Abstract
The McMurdo Dry Valleys of Antarctica are considered to be one of the most physically and chemically extreme terrestrial environments on the Earth. However, little is known about the organisms involved in nitrogen transformations in these environments. In this study, we investigated the diversity and abundance of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in four McMurdo Dry Valleys with highly variable soil geochemical properties and climatic conditions: Miers Valley, Upper Wright Valley, Beacon Valley and Battleship Promontory. The bacterial communities of these four Dry Valleys have been examined previously, and the results suggested that the extremely localized bacterial diversities are likely driven by the disparate physicochemical conditions associated with these locations. Here we showed that AOB and AOA amoA gene diversity was generally low; only four AOA and three AOB operational taxonomic units (OTUs) were identified from a total of 420 AOA and AOB amoA clones. Quantitative PCR analysis of amoA genes revealed clear differences in the relative abundances of AOA and AOB amoA genes among samples from the four dry valleys. Although AOB amoA gene dominated the ammonia-oxidizing community in soils from Miers Valley and Battleship Promontory, AOA amoA gene were more abundant in samples from Upper Wright and Beacon Valleys, where the environmental conditions are considerably harsher (e.g., extremely low soil C/N ratios and much higher soil electrical conductivity). Correlations between environmental variables and amoA genes copy numbers, as examined by redundancy analysis (RDA), revealed that higher AOA/AOB ratios were closely related to soils with high salts and Cu contents and low pH. Our findings hint at a dichotomized distribution of AOA and AOB within the Dry Valleys, potentially driven by environmental constraints.
Keywords: archaea, AOA, bacteria, AOB, ammonia oxidizers, Antarctica, Dry Valleys
INTRODUCTION
Nitrification represents the oxidative part of the nitrogen (N) cycle and refers to the two-step process where ammonia is oxidized to nitrite and subsequently to nitrate. This process is considered a central biological pathway in the global N budget and productivity of terrestrial and aquatic ecosystems. From 1890 until 2004, scientists believed that only bacteria mediated aerobic ammonia oxidation. The recent discovery of a chemoautotrophic ammonia-oxidizing archaeon, Nitrosopumilus maritimus (Könneke et al., 2005), transformed our concept of the nature of organisms involved in nitrification, highlighting the importance of ammonia-oxidizing archaea (AOA) as potential participants in global biogeochemical N transformations (Hallam et al., 2006; Brochier-Armanet et al., 2008; de la Torre et al., 2008; Pester et al., 2012). The phylogenetic uniqueness of these archaea led to the creation of a novel archaeal phylum, Thaumarchaeota, comprising all AOA (Brochier-Armanet et al., 2008). The idea that nitrification activities from AOA greatly contribute to the global N-cycle is now generally accepted, and widespread distribution of AOA has been demonstrated (e.g., Francis et al., 2005; Biller et al., 2012; Stahl and de la Torre, 2012). Quantification of the relative abundances of AOA and ammonia-oxidizing bacteria (AOB) in different habitats (e.g., Leininger et al., 2006), including in Antarctic Peninsula soils (Jung et al., 2011), indicated a general dominance of AOA over AOB. However, ammonia-oxidizing betaproteobacteria have been shown to be more abundant and potentially more active than AOA in some estuarine and coastal sediments (Santoro et al., 2008; Magalhães et al., 2009), suggesting that the relative abundances and functional importance of AOB vs. AOA could vary in natural ecosystems. Recent studies indicated that environmental drivers like substrate (i.e., NH4+) concentration, pH, oxygen availability, salinity, among others, might be responsible for differentiating AOA and AOB abundance and distribution (Martens-Habbena and Stahl, 2011; Hatzenpichler, 2012; He et al., 2012; Prosser and Nicol, 2012; Zhalnina et al., 2012; Zhang et al., 2012). Despite previous attempts to evaluate drivers of natural AOA and AOB population dynamics, there remain large gaps in our understanding of factors that control AOA vs. AOB prominence in numerous ecosystems (Hatzenpichler, 2012; Prosser and Nicol, 2012; Monteiro et al., 2014).
The distribution and abundance of biota in the McMurdo Dry Valleys of Victoria Land in Antarctica are subject to strong spatial structuring due to the extreme heterogeneity in soil geochemical properties and severe climate gradients (Wood et al., 2008; Pointing et al., 2009; Lee et al., 2012; Magalhães et al., 2012). The dominance of environmental filters, together with the trophic simplicity of the ecosystem, makes Dry Valley soils a perfect model for investigating the physicochemical drivers of microbial biodiversity and function. In this study, we examined how environmental variables may determine the diversity and abundance of AOA and AOB amoA genes in the Dry Valleys. Although N is thought to be the limiting factor in many terrestrial Antarctic ecosystems, particularly in the Dry Valleys, little is known about the abundance and diversity of organisms and genes involved in the N cycle (Barrett et al., 2007; Hopkins et al., 2008; Cary et al., 2010; Niederberger et al., 2012). Studies of microbial N processes in the Dry Valleys have primarily focused on the N-fixation pathway. These studies demonstrated an unexpectedly high diversity of diazotrophs in Dry Valley soils (Cavacini, 2001; Wood et al., 2008; Pointing et al., 2009), suggesting that both cyanobacteria and a diverse range of heterotrophic diazotrophs are important players in the total input of N to these extreme oligotrophic environments (Niederberger et al., 2012). Interestingly, previous surveys of bacterial 16S rRNA genes in the Dry Valleys recovered sequences closely related to AOB groups (i.e., Nitrosomonas; Niederberger et al., 2008; Lee et al., 2012), and a GeoChip analysis of Dry Valley soils identified genes involved in the N cycle (Chan et al., 2013). However, there is very limited research on the dynamics of AOB and archaea in these hyper-arid cold deserts. Reports of bacterial and archaeal amoA abundance and diversity are so far restricted to the considerably wetter Antarctic Peninsula (Yergeau et al., 2007; Jung et al., 2011). Recent studies reporting limited diversity and abundance of Archaea in the Dry Valleys have identified a consistently high proportion of sequences (80–99%) affiliated with Thaumarchaeota (formerly known as Crenarchaeota Marine Group 1.1b; Ayton et al., 2010; Richter et al., 2014). These findings represent cursory evidence for archaeal nitrification in the Dry Valleys.
In this study, we investigated the distribution, abundance, and diversity of AOA and AOB amoA genes in four McMurdo Dry Valleys, where soil bacterial diversity and geochemistry have been previously described (Lee et al., 2012). The previous study reported a high degree of physicochemical heterogeneity and distinct bacterial communities, likely driven by the disparate physicochemical conditions. We hypothesized that such physicochemical heterogeneities exert similar selective effects on AOA and AOB amoA genes distribution and abundance.
MATERIALS AND METHODS
DRY VALLEYS SOIL SAMPLES COLLECTION
Soils were collected from four different McMurdo Dry Valleys (Figure 1): Miers Valley (MV; 78°60’S 164°00’E), Upper Wright Valley (UW; 77°10’S, 161°50’E), Beacon Valley (BV; 77°48’S, 160°48’E), and Battleship Promontory (BP; 76°54’S 160°55’E). Miers Valley is a coastal, low altitude valley (153 m) with comparatively high C/N ratio and has been noted for sustaining diverse cyanobacterial and bacterial communities (Wood et al., 2008; Lee et al., 2012). Beacon and Upper Wright Valleys are higher altitude valleys (1500 and 1000 m, respectively), characterized by extremely low temperatures, strong desiccating winds, low C/N ratios, and high soil electrical conductivity, creating comparatively inhospitable environments for soil microorganisms (Wood et al., 2008; Lee et al., 2012). Battleship Promontory is a high altitude valley (1000 m) with transiently liquid water in snow melt ponds, leading to lower soil electrical conductivity and higher moisture content and creating favorable conditions for bacterial communities (Lee et al., 2012).
FIGURE 1.
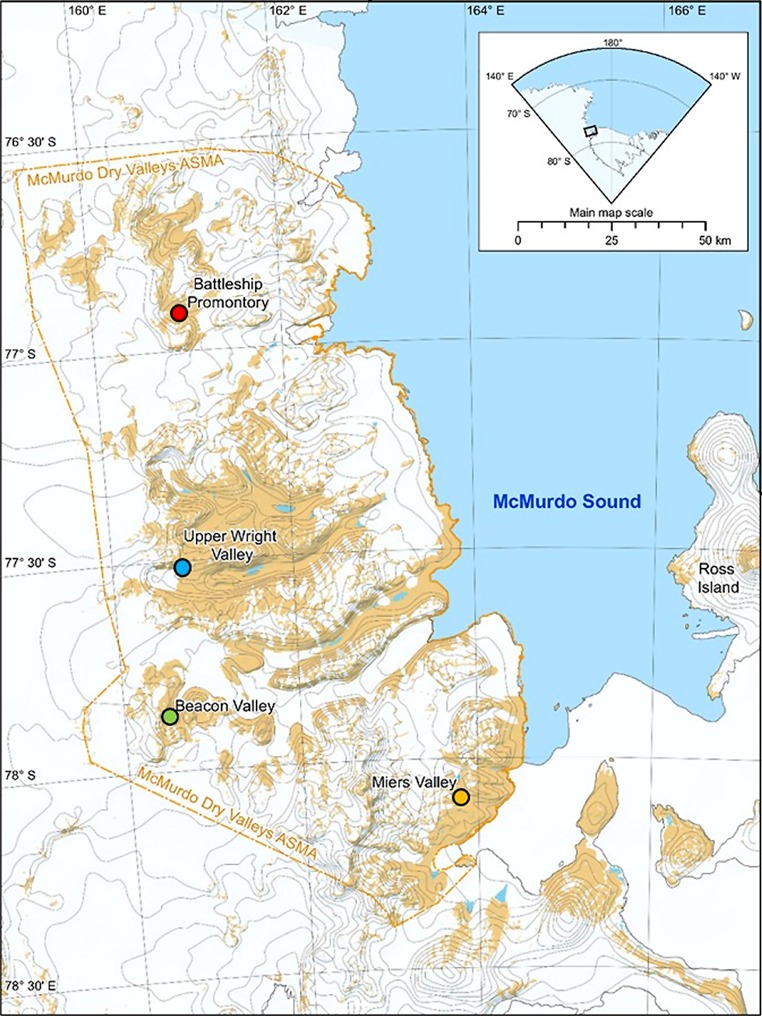
Map of the McMurdo Dry Valleys and the sampling sites.
In December 2006 (Miers Valley and Beacon Valley) and January 2008 (Battleship Promontory and Upper Wright Valley), two perpendicular transects of 50 m intersecting in the center were laid out at each sampling site, and four sampling points (A–D) were taken at the ends of each transect (Lee et al., 2012). At each sampling site, an area of 1 m2 was identified, and one scoop of soil was collected aseptically from the top 2 cm at the four corners of this 1 m2 area and combined in a sterile Whirl-Pak (Nasco International Inc., Fort Atkinson, WI, USA). All necessary and appropriate precautions were taken to avoid anthropogenic or cross-site contaminations. Samples were stored at -20°C at the earliest opportunity and transported back to University of Waikato, New Zealand, where they were stored at -80°C until analysis. Soil geochemical properties were determined as previously described (Lee et al., 2012).
DNA EXTRACTION
From each of the four samples collected at a sampling site (A–D), six replicates of DNA were extracted (total of 24 extractions per valley) from 0.6 to 0.8 g of homogenized soil using a modification of the CTAB (bromide-polyvinylpyrrolidone-bmercaptoethanol) extraction protocol (Coyne et al., 2001; Barrett et al., 2006). Recovered DNA was stored at –80°C. Reproducibility of DNA recovery, quantified using the QuBit dsDNA HS Kit (Life Technologies, Portugal), was tested among the six replicates (coefficient of variation = 14%). Three of the DNA extraction replicates were combined to obtain sufficient amounts of DNA for molecular analyses. Internal variations for each sample were assessed by processing two pooled replicates (composed of three DNA extractions) for each soil sample.
CLONING AND PHYLOGENETIC ANALYSIS
To assess AOA and AOB amoA genes diversity, two soil samples from each Dry Valley (i.e., MV-A, MV-B, BP-A, BP-B, BV-A, BV-B, UW-A, and UW-B) were randomly selected for clone library construction. Archaeal and bacterial amoA genes were amplified using Arch-amoAR/Arch-amoAF (Francis et al., 2005) and amoA-1F/534R (Rotthauwe et al., 1997) primers, respectively (Table 1). PCR products, generated using a previously described PCR protocol (Magalhães et al., 2009), were visualized with agarose gel (1.5%) electrophoresis, and a band of appropriate size was excised and purified with the QIAquick Gel Extraction Kit (Qiagen, Portugal). The resulting PCR amplicons were cloned using the TOPO TA Cloning Kit (Life Technologies, Portugal) according to manufacturer instructions. Plasmids were isolated using the GeneElute Plasmid Miniprep Kit (Sigma Aldrich, Spain), and DNA concentrations were determined with the QuBit dsDNA BS kit (Life Technologies, Portugal). Thirty colonies were selected randomly from each clone library (i.e., a total of 480 colonies for AOA and AOB amoA genes), and their insert sizes were verified by digesting 0.6–0.8 μg of DNA at 37°C for 2h with 10 U of EcoRI (Sigma-Aldrich, Portugal). A total of 420 clones with the correct insert size and containing likely AOA and AOB amoA genes were screened using restriction fragment length polymorphism analysis, where 0.6–0.8 μg of DNA was digested with 10 U of MspI (Promega, Europe) at 37°C overnight. A total of 15 clones of amoA AOA and AOB genes from each valley (total 120) were selected for sequencing in the STABVIDA Sequencing Facilities (Lisbon, Portugal).
Table 1.
Primers used in this study.
| Target gene | Primers | Sequence (5′–3′) | Reference |
|---|---|---|---|
| 16s rRNA | 341F | CCT ACG GGA GGC AGC AG | Muyzer et al. (1993) |
| 534R | ATT ACC GCG GCT GCT GG | ||
| βAOB amoA | amoA-1F | GGG GTT TCT ACT GGT GGT | Rotthauwe et al. (1997) |
| amoA-2R′ | CCT CKG SAA AGC CTT CTT C | Okano et al. (2004) | |
| Archaea amoA | Arch -amoAF | STAATGGTCTGGCTTAGACG | Francis et al. (2005) |
| Arch - amoAR | GCGGCCATCCATCTGTATGT | Francis et al. (2005) | |
| qPCR-AOA | AOAF | CCTACCACAAGCATAGT | This study |
| AOAR | GTTAACAGCACCTTACTTACT | This study | |
| qPCR-AOB | AOBF | GTCTCCATGCTCATGTTC | This study |
| AOBR | GGACCTTTGACGTAGAAGAA | This study |
Sequences were aligned with published AOA and AOB amoA sequences in GenBank using the basic local alignment search tool (BLAST). All sequences were aligned with Clustal W (Thompson et al., 1994) as implemented in Bioedit version 7.0.5 (Hall, 1999), and phylogenetic trees were constructed using MEGA version 3.1 (Kumar et al., 2001) with both maximum parsimony and neighbor-joining methods. Bootstrap analysis (1,000 replicates) was carried out. We used a 98% similarity cut off for defining operational taxonomic units (OTUs). Clone sequences from this study have been deposited in GenBank under accession numbers KF574112 to KF574224.
QUANTITATIVE REAL-TIME PCR
Quantitative PCR (qPCR) was conducted in a CFX96 real-time PCR detection system (Bio-Rad, Portugal) to determine copy numbers of bacterial 16S rRNA, bacteria amoA, and archaeal amoA genes using previously described 16S rRNA primers and new amoA primers designed for this study (Table 1). For each soil sample, qPCR was performed in triplicates for both pooled DNA replicates as 25 μl reactions with 10–20 ng of template DNA in each reaction. Each qPCR tube (white 0.2 ml PCR strips with ultra clear optical flat caps, [Bio-Rad, Portugal]) contained 12.5 μl of iQ Sybr Green Supermix (BioRad, Portugal), 2 μl of each primer (10 μM), and nuclease-free water (Promega, Portugal). For both primer sets, the thermal cycler was programmed for 5 min of denaturation at 94°C; followed by 8 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 30 s, and extension at 72°C for 30 s; followed by 27 cycles where the annealing temperature was changed to 57 °C; and a final extension step at 72°C for 10 min. Standards consisted of plasmids with AOA and AOB amoA gene inserts from clones generated for this study, as described above. Standard curves were generated in duplicate for each primer set, and amplification of standards was linear over six orders of magnitude (i.e., 0.2–0.2 × 10-6 ng of DNA). The R2 values between plasmid DNA copy numbers and the calculated threshold cycle values ranged from 0.98 to 1.00, and amplification efficiency ranged between 98 and 101% for all standard curves. Target copy numbers were calculated with an average molecular weight of 618 g mol-1, and data were standardized to copies of gene per g of sediment. Melting curves and agarose gel electrophoresis of the qPCR products were carried out following each qPCR assayed to confirm identity of the PCR products. Primers for qPCR were designed for AOA and AOB (Table 1) using AlleleID 7.6 software (Prenier Biosoft, International).
STATISTICAL ANALYSES
Relationships between gene copy numbers and soil physicochemical properties (described in Lee et al., 2012) for 16 samples (four sampling sites in each of the four Dry Valleys) were analyzed using multivariate ordination tools. Redundancy analysis (RDA) was selected as the preferred ordination method (ter Braak and Smilauer, 2002) and performed using CANOCO (version 4.5, Microcomputer Power, Ithaca, NY, USA). For RDA, environmental variables (i.e., pH, electrical conductivity, gravimetric water content, C/N, Mg, Cr, Mn, Co, Ni, and Cu) were normalized to a mean of 0 and SD of (1). Monte Carlo permutation test was used to assess the statistical significance of the relationships.
RESULTS AND DISCUSSION
AOA AND AOB DIVERSITY
Previous studies have reported high diversities and widespread distribution of AOA and AOB in natural and managed soils, marine and estuarine water and sediments, wastewater treatment bioreactors, hot springs, and many other environments (Francis et al., 2005; Beman and Francis, 2006; Leininger et al., 2006; Park et al., 2006; Dang et al., 2008; Magalhães et al., 2009; Biller et al., 2012; Stahl and de la Torre, 2012). In this study, we report AOA and AOB presence in the extreme environments of the Transantarctic Mountains. AOA and AOB amoA gene diversity recovered from these soils were extremely low, with only four AOA and three AOB amoA OTUs identified from a total of 420 clones (Figures 2 and 3). However, we cannot exclude the possibility that our primers failed to amplify some amoA genes, resulting in a lower observed AOA and AOB amoA diversity since considerable numbers of microorganisms in the Dry Valleys cannot be reliable assigned to higher taxonomic levels (Cary et al., 2010).
FIGURE 2.
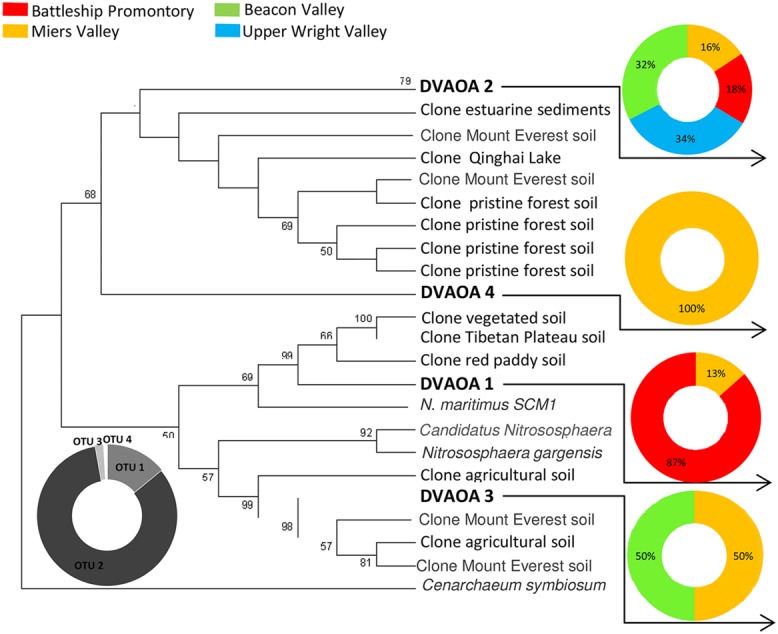
Phylogenetic relationship among Archaeal amoA sequences retrieved from Antarctica Dry Valleys and other geographic locations. The neighbor-joining tree was based on 591 nucleotide sequences and was constructed based on Kimura distances and the neighbor-joining method. Distance bootstrap values ≥50% are indicated at branch points (1000 iterations). The major clusters indicated were also supported by maximum parsimony analysis. The 210 Archaeal amoA clones obtained from this study are distributed under DVAOA1, DVAOA2, DVAOA3, and DVAOA4. Gray scale pie plot indicate the percentage of occurrence of each OTU in the total sequences, and colorful pie plots indicate the percentage of occurrence of each OTU within the four Dry Valleys.
FIGURE 3.
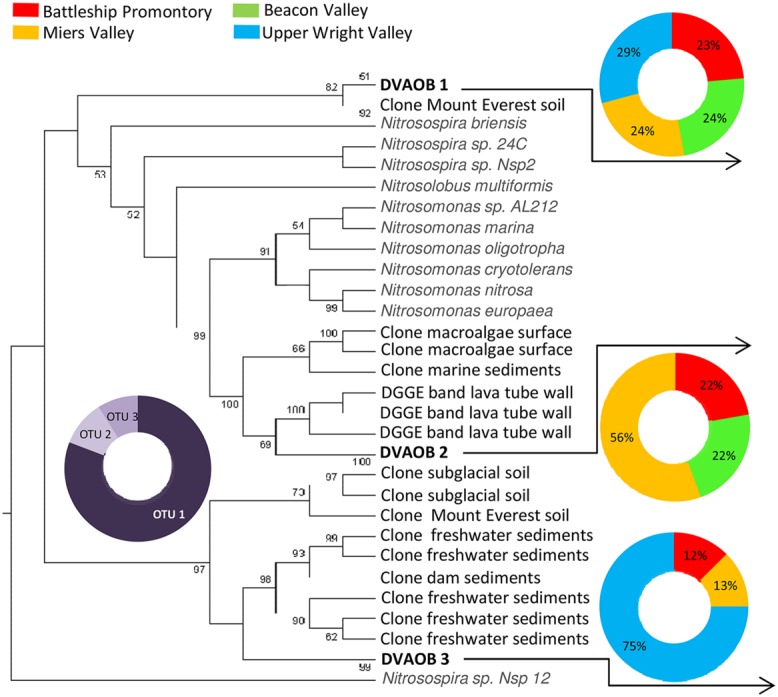
Phylogenetic relationship among Bacterial amoA sequences retrieved from Antarctica Dry Valleys and other geographic locations. The neighbor-joining tree was based on 443 nucleotide sequences and was constructed based on Kimura distances and the neighbor-joining method. Distance bootstrap values ≥50% are indicated at branch points (1000 iterations). The major clusters indicated were also supported using maximum parsimony analysis. The 210 Bacterial amoA clones obtained from this study are distributed under DVAOB1, DVAOB2, and DVAOB3. Gray scale pie plot indicate the percentage of occurrence of each OTU in the total sequences, and colorful pie plots indicate the percentage of occurrence of each OTU within the four Dry Valleys.
Archaea were only recently successfully detected in the Ross Sea region (Ayton et al., 2010) and across the McMurdo Dry Valleys (Richter et al., 2014), with reportedly low diversity and dominance of certain OTUs in these harsh environments. In Antarctic coastal mineral soils, 99% of the detected 16S rRNA sequences were affiliated with Thaumarchaeota (Ayton et al., 2010), as did more than 80% of all archaeal sequences identified in the McMurdo Dry Valleys (Richter et al., 2014). These findings hint at a high prevalence of amoA within the archaeal communities of Antarctic Dry Valley soils, which have been demonstrated by our results to have low genetic diversity (Figure 2). Taking this into account the quantification of the amoA could be a good representative of the whole archaeal community inhabiting Dry Valley soils.
Across the four AOA OTUs identified (DVAOA 1–4), 89% of the archaeal amoA clone sequences fell within DVAOA 2, which was also the only OTU recovered from all four sampling sites (Figure 2). The representative sequence of DVAOA 2 showed 95–96% nucleotide sequence similarity to sequences retrieved from a wide range of environments, such as temperate oxic lake water and anoxic sediments (Jiang et al., 2009), sediments from Plum Island Sound estuary [(Boston, Massachusetts, USA) Bernhard et al., 2010], alpine and permafrost Mount Everest bare soils (Zhang et al., 2009) and temperate pristine forest soils (Szukics et al., 2010). DVAOA2 is also less than 80% similar to the amoA gene of any known AOA isolates [71% to N. maritimus (Könneke et al., 2005)] and 79% to Candidatus Nitrososphaera gargensis (Spang et al., 2012). Only one AOA amoA OTU (DVAOA 2) was identified in the Upper Wright Valley, and Miers Valley had the highest diversity of amoA AOA, with four different OTUs present (Figure 2).
The identified AOB amoA sequences fell into two distinct phylogenetic clusters (Figure 3). All three amoA AOB OTUs (DVAOB1-3) were present in Miers Valley, whereas the other Dry Valleys contained only two of those OTUs. Similarly to the pattern for AOA amoA, the majority (81%) of amoA AOB clones fell within one OTU (i.e., DVAOB1), and the remaining clones (19%) were equally distributed between DVAOB2 and DVAOB3 (Figure 3). The representative sequences for DVAOB1 are 97% similar to a AOB amoA sequence detected in alpine soils on Mount Everest (Zhang et al., 2009). It is worth noting that a higher AOB amoA gene diversity (with sequences within four Nitrosospira and two Nitrosomonas-like clusters) was reported for the Mt. Everest soils (Zhang et al., 2009). Representative sequences for DVAOB1 and DVAOB2 were more closely related to Nitrosospira-like entries in GenBank (between 87 and 89% similarity) than Nitrosomonas-like sequences (between 70 and 78% similarity). Interestingly, previous studies have demonstrated that Nitrosomonas-like species are more tolerant of high environmental NH4+ concentrations than Nitrosospira-like species (de Bie et al., 2001; Cébron et al., 2004; Caffrey et al., 2007; Magalhães et al., 2009), which are typically associated with pristine environments with low NH4+ concentrations (Stephen et al., 1996, 1998; McCaig et al., 1999). In the extremely oligotrophic Dry Valley soils, nutrient limitation has been found to impose strong limitations on the distribution of soil microbiota (Cary et al., 2010; Lee et al., 2012; Magalhães et al., 2012). The representative sequence of DVAOB 3 showed lower similarities to amoA gene sequences from isolates (73–77% similarity). However, clustered (94–95% similarity) with sequences retrieved from a range of contrasting environments like lava cave walls biofilms (Azores, Portugal (Hathaway et al., 2014), sediments from subtropical freshwater marshes [Hongne Reserve, China (Lee and Gu, unpublished)] and subglacial soils from Robertson Glacier [Alberta, Canada (Boyd et al., 2011)].
Our findings showed that although AOA and AOB amoA gene diversities in the Dry Valleys were low, there were clear differences in the distribution of AOA and AOB among the four Dry Valleys examined.
AOA AND AOB COMMUNITY ABUNDANCE
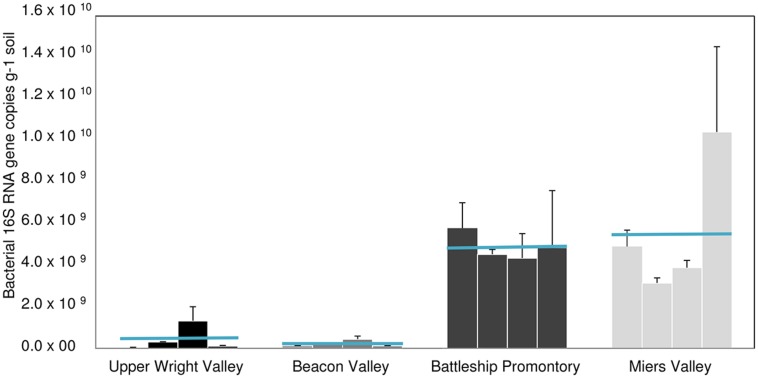
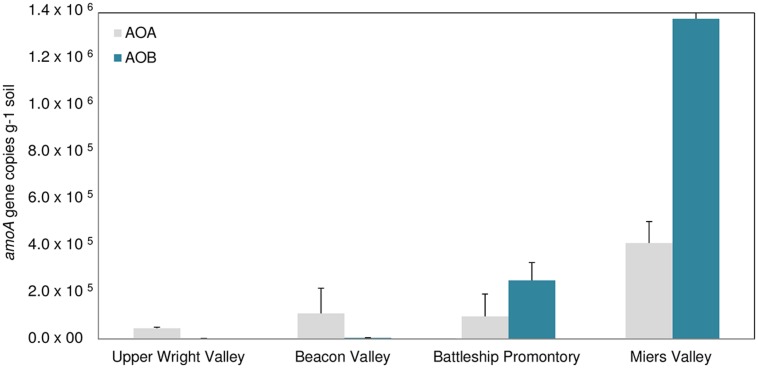
qPCR analysis of AOB amoA gene showed substantially higher abundance of AOB amoA gene at BP and MV (Figure 4A) compared to UW and BV (ANOVA, p < 0.001; Figure 4A). Coincidentally, qPCR analysis of bacterial 16S rRNA genes showed much higher abundance in BP and MV (5.6 ± 1.4 × 109 and 4.9 ± 1.4 × 109 copies g-1 soil, respectively) than in UW and BV (2.2 ± 1.4 × 108 and 4.4 ± 1.8 × 108, copies g-1 soil, respectively; Figure 5). These values fall within the range of bacterial 16S rRNA gene copy numbers previously reported for McKelvey Valley (0.06 × 108 to 2.4 × 108, Pointing et al., 2009), and for Ross Island and several McMurdo Dry Valleys, from direct cell counts (Ayton et al., 2010). AOA amoA gene qPCR results showed a different pattern, where amoA gene copy numbers did not differ significantly among samples from UW, BV, and BP, and were only significantly higher (p < 0.001) in samples from MV (Figure 4B). Soils from MV have been described as geochemically distinct (i.e., high C/N, high pH, etc.; Lee et al., 2012) from other Dry Valleys, and these conditions may favor nitrification and thus allow high diversity and abundance of bacterial and archaeal amoA. This is congruent with high bacterial and cyanobacterial diversities previously documented in samples from MV (Wood et al., 2008; Lee et al., 2012).
FIGURE 4.
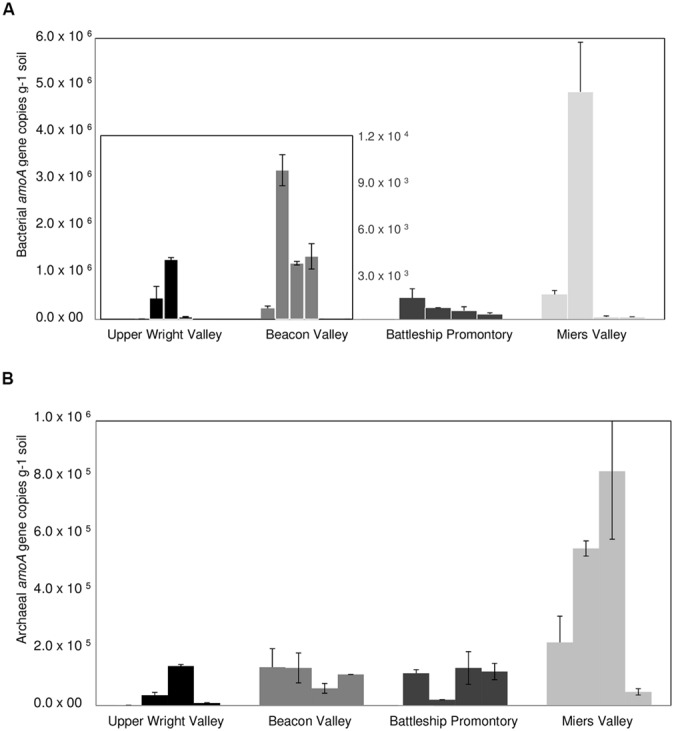
Bacterial (A) and archaeal (B)amoA gene copy numbers for locations A, B, C, and D) in Upper Wright, Beacon and Miers Valleys and in Battleship Promontory (mean ± SD of 3 qPCR replicates).
FIGURE 5.
16S rRNA gene copy numbers for locations A, B, C, and D of the four Dry Valleys (mean ± SD of 3 qPCR replicates). Blue lines show the mean value for the four sites of each valley.
Large spatial differences in AOA and AOB amoA gene abundance were detected among the four Dry Valleys (Figures 4 and 6). For example, AOB amoA gene abundances were three orders of magnitude higher in MV soils relative to UW soils (Figures 4A and 6). AOA and AOB amoA gene abundance reported here are similar to values obtained in higher altitude soils (≥5700 m) of Mount Everest, but one to three magnitudes lower than those observed for lower elevation mountain soils (<5400 m; Zhang et al., 2009). Interestingly, the amoA gene copy numbers for the Dry Valleys are significantly higher than those reported for soils from the Antarctic Peninsula (Jung et al., 2011). This may be due to the fact that we developed and optimized a new set of qPCR primers for archaeal and bacterial amoA genes found in the Dry Valleys, whereas the qPCR primers used by Jung et al. (2011) were based on existing primer sets developed for very different environments (Rotthauwe et al., 1997; Okano et al., 2004; Francis et al., 2005; de la Torre et al., 2008).
FIGURE 6.
Bacterial vs. archaeal amoA genes copy numbers in Upper Wright, Beacon and Miers Valleys and in Battleship Promontory (mean ± SD, of 3 qPCR replicates performed in each of the four sites sampled in each valley, n = 12).
Since the discovery of chemoautotrophic AOA (Venter et al., 2004; Könneke et al., 2005; Schleper et al., 2005; Treusch et al., 2005), their high relative abundances have been reported for many systems (Hallam et al., 2006; Leininger et al., 2006; Wuchter et al., 2006; Nicol et al., 2008). These studies showed a general prevalence of AOA over AOB, as was previously reported for soils from the Antarctic Peninsula (Jung et al., 2011). We observed more complex AOA vs. AOB dynamics in the Dry Valleys, with significant differences in the four Dry Valleys examined. Although AOB outnumbered AOA in MV and BP, high relative abundances of AOA amoA genes were found in UW and BV (Figure 6), where lower bacterial 16S rRNA gene abundance (Figure 5) and diversity have been described (Lee et al., 2012). Quantitative PCR analysis of amoA genes revealed clear differences in the relative abundances of AOA and AOB amoA genes among samples from the four Dry Valleys. A shift in the relative abundance of AOA and AOB has been previously reported for Mount Everest alpine and permafrost environments with greater abundance of AOA over AOB at lower (4000 m), and the reverse at higher (5800–6000 m) altitudes (Zhang et al., 2009). In addition, the results for MV and BP are in agreement with what had been observed in subglacial soils (Boyd et al., 2011), where bacteria appeared to be the predominant nitrifiers over archaea. It is also interesting to note that the AOB/AOA amoA gene ratio varied between 2.6 and 3.3 in MV and BP, but in UW and BV, where AOA amoA was more dominant, AOA/AOB ratios were much higher (33.1–24.0). Collectively, these results indicate spatial heterogeneity in the microorganisms that mediate the ammonia oxidation pathway in these extreme environments, and that the harsh conditions in UW and BV potentially impose physiological challenges for AOB and limit their distribution.
ENVIRONMENTAL CONTROLS ON AOA AND AOB DISTRIBUTION
The clearly dichotomized distribution of AOA and AOB in the four Dry Valleys examined (Figure 6) provides a rare opportunity to identify the environmental factors that control the relative abundance and diversity of these two groups of ammonia oxidizers.
As previously described, soil geochemical characteristics of McMurdo Dry Valleys varied significantly among the studied valleys (Lee et al., 2012). Large heterogeneities in bacterial diversity (Lee et al., 2012), bacterial abundance (Pointing et al., 2009), and the diversity of stress response pathways (Chan et al., 2013) among different Dry Valleys reflect significant variances in soil geochemical properties that characterize the Dry Valley ecosystem (Bockheim, 1997; Lamsal and Paudyal, 1999; Lee et al., 2012). Among the Dry Valleys examined for this study, MV stood out by virtue of the high C/N ratio, high total % C content, low conductivity, and higher pH of its soils (Lee et al., 2012), which may explain higher overall AOA and AOB amoA gene copy numbers (Figures 4A,B) and diversity (Figures 2 and 3). The low C/N ratios and high soil conductivity that characterized BV and UW soils (Lee et al., 2012) correlate with lower bacterial abundance (Figure 5) and drastically lower AOB amoA diversity (Figure 3) and gene copy numbers (ANOVA, p < 0.01; Figure 4A).
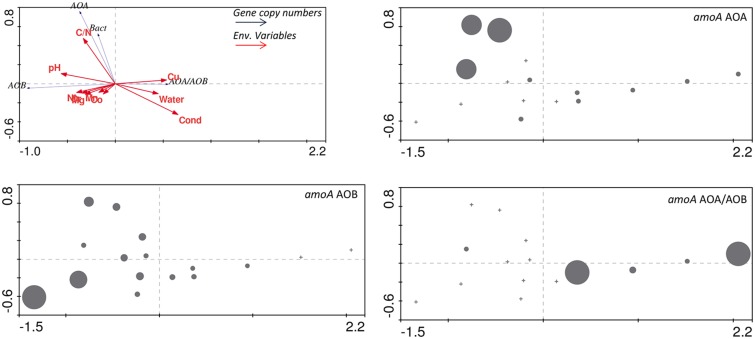
Correlations between environmental variables and 16S rRNA and amoA gene copy numbers were examined using (RDA; Figure 7). A Monte Carlo test of F-ratios (F = 2.54 and p = 0.044) identified ten environmental variables (pH, conductivity, gravimetric water content, C/N, Mg, Cr, Mn, Co, Ni, and Cu) among those reported in Lee et al. (2012) that significantly contributed to differences in gene copy numbers. The first gradient (RDA 1, horizontal, Figure 7) explained 75% of the variability in gene copy numbers and was highly correlated with the environmental variables (93.2%). The results revealed that higher abundance of AOA and AOB amoA genes are most strongly correlated with higher C/N ratio and pH in the soils (Figure 7). Higher AOA/AOB ratios were related to higher conductivity, Cu contents, and higher gravimetric water content (Figure 7). Our results suggest that AOA communities can better tolerate higher conductivity in BV and UW soils. This is in agreement with the documented prevalence of AOA over AOB in ocean waters (Santoro et al., 2010) where salinities are fairly stable at about 35 ppt. However, studies of coastal and estuarine sediments, characterized by highly fluctuating salinity levels, showed that high salinities favored numerical dominance of AOB over AOA (Santoro et al., 2008; Magalhães et al., 2009). Substrate (i.e., NH4+) concentration might also play an important role in the abundance and distribution of AOA and AOB, with AOA generally dominant in low NH4+ and oligotrophic environments (Martens-Habbena et al., 2009; Santoro et al., 2010; Martens-Habbena and Stahl, 2011; Zhalnina et al., 2012).
FIGURE 7.
Redundancy analysis ordination (RDA) plot for the biogeochemical variables and copy numbers of AOA and AOB amoA genes. Gene copy values for AOA, AOB and ratios of amoA AOA/AOB are represented as circles of diameter scaled linearly to the magnitude of the value. In the RDA ordination diagram, the angle and length of the arrow relative to a given axis reveals the extent of correlation between the variable and the canonical axis (environmental gradient).
CONCLUSION
The findings of this study represent the first report of the diversity and abundance of AOA and AOB communities in the Transantarctic Mountains, and make substantial contributions to our knowledge of the poorly understood N cycle of Antarctic Dry Valley soils. We confirmed the presence of archaeal and bacterial amoA genes in four Dry Valleys with disparate soil geochemical characteristics, demonstrating wide distributions and spatial heterogeneities of AOA and AOB, highlighting the potential role of nitrification in microbial processes in the Dry Valleys. We also identified a dichotomized distribution of AOA and AOB in the Dry Valleys that is potentially driven by environmental heterogeneities. Although more detailed studies are needed to fully understand the environmental drivers that control the relative abundance of AOA and AOB in natural environments, our results indicate that soil conductivity, may play an important role.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are sincerely grateful to Antarctica New Zealand for providing logistics support. We thank L. Torgo for their assistance in statistical analyses, F. Castro in designing the new primers, and A. Bordalo and A. P. Mucha for the loan of the real-time PCR system (CFX96, BioRad) and laboratory space for sampling processing. We also thank W. J. Wiebe for the extremely helpful comments on the manuscript. This study was conducted as part of the New Zealand Latitudinal Gradient Project (LGP) and the New Zealand Terrestrial Antarctic Biocomplexity Survey (nzTABS), a New Zealand International Polar Year program supported through a grant from the New Zealand Foundation for Research, Science and Technology to S. Craig Cary (UOWX0710). This study was funded by the Portuguese Science and Technology Foundation (FCT) through a researcher starting grant to Catarina M. Magalhães (FCT 2012) and by FCT and FEDER throughout a grant to Catarina M. Magalhães (PTDC/MAR/112723/2009–FCOMP-01-0124-FEDER-015422), and partially supported by the European Regional Development Fund (ERDF) through the COMPETE–Operational Competitiveness Program and national funds through FCT–Foundation for Science and Technology, under the project “PEst-C/MAR/LA0015/2011.” This study was also funded by PROPOLAR through a grant to Catarina M. Magalhães. CKL was supported by FRST Postdoctoral Research Fellowship (UOWX0715), the New Zealand Marsden Fund (UOW1003), and the New Zealand Antarctic Research Institute (NZARI2013-7).
REFERENCES
- Ayton J., Aislabie J., Barker G. M., Saul D., Turner S. (2010). Crenarchaeota affiliated with group 1.1b are prevalent in coastal mineral soils of the Ross Sea region of Antarctica. Environ. Microbiol. 12 689–703 10.1111/j.1462-2920.2009.02111.x [DOI] [PubMed] [Google Scholar]
- Barrett J. E., Virginia R. A., Lyons W. B., McKnight D. M., Priscu P. T., Doran P. T., et al. (2007). Biogeochemical stoichiometry of Antarctic Dry Valley ecosystems. J. Geophys. Res. 112 G01010. 10.1029/2005JG000141 [DOI] [Google Scholar]
- Barrett J. E., Virginia R. A., Wall D. H., Cary S. C., Adams B. J., Hacker A. L., et al. (2006). Co-variation in soil biodiversity and biogeochemistry in Northern and Southern Victoria Land, Antarctica. Antarct. Sci. 18 535–548 10.1017/S0954102006000587 [DOI] [Google Scholar]
- Beman J. M., Francis C. A. (2006). Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahía del Tóbari, Mexico. Appl. Environ. Microbiol. 72 7767–7777 10.1128/AEM.00946-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard A. E., Landry Z. C., Blevins A., de la Torre J. R., Giblin A. E., Stahl D. A. (2010). Abundance of ammonia-oxidizing Archaea and Bacteria along an estuarine salinity gradient in relation to potential nitrification rates. Appl. Environ. Microbiol. 76 1285–1289 10.1128/AEM.02018-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller S. J., Mosier A. C., Wells G. F., Francis C. A. (2012). Global biodiversity of aquatic ammonia-oxidizing archaea is partitioned by habitat. Front. Microbiol. 3:252 10.3389/fmicb.2012.00252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockheim J. G. (1997). Properties and classification of cold desert soils from Antarctica. Soil Sci. Soc. Am. J. 61 224–231 10.2136/sssaj1997.03615995006100010031x [DOI] [Google Scholar]
- Boyd E. S., Lange R. K., Mitchell A. C., Havig J. R., Hamilton T. L., Lafreniere M. J., et al. (2011). Diversity, abundance, and potential activity of nitrifying and nitrate-reducing microbial assemblages in a subglacial ecosystem. Appl. Environ. Microbiol. 77 4778–4787 10.1128/AEM.00376-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier-Armanet C., Boussau B., Gribaldo S., Forterre P. (2008). Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6 245–252 10.1038/nrmicro1852 [DOI] [PubMed] [Google Scholar]
- Caffrey J. M., Bano N., Kalanetra K., Hollibaugh J. T. (2007). Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1 660–662 10.1038/ismej.2007.79 [DOI] [PubMed] [Google Scholar]
- Cary S. C., McDonald I. R., Barrett J. E., Cowan D. A. (2010). On the rocks: the microbiology of Antarctic Dry Valley soils. Nat. Rev. Microbiol. 8 129–138 10.1038/nrmicro2281 [DOI] [PubMed] [Google Scholar]
- Cavacini P. (2001). Soil algae from northern Victoria Land (Antarctica). Polar Biosci. 14 45–60 [Google Scholar]
- Cébron A., Coci M., Garnier J., Laanbroek H. J. (2004). Denaturing gradient gel electrophoretic analysis of ammonia-oxidizing bacterial community structure in the lower Seine river: impact of Paris wastewater effluents. Appl. Environ. Microbiol. 70 6726–6737 10.1128/AEM.70.11.6726-6737.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y., Van Nostrand J., Zhou J., Pointing S. B., Farrell R. L. (2013). Functional ecology of an Antarctic dry valley. Proc. Natl. Acad. Sci. U.S.A. 110 8890–8895 10.1073/pnas.1300643110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne K. J., Hutchins D. A., Hare C. E., Cary S. C. (2001). Assessing temporal and spatial variability in Pfiesteria piscicida distributions using molecular probing techniques. Aquat. Microb. Ecol. 24 275–285 [Google Scholar]
- Dang H., Zhang X., Sun J., Li T., Zhang Z., Yang G. (2008). Diversity and spatial distribution of sediment ammonia-oxidizing crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea. Microbiol. 154 2084–2095 10.1099/mic.0.2007/013581-0 [DOI] [PubMed] [Google Scholar]
- de Bie M. J. M., Speksnijder A. G. C. L., Kowalchuk G. A., Schuurman T., Zwart G., Stephen J. R., et al. (2001). Shifts in the dominant populations of ammonia oxidizing β subclass Proteobacteria along the eutrophic Schelde estuary. Aquat. Microb. Ecol. 23 225–236 10.3354/ame023225 [DOI] [Google Scholar]
- de la Torre J. R., Walker C. B., Ingalls A. E., Könneke M., Stahl D. A. (2008). Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10 810–818 10.1111/j.1462-2920.2007.01506.x [DOI] [PubMed] [Google Scholar]
- Francis C. A., Roberts K. J., Beman J. M., Santoro A. E., Oakley B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 102 14683–14688 10.1073/pnas.0506625102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 41 95–98 [Google Scholar]
- Hallam S. J., Mincer T. J., Schleper C., Preston C. M., Roberts K., Richardson P. M., et al. (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:e95 10.1371/journal.pbio.0040095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway J. J. M., Sinsabaugh R. L., Dapkevicius M. L. N. E., Northup D. E. (2014). Diversity of ammonia oxidation (amoA) and nitrogen fixation (nifH) Genes in lava caves of terceira, Azores, Portugal. Geomicrobiol. J. 31 221–235 10.1080/01490451.2012.752424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R. (2012). Diversity, physiology and niche differentiation of ammonia-oxidizing archaea. Appl. Environ. Microbiol. 8 7501–7510 10.1128/AEM.01960-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.-Z., Hu H.-W., Zhang L.-M. (2012). Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil. Biol. Biochem. 55 146–154 10.1016/j.soilbio.2012.06.006 [DOI] [Google Scholar]
- Hopkins D. W., Sparrow A. D., Gregorich E. G., Elberling B., Novis P., Fraser F., et al. (2008). Isotopic evidence for the provenance and turnover of organic carbon by soil microorganisms in the Antarctic Dry Valleys. Environ. Microbiol. 11 597–608 10.1111/j.1462-2920.2008.01830.x [DOI] [PubMed] [Google Scholar]
- Jiang H., Dong H., Yu B., Ly G., Deng S., Berzins N., et al. (2009). Diversity and abundance of ammonia-oxidizing archaea and bacteria in Qinghai Lake, Northwestern China. Geomicrob. J. 26 199–211 10.1080/01490450902744004 [DOI] [Google Scholar]
- Jung M. Y., Park S. J., Min D., Kim J. S., Rijpstra W. I. C., Sinninghe Damsté J. S., et al. (2011). Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I. 1a from an agricultural soil. Appl. Environ. Microbiol. 77 8635–8647 10.1128/AEM.05787-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M., Bernhard A. E., de la Torre J. R., Walker C. B., Waterbury J. B., Stahl D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437 543–546 10.1038/nature03911 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Jakobsen I. B., Nei M. (2001). Mega 2: molecular evolutionary genetics analysis software. Bioinformatic 17 1244–1245 10.1093/bioinformatics/17.12.1244 [DOI] [PubMed] [Google Scholar]
- Lamsal K., Paudyal G. (1999). Model for assessing impact of salinity on soil water availability and crop yield. Agric. Water Manage. 41 57–70 10.1016/S0378-3774(98)00116-4 [DOI] [Google Scholar]
- Lee C. K., Barbier B. A., Bottos E. M., McDonald I. R., Cary S. C. (2012). The Inter-Valley Soil Comparative Survey: the ecology of Dry Valley edaphic microbial communities. ISME J. 6 1046–1057 10.1038/ismej.2011.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S., Urich T., Schloter M., Schwark L., Nicol G. W., Qi J., et al. (2006). Archaea predominance among ammonia-oxidizing prokaryotes. Nature 442 806–809 10.1038/nature04983 [DOI] [PubMed] [Google Scholar]
- Magalhães C., Machado A., Bordalo A. A. (2009). Temporal variability of relative abundance of ammonia oxidizing bacteria vs. archaea in the sandy at of the Douro River estuary, Portugal. Aquat. Microb. Ecol. 56 13–23 10.3354/ame01313 [DOI] [Google Scholar]
- Magalhães C., Stevens M. I., Cary S. C., Ball B. A., Storey B. C., Wall D. H., et al. (2012). At the limits of life: multidisciplinary insights on biotic diversity and environmental constraints in the Transantarctic Mountains, Antarctica. PLoS ONE 7:e44579 10.1371/journal.pone.0044578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Habbena W., Berube P. M., Urakawa H., de la Torre J. R., Stahl D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461 976–979 10.1038/nature08465 [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W., Stahl D. A. (2011). Nitrogen metabolism and kinetics of ammonia oxidizing archaea. Methods Enzymol. 496 465–487 10.1016/B978-0-12-386489-5.00019-1 [DOI] [PubMed] [Google Scholar]
- McCaig A. E., Phillips C. J., Stephen J. R., Kowalchuk G. A., Harvey S. M., Herbert R. A., et al. (1999). Nitrogen cycling and community structure of proteobacterial beta-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl. Environ. Microbiol. 65 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro M., Senéca J., Magalhães C. (2014). The history of aerobic ammonia oxidizers: from the first discoveries to today. J. Microbiol. 52 537–547 10.1007/s12275-014-4114-0 [DOI] [PubMed] [Google Scholar]
- Muyzer G., de Waal E. C., Uitterlinder A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol G. W., Leininger S., Schleper C., Prosser J. I. (2008). The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10 2966–2978 10.1111/j.1462-2920.2008.01701.x [DOI] [PubMed] [Google Scholar]
- Niederberger T. D., McDonald I. R., Hacker A. L., Soo R. M., Barrett J. E., Wall D. H., et al. (2008). Microbial community composition in soils of Northern Victoria Land, Antarctica. Environ. Microbiol. 10 1713–1724 10.1111/j.1462-2920.2008.01593.x [DOI] [PubMed] [Google Scholar]
- Niederberger T. D., Sohm J. A., Tirindelli J., Gunderson T., Capone D. G., Carpenter E. J., et al. (2012). Diverse and highly active diazotrophic assemblages inhabit ephemerally wetted soils of the Antarctic Dry Valleys. FEMS Microbiol. Ecol. 82 376–390 10.1111/j.1574-6941.2012.01390.x [DOI] [PubMed] [Google Scholar]
- Okano Y., Hristova K. R., Leutenegger C. M., Jackson L. E., Denison R. F., Gebreyesus B., et al. (2004). Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 70 1008–1016 10.1128/AEM.70.2.1008-1016.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.-D., Wells G. F., Bae H., Criddle C. S., Francis C. A. (2006). Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl. Environ. Microbiol, 72 5643–5647 10.1128/AEM.00402-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M., Rattei T., Flechl S., Gröngröft A., Richter A., Overmann J., et al. (2012). amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ. Microbiol. 14 525–539 10.1111/j.1462-2920.2011.02666.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointing S. B., Chan Y., Lacap D. C., Lau M. C. Y., Jurgens J. A., Farrell R. L. (2009). Highly specialized microbial diversity in hyper-arid polar desert. Proc. Natl. Acad. Sci. U.S.A. 107 1254–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser J. I., Nicol G. W. (2012). Archaeal and bacterial ammonia- oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 20 523–531 10.1016/j.tim.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Richter I., Herbold C. W., Lee C. K., McDonald I., Barrett J. E., Cary S. C. (2014). Influence of soil properties on archaeal diversity and distribution in the McMurdo Dry Valleys, Antarctica. FEMS Microbiol. Ecol. 89 347–359 10.1111/1574-6941 [DOI] [PubMed] [Google Scholar]
- Rotthauwe J.-H., Witzel K. P., Liesack W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63 4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro A. E., Casciotti K. L., Francis C. A. (2010). Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ. Microbiol. 12 1989–2006 10.1111/j.1462-2920.2010.02205.x [DOI] [PubMed] [Google Scholar]
- Santoro A. E., Francis C. A., de Sieyes N. R., Boehm B. B. (2008). Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 10 1068–1079 10.1111/j.1462-2920.2007.01547.x [DOI] [PubMed] [Google Scholar]
- Schleper C., Jurgens G., Jonuscheit M. (2005). Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 3 479–488 10.1038/nrmicro1159 [DOI] [PubMed] [Google Scholar]
- Spang A., Poehlein A., Offre P., Zumbrägel S., Haider S., Rychlik N., et al. (2012). The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ. Microbiol. 14 3122–3145 10.1111/j.1462-2920.2012.02893.x [DOI] [PubMed] [Google Scholar]
- Stephen J. R., Kowalchuk G. A., Bruns M. A. V., McCaig A. E., Phillips C. J., Embley T. M., et al. (1998). Analysis of β-subgroup Proteobacterial ammonia oxidizer populations in soil by denaturating gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl. Environ. Microbiol. 64 2958–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen J. R., McCaig A. E., Smith Z., Prosser J. I., Embley T. M. (1996). Molecular diversity of soil and marine 16S rRNA gene sequences related to b-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62 4147–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., de la Torre J. R. (2012). Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66 83–101 10.1146/annurev-micro-092611-150128 [DOI] [PubMed] [Google Scholar]
- Szukics U., Abell G. C. J., Hodl V., Mitter B., Sessitsch A., Hackl E., et al. (2010). Nitrifiers and denitrifiers respond rapidly to changed moisture and increasing temperature in a pristine forest soil. FEMS Microbiol. Ecol. 72 395–406 10.1111/j.1574-6941.2010.00853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Braak C. J. F., Smilauer P. (2002).CANOCO Reference Manual and CanoDraw for Windows User’s Guide: Software for Canonical Community Ordination (version 4.5). Ithaca, NY: Microcomputer Power [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). Clustal-W-Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch A. H., Leininger S., Kletzin A., Schuster S. C., Klenk H. P., Schleper C. (2005). Novel genes for nitrite reductase and amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7 1985–1995 10.1111/j.1462-2920.2005.00906.x [DOI] [PubMed] [Google Scholar]
- Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304 66–74 10.1126/science.1093857 [DOI] [PubMed] [Google Scholar]
- Wood S. A., Rueckert A., Cowan D. A., Cary S. C. (2008). Sources of edaphic cyanobacterial diversity in the Dry Valleys of Eastern Antarctica. ISME J. 2 308–320 10.1038/ismej.2007.104 [DOI] [PubMed] [Google Scholar]
- Wuchter C., Abbas B., Coolen M. J. L., Herfort L., van Bleijswijk J., Timmers P., et al. (2006). Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U.S.A. 103 12317–12322 10.1073/pnas.0600756103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau E., Kang S., He Z., Zhou J., Kowalchuk G. A. (2007). Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J. 1 163–179 10.1038/ismej.2007.24 [DOI] [PubMed] [Google Scholar]
- Zhalnina K., de Quadros P. D., Camargo F. A. O., Triplett E. W. (2012). Drivers of archaeal ammonia-oxidizing communities in soil. Front. Microbiol. 3:210 10.3389/fmicb.2012.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.-M., Hu H.-W., Shen J. P., He J.-Z. (2012). Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6 1032–1045 10.1038/ismej.2011.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. M., Wang M., Prosser J. I., Zheng Y. M., He J. Z. (2009). Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest. FEMS Microbiol. Ecol. 70 52–61 10.1111/j.1574-6941.2009.00775.x [DOI] [PubMed] [Google Scholar]