SUMMARY
The manner by which genotype and environment affect complex phenotypes is one of the fundamental questions in biology. In this study, we quantified the transcriptome—a subset of the metabolome—and, using targeted proteomics, quantified a subset of the liver proteome from 40 strains of the BXD mouse genetic reference population on two diverse diets. We discovered dozens of transcript, protein, and metabolite QTLs, several of which linked to metabolic phenotypes. Most prominently, Dhtkd1 was identified as a primary regulator of 2-aminoadipate, explaining variance in fasted glucose and diabetes status in both mice and humans. These integrated molecular profiles also allowed further characterization of complex pathways, particularly the mitochondrial unfolded protein response (UPRmt). UPRmt shows strikingly variant responses at the transcript and protein level that are remarkably conserved among C. elegans, mice, and humans. Overall, these examples demonstrate the value of an integrated multilayered omics approach to characterize complex metabolic phenotypes.
INTRODUCTION
The central dogma of molecular biology states that genetic information encoded in DNA is first transcribed by RNA polymerase, then translated by ribosomes into proteins. However, the DNA sequence of a gene provides little information for predicting when, where, and to what extent its associated RNA and protein products will be expressed. Since the advent of microarray technology, comprehensive gene expression patterns—i.e., the transcriptome— can be precisely and comprehensively quantified across large populations. Unfortunately, transcript levels generally have only modest correlation with the levels of corresponding proteins (Ghazalpour et al., 2011; Gygi et al., 1999; Schwan-häusser et al., 2011), and genetic variants similarly affecting both the transcript and peptide levels of a gene are relatively uncommon (Albert et al., 2014; Skelly et al., 2013). As proteins in most cases are more directly responsible than transcripts in the regulation of cellular pathways—and ultimately phenotypic traits—there is a critical need for efficient, large-scale, and accurately quantitative proteomics methods to complement transcriptomic data sets.
Over the past decade, the development of discovery mass spectrometry (“shotgun”) has allowed the first large-scale studies on quantitative proteomics. In this approach, protein extracts are cleaved into short peptide sequences, which are then chromatographically separated and analyzed by tandem mass spectrometry. This allows the untargeted discovery of thousands of peptides, but if the number of unique peptide fragments in a sample significantly exceeds the number of available sequencing cycles (as in whole proteome extracts), any individual peptide will be inconsistently sampled across repeat analyses. This reduces the technical reproducibility but moreover means that the number of peptides consistently quantified across all (or most) samples decreases as the study size increases (Karpievitch et al., 2012). Consequently, discovery mass spectrometry strategy has yielded mixed results in large population studies (Ghazalpour et al., 2011; Holdt et al., 2013), particularly as specific peptides of interest cannot be targeted, and the most consistently identified peptides are biased toward those of higher abundance (Callister et al., 2006). To overcome these hurdles, selected reaction monitoring (SRM) was developed, which perfects technical reproducibility and allows consistent multiplexed quantitation of target proteins by deploying a mass spectrometric measurement assay that is specific for each targeted peptide (Lange et al., 2008a). Thus, hundreds of target peptides can be consistently and accurately quantified across large populations of samples. Recent studies in yeast have shown that the proteins and transcripts of genes are typically controlled by different, distinct mechanisms (Albert et al., 2014; Picotti et al., 2013). However, these hypotheses have not been well tested in mammalian genetic reference populations (GRPs) through multilayered transcriptomic and proteomic strategies.
Large GRPs are frequently used to determine to which extent phenotypic variation is driven by genetic variants (i.e., heritability), and to subsequently identify genes driving such variation. These genes can be identified by genome-wide association studies (GWASs) or by quantitative trait locus (QTL) mapping, approaches that have been applied to various species and have led to the successful identification of dozens of major allelic variants (Andreux et al., 2012; De Luca et al., 2003; Deeb et al., 1998; Yvert et al., 2003). In mammals, the murine BXD family is the largest and most well-studied GRP, consisting of ~150 recombinant inbred strains descended from C57BL/6J (B6) and DBA/2J (D2) (Andreux et al., 2012). Using 40 strains of this population on both chow (CD) and high fat (HFD) diets, we have obtained major metabolic phenotypes and established a multilayered data set focused on 192 metabolic genes expressed in the liver. For all genes, we know the sequence variants, transcript levels, and protein levels in all cohorts. These data are further supplemented with targeted metabolite analysis in the liver and serum, generating the first large-scale multilayered quantitative picture of any cellular process in the BXD population.
RESULTS
Protein Targeting across a Genetically and Environmentally Diverse Murine Population
We first selected 192 metabolic proteins for study, with particular focus on genes regulating mitochondria and general energy metabolism. For each gene, synthetic peptides were generated based on established assays (Picotti et al., 2010) (Figure 1A) to accurately quantify each protein across all cohorts. To validate peptide measurements, we compared the coefficients of variation (CV) among technical and biological peptide replicates. Both technical and biological replicates showed a high degree of reproducibility (CV ~ 0.09 and 0.12, respectively), indicating the quantification of targeted peptides is sufficiently accurate across large, diverse populations. More importantly, the results for biological replicates reveal nearly equally high overall reproducibility (r > 0.98; Figure 1B), suggesting low biological variance within each cohort, particularly relative to cross-cohort variance (CV ~ 0.33); 82.1% of the quantified peptides are highly variable (CV > 0.20), indicating that the biological error (i.e., variation within a cohort) is much smaller than the variation induced by differences in genotypes and diet. We observed nearly complete quantification of all 192 proteins across all cohorts, with only ~60 missing peptide counts out of ~15,000 measurements (i.e., 99.6% completion; Figure 1C, top). This completion is similar to the level of completeness achieved by microarrays (Figure 1C, bottom) and is a major contrast to shotgun-acquired proteomics, which typically have completeness of ~70% (Karpievitch et al., 2012).
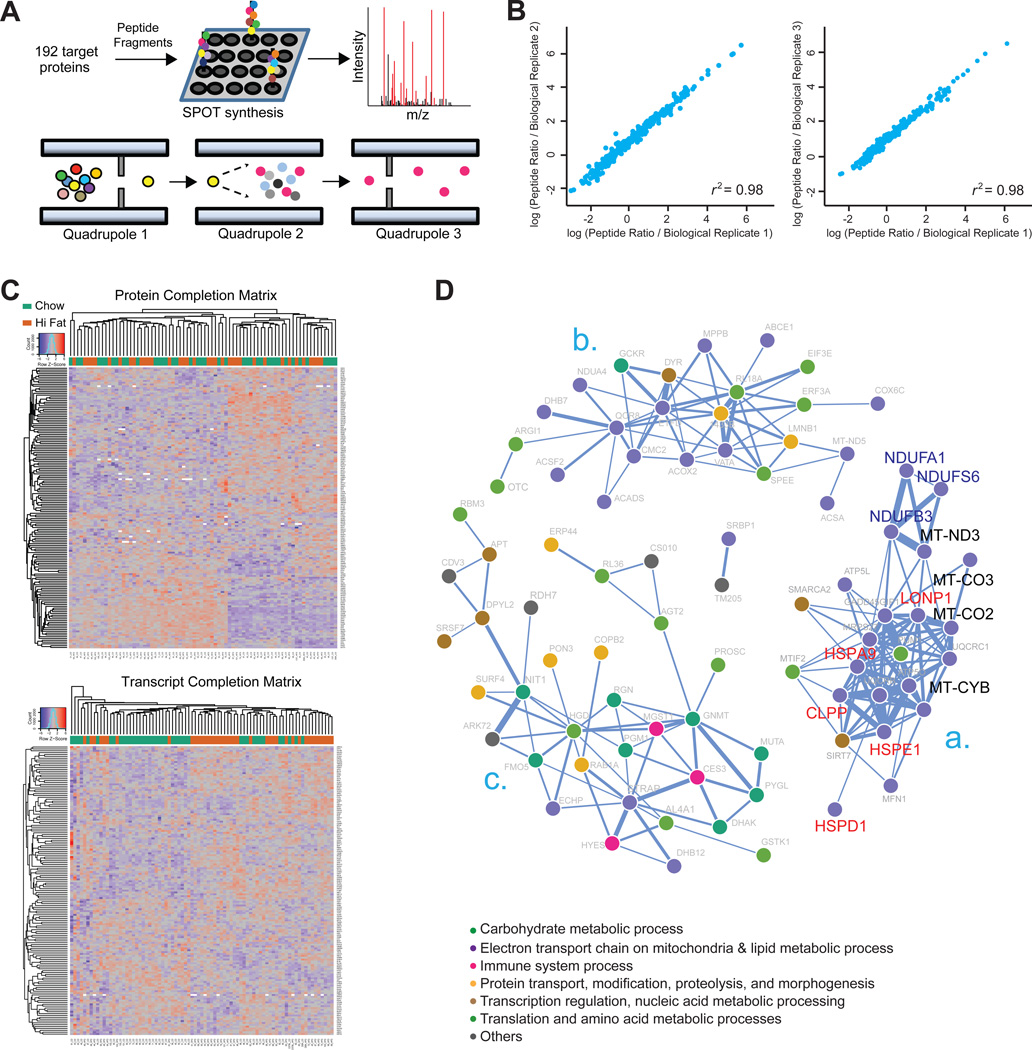
Figure 1. SRM-Based Protein Quantification and Covariation Network.
(A) SRM assay development for targeted proteomic measurements; 309 peptides corresponding to 192 genes were designed and synthesized via SPOT synthesis. Fragment ion spectra were generated on a triple-quadrupole MS with the SRM-triggered MS2 mode, and then ions were selected based on their relative intensities. Dot colors indicate different amino acids. The following abbreviation was used: m/z, mass-to-charge ratio. Mouse liver homogenate combined with the heavy reference proteome was analyzed with SRM on a triple-quadrupole MS. Different dot colors represent different peptides (Quadrupole 1) or product ions (Quadrupole 3).
(B) Biological replicates had near-perfect reproducibility (r2 ~ 0.98), shown here for all three biological replicates of the BXD60 CD cohort.
(C) Two-way cluster analysis of protein (top) and transcript (bottom) abundances in all 77 cohorts (40 CD, 37 HFD). Columns are clustered based on samples, and rows are clustered based on gene-product abundances. Protein and transcript abundances are colored in a red-blue scale. Red, high abundance; blue, low abundance; white, missing data.
(D) Protein association network based on robust Spearman correlation measures for all protein pairs. Statistically significant and strong positive associations (p < 0.01 and r > 0.6) are edges. The largest correlation clusters are labeled “a,” “b,” and “c.” Nodes are labeled with protein names and colored according to their biological process, as reported by DAVID (Huang et al., 2009).
It has been well established that transcriptomic networks of many metabolic processes covary quite well, e.g., within the electron transport chain or within the citric acid cycle (Ihmels et al., 2002). At the protein level, proteins that function in common biological processes or that localize to the same functional modules are also reported to be subject to a similar regulatory process and generally covary (Foster et al., 2006). To validate and identify which of these 192 proteins vary synchronously, we computed the robust Spearman correlation network for all protein pairs using the full SRM data set (Figure 1D). The resulting network contained 82 correlated nodes (proteins) with 211 edges in 3 main enrichment clusters. As expected, proteins within ontologies are highly correlated such as for mitochondria (Figure 1Da) and lipid metabolism (Figure 1Db). Within Figure 1Da are five of the six measured proteins involved in mitochondrial unfolded protein response (UPRmt) (HSPD1, HSPE1, HSPA9, CLPP, and LONP1, indicated in red), along with three of the four measured NADH dehydrogenase genes (NDUFA1, NDUFB3 and NDUFS6, in blue) and four of the eight measured mitochondrial-encoded proteins (MT-CYB, MT-CO2, MT-CO3, and MT-ND3, in black). Meanwhile, proteins involved in carbohydrate metabolism are enriched in the same cluster (Figure 1Dc). These results show that functionally related proteins tend to be coordinately regulated at a protein level and that coregulation of protein abundance is strongly maintained for certain processes. To validate the biological significance of these function-based covariation clusters, we further investigated one: the UPRmt network (elaborated on later).
Protein and mRNA Gene Products Generally Do Not Correlate
With the general protein measurements validated, we generated a global overview of how genotype and diet influence differential transcript and protein expression (Figure 2A; numerical values listed in Table S1 available online). At the genetic level, transcripts and proteins map to an equivalent number of significant QTLs: we detected 65 significant transcript QTLs (eQTLs—blue lines at the center) and 57 significant protein QTLs (pQTLs—red lines at the center). However, though the total number of eQTLs and pQTLs are roughly equivalent, the predominant type of regulation was very different: 74% of the eQTLs are cis-mapping (ratio of solid to dashed blue lines), versus only 31% of pQTLs (ratio of solid to dashed red lines). This indicates a closer connection between a transcript and its gene than a protein and its gene. In general, trans-mapping proteins and transcripts mapped evenly across the genome, not yielding any clear “hot spots” for these metabolic genes (sample magnification of QTLs mapping to chromosome 5 is shown in Figure 2B).
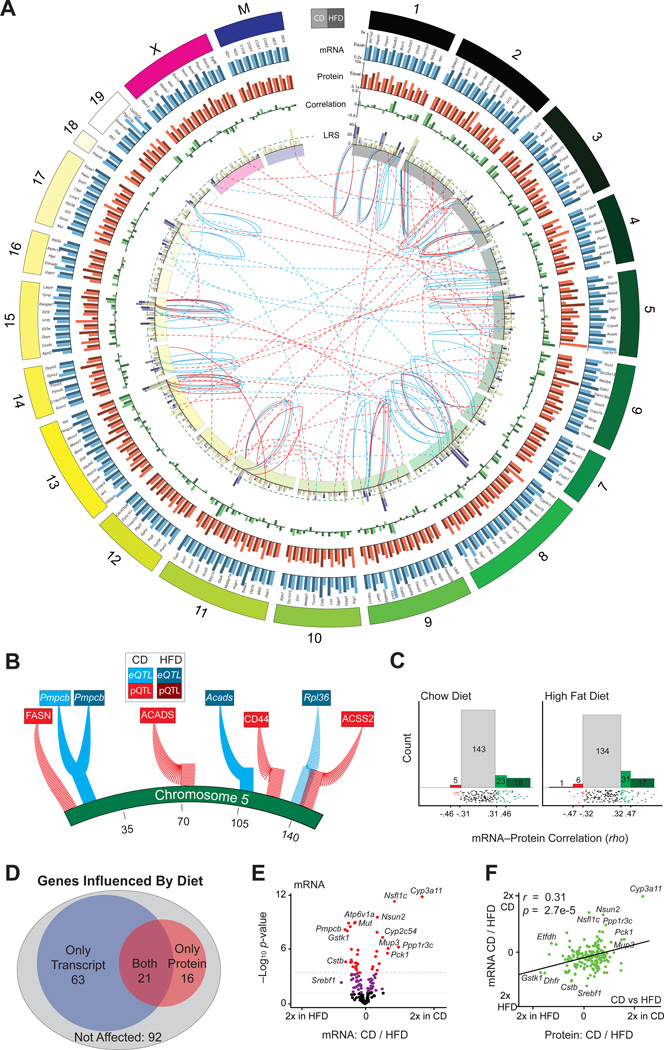
Figure 2. mRNA and Protein Overview.
(A) Circos plot of mRNA and protein data for all 192 genes, labeled on outer edge. Genes are represented by two bars: light for CD and dark for HFD. Genes are arranged by relative chromosome position; the chromosome length is according to number of genes measured. Blue bars indicate the transcript relative expression CD versus HFD; orange bars indicate the protein relative expression CD versus HFD. Bars with more unequal heights indicate diet has a larger impact. Green bars indicate the correlation between the transcript and protein for each gene within each diet. Fuchsia bars represent the strength of the peak eQTL. Yellow bars represent the strength of the peak pQTL. The two bars are overlaid with transparency. The dashed green line represents the simplified significance cutoff (LRS ≥ 18). The inner ring indicates the chromosome location. The blue central lines represent significant eQTLs, and the red central lines represent significant pQTLs. The central solid lines represent cis-QTLs, and the central dashed lines represent trans-QTLs. QTL lines stem from the LRS bar graph and terminate on the inner side of the chromosome ring at the approximate QTL location.
(B) Magnified view of eQTLs and pQTLs mapping to chromosome 5.
(C) In CD ~25% (left), and in HFD ~30% (right), of transcripts correlate nominally significantly with their protein. The lower strip charts show correlation distribution. Spearman correlation values corresponding to nominal significance (p < 0.05) and corrected significance (p < 0.0002) are displayed on the axis.
(D) Venn diagram of genes that are differentially regulated between CD and HFD as transcripts (blue), proteins (red), both (purple), or neither (gray).
(E) Volcano plot for mRNA showing the magnitude of dietary effect versus significance. ~45% vary with nominal significance (p < 0.05) between the dietary conditions. Approximately 19% vary with corrected significance (raw p < 0.0003). Some extreme genes are labeled.
(F) Plot of the effect of diet on transcripts versus the effect of diet on proteins. In general, transcripts and proteins are similarly affected by diet.
Within each diet, we found proteins and transcripts to be nominally correlated for ~25% of genes (i.e., Spearman correlation p value < 0.05; Figure 2C, diets considered separately), similar to findings in other species and populations (Foss et al., 2007; Ghazalpour et al., 2011; Schwanhäusser et al., 2011; Skelly et al., 2013). Of the correlated transcript-peptide pairs (46 in CD, 55 in HFD), 31 correlate significantly in both cohorts, whereas 70 pairs correlate in only one dietary cohort. Correspondingly, while ~50% of genes were affected by diet, transcripts were more frequently influenced (84 of 189) than proteins (37 of 192), with only 21 genes affected at both the transcript and protein levels (Figure 2D). Genes that were the most strongly affected by diet at the transcript level (Figure 2E) tend to be similarly affected at the protein level (Figure 2F)—e.g., Cyp3a11 is higher in CD—though exceptions are frequent, e.g., Srebf1 mRNA is induced by HFD, but unaffected at the protein level, while in another counterexample, the HFD cohorts have more ETFDH protein, but less of the transcript. Thus, while transcripts and proteins are moderately covarying estimations of their gene’s activity and typically have covarying responses to external factors (e.g., diet), these trends are too weak to support the measurement of any one particular transcript to serve as a proxy for the protein (or vice versa) without prior knowledge.
Most Transcript and Protein QTLs Do Not Overlap
Of the 192 target genes, 79 map to a significant eQTL or pQTL in at least one dietary condition. A strong majority of significantly mapped QTLs, ~80%, are unique to either the transcript level or protein level (Figure 3A). At the transcript level, 28 genes map to cis-eQTLs, 20 of which are in both diets, while 17 transcripts map as trans-eQTLs, none of which are observed in both diets. Together, we observed 65 significant eQTLs stemming from 45 distinct genes (Figure 3B, left; the 20 distinct genes that map cis in both diets contribute 40 significant eQTLs). The range of transcript variance within a diet was a strong predictive factor for observing an eQTL. Transcripts in the least variable quartile (range < 1.5-fold from the lowest to the highest expressing BXD strain) contained only 10% of the significant QTLs. In contrast, the second quartile (range 1.5- to 1.65-fold) contained 17%, the third quartile (range 1.65- to 2.0-fold) contained 29%, and the top quartile (range ≥ 2.0-fold) contained 41% of the significant QTLs.
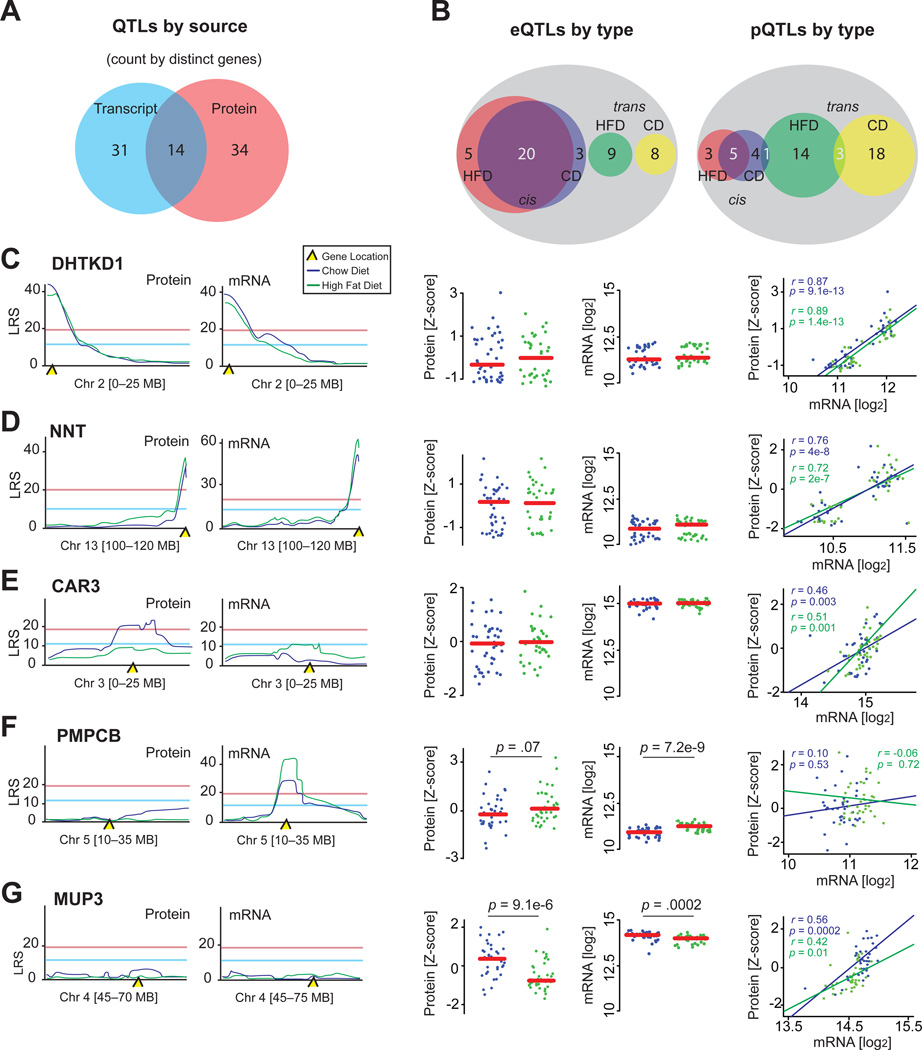
Figure 3. QTL Overview.
(A) Venn diagram separating all genes with distinct QTLs based on provenance.
(B) Venn diagrams separating eQTLs or pQTLs by dietary source and regulatory mechanism. Overlapping regions indicates genes giving an eQTL or pQTL in both diets; white numbers are counted twice for QTL count, but once for distinct gene count (e.g., there are 48 significant cis-eQTLs, which stem from 28 distinct genes).
(C) In both diets, DHTKD1 and Dhtkd1 share a common cis-QTL, are unaffected by diet, and strongly correlate.
(D) NNT and Nnt display a similar pattern.
(E) Car3 has only one significant pQTL despite an absence of dietary effect and a strong transcript-protein correlation.
(F) PMPCB does not map to a significant pQTL; however, Pmpcb maps to a significant cis-eQTL in both diets, despite a strong transcriptional upregulation by HFD. The transcript and protein levels do not correlate.
(G) Mup3 and MUP3 do not map to significant QTLs, despite having high levels of variation and a strong transcript-protein correlation.
Related to Figure S1.
At the protein level, 57 significant pQTLs stem from 48 distinct proteins (Figure 3B, right). In striking contrast to transcript regulation, only 13 distinct proteins map to cis-pQTLs, five in both diets, while 36 proteins map as trans-pQTLs. No trans-pQTLs are consistent across diets, although three proteins significantly map to separate trans-pQTLs in both diets (Cd44, Acss2, and Ndufs6), and one (Hmgcs1) maps to both cis- and trans-pQTLs. Together with transcript data, we could moderately predict pQTLs: of the 28 genes with cis-eQTLs, 12 correlated well (r > 0.40) between the transcript and protein. Of these 12 genes, five (Dhtkd1, Nnt, Tymp, Gclm, and Bckdhb) map significantly as pQTLs and eQTLs in both diets (Figures 3C, 3D, S2A, and S2B; Bckdhb in Figure 4B), and four (Acox1, Mfn1, Mri1, and Pm20d1) map as pQTLs and eQTLs in at least one diet. Interestingly, three proteins mapped as cis-pQTLs and correlated with their transcript, but did not have associated eQTLs (Hmgcs1, Car3, and Acsf2), suggesting the genetic variant driving these changes manifests more prominently at the protein level than at the transcript level (e.g., Figure 3E).
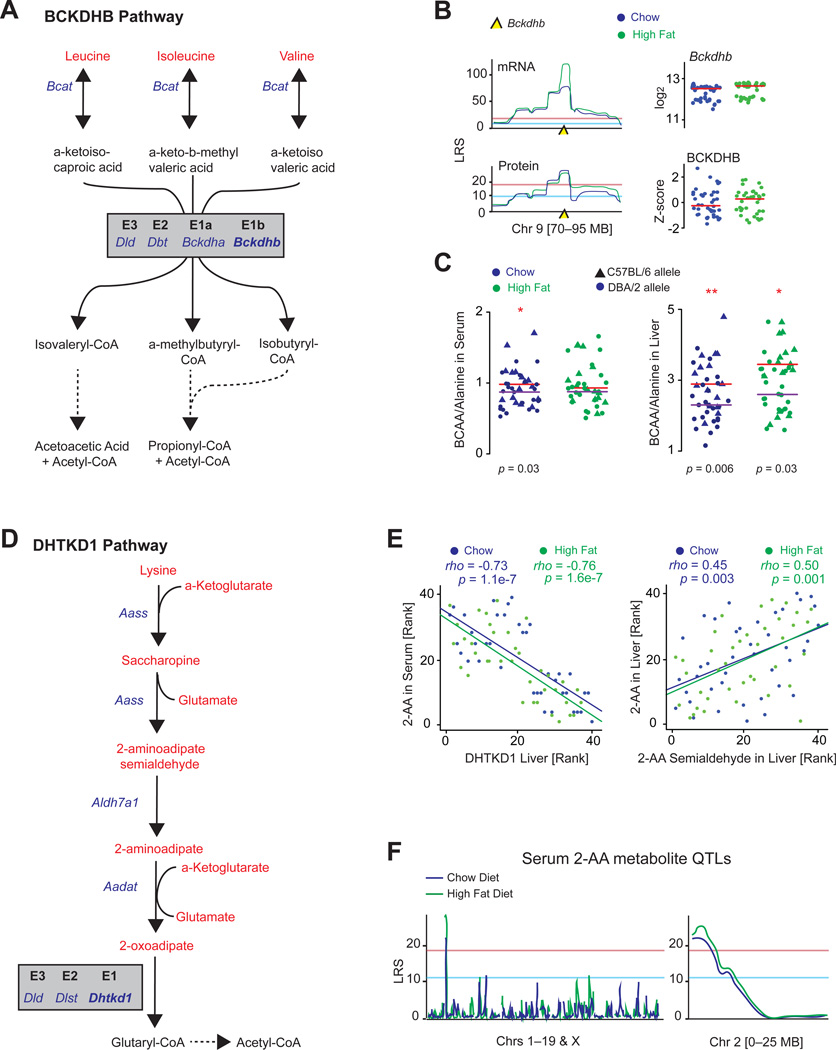
Figure 4. Metabolic Consequences of BCKDHB and DHTKD1.
(A) BCKDHB is the E1b subunit of the BCKD complex, which irreversibly converts several BCKAs.
(B) Bckdhb mRNA has the strongest eQTLs of the 192 target genes examined in the liver and has among the strongest pQTLs. Neither transcript nor protein is affected by diet.
(C) The BCAA/alanine ratio is significantly increased in animals with the dysfunctional C57BL/6 allele in CD serum and liver measurements and HFD liver measurements, in line with it acting as a risk for MSUD. Red line represents median of strains with the C57BL/6 allele, and purple lines represent the median of strains with the DBA/2 allele.
(D) DHTKD1 is the E1 subunit of the dehydrogenase complex that catalyzes the irreversible conversion of 2-oxoadipate to glutaryl-CoA.
(E) Left: serum 2-AA levels are strongly related to DHTKD1, as are liver levels (not shown). Right: other upstream metabolites in the DHTKD1 pathway correlate strongly with one another, e.g., 2-AA and 2-AA semialdehyde, shown here in liver.
(F) Serum 2-AA maps as an mQTL in both diets to proximal chromosome 2, the location of DHTKD1.
Diet also plays a clear role in the consistent identification of cis-QTLs. For the 20 genes with cis-eQTLs identified in both diets, HFD significantly affects the levels of only one (Pmpcb; Figure 3F). Conversely, for the eight genes mapping to an eQTL in only one diet, the levels of only three are affected (Ndufs2, Acads, and Aldh4a1). Similar trends are observed at the protein level: of the five cis-pQTLs found in both diets, none are affected by diet, while for the eight genes with cis-pQTLs in only one diet, only the levels of one (Hmgcs1) are affected. This relationship is similar for trans-eQTLs: of the 17 genes with trans-eQTLs, only four are influenced by diet (whereas ~50% of total transcripts are influenced by diet). However, diet does not appear to strongly influence trans-pQTLs, where 11 of the 39 are affected by diet, a similar proportion to the overall effect, recalling that ~20% of proteins have levels influenced by diet (Figure 2D). Together this suggests diet can change the genetic factors regulating these genes without changing their overall levels.
While high expression variability is associated with stronger QTLs, it is not a definitive predictor, e.g., Mup3 is highly variable, strongly affected by diet, and consistently expressed between mRNA and protein, but does not map to a QTL in any measurement (Figure 3G). Therefore, while the ability to predict peptide levels based on transcript measurements on a systems scale is quite powerful (the ~25% to ~37% of correlated transcript-peptide pairs is much better than the ~5% expected by chance), the probability to fail of any one particular gene is quite high. These chances can be adjusted somewhat—perturbations dramatically affecting transcript levels are more likely to manifest at the protein level and vice versa—but even so, prior research must be established before gene expression can be confidently considered a proxy for protein levels in targeted genetic studies. This also indicates that applying quantitative proteomic data to pathways established at the transcript level can indicate new links that were previously obscured.
Functional Relationships of pQTLs to Phenotypes
To characterize the cellular function and potential physiological relevance of the pQTLs, we first collated all Entrez (Maglott et al., 2005) and UniProt (Magrane and Consortium, 2011) entries for genes with the most significant pQTLs (LRS ≥ 20) (Table 1). As the BXDs have extensive historical phenotype and metabolite data available on GeneNetwork (Wang et al., 2003), we performed a phenome-wide association study to determine if any collected phenotype data mapped at least suggestively (likelihood ratio statistic [LRS] ≥ 12) as clinical QTLs (cQTL) to the same loci. A handful of phenotypic connections in the BXDs were supported by literature, including a link between Nnt and insulin (Wong et al., 2013) and between Car3 and subcutaneous adipose mass (Mitterberger et al., 2012). However, for the majority of pQTLs, no established cQTLs mapped to the same loci. We thus selected the two genes with the most significant and novel pQTLs in both diets for follow-up analysis and validation— Bckdhb and Dhtkd1.
Table 1.
pQTLs Link to Phenotypes
| Protein | Diet | LRS | QTL Area [Mb] | Gene | Function | Phenotype |
|---|---|---|---|---|---|---|
| DHTKD1 | CD | 43 | Chr2: 3–8 | cis | lysine catabolism | Reye syndrome, glucose regulation |
| DHTKD1 | HFD | 41 | Chr2: 3–8 | cis | lysine catabolism | Reye syndrome, glucose regulation |
| NNT | CD | 37 | Chr13: 119–120 | cis | NADPH regeneration | insulin secretion |
| GCLM | HFD | 34 | Chr3:118–124 | cis | glutathione synthesis | detoxification, ischemia, anemia |
| NNT | HFD | 31 | Chr13: 119–120 | cis | NADPH regeneration | insulin secretion |
| PRPS1 | CD | 31 | Chr6: 61–66 | 73 | nucleotide synthesis | arts syndrome |
| GPX1 | CD | 30 | Chr11: 119–122 | 70 | glutathione peroxidase | detoxification, cardiomyopathy |
| PM20D1 | HFD | 29 | Chr1: 134–137 | cis | zinc binding (putative) | tuberculosis sensitivity |
| MT-ND3 | CD | 28 | Chr3: 11–18 | 21 | complex I subunit | Leigh syndrome |
| CMAS | CD | 28 | Chr7: 95–105 | 70 | sialic acid adhesion | fibrosarcoma, dermatomyositis |
| MRI1 | CD | 28 | Chr8: 88–90 | cis | tyrosine phosphorylation | cell invasion regulation |
| NDUFS6 | CD | 27 | Chr3: 11–18 | 21 | complex I subunit | lactic acidosis |
| PUM2 | HFD | 27 | Chr10: 106–109 | 7 | RNA binding | development, cell differentiation |
| BCKDHB | CD | 26 | Chr9: 83–85 | cis | BCAA degradation | maple syrup urinary disease |
| BCKDHB | HFD | 24 | Chr9: 83–83 | cis | BCAA degradation | maple syrup urinary disease |
| TYMP | CD | 24 | Chr15: 89–90 | cis | thymidine phosphorylation | angiogenesis, mtDNA regulation |
| RPL18A | CD | 24 | Chr3: 25–31 | 33 | ribosome 60S subunit | hepatitis susceptibility |
| CAR3 | CD | 24 | Chr3: 14–21 | cis | carbon dioxide hydration | laryngeal disease, adipogenesis |
| DAK | HFD | 24 | Chr4: 128–130 | 34 | dihydroxyacetone kinase, FMN | glycerolipid metabolism |
| NDUFA4 | HFD | 24 | Chr11: 33–40 | 5 | complex I subunit | glucose regulation |
| ACADS | CD | 23 | Chr5: 67–76 | 42 | beta oxidation, short chain | SCAD deficiency |
| HMGCS1 | HFD | 23 | Chr4: 80–84 | 10 | cholesterol synthesis | cholesterolemia |
| FASN | CD | 23 | Chr5: 137–139 | 38 | fatty acid synthesis | obesity |
| GCLM | CD | 23 | Chr3: 118–124 | cis | glutathione synthesis | ischemia, anemia |
| ABCE1 | CD | 23 | Chr13: 114–121 | 28 | ribonuclease L inhibitor | viral load, RNA stability |
| ABCB8 | CD | 23 | Chr5: 23–25 | cis | mito membrane transporter | iron homeostasis |
| ARG1 | HFD | 22 | Chr11: 64–66 | 13 | urea cycle | argininemia, hyperammonemia |
| DCN | HFD | 22 | Chr11: 51–52 | 11 | collagen assembly | cell growth and proliferation |
| AKR7A2 | HFD | 22 | Chr1: 23–24 | cis | succinic semialdehyde redox | detox of aldehydes and ketones |
| AFP | HFD | 21 | Chr6: 86–88 | 29 | fetal serum albumin (putative) | copper, fatty acid, bilirubin binding |
| COPB1 | CD | 21 | Chr16: 8–15 | 56 | Golgi complex transporter | lipid storage, LDL levels |
| ACSS2 | HFD | 21 | Chr11: 115–121 | 101 | acetate activation | lipid synthesis, energy generation |
| TYMP | HFD | 21 | Chr15: 89–90 | cis | thymidine phosphorylation | angiogenesis, mtDNA regulation |
| COX6C | CD | 20 | Chr7: 141–144 | 14 | cytochrome C electron transfer | energy generation |
| MYBBP1A | HFD | 20 | Chr14: 112–116 | 1 | regulation of ribosomal DNA | nucleolar stress response |
| GSTK1 | HFD | 20 | Chr4: 67–70 | 3 | glutathione regulation | cellular detoxification |
| EPAS1 | HFD | 20 | Chr10: 106–109 | 7 | O2-responsive transcription factor | erythryocytosis, hypoxia response |
| ACSF2 | CD | 20 | Chr11: 116–118 | cis | fatty acid metabolism | tuberculosis sensitivity |
| SRSF7 | HFD | 20 | Chr15: 40–48 | 11 | spliceosome, mRNA export | immune response |
A list of all genes with highly significant (LRS ≥ 20) pQTLs. For each gene, the significant peak QTL width considers a 5 LRS drop-off on either side of the peak. A count of all genes under the wide peak is displayed, unless it is a cis-pQTL as the candidate gene can be easily identified (itself). The cellular function and overt physiological effect described by literature for each gene is given.
Bckdhb is a subunit of the branched chain amino α-keto acid dehydrogenase (BCKD, EC 2.7.11.4) complex (Figure 4A) and maps to two of the most significant eQTLs and among the most significant consistent pQTLs (Figure 4B; Tables 1 and S1). Bckdhb transcript and protein levels are highly variable across strains and are unaffected by diet (Figure 4B), but no cQTL mapped to this locus. In humans, variant alleles of BCKDHB are known to cause Type IB maple syrup urinary disease (MSUD), an inborn error of metabolism characterized by a buildup of branched chain amino acids (BCAAs) and branched chain α-keto acids (BCKAs). MSUD manifests in newborns and is associated with neurological problems and delayed development, and if untreated it can be fatal (Chuang and Shih, 2001). This pathway is a multi-tissue system driven particularly by the muscle and liver, and consequently it is diagnosed by significant increases of BCAAs and BCKAs in both the serum and urine, a decrease in alanine, and/or by detection of L-alloisoleucine (Chuang and Shih, 2001; Haymond et al., 1978). Genetically engineered mouse models of MSUD have similar phenotypic progression as humans, including the same metabolite buildups and related phenotypes such as movement difficulties and reduced body growth (Koepp et al., 1974; Wu et al., 2004). We thus examined the BXDs with respect to these traits. While L-alloisoleucine was not detected, strains with the less functional B6 allele of Bckdhb had a marked increase in the BCAA/alanine ratio in the serum and liver (Figure 4C, left and right, respectively). However, no association with movement, growth or body weight was present in the BXDs (data not shown), suggesting that this Bckdhb variant carries only the subclinical metabolic effects of MSUD. This observation is also in line with the human disease, where MSUD symptoms are only apparent when BCKD activity is < ~30% of normal (Skvorak, 2009). In contrast, strains with the B6 allele of Bckdhb have ~50% the enzymatic activity compared to those with the D2 allele. However, this raises the possibility that a sustained BCAA loading test in the BXDs may lead to clinical manifestation of the intermittent form of MSUD.
To continue scanning for physiological links with the proteins examined, we also investigated the strongest pQTL in both diets. This gene, Dhtkd1, encodes a mitochondrial protein operating as the E1 subunit of a dehydrogenase complex involved in lysine metabolism (EC 1.2.4.2; Figure 4D). In humans, variants in DHTKD1 have been linked to urinary buildup of 2-aminoadipate (2-AA) and 2-oxoadipate, yet the few studies on the physiological consequences of these variants are conflicted (Danhauser et al., 2012; Houten et al., 2013; Xu et al., 2012). After measuring the metabolites involved in this pathway in the BXD liver and serum, we observed a striking correlation between DHTKD1 levels and 2-AA (Figure 4E, left) as well as between metabolites in the pathway itself (Figure 4E, right and Figure S2A). Moreover, 2-AA mapped significantly in both diets as an mQTL to the locus of Dhtkd1 itself, indicating the BXDs share an inborn error of metabolism similar to humans (Figure 4F). Due to the strength and consistency of this correlation across diets, we hypothesized this variant may lead to phenotypic consequences and inform for human validation.
DHTKD1: A Regulator of Glucose Homeostasis
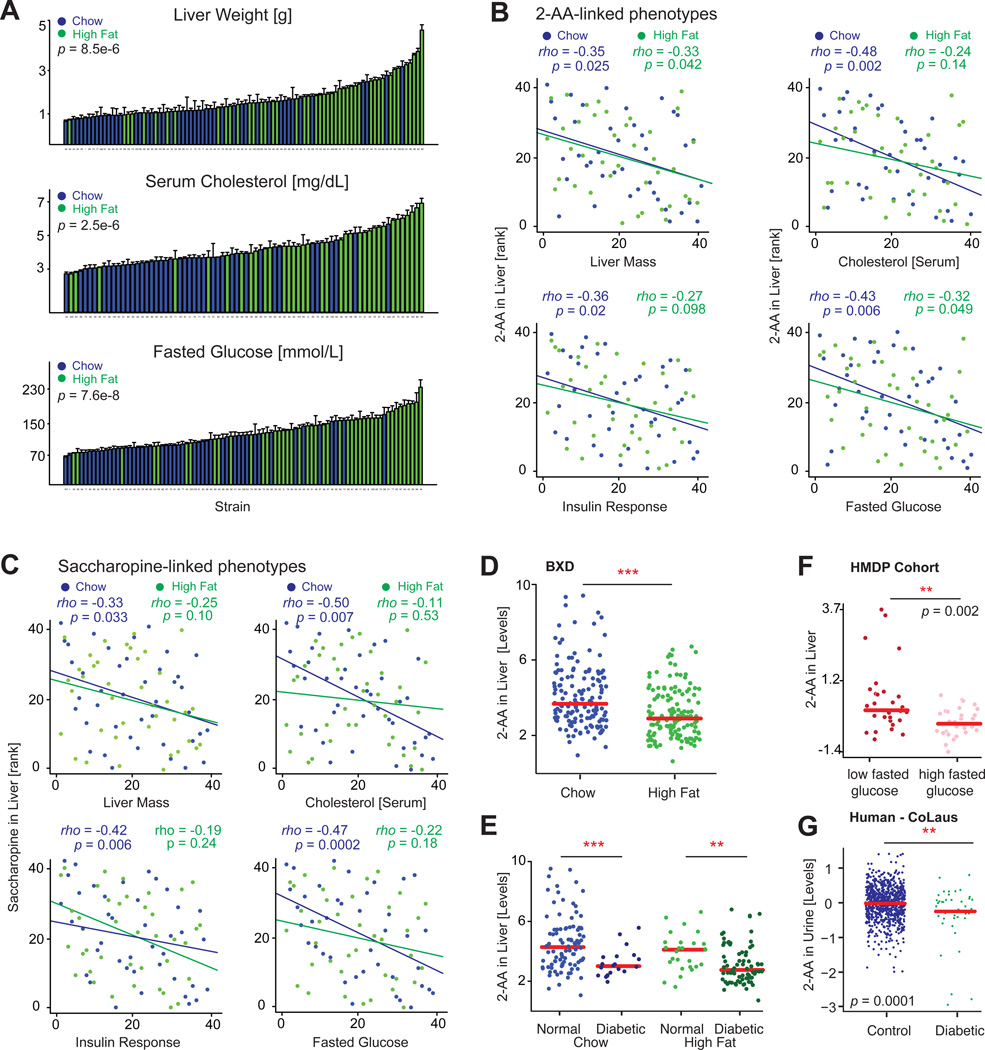
Inhibition of DHTKD1 in human liver cells is known to diminish mitochondrial activity (Xu et al., 2013), and external administration of 2-AA improves insulin secretion in mice (Wang et al., 2013). We thus examined whether the natural genetic variants in Dhtkd1 may also influence glucose homeostasis in the BXDs. The BXDs have varying responses to HFD-induced diabesity and include some resistant strains, but most strains are affected, leading to general increases in liver weight, serum cholesterol, glucose, and overall body weight (Figures 5A and S2B). Strikingly, 2-AA is negatively associated with liver mass and fasted glucose levels in both diets, and with serum cholesterol and insulin in CD (Figure 5B). Other metabolites in this pathway were also connected to the same phenotypes and with negative correlation [e.g., saccharopine (Figure 5C) and α-ketoglutarate (Figure S2C)]. Interestingly, 2-AA levels are significantly decreased in the HFD cohorts (Figure 5D), despite the genetic regulation of 2-AA by Dhtkd1 being equivalent between CD and HFD cohorts (Figures 4E and 4F). However, this decrease is not directly linked to diet, but instead with the animals’ progression toward diabetes. Based on HOMA insulin resistance indices (Lee et al., 2008), most HFD-fed BXD animals (108 of 180) became glucose intolerant and insulin resistant over the six-month period of the dietary challenge (Figure S2D). Conversely, only 18% of CD animals (29 of 165) have poor insulin sensitivity. Strikingly, the changes in 2-AA levels go in tandem with the HOMA index: animals with worse insulin sensitivity have lower 2-AA, and animals with good insulin sensitivity have equivalent levels of 2-AA, with no effects linked directly to diet (Figure 5E). With this in mind, we further examined 2-AA in two independent population studies. In the Hybrid Mouse Diversity Panel, a recent profile of liver metabolomes in 96 cohorts in the fasted state contained 2-AA measurements (Ghazalpour et al., 2014). In this diverse population, which includes~30 BXD strains (with only one strain overlapping with those in our study), 2-AA is diminished in cohorts with higher fasted glucose (Figure 5F), and the two are again negatively correlated (Figure S2E). Likewise, we observed the same trend in a human population study (Cohorte Lausannoise [CoLaus] study [Firmann et al., 2008]), in which we recently analyzed the spot urine samples of 835 middle-aged or elderly individuals (Rueedi et al., 2014). In this cohort, diabetic patients (Figure S2E) also displayed a marked decrease in 2-AA levels (Figure 5F). Together, these findings indicate that Dhtkd1 plays a major role in the regulation of 2-AA in both mice and humans and that the modulation of this pathway may influence the development of diabetes.
Figure 5. Physiological Consequences of DHTKD1 Variants.
(A) Liver size, serum cholesterol, and fasted glucose levels increase after HFD across the BXDs. Error bars represent mean + SEM.
(B) In CD livers, 2-AA is associated negatively with liver mass, serum cholesterol, insulin, and glucose. In HFD livers, 2-AA is associated negatively with liver mass and fasting glucose.
(C) Liver levels of saccharopine, a metabolite upstream of 2-AA, are also associated negatively with the same phenotypes, although only in CD.
(D) 2-AA levels in the liver are significantly decreased in HFD-fed BXD cohorts.
(E) However, when correcting for diabetes status (HOMA-IR > 10 as diabetic, or HOMA-IR < 5 as healthy), there was no difference between CD and HFD—instead, only between diabetic and nondiabetic cohorts.
(F) Using publicly available data from a recent metabolomic profiling of livers from the HMDP (Ghazalpour et al., 2014), we can observe that the inverse relationship between 2-AA and fasted glucose is highly consistent in mouse populations. p = 0.01 if the four high nonoutliers in the low group are suppressed.
(G) Also in a human population study with urine metabolomics, diabetic patients had markedly lower levels of 2-AA.
Related to Figure S2.
The Mitochondrial Unfolded Protein Response
The UPRmt is a mitochondrial stress response pathway that is activated by proteostatic stress, such as by accumulation of unassembled or unfolded proteins in the mitochondria (Zhao et al., 2002), by the presence of an imbalance between mitochondrial and nuclear encoded proteins (Houtkooper et al., 2013), or by electron transport chain defects (Durieux et al., 2011; Runkel et al., 2013). The activation of UPRmt in turn leads to the transcription and translation of nuclear-encoded protective genes such as mitochondrial chaperones and proteases to reestablish mitochondrial proteostasis (reviewed in Haynes et al., 2013; Jovaisaite et al., 2014; Wolff et al., 2014). The bulk of research on UPRmt has taken place using C. elegans and mammalian cell lines, thus little is known about when or how UPRmt occurs in vivo in mammals. Furthermore, as the UPRmt is a stress response tied to maintaining mitochondrial protein balance, we hypothesized that its protein correlation networks may be different than those generally examined at the transcriptional level.
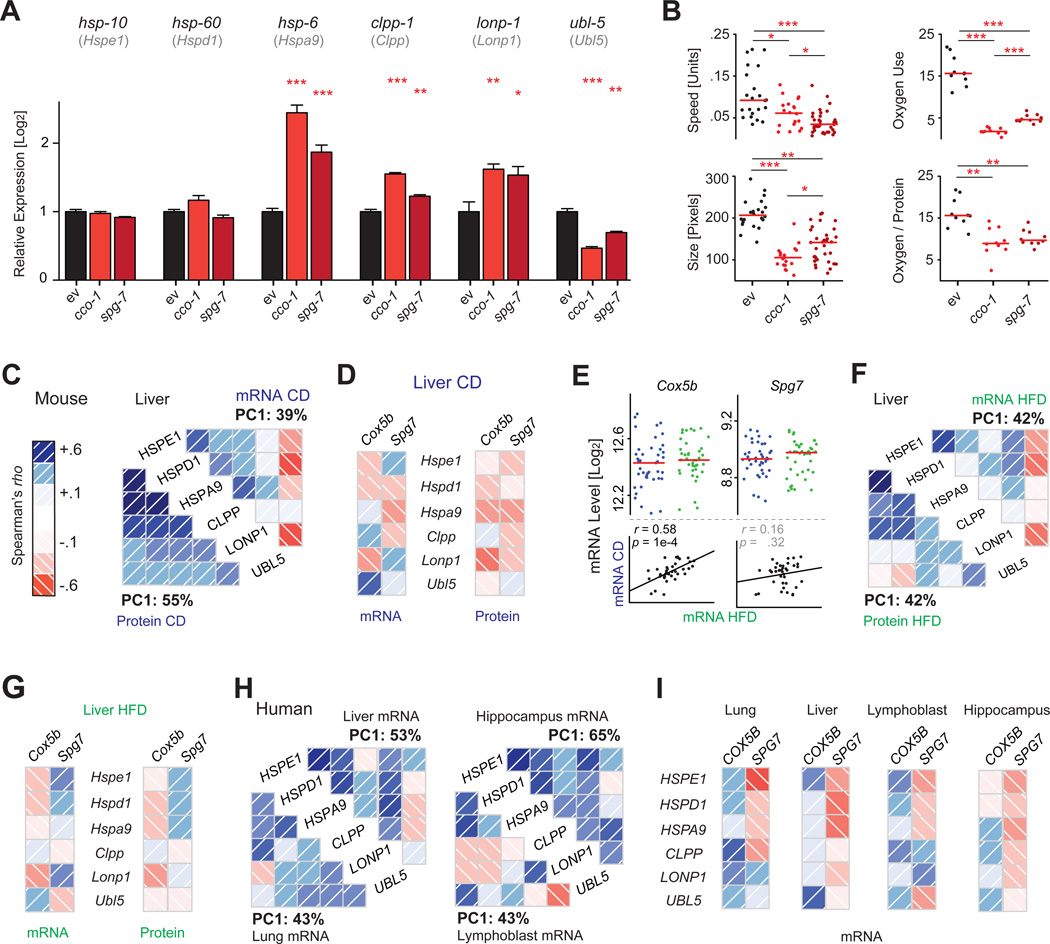
In the worm, two “classical” approaches have been typically used to induce UPRmt: the loss of function of cco-1, a nuclear encoded component of the electron transport chain (Durieux et al., 2011), or the loss of function of spg-7, a mitochondrial protein quality-control protease (Yoneda et al., 2004). We confirmed that the knockdown of either gene by RNAi triggers the UPRmt response in C. elegans, by strong induction of the mitochondrial chaperone hsp-6 and of the proteases lonp-1 and clpp-1 (Figure 6A). Moreover, we linked this UPRmt activation to specific phenotypes—a major reduction in size and mobility, as well as a decrease in oxygen consumption—which are consequences of mitochondrial stress (Figure 6B). However, it has not been previously shown whether this coordinated regulation of UPRmt genes is conserved in mammals in vivo.
Figure 6. The Mitochondrial Unfolded Protein Response.
(A) UPRmt induction in C. elegans triggered by interference with ETC (RNAi of cco-1) or mitochondrial proteostasis (RNAi for spg-7). These triggers result in upregulation of UPRmt effectors hsp-6, clpp-1, and lonp-1 and a reduction in ubl-5. The orthologous mouse genes are indicated below the respective C. elegans gene symbol. Error bars represent mean + SEM.
(B) UPRmt induction in C. elegans decreases movement, size, and oxygen consumption.
(C) UPRmt genes and proteins form a network of coordinately expressed mRNAs and proteins in vivo in mice, which is stronger at the protein than at the mRNA level.
(D) Cox5b and Spg7 (orthologs of C. elegans cco-1 and spg-7) are generally negatively associated with the levels of all UPRmt genes in CD cohorts, particularly at the protein level, in line with observations in the worm.
(E) While the levels of Cox5b and Spg7 are not affected by diet, expression is consistent by strain across the two diets only for Cox5b.
(F) The UPRmt network in HFD livers is similar to that observed in CD, but somewhat weaker. Ubl5 remains a striking negative correlate at the mRNA level.
(G) In HFD, Cox5b remains a negative correlate of UPRmt transcripts and proteins, while Spg7 does not.
(H) The features of the UPRmt network are also conserved in 427 human liver biopsies (Schadt et al., 2008), 405 lung biopsies (Ding et al., 2004), 180 lymphoblast lines (Monks et al., 2004), and 43 hippocampi (Berchtold et al., 2008).
(I) In humans, SPG7 is a consistent negative correlate of the UPRmt transcripts.
In the BXDs, we investigated the expression of six members of the UPRmt pathway, which are well conserved from C. elegans: mitochondrial chaperones (Hspd1, Hspe1, Hspa9), proteases (Clpp, Lonp1), and a transcriptional regulator involved in UPRmt (Ubl5). These UPRmt genes are also coordinately regulated at both mRNA and protein level in the BXDs, but with much stronger connections among proteins (Figure 6C). Moreover, the UPRmt network correlates negatively with Cox5b and Spg7 (mouse orthologs of worm cco-1 and spg-7, respectively), indicating that low abundance of these genes amplifies UPRmt in mammals as in C. elegans (Figure 6D). The network is also influenced in part by diet. While Cox5b expression patterns are similar between CD and HFD, Spg7 covariation is disjointed between the dietary cohorts (Figure 6E). This may explain why, despite a similar overall UPRmt response in both diets (Figures 6D and 6F), Spg7 trends positively in HFD cohorts, while Cox5b remains consistent (Figure 6G). Using four large transcriptional studies of human tissue biopsies, we observe similar transcriptional links, particularly including a strong network between all the UPRmt genes (Figure 6H). In humans, SPG7 is a consistent negative correlate of this network, in contrast to COX5B, which generally has positive covariation with the UPRmt response (Figure 6I). Thus, while many of the overall regulators of UPRmt remain coregulated across species—worm, mouse, and human—particular nuances of its activation pathways appear to be variable dependent on species, environment, tissue, and likely other factors.
DISCUSSION
Due to major differences in transcript and protein regulation, it has become increasingly clear that systems proteomics is essential for the analysis of complex systems such as metabolism (Khan et al., 2013; Skelly et al., 2013). Traditionally this was attempted by semiquantitative immunoassays such as ELISA or western blotting, yet these techniques allow only a handful of proteins to be measured in parallel and are moreover limited by the scarcity of quantitative immunoassays and the dubious quality of many antibodies (Marx, 2013). Recent shotgun proteomics experiments on diverse populations have provided fundamental data on protein variance in populations of yeast (Albert et al., 2014; Skelly et al., 2013), mammals (Ghazalpour et al., 2011), and humans (Hwang et al., 2010). However, the inability to measure target proteins has limited the application of this approach in the study of disease pathways. New developments in mass spectrometry have led to SRM, a targeted quantitative systems proteomics technique in which choice proteins can be quantified based on a priori information. This technique has been recently applied in a moderate-sized yeast population study (Picotti et al., 2010), but until now it has not been applied to study genetic regulation in multicellular species or for a choice pathway.
In this study, we quantified 192 metabolism genes at the transcript and protein levels in livers from 77 cohorts of the BXD GRP under two different dietary conditions. Along with sequence variants, basic metabolomics, and phenotype data, this combined multilayered population data set enabled us to accurately estimate abundance changes in gene products due to genotype, diet, and gene-by-environment interactions (GXE). By complementing these layers, we were able to tentatively link dozens of pQTLs with phenotypes. Of the ~50 of significant pQTLs identified, we established links between four genes leading to phenotypic consequence: two novel and two confirmatory. For these genes, Nnt, Car3, Dhtkd1, and Bckdhb, coding differences between B6 and D2 alleles were linked to robust differences at both the transcript level and protein level, leading to shared cis-regulatory eQTLs and pQTLs and then to phenotypic consequences. While Nnt has been previously attributed as causal to differences in insulin secretion in the BXDs (Wong et al., 2013), the other pQTLs are novel, including Car3, which was previously linked to adiposity only in knockout mice (Mitterberger et al., 2012). For Bckdhb, strains carrying the B6 allele have a ~50% reduction in enzyme levels, which leads to a buildup of BCAA levels and a decrease in alanine levels, as in the intermittent form of MSUD. In humans, MSUD is a rare inborn error of metabolism, and while BXDs do not suffer any overt physiological consequences of BCKDHB deficiency in the basal state, certain dietary stresses may magnify the symptoms and make the BXDs appropriate to model MSUD.
For Dhtkd1, the BXD strains map to highly significant and overlapping eQTLs and pQTLs with an approximately bimodal distribution with 2-fold variance between strains with the B6 (low) or the D2 allele (high) of the gene and with no effect from diet. Furthermore, the metabolite 2-AA maps as a significant mQTL to the Dhtkd1 locus in both diets, despite a marked decrease of 2-AA levels in HFD cohorts. Interestingly the decrease in 2-AA is not directly due to the HFD, but rather it is due to the increased penetrance of insulin resistance after HFD. Moreover, prior works show that external administration of 2-AA can improve insulin sensitivity and glucose response in mice (Wang et al., 2013), indicating that variants in Dhtkd1 and 2-AA levels may be driving the pathogenesis of diabetes, rather than being mere passive biomarkers of the disease. Strikingly, we replicated this association between 2-AA and high glucose levels in a recent and completely independent metabolomic data set collected in the Hybrid Mouse Diversity Panel (Ghazalpour et al., 2014). Furthermore, and clinically relevant, we demonstrated in a human population-based sample, Co-Laus, that diabetic patients likewise have lower urinary levels of 2-AA (Rueedi et al., 2014). Thus, 2-AA levels are consistently linked to the diabetes status in both mice and humans, and in the BXDs we can unequivocally attribute a causal part of this variance to variants in Dhtkd1. The BXD strains were necessary to bring the hypothesis to power a human study and also confirm that the mouse link has relevance for human disease. Together, these findings further validate the BXD mouse population as a model for human metabolic diseases (Andreux et al., 2012), and they indicate a critical need for multilayered measurements to effectively analyze complex systems, particularly within the context of the study of GXE.
It is worth stressing that novel regulatory mechanisms can be found either through QTL analysis and their equivalent from GWAS, SNP analysis, or through network analyses, which are a complementary and powerful approach to dissect complex traits. The network approach is particularly viable when backed by high-depth multilayered data sets such as illustrated by our example of UPRmt. UPRmt is a reparative pathway activated by mitochondrial proteotoxic stress that has been primarily studied in the C. elegans and in cultured cells, but little is known about whether it occurs in vivo in mammals. We examined six genes that are known to be major regulators of C. elegans UPRmt and that are conserved in mammals. These six genes form a robust coexpression network in both diets at the transcriptional and proteomic levels, with the proteomic connections being stronger, befitting the role of UPRmt as a sensor and regulator of protein stress. One observation that stood out in the analysis of the UPRmt was the striking “contradiction” between the Ubl5 transcript and UBL5 protein correlations to the UPRmt network. Ubl5 is a transcriptional regulator known to induce UPRmt; yet, in both worms and mice, its transcript levels decrease when UPRmt is activated. Conversely, the UBL5 protein is increased with UPRmt activation in the BXDs, an observation also previously reported in C. elegans (Benedetti et al., 2006). This discordance in protein and transcript regulation suggests the existence of posttranscriptional mechanisms or a negative feedback loop, which could not be detected at the transcript or protein level alone. While there remains a great deal of this pathway left to be explored, it is clear that accurate, systems-scale protein measurements are essential to effectively model complex protein response networks like UPRmt.
Today, there is an unprecedented capacity for accurate measurement quantification of nearly all aspects of biology. Surprisingly, perhaps, proteins remain one of the most difficult aspects of biology to precisely measure on a systems scale, though it has been long clear that transcripts only serve as a weak proxy for protein levels (Gygi et al., 1999). Our data show that the integration of systems proteomics data sets with different layered systems measurements provides unprecedented insights into the mechanistic regulation of complex systems, such as exemplified by metabolism, which can eventually lead to the improved diagnosis and treatment of metabolic disease.
EXPERIMENTAL PROCEDURES
Animals
Forty strains of the BXD population—40 on CD, 37 on HFD—with ~10 male animals from each strain were separated into 2 cohorts of 5 for each diet. Food access was ad libitum for CD—Harlan 2018 (6% kCal/fat, 74% carbohydrate, 20% protein)—and for HFD (Harlan 06414 [60% kCal/fat, 20% carbohydrate, 20% protein]). HFD cohorts received the diet from week 8 until sacrifice. Each cohort was communally housed until week 23, after which animals were single caged until tissues collection at 29 weeks after an overnight fast. Tissues were collected from 183 CD and 168 HFD animals, with at least 3 biological replicates for all cohorts. All research was approved by the Swiss cantonal veterinary authorities of Vaud under licenses 2257.0 and 2257.1.
Sample Preparation and Analysis
For liver analyses, three ~100 mg pieces were taken from cold storage for each individual and then weighed and sorted for mRNA, protein, and metabolite measurements. For microarray, mRNA from three to five individuals per cohort were prepared separately and then pooled equally after nanodrop quantification and run on the Affymetrix Mouse Gene 1.0 ST array platform. For proteomics, protein was prepared from one to three biological replicates per cohort. For metabolite measurements, all individuals in all cohorts were measured individually after polar extraction of samples.
SRM Assay Development and Protein Quantification
Generation of peptide library and development ofSRM assays were performed as described (Picotti et al., 2010). Identical SRM assays for all 192 target proteins were run on all 77 cohorts. SRM traces were manually checked according to established criteria (Lange et al., 2008a). For relative quantification of each protein across the set of different cohorts, the raw intensity of transitions of the native and (13C6, 15N4)-Arginine, (13C6, 15N2)-Lysine peptides were considered. The technical reproducibility of SRM-based quantification was validated by measuring the individual samples with three independent mass spectrometry injections.
General Informatic Analyses
Outliers were Windsorized prior to all analyses. SNP analysis was also performed using GeneNetwork, which contains the full sequence data of C57BL/6J and DBA/2J, as well as high density genotype information for all the BXD lines. The correlations are Pearson’s r or Spearman’s rho, depending on the absence or presence of outliers. Student’s t test was used for comparing two groups in normalized data (all protein and mRNA are normalized). Bonferroni’s correction for multiple testing was performed, and cutoffs for both nominal significance (p < 0.05) and corrected significance (p < 0.05/n tests) are displayed. Except for QTL plots, graphs and analyses were performed in R.
Metabolite Measurements
Murine metabolite measurements for this study were performed on serum and livers from the BXDs using ultra performance liquid chromatrography-mass spectrometry and flow-injection time-of-flight mass spectrometry, respectively. Human urinalysis was performed on 835 Caucasian adults in the CoLaus study, including 43 diabetics and 792 nondiabetics, males and females. The CoLaus study was approved by the Institutional Ethics Committee of the University of Lausanne. All study participants gave written consent. Full details of the human NMR metabolomics data are in a separate parallel publication (Rueedi et al., 2014).
Data Repository
All BXD data can be found at http://www.GeneNetwork.org. To download the clinical phenotype data, change the type to “Phenotypes” and enter “LISP2” to find all associated results. Select all and export to recover the data, or analyze it online. Microarray data and protein measurements can be found on the same resource; change the type to “Liver Proteome” or “Liver mRNA” and navigate to the named EPFL data sets under CD or HFD (e.g., EPFL/LISP BXD CD Liver Affy Mouse Gene 1.0 ST [Apr13] RMA). To download these data, please click the “INFO” button at the right-hand side of the search page and download the raw data set in the upper right box of the screen.
Extended Experimental Procedures and Supplemental References are available in the Supplemental Information.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. van Cruchten and W. Smit for the serum analysis of 2-AA. The CoLaus study received financial contributions from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (SNSF; 33CSCO-122661). The authors thank P. Vollenweider, G. Waeber, V. Mooser, and D. Waterworth, co-principal investigators of the CoLaus study. We also thank L. Gillet, H. Röst, and P. Picotti for insightful discussions and R. Huttenhain, A. Maiolica, P. Kouvonen, T. Sajic, A. Leitner, and A. Bensimon for technical support. Special thanks to M. Bochud, Y. Barreau, M. Firmann, V. Mayor, A. Bastian, B. Ramic, M. Moranville, M. Baumer, M. Sagette, J. Ecoffey, and S. Mermoud for data collection. Y.W. was supported by the ERC (Proteomics v3.0; AdG-233226 to R.A.) and the LiverX program of SystemsX, the Swiss Initiative for Systems Biology and E.G.W. by a fellowship from the Fondation Romande pour la Recherche sur le Diabéte. J.A. is the Nestlé Chair in Energy Metabolism. Research was supported by the EPFL, ETHZ, ERC (Sirtuins; AdG-231138 and Proteomics v3.0; AdG-233226), Velux Stiftung, LiverX, and AgingX programs of the Swiss Initiative for Systems Biology (51RTP0-151019 and 2013/153), SNSF (31003A-140780, 31003A-143914, 3100A0-107679, and CSRII3-136201), and the NIH (R01AG043930).
Footnotes
ACCESSION NUMBERS
All array data can also be found on the Gene Expression Omnibus (GEO) under the accession number GSE60149.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, two figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.07.039.
AUTHOR CONTRIBUTIONS
Y.W. and E.W. contributed equally to this work, planning and performing most of the specific experiments, writing the manuscript, and preparing the figures. R.A. and J.A. conceived, designed, and supervised the general project and edited the manuscript. E.W. performed all BXD experiments and transcriptomics. Y.W. and P.F. ran all proteomics assays. S.D. and N.Z. performed liver metabolomics. A.M. and V.J. performed the C. elegans experiments. S.M.H. and C.A.A. performed serum metabolomics. W.W. provided bioinformatic expertise, and Z.K. analyzed the CoLaus study.
REFERENCES
- Albert FW, Treusch S, Shockley AH, Bloom JS, Kruglyak L. Genetics of single-cell protein abundance variation in large yeast populations. Nature. 2014;506:494–497. doi: 10.1038/nature12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy MF, Henry H, Schoonjans K, Williams RW, Auwerx J. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc. Natl. Acad. Sci. USA. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callister SJ, Barry RC, Adkins JN, Johnson ET, Qian WJ, Webb-Robertson BJ, Smith RD, Lipton MS. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res. 2006;5:277–286. doi: 10.1021/pr050300l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang DT, Shih VE. Maple syrup urine disease (branched-chain ketoaciduria) In: Scriver CR, et al., editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 1971–2005. [Google Scholar]
- Danhauser K, Sauer SW, Haack TB, Wieland T, Staufner C, Graf E, Zschocke J, Strom TM, Traub T, Okun JG, et al. DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. Am. J. Hum. Genet. 2012;91:1082–1087. doi: 10.1016/j.ajhg.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Roshina NV, Geiger-Thornsberry GL, Lyman RF, Pasyukova EG, Mackay TF. Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat. Genet. 2003;34:429–433. doi: 10.1038/ng1218. [DOI] [PubMed] [Google Scholar]
- Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- Ding L, Quinlan KB, Elliott WM, Hamodat M, Paré PD, Hogg JC, Hayashi S. A lung tissue bank for gene expression studies in chronic obstructive pulmonary disease. COPD. 2004;1:191–204. doi: 10.1081/copd-120039810. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firmann M, Mayor V, Vidal PM, Bochud M, Pécoud A, Hayoz D, Paccaud F, Preisig M, Song KS, Yuan X, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc. Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss EJ, Radulovic D, Shaffer SA, Ruderfer DM, Bedalov A, Goodlett DR, Kruglyak L. Genetic basis of proteome variation in yeast. Nat. Genet. 2007;39:1369–1375. doi: 10.1038/ng.2007.22. [DOI] [PubMed] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett BJ, Shih D, Che N, Orozco L, Pan C, Hagopian R, He A, Kayne P, Yang WP, et al. Genetic regulation of mouse liver metabolite levels. Mol. Syst. Biol. 2014;10:730. doi: 10.15252/msb.20135004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haymond MW, Ben-Galim E, Strobel KE. Glucose and alanine metabolism in children with maple syrup urine disease. J. Clin. Invest. 1978;62:398–405. doi: 10.1172/JCI109141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Fiorese CJ, Lin YF. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol. 2013;23:311–318. doi: 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, von Delft A, Nicolaou A, Baumann S, Kostrzewa M, Thiery J, Teupser D. Quantitative trait loci mapping of the mouse plasma proteome (pQTL) Genetics. 2013;193:601–608. doi: 10.1534/genetics.112.143354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Te Brinke H, Denis S, Ruiter JP, Knegt AC, de Klerk JB, Augoustides-Savvopoulou P, Häberle J, Baumgartner MR, Coş-kun T, et al. Genetic basis of hyperlysinemia. Orphanet J. Rare Dis. 2013;8:57. doi: 10.1186/1750-1172-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, Smoke CC, Meyer C, Højlund K, Yi Z, Mandarino LJ. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes. 2010;59:33–42. doi: 10.2337/db09-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- Jovaisaite V, Mouchiroud L, Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J. Exp. Biol. 2014;217:137–143. doi: 10.1242/jeb.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpievitch YV, Dabney AR, Smith RD. Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinformatics. 2012;13(Suppl 16):S5. doi: 10.1186/1471-2105-13-S16-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z, Ford MJ, Cusanovich DA, Mitrano A, Pritchard JK, Gilad Y. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science. 2013;342:1100–1104. doi: 10.1126/science.1242379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp P, Rybak C, Rüdiger HW, Wendel U. Maple syrup urine disease variant: report on an infant. Z. Kinderheilkd. 1974;116:177–184. doi: 10.1007/BF00464456. [DOI] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 2008a;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Muniyappa R, Yan X, Chen H, Yue LQ, Hong EG, Kim JK, Quon MJ. Comparison between surrogate indexes of insulin sensitivity and resistance and hyperinsulinemic euglycemic clamp estimates in mice. Am. J. Physiol. Endocrinol. Metab. 2008;294:E261–E270. doi: 10.1152/ajpendo.00676.2007. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011 doi: 10.1093/database/bar009. bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. Finding the right antibody for the job. Nat. Methods. 2013;10:703–707. doi: 10.1038/nmeth.2570. [DOI] [PubMed] [Google Scholar]
- Mitterberger MC, Kim G, Rostek U, Levine RL, Zwerschke W. Carbonic anhydrase III regulates peroxisome proliferator-activated receptor-γ2. Exp. Cell Res. 2012;318:877–886. doi: 10.1016/j.yexcr.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, Phillips JW, Sachs A, Schadt EE. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wen-schuh H, Aebersold R. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- Picotti P, Clément-Ziza M, Lam H, Campbell DS, Schmidt A, Deutsch EW, Rö st H, Sun Z, Rinner O, Reiter L, et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueedi R, Ledda M, Nicholls AW, Salek RM, Marques-Vidal P, Morya E, Sameshima K, Montoliu I, Da Silva L, Collino S, et al. Genome-wide association study of metabolic traits reveals novel gene-metabolite-disease links. PLoS Genet. 2014;10:e1004132. doi: 10.1371/journal.pgen.1004132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel ED, Liu S, Baumeister R, Schulze E. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet. 2013;9:e1003346. doi: 10.1371/journal.pgen.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Skelly DA, Merrihew GE, Riffle M, Connelly CF, Kerr EO, Johansson M, Jaschob D, Graczyk B, Shulman NJ, Wakefield J, et al. Integrative phenomics reveals insight into the structure of phenotypic diversity in budding yeast. Genome Res. 2013;23:1496–1504. doi: 10.1101/gr.155762.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvorak KJ. Animal models of maple syrup urine disease. J. Inherit. Metab. Dis. 2 009;32:229–246. doi: 10.1007/s10545-009-1086-z. [DOI] [PubMed] [Google Scholar]
- Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, Ghorbani A, O’Sullivan J, Cheng S, Rhee EP, et al. 2-aminoadipic acid is a biomarker for diabetes risk. J. Clin. Invest. 2013;123:4309–4317. doi: 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Weissman JS, Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Wong N, Morahan G, Stathopoulos M, Proietto J, Andrikopoulos S. A novel mechanism regulating insulin secretion involving Herpud1 in mice. Diabetologia. 2013;56:1569–1576. doi: 10.1007/s00125-013-2908-y. [DOI] [PubMed] [Google Scholar]
- Wu JY, Kao HJ, Li SC, Stevens R, Hillman S, Millington D, Chen YT. ENU mutagenesis identifies mice with mitochondrial branched-chain aminotransferase deficiency resembling human maple syrup urine disease. J. Clin. Invest. 2004;113:434–440. doi: 10.1172/JCI19574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WY, Gu MM, Sun LH, Guo WT, Zhu HB, Ma JF, Yuan WT, Kuang Y, Ji BJ, Wu XL, et al. A nonsense mutation in DHTKD1 causes Charcot-Marie-Tooth disease type 2 in a large Chinese pedigree. Am. J. Hum. Genet. 2012;91:1088–1094. doi: 10.1016/j.ajhg.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Zhu H, Gu M, Luo Q, Ding J, Yao Y, Chen F, Wang Z. DHTKD1 is essential for mitochondrial biogenesis and function maintenance. FEBS Lett. 2013;587:3587–3592. doi: 10.1016/j.febslet.2013.08.047. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackel-prang R, Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.