https://genome.ucsc.edu/ENCODE/downloads.html under the “Genome Segmentations” link within the ENCODE Analysis Hub. Similarly, segmentations for the Roadmap data are available from the University of California Santa Cruz (UCSC) Table Browser within the “Roadmap Epigenomics Data Complete Collection at Wash U VizHub” under the “Roadmap ChromHMM” track. The primary difference between these two annotations is resolution. While segments in published ChromHMM segmentation25 have a mean of 4,862 bp and a median of 800 bp,25 Segway segments have a mean length of 168 bp and a median of 124 bp.24 Although these assays could use a large number of labels, the number of states with easily meaning (such as transcription start sites, promoters, enhancers, or untranslated regions [UTR]) becomes equivocal after approximately 25 states because the clustering of histone modifications cannot readily be linked with a known chromatin state. However, an advantage of machine learning techniques is the capacity to elucidate new putatively functional epigenetic states, in addition to summarizing complex data for easier interpretation (see Fig. 2). It should be acknowledged that our understanding of the true underlying biology is incomplete, and given the paucity of gold standard data, assigning broader categories reflecting more general biological properties is usually advisable for functional follow-up.
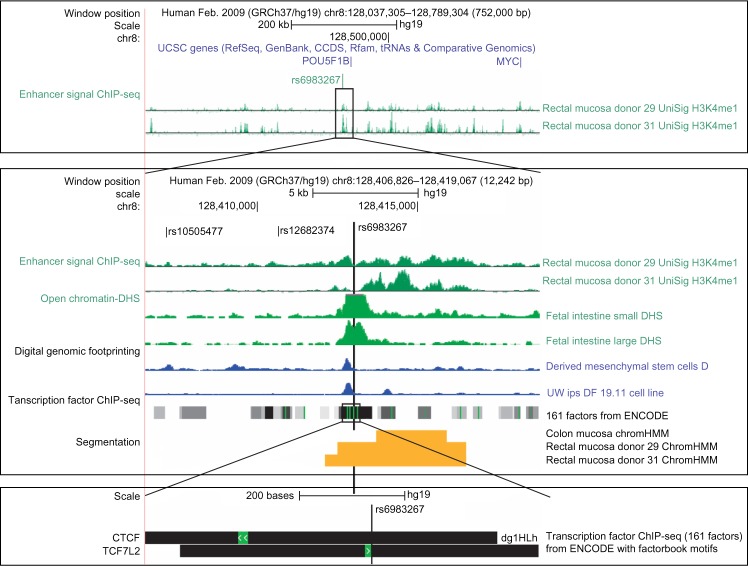
Figure 2.
Bioinformatics annotation of a promoter element using UCSC Genome Browser.
Notes: Figure 2 Bioinformatics annotation using the UCSC Genome Browser for the 8q24 index SNP rs6983267. In the first panel, the index SNP is shown to be located in an intergenic region over 200 kb upstream of MYC. The Roadmap tracks in the second panel zoom in on 8q24 showing the genomic position chr8:128,406,826–128,419,067. The chromosomal positions of three variants in LD (r2 > 0.8) with rs6983267 are shown in the first custom bed file track. The next two plots are biological replicates of normal rectal enhancer ChIP-seq signal. The following two show DHS peak enrichment in fetal intestine. Shown below the DHS plots are genomic footprinting tracks and below that is aggregate ChIP-seq signal from 161 TFs. Genomic segmentation by ChromHMM in normal colon and rectal tissue labels the region containing rs6983267 as an enhancer. In the third panel we see strong TCF7L2 and CTCF binding across the rs6983267 variant. While rs6983267 has a strong bioinformatic functional annotation, the other two variants in strong LD, rs10505477 and rs12682374, do not align to predicted regulatory elements. In vitro evaluation of this region has shown that the rs68983267 is a functional variant that disrupts the binding of TCF7 and expression of the target gene MYC.56,84
In addition to classifying segments of the genome, a new score, referred to as the combined annotation dependent depletion (CADD),26 has been developed to annotate the genome at single nucleotide resolution using a support vector machine (SVM) on features identified from previously mentioned genome-wide annotations. In contrast to Segway and ChromHMM, the C-score from CADD is one of the first attempts to assign a measure of “deleteriousness” to all possible alleles across the human genome. CADD shows great promise in the identification of Mendelian alleles (as in Thalassemia or Kabuki syndrome), with total penetrance and large effects on fitness. However, it is unclear how sensitive CADD is in detecting functional alleles with more subtle effects. By definition, deleterious alleles are functional, but functional alleles might not always cause a significant reduction in evolutionary fitness. In particular, alleles associated with late-onset conditions may not exhibit evolutionary constraint as they have modest, quantitative effects in disease risk. Thus, the negative predictive value of modest CADD (C-score < 5) scores will need to be evaluated in panels of known functional variants for common, late-onset diseases.
Chromatin interaction annotations
In general, chromatin immunoprecipitation (ChIP) is a powerful technique for studying protein–DNA interactions. In addition to detecting histone modifications and TF binding, various ChIP methods can be used to detect long-range interactions, such as the interaction of distal enhancers with target gene promoters. A detailed review of ChIP-based methods has previously been published by Fullwood and Ruan.27 In short, chromosome conformation capture (3C)28 methods are able to detect chromatin architecture, but are expensive, burdensome to implement, are unable to provide insight at a genome-wide scale, and are difficult to interpret because of a large noise-to-signal ratio. In addition, these data are often nonspecific, resulting from the stochastic properties of chromatin interactions. Indeed, two points closely positioned on the chromosome are more likely to collide at random than those at greater physical distance.29 This randomness can lead to the detection of a large amount of nonfunctional interactions relative to true functional signal in all chromatin capture techniques. Higher-throughput methods such as 4C,30,31 which can detect one regulatory element interacting with other targets, or 5C,32 detection of many regulatory elements interacting with targets, are still not implemented at a genome-wide scale.
Adding ChIP to chromosome capture methods helps to reduce noise by requiring the presence of a particular protein. However, chromosome capture methods still restrict the scope of detection because they depend on specific sites, and the addition of ChIP further limits the analysis to a single factor. Under the assumption that functional interactions between chromosomal regions are tethered, it was proposed that the use of sonication rather than the restriction enzyme digestion can help reduce noise by removing weak interactions associated with the random proximity of two regions. Chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) is the first of these techniques to move toward genome-wide scale. Unlike other chromatin capture techniques, ChIA-PET introduces a linker sequence to the DNA fragments that are anchored together by a protein. Introduction of the linker allows this approach to be conducted genome-wide. The addition of both ChIP and sonication to ChIA-PET is a powerful method to detect long-range interactions across the genome associated with particular TFs.33
Expressed quantitative trait loci (eQTL) annotation
The majority of cancer GWAS do not identify plausible coding variants obviously connected to a single gene. ENCODE chromatin conformation capture experiments in modest subsets of the genome suggest that many intergenic regulatory elements skip over the closest promoter to interact with more distal genes.15,34 As a result, ad hoc identification of the causal gene(s) (eg, reporting the gene that is physically closest to the index SNP) that underlie an association signal remains a significant problem that can limit the biological interpretability of disease association study results. However, eQTLs are by definition associated with a specific gene, and trait-associated variants that are also eQTLs can generate candidate causal genes for further functional studies. The Genotype-Tissue Expression (GTEx) database is a useful resource for accessing eQTL data and can be accessed through http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi. HaploReg19 is an additional tool for identifying variants that are positioned in eQTLs for various traits. Pairing eQTL analysis with association results to identify target genes is a powerful tool for functional follow-up because nonspecific enhancer activity assays using weak promoters are generally meaningful, but less compelling than activity assays measuring the specific interactions between an enhancer and the natural target promoter. As such, knowledge of the target gene allows for stronger experimental design before entering the lab.
Resources for Annotation of Variants
In addition to the difficulty of predicting regulatory effects of sequence variation, follow-up of cancer risk loci is complicated by the statistical correlation between nearby variants, a phenomenon referred to as linkage disequilibrium (LD). Since genetic architecture differs among populations, the resolution of GWAS results is defined by the LD structure of the studied population. Since the vast majority of cancer loci were originally discovered in populations of European descent, there are often a very large number of potential functional variants tagged by the index variant. As such, the first step in bioinformatic follow-up of functional loci is to identify all variants tagged by the index variant in the population from which the variant was discovered.
There are now many databases that catalog human variation across populations including 1000 Genomes Project,35 UK10 K project (www.uk10k.org), and a similar effort initiated in Saudi Arabia to sequence the genomes of up to 100,000 individuals (Saudi human genome program http://rc.kfshrc.edu.sa/sgp/). These catalogs can be used to identify variants that are in LD with the initial index SNP, and LD can be retrieved through various resources including Haploview36 or HaploReg19 from the Broad Institute, and SNAP.23 In addition to obtaining LD information, Haploview36 and SNAP23 allow visualization of LD across different populations through two different regional plots. Among these resources, HaploReg provides LD information for the following super populations: Europeans (EUR), Africans and African Americans (AFR), East Asians (ASN), and Hispanics (AMR) from the 2011 release of the 1000 Genomes Project. If a more specific population is desired, then Haploview is the most appropriate among these three. SNAP calculates LD in CEU, YRI, CHB/JPT populations but is restricted to 500 kb. These resources should be consulted for the identification of the bin of tagged SNPs for a given GWAS association. Early maps of common variation (the HapMap) may only have cataloged a portion of the variants within a bin, so it is advisable to consult the more recent sequencing data in order to more comprehensively identify the panel of candidate functional variants within a bin.
Annotation of variant files
As described above, there are many different datasets that can be used to annotate variants. In addition, there are many resources that have been developed to take lists of candidate functional SNPs (variant files) and integrate a large set of functional annotations for all SNPs in the file. Annotation files are typically formatted as bed track files (http://genome.ucsc.edu/FAQ/FAQformat.html#format1) for the desired functional annotations and in variant call format (VCF) (http://samtools.github.io/hts-specs/VCFv4.1.pdf) for the variants to be annotated. The functional annotation bed files can be downloaded directly from the UCSC Table Browser37 or through the MySQL site (genome-mysql.cse.ucsc.edu). There are many tools designed to annotate, visualize, or manipulate VCF files such as VCFtools,38 Annovar,39 and AnnTools.40 Other programs, such as HaploReg,19 can take lists of SNP identifiers to annotate variants with regulatory annotations in the attempt to streamline bioinformatics follow-up of GWAS loci. HaploReg takes the input variant(s), generates a list of correlated variants with the input variants, and then annotates all listed variants with pre-loaded ENCODE, Roadmap, or expression data. Recently, a bioinformatics tool called Enlight (http://enlight.usc.edu/) was developed to help identify causal variants by taking regional plots from GWAS results and overlaying biofeatures such as epigenetic modification, DHS, and TFBS. One limitation of both HaploReg and Enlight is that the meta-collection available from these sites reflects only a limited proportion of the available functional annotations, which may not be relevant to the disease of interest, and may or may not be the most updated version of the dataset. Furthermore, to make these resources usable, the LD calculation may not be in the most relevant population, and the calculation may not encompass a large enough region of the genetic architecture at that particular locus.
A relatively new annotation tool, GWAS3D (http://jjwanglab.org/gwas3d), is a web server that combines chromatin interaction data (5C, Hi-C, ChIA-PET) with histone modification, TF binding (ChIP-seq), and ChromHMM segmentation in 16 cell types from ENCODE to predict the probability that a genetic variant affects regulatory pathways. To predict functional variants, GWAS3D then looks to differences in binding affinity for the predicted regulatory factors. This information is then used to annotate GWAS signals, taking into consideration both LD structure of super populations and effect estimates, to aid in the interpretation of results. Input data can be VCF or PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) files, and the implementation is straightforward. A potential limitation of this resource is that it is not currently made tissue specific. As such, it is possible that this resource could miss regulatory elements that are tissue specific and related only to certain cancers. However, this restriction is reflective of the scarcity of chromosomal interaction data for many tissues and cell types.
Cancer-specific annotations
The annotations described thus far have focused on the annotation of noncoding germline variation to predict causal variants within susceptibility loci. These annotations can be generalized to functional annotation of associations for any disease or trait. However, there are also a few cancer-specific resources. COSMIC41 and TCGA (https://tcga-data.nci.nih.gov/tcga/) focus on acquired somatic variants in cancer and how to detect those that play a significant role in tumor development, proliferation, or treatment response.42,43 Genes that are frequently amplified, deleted, dysregulated, or somatically mutated in the cancer of interest can be prioritized as potential regulatory targets for nearby index SNP associations. Similar to eQTL and chromosome capture analyses, knowledge of the target gene for a candidate regulatory enhancer can be used to design stronger functional analysis experiments, as shown for the MYC region and rs6983267 in colon cancer.44,45 In order to identify novel driver mutations, several large-scale sequencing projects of tumor tissue have commenced, such as TCGA. Many of the available annotations for identifying driver mutations relate somatic mutations to expression data, or leverage existing knowledge about functional mutations, cancer genes, or associated pathways. A comprehensive list of available programs and algorithms for distinguishing driver mutations was provided in a recent publication by Zhang et al.46 Many of the tools focus on annotation of the variants themselves using measurements of deleteriousness (eg, PolyPhen2). However, these tools also include methods that compare the genes harboring aberrations to known cancer genes in order to predict novel drivers. More sophisticated algorithms consider mutational profiles or background rates for passenger mutations or use machine learning methods to train a classifier on a known set of driver and passenger mutations, which can be collected from resources like the COSMIC database,41 and then apply those predictors to a new sequencing dataset (eg, Driver Mutation Identification [DMI],47 Cancer-Specific High-throughput Annotation of Somatic Mutations [CHASM],48 and Screening for Non-Acceptable Polymorphism [SNAP]).49 Many of the annotations that specifically try to identify driver mutations are based on the hypothesis that a driver gene would accumulate a greater number of mutations than passenger genes. Major limitations of these tools include differences in predictions based on the method used, and that passenger mutations can assume the role of driver mutations given a change in the environment, such as targeted chemotherapy.46 In addition, it is now appreciated that there is a great deal of tumor heterogeneity, both intratumor (within a single tumor) and intertumor (between tumors) that make it difficult to generalize parameters trained on a limited number of examples.
Visualization of variant annotations
Databases used to visualize annotated variants include the UCSC Genome Browser,50 the UCSC Cancer Genome Browser,51 and as described earlier, Enlight. The UCSC Cancer Genome Browser currently displays a growing catalog of data, including 201 datasets from 22 TCGA (The Cancer Genome Atlas) cancers as well as data from Cancer Cell Line Encyclopedia and Stand Up To Cancer. This database allows samples to be examined by common clinical features such as therapeutic response or by genomic signatures that predict drug response. Genomic data can be viewed by genes, which allows users to easily see functional changes to the genome as well as examine trends across pathways of genes. Several statistical tools are available to test quantitative differences in expression as well. Additionally, the Tumor Image Viewer, based on Google Maps, allows users to interactively view slides of tumor tissue samples. The UCSC Genome Browser contains the ENCODE, Roadmap, and many other functional data-sets, which can be used to identify variants that could impact splicing, transcription, translational, and post-translational regulation of gene expression. After identifying a list of strong functional candidates it is often useful to upload a bed file containing potentially functional variants underlying GWAS signal to visualize their proximity to genes and their positioning relative to segments of the genome likely to have regulatory potential.
Proposed Bioinformatics Framework
Given the volume of annotations and resources presently available, it is useful to establish a bioinformatics framework to guide functional evaluation of noncoding variants associated with cancer risk. As such, a framework is presented below as a general approach for conducting such analyses.
To follow up an index SNP that has been associated with cancer, it is first necessary to generate a list of variants tagged by the index SNP in the studied population. There are now a number of public resources available to achieve this (such as HaploReg). After obtaining a list of correlated variants, it is useful to create a custom bed file to visualize these variants in the UCSC Genome Browser. In Figure 2 the bin of variants associated with an index SNP (rs6983267) for colorectal cancer risk (P = 5 × 10−11)52 is smaller than most containing only three SNPs. This bin of variants is positioned 300 kb upstream of the protooncogene MYC. It is not unusual for variants to be located in intergenic regions that are tens of thousands of base pairs away from the closest gene (see Table 2) making it difficult to predict the potential target gene. The cancer genome browser (https://genome-cancer.ucsc.edu/)51 and the Online Mendelian Inheritance in Man (OMIM)53 are useful resources for identifying biologically plausible genes. For instance, an OMIM search of MYC reveals that its gene product plays a key role in cell proliferation, differentiation, transformation, and apoptosis. Although this locus is now well established as an enhancer for MYC, in many instances the target is less clear. For example, two different breast cancer loci have been followed up in laboratory, and functional variants, rs691357854 and rs4784227,55 were found to impact reporter protein expression but have unknown targets. Further chromosome capture in relevant tissue could provide substantial insight to help translate these loci into better clinical targets.
Table 2.
Examples of variants with laboratory follow-up.
| VARIANT | REGULATORY REGION | GENE | TRAIT | PUBLICATION | YEAR | BEFORE GWAS? |
|---|---|---|---|---|---|---|
| rs6983267 | Distal Enhancer 300kb downstream | MYC | Colorectal Cancer | Pomerantz et al., Nat Genet56 | 2009 | Follow-up |
| rs16888589 | Enhancer 20kb downstream | EIF3H | Colorectal Cancer | Pittman et al., PLoS Gen85 | 2010 | Follow-up |
| rs10822013 | Intronic | ZNF365 | Breast Cancer | Cai et al., Hum Mol Genet86 | 2011 | Follow-up |
| rs2735940 | Promoter | TERT | Telomere Length | Matsubara et al, Bichem Biophys Res Commun | 2006 | Pre-GWAS |
| rs2735940 | Promoter | TERT | Acute Lymphoblastic Leukemia (ALL) | Sheng et al., Carcinogenesis69 | 2013 | Follow-up |
| rs2736108 | Promoter | TERT | Ovarian and Breast Cancer | Beesley et al., PLoS One87 | 2011 | Follow-up |
| rs1512268 | Enhancer 14kb downstream | NKX3.1 | Prostate Cancer | Akamatsu et al., Hum Mol Genet88 | 2010 | Follow-up |
| rs6913578 | Regulates unknown target | Unknown | Breast Cancer | Cai et al., Cancer Res54 | 2011 | Follow-up |
| rs4784227 | Possible enhancer of TOX3, 18.4 kb upstream | Unknown | Breast Cancer | Long et al., PLoS Genet55 | 2010 | Follow-up |
| rs2239632 | Promoter | CEBPE | ALL | Ryoo et al., J Hum Genet89 | 2013 | Follow-up |
| rs11730582 | Promoter | OPN | Gastric Cancer | Zhao et al., BMC Cancer90 | 2012 | Follow-up |
| rs11730582 | Promoter | OPN | Melanoma | Schultz et al., Mol Carcinog91 | 2009 | Follow-up |
| rs4590952 | Intronic | KITLG | Testicular Cancer | Zeron-Medina et al., Cell92 | 2013 | Follow-up |
| rs944289 | Enhancer | PTCSC3 (lincRNA) | Papillary thyroid carcinoma (PTC) | Jendrzejewski et al., Proc Natl Acad Sci93 | 2012 | Follow-up |
| rs1859961 | Distal Enhancer 1Mb upstream | SOX9 | Prostate Cancer | Zhang et al., Genome Res94 | 2012 | Follow-up |
| rs12194974 | Promoter | LIN28B | Ovarian Cancer | Permuth-Wey et al., Cancer Res95 | 2011 | Follow-up |
| rs8506 | miRNA binding in exon | lincRNA-NR_024015 | Gastric Cancer | Fan et al., PLoS One96 | 2014 | Follow-up |
| rs10993994 | Promoter | MSMB | NA | Buckland et al., Hum Mutat70 | 2005 | Pre-GWAS |
| rs10993994 | Promoter | MSMB | Prostate Cancer | Chang et al., Hum Mol Genet71 | 2009 | Follow-up |
| rs10993994 | Promoter | MSMB | Prostate Cancer | Lou et al., Proc Natl Acad Sci72 | 2009 | Follow-up |
Bioinformatics follow-up of loci positioned in intronic or intergenic regions can be conducted within the UCSC Genome Browser, or another annotation tool. After aligning variants of interest to the reference genome, it is useful to annotate the region with biofeatures (ChIP-seq and DHS) assayed in relevant tissues and cell lines from Roadmap and ENCODE, respectively. Figure 2 illustrates that unlike the other two tagged variants (rs10505477 and rs12682374), the index SNP (rs6983267) aligns to a region of open chromatin with promoter histone modifications in normal colon and rectal mucosa. Furthermore, rs6983267 is located within a TFBS for TCF4 making this a likely functional candidate for laboratory follow-up. Figure 3 illustrates that although the ChIP-seq signal was quite broad, DHS footprinting identifies a much smaller region that demarks the true underlying binding site for TCF4 and that rs6983267 falls in the center of that footprint. ChromHMM segmentation in normal colon and rectal mucosa illustrates the ease of interpretation of the annotation, labeling the region harboring rs6983267 as an enhancer in rectal mucosa. However, the variant misses this region in colon tissue. This variant has now been confirmed as causal in the laboratory, with the risk allele decreasing the binding affinity of TCF4.56 For variants within UTRs one can look for miRNA binding sites using the TargetScanS 5.2 track within the UCSC Genome Browser. Variants falling near splice sites can be annotated using a number of different tools including BDGP, ESE finder,57 RescueESE,58 ESRSearch,59 PESX,60 or Ensembl.61
Limitations of Existing Resources
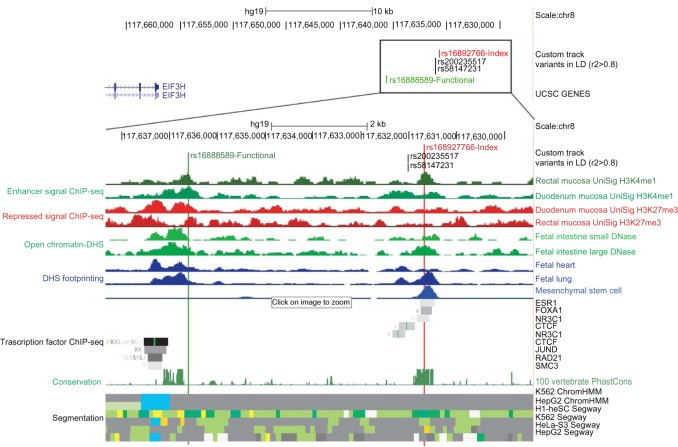
In noncoding regions, the majority of functional predictions to date inform whether a polymorphism is positioned in a region that is likely to be biologically relevant based on the presence of one or more biochemical signals. As such, with the exception of CADD and PWM motif analysis, noncoding annotations do not predict the effects of a specific allele. Furthermore, even when a variant has a particularly strong bioinformatics annotation relative to other tagged SNPs, laboratory follow-up of multiple variants is often required. For instance, Figure 4 outlines an example where the variant with the strongest bioinformatics annotation (rs16892766) is not the same variant shown to have allelic effects on gene expression in vitro. The index tagSNP rs16892766, which is strongly associated with colorectal cancer (1 × 10−18),62–67 is located in an intergenic region over 140 kb telomeric of the promoter for the closest gene EIF3H. The bioinformatics annotation of this locus shown in Figure 4 suggests that rs16892766 is positioned in a poised enhancer in colon and rectal tissues and falls within a DHS footprint that corresponds to ChIP-seq binding sites for ESR1, FOXA1, and NR3C1. Alternatively, rs16888589 is in strong LD with rs16892766 (r2 = 0.89 in 1000 Genomes Project EUR population), has similar enhancer signals, but does not fall within a binding site for an assayed TF. Fine-mapping of both variants through in vitro and in vivo analysis suggested that in human colorectal cancer cell lines, the ancestral allele rs16888589-A significantly repressed gene expression while there were no allelic effects observed with rs16892766. The authors also reported that rs16888589 interacts with the promoter of EIF3H, as shown through chromatin capture techniques. Interestingly, increased expression of this gene was shown to increase colorectal cancer growth and invasiveness. Thus, despite the relatively stronger bioinformatic annotation for the index SNP, another predicted functional SNP, rs16888589, was shown to be the likely causal variant. This illustrates the importance of laboratory follow-up for multiple variants, particularly when there is more than one putative functional variant predicted.
Figure 4.
Limitation of bioinformatics annotation.
Notes: Figure 4 outlines an example where the variant with the strongest annotation (shown in red) is not the underlying functional variant (shown in green). The rs16892766 locus is located in an intergenic region over 20 kb downstream of EIF3H. Although both rs16888589 and rs16892766 fall in a poised enhancer based on histone modification, DHS, ChromHmm segmentation, and footprinting evidence, only the index rs16892766 falls in the highly conserved ChIP-seq binding site for the three shown TFs. Laboratory follow-up of this variant revealed that the true underlying causal variant is rs16888589 and not the index, although the index had the stronger bioinformatic annotation illustrating the importance of laboratory follow-up, particularly when there exists more than one predicted functional variant.
Although higher throughput functional assays might increase the production of training data to improve predictions on the functionality of noncoding variants, it is possible that a great deal of pre-GWAS knowledge of functional variation is lost in the literature. Table 2 outlines several examples of variants that were confirmed to be functional through in vitro analysis after the initial association was detected in GWAS. However, this table also reveals that several known functional variants were subsequently investigated as a follow-up of a GWAS finding without reference to the original experiment. For instance, the TERT promoter variant, rs2735940, was shown to have functional effects on gene expression in 2006,68 and then again after a recent GWAS finding for acute lymphoblastic leukemia in 2013.69 Similarly, the MSMB promoter variant, rs10993994, which is associated with prostate cancer, has recently been investigated in the laboratory three times, while the original experiment showing the allelic effects of rs10993994 was conducted in 2005.70–72 A significant challenge for extracting functional evidence from the literature is that prior to GWAS, promoters (rarely enhancers) may have been investigated because the gene was interesting for a trait that may be unrelated to the subsequent disease or trait association detected through GWAS. These examples indicate that there is a need to more effectively link the laboratory investigations of allelic variants to GWAS findings in order to avoid redundant efforts.
Future Directions
Advances in sequencing technology have enabled genetic association studies of complex diseases to test rare variant hypotheses in both coding and noncoding regions. Unlike traditional genome-wide approaches, which make use of markers to capture bins of correlated SNPs, sequencing-based approaches identify a much larger number of rare variants, significantly reducing the power of a single variant approach. To address this loss in power, aggregate association tests such as burden and variance-component analyses have been developed to effectively collapse the genome into smaller units of analysis, and decrease the multiple testing penalty. Each method collapses a large number of infrequent observations into a much smaller set of testing units. In the exome, a gene serves as a natural functional unit for combining rare variants. However, segmenting intergenic space into functional units is more difficult because enhancers work in concert to regulate one or more target genes. Although chromatin capture technology can provide substantial insight into how distal regions interact, these assays are only available for a limited number of tissues and cell types because of the associated cost, and there remains a large number of nonfunctional predicted interactions. Since rare variants are intermittently scattered across intergenic space, aggregation within a single enhancer unit, as defined through segmentations like ChromHMM or Segway, is not feasible since each element is likely to contain only a single variant. Furthermore, the resolution of ChIP-seq is on the order of thousands of nucleotides and is therefore far too broad to distinguish between variants closely positioned on the chromosome. Although the additional requirement of DHS overlap can refine this signal to a few hundred nucleotides, the underlying binding motif for functional machinery (TFs) is about 10 nucleotides (5–31 nucleotides, with mean 9.9 nucleotides for eukaryotes).73 Furthermore, these annotations do not provide insight into whether the minor allele of the polymorphism would impact the binding of the TF. As such, in aggregate testing methods, it is difficult to prioritize likely functional variants over those less likely to be functional in the same testing unit. Since this is an important component for improving power in aggregate testing, this is a significant limitation that will need to be addressed.
Moving forward it will be important to develop more robust and high-throughput methods to identify both functional and nonfunctional allelic changes in motif sequences and their relative impact on the expression of a target. The generation of these data will lead to improved training data-sets, as is currently available for nsSNPs. However, it will also be important to create a better reporting system to link this information into a publicly available resource to avoid redundant efforts and maximize the utility of these more onerous experiments. Although there is now a wealth of information on the functional potential of noncoding regions, there remains much to be discovered on how these regions interact, and the impact of regulatory variation.
Given the time and expense associated with laboratory follow-up, in silico prediction remains a critical step for generating testable hypotheses. The recognition of these functional, regulatory polymorphisms is becoming feasible as datasets annotating evidence for functional noncoding regions across the genome accrue. Hopefully, this will facilitate the identification of the actual variant(s) in each region associated with disease. In the short term, genetic risk models based on functional variants rather than index tagSNPs should yield more precise genetic risk models, as well as facilitate the translation of GWAS associations into therapeutic targets. In the longer term, as methods for the prediction of regulatory function improve, these methods might be used to identify less frequent regulatory polymorphisms associated with cancer, even when such variants are too rare for statistical correlations in reasonably sized populations, and thereby account for some of the missing heritability of these diseases.
Footnotes
ACADEMIC EDITOR: JT Efird, Editor in Chief
FUNDING: This work was supported by the National Institutes of Health [U19 CA148107] and the National Cancer Institute [UO1 CA164930]. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants. Provenance: the authors were invited to submit this paper.
Author Contributions
SR wrote the first draft of the manuscript; CC contributed to the writing of the manuscript; PA made critical revisions and approved the final version. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Hindorff LA, MacArthur J, European Bioinformatics Institute. Morales J, European Bioinformatics Institute et al. A catalog of published genome-wide association studies. 2013. Available at www.genome.gov/gwastudies.
- 2.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawabata T, Ota M, Nishikawa K. The protein mutant database. Nucleic Acids Res. 1999;27(1):355–7. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giardine B, Riemer C, Hefferon T, et al. PhenCode: connecting ENCODE data with mutations and phenotype. Hum Mutat. 2007;28(6):554–62. doi: 10.1002/humu.20484. [DOI] [PubMed] [Google Scholar]
- 6.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gromiha MM, An J, Kono H, et al. ProTherm, version 2.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 2000;28(1):283–5. doi: 10.1093/nar/28.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prabakaran P, An J, Gromiha MM, et al. Thermodynamic database for protein-nucleic acid interactions (ProNIT) Bioinformatics. 2001;17(11):1027–34. doi: 10.1093/bioinformatics/17.11.1027. [DOI] [PubMed] [Google Scholar]
- 9.ENCODE Project Consortium. Bernstein BE, Birney E, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chadwick LH. The NIH roadmap epigenomics program data resource. Epigenomics. 2012;4(3):317–24. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu H, Fang X. A brief review on the human encyclopedia of DNA elements (ENCODE) project. Genomics Proteomics Bioinformatics. 2013;11(3):135–41. doi: 10.1016/j.gpb.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12(4):283–93. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vernot B, Stergachis AB, Maurano MT, et al. Personal and population genomics of human regulatory variation. Genome Res. 2012;22(9):1689–97. doi: 10.1101/gr.134890.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathelier A, Zhao X, Zhang AW, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42(Database issue):D142–7. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32(Web Server issue):W249–52. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karchin R, Diekhans M, Kelly L, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21(12):2814–20. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 21.Capriotti E, Calabrese R, Fariselli P, Martelli PL, Altman RB, Casadio R. WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics. 2013;14(suppl 3):S6. doi: 10.1186/1471-2164-14-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21(14):3176–8. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 23.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9(5):473–6. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107(1):30–9. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 29.Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3(1):17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 30.Simonis M, Klous P, Splinter E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38(11):1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Z, Tavoosidana G, Sjölinder M, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38(11):1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 32.Dostie J, Richmond TA, Arnaout RA, et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16(10):1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goh Y, Fullwood MJ, Poh HM, et al. Chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) for mapping chromatin interactions and understanding transcription regulation. J Vis Exp. 2012;62(ii):3770. doi: 10.3791/3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.1000 GenomesProject Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.Karolchik D, Hinrichs AS, Furey TS, et al. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–6. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makarov V, O’Grady T, Cai G, Lihm J, Buxbaum JD, Yoon S. AnnTools: a comprehensive and versatile annotation toolkit for genomic variants. Bioinformatics. 2012;28(5):724–5. doi: 10.1093/bioinformatics/bts032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331(6024):1553–8. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 44.Freedman ML, Monteiro AN, Gayther SA, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43(6):513–8. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahmadiyeh N, Pomerantz MM, Grisanzio C, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A. 2010;107(21):9742–6. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Liu J, Sun J, Chen C, Foltz G, Lin B. Identifying driver mutations from sequencing data of heterogeneous tumors in the era of personalized genome sequencing. Brief Bioinform. 2014;15(2):244–55. doi: 10.1093/bib/bbt042. [DOI] [PubMed] [Google Scholar]
- 47.Tan H, Bao J, Zhou X. A novel missense-mutation-related feature extraction scheme for ‘driver’ mutation identification. Bioinformatics. 2012;28(22):2948–55. doi: 10.1093/bioinformatics/bts558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carter H, Samayoa J, Hruban RH, Karchin R. Prioritization of driver mutations in pancreatic cancer using cancer-specific high-throughput annotation of somatic mutations (CHASM) Cancer Biol Ther. 2010;10(6):582–7. doi: 10.4161/cbt.10.6.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24(20):2397–8. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karolchik D, Barber GP, Casper J, et al. The UCSC genome browser database: 2014 update. Nucleic Acids Res. 2014;42(Database issue):D764–70. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldman M, Craft B, Swatloski T, et al. The UCSC cancer genomics browser: update 2013. Nucleic Acids Res. 2013;41(Database issue):D949–54. doi: 10.1093/nar/gks1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters U, Jiao S, Schumacher FR, et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology. 2013;144(4):799–807.e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johns Hopkins University (Baltimore, MD) McKusick-Nathans Institute of Genetic Medicine. Online Mendelian Inheritance in Man. 2014. OMIM Web site: http://omim.org/2014.
- 54.Cai Q, Wen W, Qu S, et al. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of Chinese, Japanese, and European ancestry. Cancer Res. 2011;71(4):1344–55. doi: 10.1158/0008-5472.CAN-10-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long J, Cai Q, Shu XO, et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia breast cancer consortium. PLoS Genet. 2010;6(6):e1001002. doi: 10.1371/journal.pgen.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41(8):882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–71. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci U S A. 2004;101(44):15700–5. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297(5583):1007–13. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 60.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol Cell Biol. 2005;25(16):7323–32. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hubbard TJ, Aken BL, Beal K, et al. Ensembl 2007. Nucleic Acids Res. 2007;35(Database issue):D610–7. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomlinson IP, Carvajal-Carmona LG, Dobbins SE, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7(6):e1002105. doi: 10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39(5):645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 64.Cui R, Okada Y, Jang SG, et al. Common variant in 6q26-q27 is associated with distal colon cancer in an ASIAn population. Gut. 2011;60(6):799–805. doi: 10.1136/gut.2010.215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 66.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40(3):316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 67.Schumacher FR, Berndt SI, Siddiq A, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20(19):3867–75. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsubara Y, Murata M, Yoshida T, et al. Telomere length of normal leukocytes is affected by a functional polymorphism of hTERT. Biochem Biophys Res Commun. 2006;341(1):128–31. doi: 10.1016/j.bbrc.2005.12.163. [DOI] [PubMed] [Google Scholar]
- 69.Sheng X, Tong N, Tao G, et al. TERT polymorphisms modify the risk of acute lymphoblastic leukemia in chinese children. Carcinogenesis. 2013;34(1):228–35. doi: 10.1093/carcin/bgs325. [DOI] [PubMed] [Google Scholar]
- 70.Buckland PR, Hoogendoorn B, Coleman SL, Guy CA, Smith SK, O’Donovan MC. Strong bias in the location of functional promoter polymorphisms. Hum Mutat. 2005;26(3):214–23. doi: 10.1002/humu.20207. [DOI] [PubMed] [Google Scholar]
- 71.Chang BL, Cramer SD, Wiklund F, et al. Fine mapping association study and functional analysis implicate a SNP in MSMB at 10q11 as a causal variant for prostate cancer risk. Hum Mol Genet. 2009;18(7):1368–75. doi: 10.1093/hmg/ddp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lou H, Yeager M, Li H, et al. Fine mapping and functional analysis of a common variant in MSMB on chromosome 10q11.2 associated with prostate cancer susceptibility. Proc Natl Acad Sci U S A. 2009;106(19):7933–8. doi: 10.1073/pnas.0902104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stewart AJ, Hannenhalli S, Plotkin JB. Why transcription factor binding sites are ten nucleotides long. Genetics. 2012;192(3):973–85. doi: 10.1534/genetics.112.143370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cingolani P, Platts A, Wang Le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso−3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382–8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE data in the UCSC genome browser: year 5 update. Nucleic Acids Res. 2013;41(Database issue):D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS. RNAdb 2.0–an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35(Database issue):D178–82. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karolchik D, Hinrichs AS, Kent WJ. The UCSC genome browser. Curr Protoc Bioinformatics. 2012 doi: 10.1002/0471250953.bi0104s40. Chapter 1:Unit1.4. [DOI] [PubMed] [Google Scholar]
- 82.GTEx Consortium The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Felsenstein J, Churchill GA. A hidden Markov model approach to variation among sites in rate of evolution. Mol Biol Evol. 1996;13(1):93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- 84.Tuupanen S, Turunen M, Lehtonen R, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41(8):885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 85.Pittman AM, Naranjo S, Jalava SE, et al. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet. 2010;6(9):e1001126. doi: 10.1371/journal.pgen.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cai Q, Long J, Lu W, et al. Genome-wide association study identifies breast cancer risk variant at 10q21.2: results from the Asia breast cancer consortium. Hum Mol Genet. 2011;20(24):4991–9. doi: 10.1093/hmg/ddr405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beesley J, Pickett HA, Johnatty SE, et al. Functional polymorphisms in the TERT promoter are associated with risk of serous epithelial ovarian and breast cancers. PLoS One. 2011;6(9):e24987. doi: 10.1371/journal.pone.0024987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Akamatsu S, Takata R, Ashikawa K, et al. A functional variant in NKX3.1 associated with prostate cancer susceptibility down-regulates NKX3.1 expression. Hum Mol Genet. 2010;19(21):4265–72. doi: 10.1093/hmg/ddq350. [DOI] [PubMed] [Google Scholar]
- 89.Ryoo H, Kong M, Kim Y, Lee C. Identification of functional nucleotide and haplotype variants in the promoter of the CEBPE gene. J Hum Genet. 2013;58(9):600–3. doi: 10.1038/jhg.2013.62. [DOI] [PubMed] [Google Scholar]
- 90.Zhao F, Chen X, Meng T, Hao B, Zhang Z, Zhang G. Genetic polymorphisms in the osteopontin promoter increases the risk of distance metastasis and death in Chinese patients with gastric cancer. BMC Cancer. 2012;12:477. doi: 10.1186/1471-2407-12-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schultz J, Lorenz P, Ibrahim SM, Kundt G, Gross G, Kunz M. The functional -443T/C osteopontin promoter polymorphism influences osteopontin gene expression in melanoma cells via binding of c-Myb transcription factor. Mol Carcinog. 2009;48(1):14–23. doi: 10.1002/mc.20452. [DOI] [PubMed] [Google Scholar]
- 92.Zeron-Medina J, Wang X, Repapi E, et al. A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell. 2013;155(2):410–22. doi: 10.1016/j.cell.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jendrzejewski J, He H, Radomska HS, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A. 2012;109(22):8646–51. doi: 10.1073/pnas.1205654109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang X, Cowper-Sal lari R, Bailey SD, Moore JH, Lupien M. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Res. 2012;22(8):1437–46. doi: 10.1101/gr.135665.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Permuth-Wey J, Kim D, Tsai YY, et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011;71(11):3896–903. doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fan QH, Yu R, Huang WX, Cui XX, Luo BH, Zhang LY. The has-miR-526b binding-site rs8506G.a polymorphism in the lincRNA-NR_024015 exon identified by GWASs predispose to non-cardia gastric cancer risk. PLoS One. 2014;9(3):e90008. doi: 10.1371/journal.pone.0090008. [DOI] [PMC free article] [PubMed] [Google Scholar]