Abstract
Background
Mounting evidence shows that microRNAs may be useful as prognostic biomarkers of gastric cancer. The aim of this meta-analysis was to summarize the predictive role of miR-21 for survival in patients with gastric cancer and to verify the association between expression of miR-21 and clinical characteristics.
Material/Methods
All the eligible studies were searched by PubMed and EMBASE and clinical characteristics and survival results were extracted. Then a meta-analysis was carried out to clarify the prognostic role of the miR-21 expression in different subgroups.
Results
We included 8 studies dealing with gastric cancer in this meta-analysis. For overall survival, the pooled hazard ratio of higher miR-21 expression in tumor tissue was 2.00 (95% CI: 1.39–2.88, P<0.01), which could significantly predict poorer survival in gastric cancer patients. Importantly, subgroup analysis suggested that higher expression of miR-21 correlated with tumor differentiation 0.42 (95% CI: 0.25–0.70 p<0.01), lymph node metastasis 6.39(95% CI: 3.11–13.14, P<0.01), and TNM stage 0.38 (95% CI: 0.21–0.67, P<0.01).
Conclusions
This meta-analysis indicates that miR-21 detection has a prognostic value in patients with gastric cancer. In addition, overexpression of miR-21 is associated with worse tumor differentiation, lymph node metastasis, and TNM stage.
MeSH Keywords: MicroRNAs, Prognosis, Stomach Neoplasms
Background
Gastric cancer is the fourth most commonly diagnosed cancer and the second leading cause of cancer death worldwide. Incidence rates (per 100 000) vary from 3.3 in men and 2.0 in women in Egypt to 65.9 in men and 25.9 in women in Korea [1,2]. In China, there are nearly 460 000 new cases of gastric cancer and 350 000 deaths each year [3]. Despite many advances in GC therapy, the prognosis for patients with gastric cancer remains poor and the 5-year survival rate is 5–20%. Therefore, there is an increasingly urgent need to find prognostic markers that can accurately display biological features of GC and predict the clinical results to heighten the clinical management of GC patients.
MicroRNAs (miRNAs) are a large family of about 22-nucleotide-long RNAs that play important roles by targeting mRNAs as key post-transcriptional regulators of gene expression [4–7]. Mounting evidence suggests that some miRNAs may function as oncogenes or tumor suppressors by regulating cell proliferation and other related biological behaviors [8–10].
MicroRNA-21 (miR-21) is one of the most widely studied oncogenic miRNAs [4,11]. Many studies have investigated the role of miR-21 in the prognosis of GC, with varied results. This study aimed to summarize the evidence for the usefulness of miR-21 in predicting the clinical outcomes of GC patients. We also evaluated the overall risk of raised miR-21 for survival in patients with GC.
Material and Methods
Search strategy
We searched PubMed, Web of Science, and EMBASE from 1966 to 30 February 2014 to identify relevant studies. The search used the following keywords: ‘microRNA- 21’ or ‘miR-21’ and ‘gastric cancer’ or ‘stomach cancer’ and ‘miRNAs and gastric cancer prognosis.’ Studies were considered eligible if they met the following criteria: (I) studied patients with gastric cancer; (II) measured expression of miR-21 in tissue or serum; and (III) investigated the survival outcome or the correlation between miR-21 expression and the clinical variables. Articles were excluded based on the following criteria: (I) review articles or letters, (II) non-English articles, (III) lacked key information for calculation with methods developed by Parmar, Williamson, and Tierney [12–14].
Data extraction
Eligible articles were reviewed independently by 2 investigators (Zhenqiang Wang and Qiang Cai). Any disagreement on a controversial study was resolved by discussion and consensus. Multivariate Cox hazard regression analysis reported in the article was included in our analysis; if these data were not available, we extracted univariate Cox hazard regression analysis or log-rank p value and Kaplan-Meier survival curves of survival outcomes instead. The above primary information was extracted by 2 investigators working independently. Additional data extracted from the studies included first author, publication year, study size, patient age and sex, TNM stage, lymph node metastasis, histological classification, methods used to detect miR-21, positive miR-21 definition, the conclusion, and other clinical characteristics.
Statistical methods
All these HRs and 95% confidence interval (CI) were calculated following Tierney’s method [13] and the logHR and SE (logHR) (SE) were used for aggregation of the survival results, but these statistical variables were not directly stated in most studies. We calculated the necessary statistics on the basis of available numerical data with methods developed by Parmar, Williamson, and Tierney [13,14]. We also investigate the correlation between miR-21 expression and the clinical variables, including TNM stage, lymph node metastasis, and histological type. According to clinical characteristics, Stage I and Stage II were combined and Stage III and Stage IV were combined. Odds ratio (OR) was used as the measure.
Heterogeneity of combined HRs was conducted using Cochran’s Q test and Higgins I-squared statistic. Heterogeneity was defined as p<0.10 or I2>50%. A random effects model was used if heterogeneity was observed (P<0.10), and the fixed effects model was used in the absence of between-study heterogeneity (P≥0.1) [15]. Publication bias was evaluated using the funnel plot. Forrest plots were used to estimate the effect of miR-21 expression on survival outcome and the correlation between miR-21 expression and the clinical variables. All of the above calculations were performed using RevMan 5.2 (Cochrane collaboration, Oxford, UK) software.
Results
Characteristics of the eligible studies
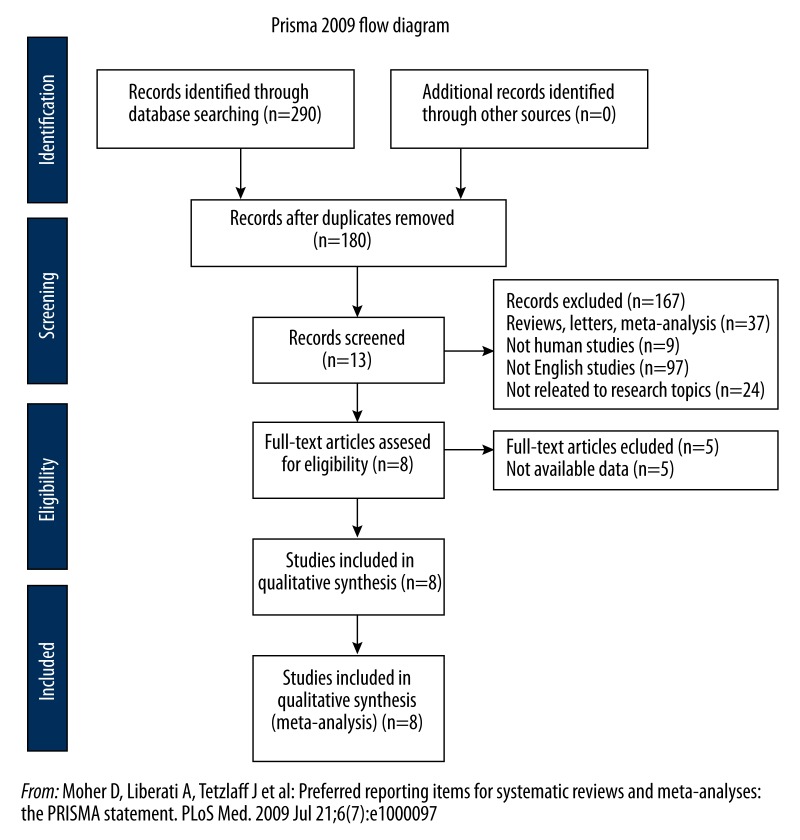
We initially searched 180 studies in PubMed and EMBASE. With the review of these abstracts, 13 potentially relevant studies were identified as eligible for full-text review. Five studies were excluded because they lacked the data necessary for calculation. Finally, 8 studies were identified according to the inclusion criteria of the meta-analysis (Figure 1). These eligible studies were published from 2008 to 2013 and included a total of 523 patients. Among all eligible studies, 5 studies [16–20] were used to analyze the correlation between miR-21 and overall survival rate and 5 studies [17,19,21–23] were chosen to demonstrate the connection of miR-21 expression and clinical features. The main features of eligible studies are summarized in Table 1.
Figure 1.
Selection of studies.
Table 1.
Clinical characteristics of included studies.
| Author | Publication year | Origin of population | Number of patient | Sample | TNM stage | miR-21 assay | Cut-off | Survival analysis | Hazard ratios |
|---|---|---|---|---|---|---|---|---|---|
| Soshi Osawa | 2011 | Japan | 37 | Tumor tissue | I–IV | qRT-PCR | Median | OS | Reported |
| Jianning Song | 2013 | China | 103 | Serum | I–IV | qRT-PCR | Median | OS | Reported |
| Jingting Jiang | 2011 | China | 56 | Tumor tissue | I–IV | qRT-PCR | Median | OS | Reported |
| Yuejuan Xu | 2012 | China | 86 | Tumor tissue | I–IV | qRT-PCR | Median | OS | Reported |
| Shin-hsuan Chan | 2008 | China | 37 | Tumor tissue | I–IV | qRT-PCR | 2-fold | – | Reported |
| Kazuo Motoyama | 2010 | Japan | 105 | Tumor tissue | I–IV | qRT-PCR | Median | – | – |
| Baogui Zhang | 2011 | China | 30 | Tumor tissue | I–IV | qRT-PCR | Median | – | – |
| Kamastu.S | 2013 | Japan | 69 | Tumor tissue | I–IV | qRT-PCR | Median | OS | Reported |
High expression of miR-21 predicts poor survival
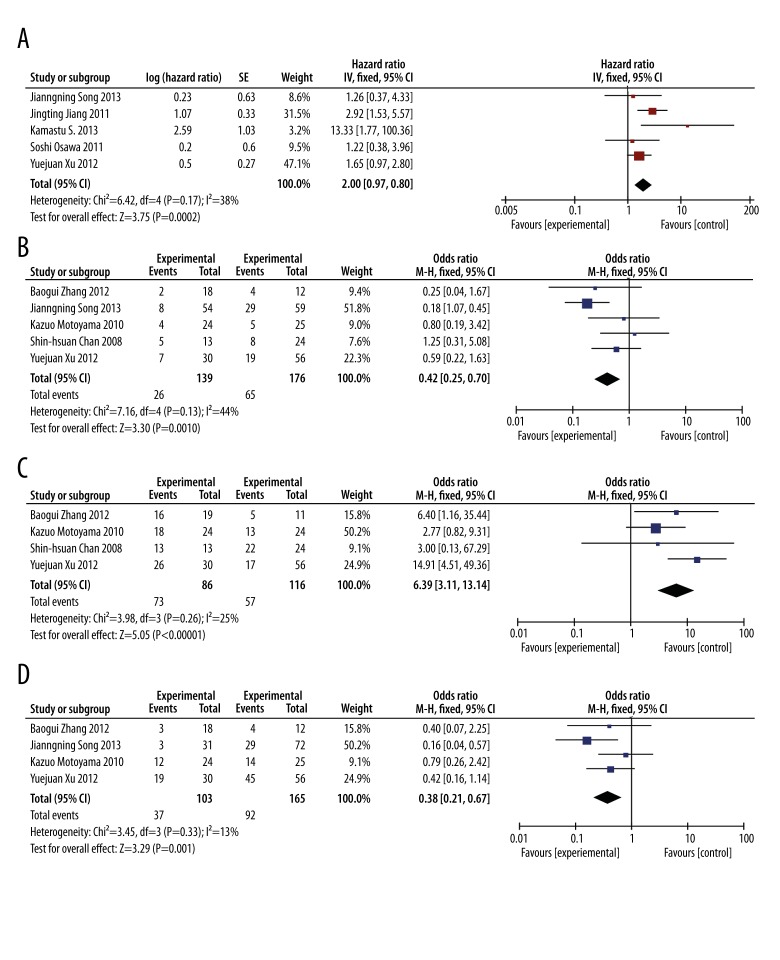
The meta-analysis of 5 studies for OS showed significant prognostic effect on miR-21 detected in tumor samples and serum. The method for detecting miR-21 expression was quantitative real-time PCR (qRT-PCR). In each study, the cut-off values of miR-21 appeared to be different. As shown in Figure 2 A, higher expression levels of miR-21 significantly predicted poorer survival, with the pooled HR of 2.00 (95% CI: 1.39–2.88, P<0.01) and heterogeneity of the 5 studies was P=0.17, I2=38%. Observed HR>1 indicated a worse outcome for the positive group relative to the negative group and would be considered statistically significant if the 95% CI did not overlap 1. Based on data displayed in Figure 2A, we conclude that high expression of miR-21 indicates poor survival in gastric cancer patients.
Figure 2.
The association between high miR-21 expression and survival of patients with gastric cancer (A). Estimated odds ratios (ORs) Summary for correlation of (B) miR-21 expression and histological differentiation, (C) miR-21 expression and lymph node status, and (D) miR-21 expression and TNM stage.
Correlation between miR-21 expression and clinical characteristics
There were 5 studies that reported correlations between miR-21 expression and some clinical characteristics (TNM stage, lymph node metastasis, and histological type); these were pooled to calculate the ORs. In our study, we found that miR-21 expression was significantly correlated with tumor differentiation, lymph node metastasis, and TNM stage. The ORs and the corresponding CIs of studies reporting tumor differentiation, lymph node metastasis, and TNM stage were 0.42 (95% CI: 0.25–0.70 p<0.01), 6.39 (95% CI: 3.11–13.14, P<0.01), 0.38 (95% CI: 0.21–0.67, P<0.01), respectively (Figure 2B–2D).
Assessment of publication bias
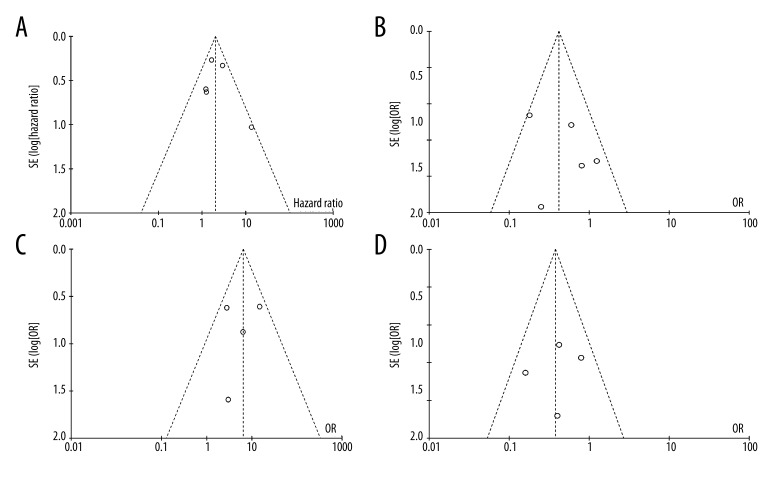
The funnel plot test was used to examine publication bias (Figure 3A–3D). No significant publication biases were found in the results of meta-analyses of miR-21 prediction value for OS using tumor and serum samples. There was also no publication bias in the studies for clinical characteristics.
Figure 3.
Funnel plots of studies included in the meta-analysis: (A) overall survival, (B) histological differentiation, (C) lymph node status, and (D) TNM stage.
Sensitivity analysis
Sensitivity analyses were performed to assess the contribution of each study to the pooled estimate by excluding individual studies one at a time and recalculating the pooled HR or OR estimates for the remaining studies. Eliminating any one study in each group did not substantially change the pooled point estimate, which indicates the reliability of our results.
Discussion
The increased use of miRNAs in cancer research has substantially changed the understanding of gene regulation. MiRNAs play a key role in tumor development by regulating the expression of oncogenes and tumor suppressors or by directly exerting their functions as oncogenes or tumor suppressors [24–26]. The prognostic values of miR-21 have been shown in the recent meta-analysis in head and neck squamous cell carcinoma (HNSCC) [27] and non-small cell lung cancer (NSCLC) [28]. Although the potential diagnostic effect of miR-21in gastric cancer had already been reported [29], the prognostic role of miR-21 in gastric cancer is still unclear. In addition, the relation between gastric cancer patients’ clinical characteristics and expression level of miR-21still needs to be defined. This systematic review and meta-analysis showed that elevated miR-21 did predict poor survival in patients with GC. Although the number of relevant prognostic studies is limited, the available data show that miR-21 is associated with the OS of gastric cancer. As far as we know, this is the first meta-analysis of the prognostic value of miR-21 for GC.
In this meta-analysis, we calculated pooled HRs from 454 patients with gastric cancer from 5 studies. Our result of pooled HR for OS was 2.00 (95% CI: 1.39–2.88, P<0.01). The interval of HR did not overlap 1, which demonstrated that the miR-21 expression did have prognostic significance on GC. In the correlation study of miR-21 expression with patients’ clinical characteristics, ORs for TNM stage, lymph node metastasis, and histological type were significant. The over-expression of miR-21 is positively related to poor histological type, lymph node metastasis, and high-grade of TNM stage, which means gastric cancer patients with the clinical characteristics mentioned above were more likely to have shorter survival time and poorer prognosis after the diagnosis was made.
The association between miR-21 expression and cancer outcome may be partly caused by the biological function of the miR-21. Recent studies have provided supporting evidence for an oncogenic role of miR-21 and shed light on the molecular mechanisms by which it regulates cellular processes. Elevated miR-21 can increase cell proliferation, colony formation, and migration and miR-21 had been reported to regulate the metastatic behavior in both mice and humans by activating various signaling pathways [22,30].
Although we tried to avoid biases in performing this meta-analysis, there were still several limitations to our study. Firstly, appreciation of the oncogenic role of miR-21 in GC has grown in recent years, but only limited research has been done on its relationship to prognosis and clinical characteristics. Results regarding GC outcomes were less powerful because only 5 studies were included for OS meta-analysis, with a relatively small sample size of 351 patients. Secondly, funnel plots showed no evidence of publication bias, but these studies are all from China and Japan, indicating possible geographic publication bias. Thirdly, this meta-analysis could not provide a clear definition about “how high is high” and a clear definition is needed of the cut-off value of miR-21 level for increased survival risk. To date, most researchers use median or mean value in their laboratory or hospital as the cut-off value but the actual value was different. Lack of abundant miR-21 expression data by qRT-PCR in the global population makes it difficult to set a standard cut-off.
Conclusions
High expression of miR-21 was significantly associated with poor survival in patients with gastric cancer. In addition, overexpression of miR-21 is associated with worse tumor differentiation, lymph node metastasis, and TNM stage. Given its limitations, the results of the present analysis should be interpreted with caution. Further clinical investigations are needed to determine the association between miR-21 and cancer prognosis, as well as the efficiency of therapies. More clinical investigations should be conducted before miR-21 can be implemented in the routine clinical management of cancer.
Footnotes
Conflict of Interest
The authors have declared that no competing interests exist.
Source of support: Departmental sources
References
- 1.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Chen W-q, Zheng R-s, Zhang S-w, et al. Report of incidence and mortality in China Cancer Registries, 2008. Chin J Cancer Res. 2012;24:171–80. doi: 10.1007/s11670-012-0171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 8.Chen C-Z. MicroRNAs as Oncogenes and Tumor Suppressors. N Engl J Med. 2005;353:1768–71. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 9.Nohata N, Hanazawa T, Kinoshita T, et al. MicroRNAs function as tumor suppressors or oncogenes: Aberrant expression of microRNAs in head and neck squamous cell carcinoma. Auris Nasus Larynx. 2013;40:143–49. doi: 10.1016/j.anl.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Babashah S, Soleimani M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer. 2011;47:1127–37. doi: 10.1016/j.ejca.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–44. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Williamson PR, Smith CT, Hutton JL, Marson AG. Aggregate data meta analysis with time to event outcomes. Stat Med. 2002;21:3337–51. doi: 10.1002/sim.1303. [DOI] [PubMed] [Google Scholar]
- 13.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–34. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osawa S. MicroRNA profiling of gastric cancer patients from formalin-fixed paraffin-embedded samples. Oncol Lett. 2011;2(4):613–19. doi: 10.3892/ol.2011.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Sun J, Xu J, et al. miR-21 Is a Promising Novel Biomarker for Lymph Node Metastasis in Patients with Gastric Cancer. Gastroenterol Res Pract. 2012;2012:640168. doi: 10.1155/2012/640168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blagosklonny MV, Jiang J, Zheng X, et al. Prognostic Significance of miR-181b and miR-21 in Gastric Cancer Patients Treated with S-1/Oxaliplatin or Doxifluridine/Oxaliplatin. PLoS One. 2011;6(8):e23271. doi: 10.1371/journal.pone.0023271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song J, Bai Z, Zhang J, et al. Serum microRNA-21 levels are related to tumor size in gastric cancer patients but cannot predict prognosis. Oncol Lett. 2013;6:1733–37. doi: 10.3892/ol.2013.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu S, Ichikawa D, Tsujiura M, et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013;33:271–76. [PubMed] [Google Scholar]
- 21.Chan SH, Wu CW, Li AF, et al. miR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 2008;28:907–11. [PubMed] [Google Scholar]
- 22.Zhang BG, Li JF, Yu BQ, et al. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep. 2012;27:1019–26. doi: 10.3892/or.2012.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motoyama K, Inoue H, Mimori K, et al. Clinicopathological and prognostic significance of PDCD4 and microRNA-21 in human gastric cancer. Int J Oncol. 2010;36:1089–95. doi: 10.3892/ijo_00000590. [DOI] [PubMed] [Google Scholar]
- 24.Nicoloso MS, Spizzo R, Shimizu M, et al. MicroRNAs – the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9(4):293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 25.Cho W. MicroRNAs in cancer – from research to therapy. Biochim Biophys Acta. 2010;1805(2):209–17. doi: 10.1016/j.bbcan.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Cho W. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42(8):1273–81. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, Cabay RJ, Jin Y, et al. MicroRNA Deregulations in Head and Neck Squamous Cell Carcinomas. J Oral Maxillofac Res. 2013;4(1):e2. doi: 10.5037/jomr.2013.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma XL, Liu L, Liu XX, et al. Prognostic role of microRNA-21 in non-small cell lung cancer: a meta-analysis. Asian Pac J Cancer Prev. 2012;13(5):2329–34. doi: 10.7314/apjcp.2012.13.5.2329. [DOI] [PubMed] [Google Scholar]
- 29.Zeng Z, Wang J, Zhao L, et al. Potential role of microRNA-21 in the diagnosis of gastric cancer: a meta-analysis. PLoS One. 2013;8(9):e73278. doi: 10.1371/journal.pone.0073278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamanaka S, Olaru AV, An F, et al. MicroRNA-21 inhibits Serpini1, a gene with novel tumour suppressive effects in gastric cancer. Dig Liver Dis. 2012;44(7):589–96. doi: 10.1016/j.dld.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]