Abstract
Doxycycline hyclate (DOX) is a highly photosensitive drug, a feature that limits the stability of the corresponding dosage forms. The main objectives of this work were the preparation and characterization of an inclusion complex of DOX with β-cyclodextrin (βCD) and to investigate if this approach could improve the photostability of the drug. Guest-host interactions were investigated using nuclear magnetic resonance, which were afterwards combined with molecular modeling methods to study the complex formation and its three-dimensional structure was proposed. A freeze-drying method was applied to obtain the complex in the solid state, which was further confirmed by thermal and spectroscopic techniques. To evaluate the complexation effect on DOX integrity, the photostability of the inclusion complex was studied, with a significant decrease in the photodegradation of DOX being found in aqueous solution upon complexation. Finally, the photoprotection produced by the complexation was evaluated by means of an antimicrobial assay. Overall, the presented results suggest that the formulation of DOX complexed with βCD constitutes an interesting approach for the preparation of pharmaceutical dosage forms of DOX with enhanced stability properties.
KEY WORDS: β-cyclodextrin, doxycycline hyclate, microbiological assay, molecular modeling, photostability
INTRODUCTION
Doxycycline (DOX, Fig. 1a) is a drug that has been widely used in the treatment of infectious diseases and as an additive in animal nutrition to facilitate growth (1,2). Often, the choice of DOX over other tetracyclines in the treatment of infections is due to its adequate oral absorption and extended half-life, which allows fewer daily doses (3). DOX is useful for the treatment of respiratory tract infections produced by atypical microorganisms, which are becoming increasingly resistant to other classes of drugs (1). Moreover, DOX is considered the tetracycline of choice in patients with poor renal function due to its limited clearance by kidneys (4). It is active against protozoa and can be administered in combination with quinine in the management of chloroquine resistant Plasmodium falciparum infection, and solutions of DOX are also used for the treatment of malignant effusion conditions that are often associated with lung, breast, and ovary lymphomas (3).
Fig. 1.
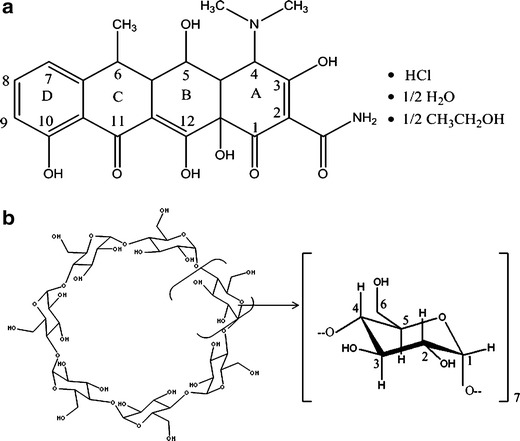
Chemical structure and proton atom numbering of: a doxycycline hyclate and b βCD
DOX exists in three forms: hydrochloride, monohydrate, and hyclate, with DOX hyclate being a hydrochloride hemihydrate hemiethanolate species (Fig. 1a), that exist as a yellow hygroscopic crystalline powder which is freely soluble in water (3). Unfortunately, as DOX is highly susceptible to light, it is necessary to use different formulation strategies in order to enhance its photostability. A commonly applied approach to increase the stability of drugs is the formation of inclusion complexes (IC) with macromolecules, of which molecular encapsulation with cyclodextrins (CD) constitutes a promising alternative for the development of new pharmaceutical dosage forms (5–7). Many natural and modified CD are available for therapeutic purposes, among which β-cyclodextrin (βCD, Fig. 1b) constitutes the most widely used macromolecule due to its low industrial cost. Complexation of drugs with CD has been used to increase their solubilities and stabilities, as well as to modulate the organoleptic properties of pharmaceutical formulations (8–10). The effect of IC formation on the photostability of different drugs has also been previously studied, with reports showing that complexation with CD can significantly affect the light absorption properties of the included compound (11). However, there are marked differences in the stabilizing effects among different CD, a feature that depends on the shape and volume of the cavity available for inclusion, and in some cases the complexation has a catalyzing rather than a stabilizing effect on the photodegradation process (12). For this reason, state of the art computer based molecular modeling techniques combined with experimental techniques, potentiates the ability to elucidate and further predict the IC three-dimensional structures (9,13–15).
In previous reports, various authors have studied the inclusion behavior of DOX with methyl and hydroxypropyl βCD (16–18), with some of these complexes exhibiting promising properties for the development of new DOX formulations intended for aerosol administration (19) and ophthalmic delivery (20–22).
Based on above commented aspects, the objective of the present study was to investigate further the possibility of obtaining an IC between DOX and βCD in aqueous solution and in solid state, and to evaluate the effect of complexation on the photostability of this drug. After the IC complex was prepared, several experimental and theoretical techniques were applied to study in detail the three-dimensional array of the resulting complex in aqueous solution and to characterize the solid product obtained by a freeze-drying method. In order to assess the efficacy of the inclusion to protect DOX from degradation, photolytic and antimicrobial assays were also performed.
MATERIALS AND METHODS
Materials
Doxycycline hyclate was provided courtesy of União Química Indústria Farmacêutica (Brazil), and βCD (MW = 1,135) was kindly supplied by Roquette (France). The D2O 99.9 atom% D was used for the NMR experiments (Sigma-Aldrich®), with all other materials and solvents being of analytical reagent grade. A Milli-Q Water Purification System (Millipore®, Bedford, Massachusetts, USA) generated the water used in these studies.
NMR Studies
NMR experiments were performed on a Bruker® Avance II High Resolution Spectrometer, equipped with a Broad Band Inverse probe (BBI) and a Variable Temperature Unit (VTU). The spectra were measured at 298 K and the value of the pD found for the samples was 2.3. The operating frequency for protons was 400.16 MHz, with the chemical shift of the residual solvent at 4.8 ppm being used as the internal reference. Induced changes in the 1H chemical shifts for DOX and βCD (Δδ), which originated due to its complexation, were calculated using the following equation (Eq. 1):
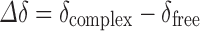 |
1 |
The geometry of the inclusion complex was studied by two-dimensional rotating frame Overhauser experiments (2D ROESY), with spinlock for mixing phase sensitive, using 180x 180-x pulses for polarization transfer. The spectra were measured with a relaxation delay of 2 s, p15 pulse for ROESY spinlock of 20 ms and l4, spinlock loop, (p15/p25 × 2) = 400. Before Fourier transformation, the matrix was zero filled to 4,096 (F2) by 2,048 (F1), and Gaussian apodization functions were applied in both dimensions.
Molecular Modeling Studies
The initial structure of DOX was constructed using the Gabedit software (23), after which a systematic conformational search was carried out in order to obtain the minimum energy conformation. The minimization protocol was conducted applying a semiempirical method (AM1), with the final minimum energy conformation being optimized using an ab initio (HF-6-31G) method as implemented in Gaussian03 (24). The initial structure of βCD was obtained from the Cambridge Structural Database (code BCDEXD10), which was used for docking studies as downloaded.
Molecular docking assays were performed using software packages designed by OpenEye Scientific Software (25), with the protocol including three main stages: (a) conformer library generation, (b) rigid exhaustive docking and (c) visualization and analysis of the results. In the first stage (a), the minimum energy conformation of DOX was used to generate a library of conformers using OMEGA software (26,27), by applying the MMFF94 force field and an energy threshold of 40 Kcal/mol. For the second stage (b), this library of conformers was subjected to molecular docking using the FRED3 and the OEDocking software suites (28–30) and performing an exhaustive evaluation of the docking poses by means of the Chemgauss3 scoring function. In the third stage (c), a clustering analysis was performed in order to identify the main binding conformations, which were afterwards subjected to molecular dynamics (MD) simulations.
To obtain the MD trajectories, the complexes predicted by molecular docking were parameterized using the general amber force field (GAFF) and the GLYCAM06 force fields, for DOX and βCD, respectively (31,32). The parameterized complexes were afterwards solvated with a pre-equilibrated TIP3P octahedric water box, considering a minimum distance between the edge of the box and the simulated system of 10 Å. After this setup was applied, an initial minimization of the solvent (keeping βCD restrained) followed by a minimization of the whole system were carried out by combining the steepest descent (5,000 steps) and conjugate gradient methods (5,000 steps). The minimized systems were then heated to 300 K for 200 ps under constant volume conditions, and this was followed by an equilibration run (1 ns) under constant pressure conditions. After the equilibration phase was finished, production runs were performed for 10 ns, using a timestep of 2 fs and with the SHAKE algorithm being applied to restrain bonds involving hydrogen atoms. The MD procedures were performed using the Amber12 software package (33).
Energetic decomposition analyses were performed using the Molecular Mechanics Poisson-Bolzmann Surface Area (MM-PBSA) modules of Amber12 (34), which were applied over the entire trajectory (10 ns), with individual snapshots being sampled every 10 frames. The corresponding structures and figures were constructed using VMD (35) and VIDA software (36).
Molecular dynamics trajectories were obtained using CUDA designed code (pmemd.cuda), with computational facilities provided by the GPGPU Computing group of the Facultad de Matemática, Astronomía y Física (FAMAF), Universidad Nacional de Córdoba, Argentina.
Preparation of the DOX:βCD System in Solid State
Freeze-dried system (FD): prepared by dissolving equimolar amounts DOX and βCD in distilled water. The resulting solutions were placed in an ultrasonic bath for 1 h and afterwards frozen at −40°C prior to starting the freeze-drying procedure (Freeze Dri 4.5 Labconco Corp., Kansas City, MI).
Physical mixture system (PM): prepared by mixing DOX and βCD uniformly in a mortar at a 1:1 M ratio.
Fourier Transform Infrared Spectroscopy (FT-IR)
The FT-IR spectra of DOX, βCD, the PM system, and the FD system were collected using identical protocols on potassium bromide discs on a Nicolet 5 SXC FT-IR Spectrometer. All spectra were acquired and processed using EZ OMNIC E.S.P v.5.1 software.
Differential Scanning Calorimetry (DSC) and Thermogravimetric Analysis (TGA)
DSC measurements of the pure materials and the binary systems were carried out using a DSC TA 2920 (TA Instruments, Inc., New Castle, USA). The thermal behavior was studied by heating 1–3 mg of samples in aluminum-pinhole pans, from 30 to 280°C, at a rate of 10°C min−1 under nitrogen gas flow. An empty pinhole pan was used as reference, and Indium (99.98%, mp 156.65°C, Aldrich, Milwaube, USA) utilized as the standard for calibrating the temperature.
The TGA curves of the different samples were recorded on a TG TA 2950 (TA Instruments, Inc., New Castle, USA), using the same conditions as in the DSC studies, but over a temperature range of 30 to 350°C. The TG temperature axes were calibrated with the Curie point of Ni (353°C). In both cases, data were obtained and processed using TA Instruments Universal Analysis 2000 software.
Photostability Studies
Stock solutions of DOX (80 μg/ml), prepared in water in the absence or presence of βCD (1:1 mol/mol), were used to study the photodegradation of the drug. These solutions were positioned 20 cm away from a UV light source at 30°C, with the corresponding solutions being sampled at specified time intervals (3.0 and 6.0 h) and the concentration of DOX determined by a previously validated HPLC assay (37). All results were expressed as percentages of the remaining DOX. The HPLC equipment consisted of a Waters system including: a Waters 1525 pump, a Model 2487 dual wavelength absorbance detector set at 360 nm, and a Rheodyne Breeze 7725 manual injector, with a Luna CN (4.6 × 250 mm, 5 μm) column. The mobile phase was water:acetonitrile:trifluoroacetic acid (60:40:0.1), at a flow rate of 1.0 ml/min. Assays were performed at 25°C, by injecting 20 μL of solution in each chromatographic run. Under the reported conditions, the retention time for DOX was 4.7 min.
Microbiological Studies
A Shaker Marconi model MA420 incubator was used for the incubation of microorganisms, with gases for the bacterial culture being generated with an ECB Digital 1.2 (Odontobrás) device. For the quantitative determinations, a Beckman spectrophotometer model DU® 530 was utilized, with Brain Heart Infusion (BHI) agar culture medium being used for maintaining the viability of the microorganisms and BHI broth used for the tests. The culture media were prepared as indicated by manufacturer’s guidelines, with media being sterilized by autoclaving (conditions: 121°C, 1 atm) for 15 min.
The cultures of Escherichia coli were subcultured with a platinum spatula for BHI broth and maintained for their development in microbial gases at 35 ± 2°C for 24 h prior to the experiment. Inoculums were standardized by measuring their transmittance at 530 nm in a spectrophotometer, with values of 25 ± 2% being obtained. The standardized culture medium was prepared on the day of use.
DOX stock (100.0 μg/mL) and standard solutions (4.0, 6.0, and 9.0 μg/mL) were prepared in water. To perform the turbidimetric assay, 200 μL of DOX standard solutions were added to 10.0 mL of sterile broth. The tubes were then incubated in a water bath shaker at 35 ± 2°C for 4 h, after which the growth of microorganisms was terminated by the addition of 0.5 mL of 12% formaldehyde, and the absorbance was measured at 530 nm. Assays were performed in triplicate.
To assess DOX photodegradation, solutions of free DOX and DOX:βCD IC were subjected to UV light irradiation at 30°C for 7 days, before quantifying the antimicrobial potencies of the irradiated solutions using the abovementioned turbidimetric assay.
RESULTS AND DISCUSSION
Structural Characterization of DOX:βCD IC in Aqueous Solution
NMR Studies
Table I shows the chemical shift displacements for DOX and βCD protons in their free and complexed states. It can be observed that in the presence of the macromolecule, almost all the proton resonances of DOX experienced shifts to higher ppm values, with the exception of H4, which exhibited a slight displacement toward lower ppm values (Δδ = −0.018). In addition, marked upfield displacements for H3, H5, and H6 protons of the host molecule were observed (Δδ = −0.0981, −0.1068, and −0.0427, respectively), which could have been due to the inclusion of groups that are rich in π electrons into the βCD hydrophobic cavity, such as the aromatic ring of DOX. These results suggest the formation of an IC between DOX and βCD in aqueous solution.
Table I.
Chemical Shifts for the Protons of DOX and βCD in the Free and Complex Forms
| Proton | βCD (ppm) | DOX:βCD (ppm) | Δδ (ppm) |
| H1 | 5.1018 | 5.0428 | −0.059 |
| H2 | 3.6812 | 3.6340 | −0.0472 |
| H3 | 3.9967 | 3.8986 | −0.0981 |
| H4 | 3.615 | 3.5576 | −0.0574 |
| H5 | 3.9095 | 3.8028 | −0.1068 |
| H6 | 3.9095 | 3.8668 | −0.0427 |
| Proton | DOX (ppm) | DOX:βCD (ppm) | Δδ (ppm) |
| H 4 | 4.2864 | 4.2846 | −0.0018 |
| H 4A | 2.8212 | 2.8225 | 0.0153 |
| H 5A | 2.5518 | 2.5750 | 0.0490 |
| H 6 | 2.6830 | 2.7497 | 0.0667 |
| H 7 | 6.9868 | 7.0097 | 0.0326 |
| H 8 | 7.5124 | 7.5444 | 0.0320 |
| H 9 | 6.8001 | 6.8047 | 0.0059 |
| H CH3 | 1.4630 | 1.5153 | 0.0523 |
Italics are presented DOX protons to distinguish them from βCD protons
Molecular Modeling Studies
To study the corresponding three-dimensional geometry of the DOX:βCD system, molecular modeling studies were performed. The docking of DOX was performed by considering its ionization state and tautomeric equilibrium at pH 2.3, in agreement with the value observed in the experimental studies. From these docking results, two main binding modes were observed (Fig. 2a, b).
Fig. 2.
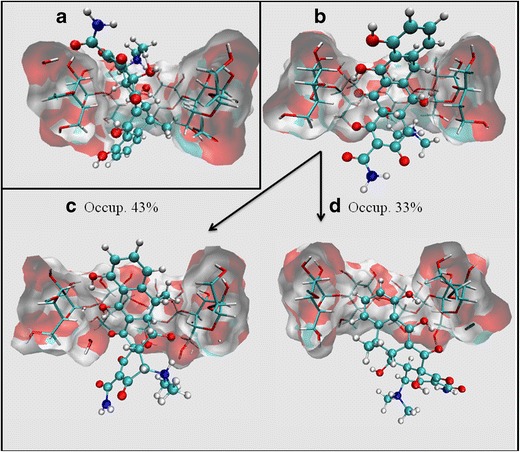
Three-dimensional structures of the binding modes between DOX and βCD as predicted by molecular docking: a pose-1, b pose-2. Representative structures obtained by applying a clustering algorithm on the molecular dynamics trajectory of DOX pose-2. c Main cluster conformation (cluster-1) (occupation = 43%); d secondary cluster conformation (cluster-2) (occupation = 33%)
In the first binding mode (pose-1, Fig. 2a), ring A was oriented toward the wide rim of βCD, while rings B, C, and D were deeply buried into the βCD cavity. In the second binding mode (pose-2, Fig. 2b), ring A was oriented toward the narrow rim of βCD, while the ring system comprised of B, C, and D was completely included in the hydrophobic cavity of the host molecule.
In order to study which of the two binding poses was most stable, molecular dynamics studies were performed to obtain simulation trajectories that included the temperature and solvent effects in the complex conformation. The simulation of both complexes was adequately equilibrated, with stable kinetic, potential, and total energy values being observed. After the corresponding trajectories were obtained, energetic decomposition analyses were performed (Table II), which revealed that pose-2 was more stable than pose-1, since higher contributions from both electrostatic and Van der Waals intermolecular interactions were observed. Then, after accounting for the corresponding solvation energies, it was found that the binding of DOX in pose-2 was ~5 Kcal/mol more stable than pose-1 (−22.84 and −17.96, respectively).
Table II.
Energetic Component Analysis Obtained by MM-PBSA for DOX:βCD Complex Analyzed by Molecular Dynamics Simulations
| Component | pose-1 (Kcal/mol) | pose-2 (Kcal/mol) |
|---|---|---|
| Electrostatic energy | −30.74 | −33.05 |
| Van der Waals | −9.81 | −15.36 |
| Total gas phase energy | −40.56 | −48.41 |
| Nonpolar solvation | −2.77 | −3.12 |
| Electrostatic solvation | 25.67 | 28.70 |
| Total solvation | 22.59 | 25.57 |
| Estimated binding energy | −17.96 | −22.84 |
Figure 2c and d show the two most populated clusters of conformations obtained from the simulated trajectory of pose-2. As can be seen from the clustering analyses, DOX exhibited a dynamic behavior in its inclusion into βCD, with the most populated conformation corresponding to the disposition in which the fused ring system was deeply buried into the host molecule hydrophobic cavity (43% of the trajectory, Fig. 2c), while the most hydrophilic moieties of the molecule (ternary amine and amide groups) were exposed to the solvent through the narrow rim of βCD. This alternation in the positioning of the included ligand was also verified by measuring the distance from the methyl group bound to ring C to H3 and H6 of βCD (Fig. 3).
Fig. 3.
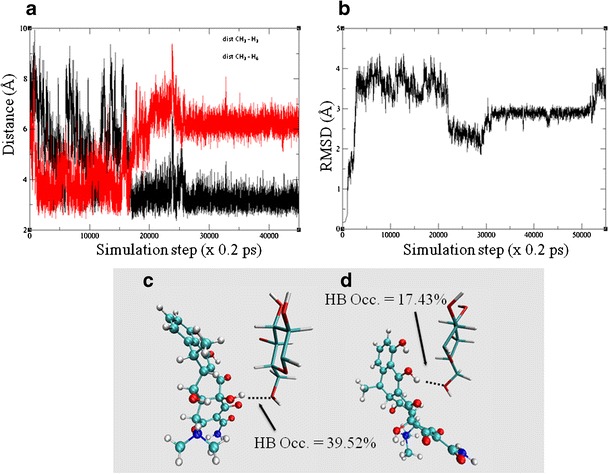
a Plot of distance between the methyl groups bound to ring C in DOX and the βCD protons lying inside the hydrophobic cavity (H3 and H6). b RMSD vs. time for the molecular dynamics trajectory of DOX:βCD pose-2 (reference: BCDEXD10). c–d The hydrogen bond analysis and occupance (%) between DOX and βCD depicted for: cluster-1 (c) and cluster-2 (d)
The orientations observed in the molecular dynamics trajectories were consistent with the ∆δ calculated from NMR studies, in which a marked shielding was observed in the protons located in the interior of the hydrophobic cavity (−0.0981 and −0.10675 for H3 and H5, respectively), and that originated due to the proximity of the electron rich fused ring system. In addition, a significant shielding was observed in H6 (−0.0427), which originated in the shielding effect of DOX (mainly in the conformation depicted in Fig. 2d, second cluster).
The ∆δ observed for the protons located outside the hydrophobic cavity (H1, H2, and H4, Table I) may have been attributed to the conformational change induced on βCD by DOX upon complexation. In order to quantitatively describe this phenomenon, the RMSD of βCD respect to the native structure (code BCDEXD10) was calculated (Fig. 3b). As can be observed, a significant conformation change occurred in the βCD three-dimensional structure when DOX was included in the hydrophobic cavity, with two stable conformations corresponding to both the clusters shown in Fig. 3a and b.
Hydrogen bond analyses were performed to identify the main intermolecular interactions originating the electrostatic energetic component. As presented in Fig. 3c and d for the conformation corresponding to cluster-1, the main hydrogen bond was established between the OH bound to C12a and O6 from βCD (HB Occ. = 39.52%). In the case of cluster-2, the main hydrogen bond was established between the OH bound to C5 of DOX and O6 from βCD (HB Occ. = 17.43%). The downfield displacement observed in the NMR studies for the H4A and H5A protons (deshielding effect) might have been originated by the presence of hydrogen bond interactions between the hydroxyl group bond to C5 of DOX and the hydroxyl groups of βCD.
ROESY Experiments
The spatial interaction between the host and guest molecules predicted by molecular modeling was confirmed on carrying out a 2D ROESY experiment. An expansion of the ROESY spectrum obtained is shown in Fig. 4, where interactions of the aromatic protons of DOX (ring D) may be observed with both internal protons of βCD (H3 and H5). Moreover, correlations were revealed between the aromatic protons of DOX and proton H6 located at the narrow side of βCD, in agreement with the observation that ring D was deeply buried into the βCD hydrophobic cavity. These results are consistent with molecular modeling studies and confirm the formation of the inclusion complex between DOX and βCD in aqueous solution.
Fig. 4.
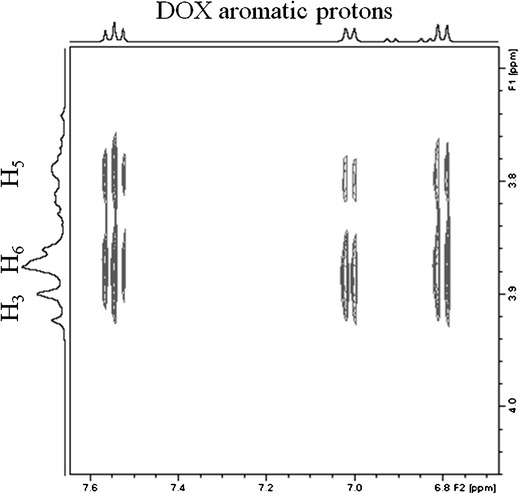
Expansion from the 2D ROESY spectrum of the DOX:βCD system
Characterization of IC in the Solid State
Fourier Transform Infrared Spectroscopy (FT-IR)
The infrared spectra of the pure components and the PM and FD systems are presented in Fig. 5. As may be observed, the spectrum for the PM system consisted of an overlay of the pure compound spectra, whereas the spectrum for the FD system exhibited differences from those of pure components. The absence or reduction of the intensities of the DOX bands at 3,449, 3,330, 1,534, 1,479, 1,459, 1,219, 1,129, 881, and 732 cm−1 in the FD system was consistent with the vibrational constrictions imposed on the included molecule in the βCD cavity. Based on this evidence, it is possible to confirm the existence of intermolecular interactions between DOX and βCD in the FD system.
Fig. 5.
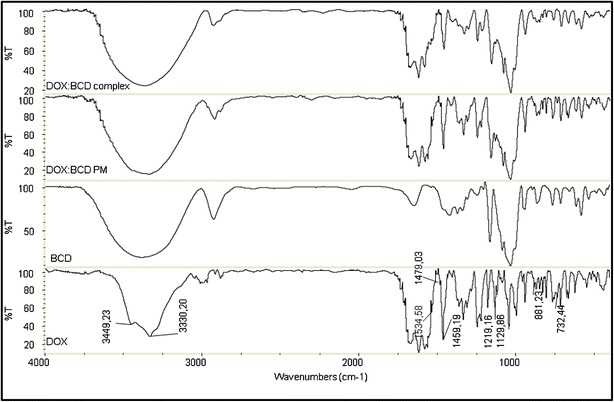
IR spectra of DOX, βCD, physical mixture system (DOX:βCD PM), and freeze-dried system (DOX:βCD complex)
Differential Scanning Calorimetry and Thermogravimetric Analysis
The thermal behavior of the DOX, βCD, PM, and FD systems is shown in Fig. 6. The endothermic peak (25–100°C) observed in the DSC curve of βCD corresponded to its dehydration, with decomposition taking place above 300°C as evidenced by the mass loss observed in the TGA curve (Δm = 11 and 77%, respectively). The DSC curve of DOX had an endothermic peak at 168°C, which corresponded to its melting and was followed by an exothermic peak at 223°C originated by its decomposition. The TGA curve of DOX presented two weight loss events, the first of which was in the 125–175°C range (Δm = 7%) whereas the second occurred near 200°C, (Δm = 36%).
Fig. 6.
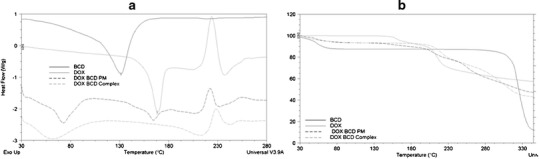
DSC (a) and TG (b) curves of DOX, βCD, physical mixture system (DOX:βCD PM), and freeze-dried system (DOX:βCD complex)
The analysis of the DSC and TGA curves corresponding to the PM system revealed identical thermal events than those observed for the pure compounds. In the case of the FD system, the TGA curve exhibited three weight loss events, the first being in the 25–100°C range (Δm = 7%), the second event occurring between 175 and 225°C (Δm = 15%), and the third at 228°C (Δm = 35%), while the mass loss observed for DOX and PM between 125 and 175°C was not present in the IC curve. In addition, the DSC curve confirmed that the endothermic peak of pure DOX at 168°C was not present in this system, indicating that the melting event did not take place, which might have been due to changes in the crystalline form of the solid or to an inclusion complex formation. This behavior was consistent with the formation of an inclusion complex between DOX and βCD in the solid state when the system was prepared by the freeze-dried method.
Photostability Studies
In order to enhance the photostability of DOX, the effect of βCD inclusion was assayed in aqueous solution. Then, the photodegradation of pure DOX and DOX:βCD complex was analyzed by an HPLC method after its exposure to UV light for 3 and 6 h. Free DOX showed a fast degradation as a result of UV light, with recovery percentages being 84 ± 2% and 68 ± 1%, at 3 and 6 h, respectively, which is consistent with the reported photolability of this drug. On the other hand, the DOX:βCD complex showed an increased stability, with recovery percentages of DOX of 99 ± 1% and 96.8 ± 0.9%, at 3 and 6 h, respectively. These results demonstrate that the inclusion of DOX into the βCD cavity had a photoprotective effect on this drug stability.
Microbiological Studies
The objective of this study was to compare the activity between the IC and free DOX, by applying a turbidimetric microbiological technique. In addition, taking into account that the photodegradation of DOX produced a significant loss of potency, a second objective of this study was to compare the activity of the IC and free DOX after the exposure of both products to UV light. Three different concentrations of DOX and IC solutions were tested, while pure βCD solutions were analyzed as control. After incubation, the tubes containing solutions of DOX showed a comparable turbidity to the culture medium with the tubes containing solutions of IC (Table III), indicating that the in vitro antibacterial activity of DOX present in the IC was not lowered as a consequence of the complexation. The βCD solutions used in the test, after the incubation period, showed absorbance values equal or higher than those of the positive control, confirming that the βCD did not have any antimicrobial activity.
Table III.
Absorbance Values of DOX in Free and Complex Forms
| DOX Conc. (μg/mL) | Without degradation in UV light | With degradation in UV light | ||||||
|---|---|---|---|---|---|---|---|---|
| Free form | Complex form | Free form | Complex form | |||||
| A Aa | CV (%) | A Aa | CV (%) | A Aa | CV (%) | A Aa | CV (%) | |
| 4 | 0.807 | 2.14 | 0.806 | 1.24 | 1.139 | 2.26 | 0.885 | 0.28 |
| 6 | 0.584 | 4.11 | 0.53 | 4.27 | 1.045 | 3.12 | 0.684 | 2.88 |
| 9 | 0.504 | 3.48 | 0.412 | 7.18 | 0.609 | 0.78 | 0.493 | 0.88 |
aAbsorbance Average
Finally, DOX and IC solutions were exposed to UV light for 7 days before being analyzed by the turbidimetric method. Table III shows the absorbance values of DOX and IC subject to UV light irradiation, with the tubes containing solutions of free DOX showing a higher turbidity of the culture medium than the tubes containing solutions of IC, thus confirming the photoprotective effect of βCD on DOX observed during the photostability studies.
CONCLUSION
The preparation of an IC between DOX and βCD in the solid state by a freeze-drying method was confirmed by spectroscopic and thermal techniques. In addition, it was possible to combine molecular modeling studies with experimental spectroscopic studies to elucidate the molecular basis of the interactions between the guest and host molecules. The IC exhibited an enhanced photostability compared to the free DOX, which in turn originated a better antimicrobial activity when both species were exposed to UV light. These observations suggest that the formulation of DOX with βCD may have overcome the stability problems currently described for the drug, and also provides an interesting alternative for the preparation of pharmaceutical dosage forms of DOX.
ACKNOWLEDGMENTS
The authors acknowledge CNPq (Brasília, Brazil), CAPES, PADC-FCF, FUNDUNESP (São Paulo, Brazil), the Secretaría de Ciencia y Técnica de la Universidad Nacional de Córdoba (SECyT), and the Consejo Nacional de Investigaciones Científicas y Tecnológicas de la Nación Argentina (CONICET) for financial support. We also thank Ferromet S.A. (agent of Roquette in Argentina) for its donation of β-cyclodextrin. We are grateful to Dr. Gloria Bonetto for NMR measurements and for her helpful discussion of the 1H NMR spectra. The authors also acknowledge the GPGPU Computing Group and Dr. Nicolás Wolovick from the Facultad de Matemática, Astronomía y Física (FAMAF), Universidad Nacional de Córdoba, Argentina, for providing access to computing resources.
Contributor Information
Ana Carolina Kogawa, Email: ac_kogawa@yahoo.com.br.
Ariana Zoppi, Email: ariana@fcq.unc.edu.ar.
Mario Alfredo Quevedo, Email: alfredoq@fcq.unc.edu.ar.
Hérida Regina Nunes Salgado, Email: salgadoh@fcfar.unesp.br.
Marcela Raquel Longhi, Phone: 54-0351-5353865, Email: mrlcor@fcq.unc.edu.ar.
REFERENCES
- 1.Brunton LL, Lazo JS, Parker KL. Goodman and Gilman’s: the pharmacological basis of therapeutics. 11. Rio de Janeiro: McGraw Hill Interamericana do Brasil; 2006. [Google Scholar]
- 2.Kogawa AC, Salgado HRN. Doxycycline hyclate: a review of properties, applications and analytical methods. Int J Life Sci Pharma Res. 2012;2(4):11–25. [Google Scholar]
- 3.Sweetman SC, Blake PS. Martindale: the complete drug reference. 36. London: Pharmaceutical Press; 2009. [Google Scholar]
- 4.Clinical Pharmacology Online, 2009. Gold Standard, an Elsevier Company, Tampa, FL. Available from URL: www.clinicalpharmacology.com. Cited 10 Jan 2013.
- 5.Brewster ME, Loftsson T. Cyclodextrins as pharmaceutical solubilizers. Adv Drug Deliv Rev. 2007;59(7):645–666. doi: 10.1016/j.addr.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Loftsson T, Brewster ME. Pharmaceutical applications of cyclodextrins: basic science and product development. J Pharm Pharmacol. 2010;62(11):1607–1621. doi: 10.1111/j.2042-7158.2010.01030.x. [DOI] [PubMed] [Google Scholar]
- 7.Loftsson T, Brewster ME. Cyclodextrins as functional excipients: methods to enhance complexation efficiency. J Pharm Sci. 2012;101(9):3019–3032. doi: 10.1002/jps.23077. [DOI] [PubMed] [Google Scholar]
- 8.Delrivo A, Zoppi A, Longhi MR. Interaction of sulfadiazine with cyclodextrins in aqueous solution and solid state. Carbohydr Polym. 2012;87(3):1980–1988. doi: 10.1016/j.carbpol.2011.10.025. [DOI] [Google Scholar]
- 9.Zoppi A, Garnero C, Linck YG, Chattah AK, Monti GA, Longhi MR. Enalapril: β-CD complex: stability enhancement in solid state. Carbohydr Polym. 2011;86(2):716–721. doi: 10.1016/j.carbpol.2011.05.008. [DOI] [Google Scholar]
- 10.Zoppi A, Delrivo A, Aiassa V, Longhi MR. Binding of sulfamethazine to β-cyclodextrin and methyl-β-cyclodextrin. AAPS PharmSciTech. 2013;14(2):727–735. doi: 10.1208/s12249-013-9958-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loftsson T, Jarho P, Másson M, Järvinen T. Cyclodextrins in drug delivery. Expert Opin Drug Deliv. 2005;2(2):335–351. doi: 10.1517/17425247.2.1.335. [DOI] [PubMed] [Google Scholar]
- 12.Tønnesen HH. Formulation and stability testing of photolabile drugs. Int J Pharm. 2001;225(1–2):1–14. doi: 10.1016/S0378-5173(01)00746-3. [DOI] [PubMed] [Google Scholar]
- 13.Onnainty R, Schenfeld EM, Quevedo MA, Fernández MA, Longhi MR, Granero GE. Characterization of the hydrochlorothiazide: β-cyclodextrin inclusion complex. Experimental and theoretical methods. J Phys Chem B. 2013;117(1):206–217. doi: 10.1021/jp311274c. [DOI] [PubMed] [Google Scholar]
- 14.Zoppi A, Quevedo MA, Longhi MR. Specific binding capacity of beta-cyclodextrin with cis and trans enalapril: physicochemical characterization and structural studies by molecular modeling. Bioorg Med Chem. 2008;16(18):8403–8412. doi: 10.1016/j.bmc.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 15.Zoppi A, Quevedo MA, Delrivo A, Longhi MR. Complexation of sulfonamides with beta-cyclodextrin studied by experimental and theoretical methods. J Pharm Sci. 2010;99(7):3166–3176. doi: 10.1002/jps.22062. [DOI] [PubMed] [Google Scholar]
- 16.Bakkour Y, Vermeersch G, Morcellet M, Boschin F, Martel B, Azaroual N. Formation of cyclodextrin inclusion complexes with doxycyclin-hyclate: NMR investigation of their characterisation and stability. J Incl Phenom. 2006;54(1–2):109–114. doi: 10.1007/s10847-005-5108-7. [DOI] [Google Scholar]
- 17.Benghodbane S, Khatmi D. A theoretical study on the inclusion complexation of doxycycline with Crysmeb. C R Chim. 2012;15(5):371–377. doi: 10.1016/j.crci.2011.11.007. [DOI] [Google Scholar]
- 18.Benghodbane S, Khatmi D. Quantum chemical calculations based on ONIOM and the DFT methods in the inclusion complex: doxycycline/2-O-Me-β-cyclodextrin. J Incl Phenom Macro. 2013;77(1-4):231–40.
- 19.Gueders MM, Bertholet P, Perin F, Rocks N, Maree R, Botta V, et al. A novel formulation of inhaled doxycycline reduces allergen-induced inflammation, hyperresponsiveness and remodeling by matrix metalloproteinases and cytokines modulation in a mouse model of asthma. Biochem Pharmacol. 2008;75(2):514–526. doi: 10.1016/j.bcp.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 20.He Z, Wang Z, Zhang H, Pan X, Su W, Liang D, et al. Doxycycline and hydroxypropyl-β-cyclodextrin complex in poloxamer thermal sensitive hydrogel for ophthalmic delivery. Acta Pharm Sin B. 2011;1(4):254–260. doi: 10.1016/j.apsb.2011.10.004. [DOI] [Google Scholar]
- 21.Zhang H, Chen M, He Z, Wang Z, Zhang M, Wan Q, et al. Molecular modeling-based inclusion mechanism and stability studies of doxycycline and hydroxypropyl-β-cyclodextrin complex for ophthalmic delivery. AAPS PharmSciTech. 2013;14(1):10–8. [DOI] [PMC free article] [PubMed]
- 22.Wang Z, He Z, Zhang L, Zhang H, Zhang M, Wen X, et al. Optimization of a doxycycline hydroxypropyl-β-cyclodextrin inclusion complex based on computational modeling. Acta Pharm Sin B. 2013;3(2):130–139. doi: 10.1016/j.apsb.2013.01.002. [DOI] [Google Scholar]
- 23.Allouche AR. Gabedita - A graphical user interface for computational chemistry softwares. J Comput Chem 2011;32(1):174–182. [DOI] [PubMed]
- 24.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Software: GAUSSIAN 98. 1998.
- 25.OpenEye Scientific.Software, http://www.eyesopen.com.
- 26.Hawkins PCD, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the protein databank and Cambridge structural database. J Chem Inf Model. 2010;50(4):572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omega.2.4.3. OpenEye Scientific Software, Santa Fe, NM http://www.eyesopen.com.
- 28.Fred.3.0.0. OpenEye Scientific Software, Santa Fe, NM http://www.eyesopen.com.
- 29.McGann M. FRED pose prediction and virtual screening accuracy. J Chem Inf Model. 2011;51(3):578–596. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- 30.McGann M. FRED and HYBRID docking performance on standardized datasets. J Comput Aided Mol Des. 2012;8(26):1–10. doi: 10.1007/s10822-012-9584-8. [DOI] [PubMed] [Google Scholar]
- 31.Kirschner KN, Yongye AB, Tschampel SM, González-Outeiriño J, Daniels CR, Foley BL, et al. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J Comput Chem. 2008;29(4):622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general Amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 33.Case DA, Cheatham Iii TE, Darden T, Gohlke H, Luo R, Merz KM, Jr, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn B, Gerber P, Schulz-Gasch T, Stahl M. Validation and use of the MM-PBSA approach for drug discovery. J Med Chem. 2005;48(12):4040–4048. doi: 10.1021/jm049081q. [DOI] [PubMed] [Google Scholar]
- 35.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 36.Connors KA. The stability of cyclodextrin complexes in solution. Chem Rev. 1997;97(5):1325–1357. doi: 10.1021/cr960371r. [DOI] [PubMed] [Google Scholar]
- 37.Kogawa AC, Salgado HRN. Quantification of doxycycline hyclate in tablets by HPLC–UV method. J Chromatogr Sci. 2013;51(10):919–25. [DOI] [PubMed]


