Abstract
Long non-coding RNAs (lncRNAs) can be important regulators of various biological processes such as RNA-directed DNA methylation (RdDM). In the RdDM pathway, recruitment of the DNA methylation complex is mediated through complementary pairing between scaffold RNAs and Argonaute-associated siRNAs. Scaffold RNAs are chromatin-associated lncRNAs transcribed by RNA polymerase Pol V or Pol II, while siRNAs originate from Pol IV- or Pol II-dependent production of lncRNAs. In contrast to the vast literature on co-transcriptional and post-transcriptional processing of mRNAs, information is limited for lncRNA regulation that enables their production and function. Recently Arabidopsis RRP6L1, a plant paralog of the conserved nuclear RNA surveillance protein Rrp6, was shown to mediate RdDM through retention of lncRNAs in the chromatin, thereby revealing that accumulation of functional lncRNAs requires more than simply RNA polymerases. By focusing on the canonical RdDM pathway, here we summarize recent evidence that indicate co-transcriptional and/or post-transcriptional regulation of lncRNAs, and highlight the emerging theme of lncRNA regulation by RNA processing factors.
Keywords: Long non-coding RNA, Pol IV, Pol V, RNA retention, RNA-directed DNA methylation, Rrp6, splicing, transcriptional silencing
DNA methylation is a major epigenetic mark that can confer transcriptional silencing. In plants and animals, complementary pairing between lncRNAs and small interfering RNAs (siRNAs) mediates de novo DNA methylation in a sequence-specific manner.1-5 RdDM mainly target heterochromatic regions that are enriched with transposons (TEs) and other DNA repeat sequences.6,7 Paradoxical to its silencing function, RdDM requires low levels of initial transcription at its target loci to produce scaffold lncRNAs and the precursors of siRNAs.
lncRNAs as Precursors of siRNAs
In the canonical RdDM pathway that involves 24nt siRNAs, production of siRNAs predominantly depends on Pol IV.8,9 Pol IV is thought to transcribe its target loci to produce single-stranded non-coding RNAs that function as siRNA precusors, although these transcripts have yet to be identified, possibly due to their very low abundance and/or instability. Pol IV-dependent transcripts can be considered as lncRNAs because they function as precursors of siRNAs and are therefore apparently longer than and distinct from siRNAs. Pol IV-dependent lncRNAs are transcribed by the RNA-dependent RNA polymerase RDR2 into double-stranded RNAs, which are subsequently cleaved by DCL3, resulting in 24nt siRNAs that are stabilized by HEN1-catalyzed 3′-O-methylation and loaded onto AGO4 or its close paralogs (Fig. 1).1-4 RDR2 physically interacts with Pol IV and requires the chromatin remodeler CLSY1 during synthesis of double-stranded RNAs,10,11 indicating that Pol IV-dependent lncRNAs may be retained on the chromatin to serve as templates of RDR2 transcription. Thus the co-transcriptional processing of Pol IV-dependent lncRNAs probably is different from that of mRNAs.
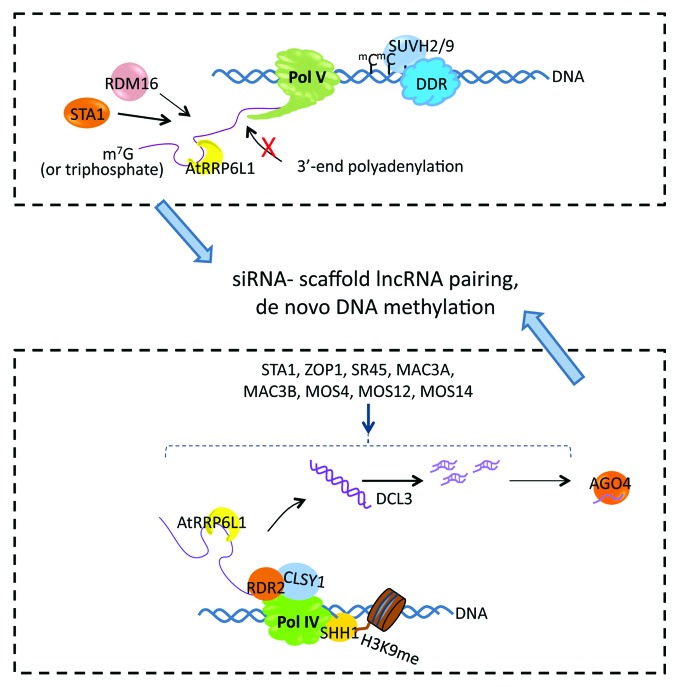
Figure 1. Proposed involvement of RNA processing factors in the accumulation and function of Pol IV- and Pol V-dependent lncRNAs in the RdDM pathway. Upper panel, Pol V occupancy in the chromatin requires the chromatin-remodeling complex DDR as well as SUVH2 and SUVH9 that are methyl-DNA binding proteins. Pol V lncRNAs may have triphosphates or caps at their 5′ ends and are devoid of 3′-end polyadenylation. AtRRP6L1 helps retain Pol V lncRNAs in the chromatin for a scaffold function. STA1 and RDM16, two splicing factors that seem to function separately, affect the levels of Pol V lncRNAs via unclear mechanisms. Lower panel, Pol IV can be recruited to its target loci by physical interaction with SHH1 that binds methylated histone H3 lysine 9 (H3K9me). RDR2 associates with Pol IV and requires CLSY1 for synthesis of double-stranded RNAs using Pol IV-transcribed lncRNAs as templates. AtRRP6L1 may help retain Pol IV lncRNAs in the Pol IV transcription complex to facilitate RDR2 transcription. Accumulation of Pol IV-dependent siRNAs is reduced to varying extents by mutations in pre-mRNA splicing factors, including STA1, ZOP1, SR45, MAC3A, MAC3B, MOS4, MOS12, and MOS14. The complementary pairing between scaffold lncRNAs and siRNAs leads to de novo DNA methylation in this canonical RdDM pathway.
Recruitment of Pol IV to its target loci can be mediated by DTF1/SHH1, which interacts with Pol IV and binds methylated histone H3 lysine 9 (Fig. 1).11-13 At some RdDM loci, Pol IV recruitment and siRNA production require the histone deacetylase HDA6 and the DNA methyltrasferase MET1,14 consistent with the notion that pre-existing chromatin marks contribute to Pol IV recruitment. While Pol IV is required for biogenesis of most 24nt siRNAs, Pol II generates 24nt siRNAs from inverted repeat sequences, where Pol II transcription produces lncRNAs that form double-stranded hairpin structures and can thus be directly cleaved by DCL3.15
Scaffold lncRNAs
Pol V-dependent lncRNAs and some Pol II-dependent lncRNAs are thought to function as chromatin-associated scaffold molecules to recruit Argonaute-bound siRNAs and consequently other components of the methylation complex. Production of scaffold lncRNAs is independent of either siRNA production or DNA methylation that is established through the RdDM pathway.17,18 Pol V transcription of lncRNAs requires the chromatin-remodeling complex DDR, which consists of DRD1, DMS3, and RDM1 (Fig. 1).16,17,19 Genome-wide chromatin occupancy of Pol V depends redundantly on SUVH2 and SUVH9, two methyl-DNA binding proteins that physically interact with several chromatin-remodeling proteins including the DDR complex.20,21 SUVH2 and SUVH9 appear to recognize DNA methylation that is dependent on MET1,21 which is the methyltransferase responsible for maintenance of DNA methylation in the symmetric CG context.1,22 Thus the initiation of Pol V-dependent lncRNA production is potentially determined by pre-existing epigenetic marks.
Pol V-dependent lncRNAs from the same locus displayed different 5′ ends, indicating that the initiation of Pol V transcription may be independent of potential promoters.17 Similar to Pol II-transcribed mRNAs, some Pol V-dependent lncRNAs possess 7-methylguanosine caps at the 5′ ends,17 indicating that Pol V-dependent lncRNAs can be subjected to RNA processing activities at least at the early stages of transcription. On the other hand, processing of lncRNAs does not resemble that of mRNAs in every way, because Pol V-dependent lncRNAs, unlike mRNAs, are devoid of polyadenylation at the 3′ ends.17
Retention of lncRNAs in the Chromatin for Proper Function
Recent evidence revealed that Pol V-dependent lncRNAs, and possibly Pol IV-dependent lncRNAs as well, are subjected to AtRRP6L1-mediated transcript retention in the chromatin (Fig. 1).23 AtRRP6L1 is a plant homolog of yeast Rrp6, which plays a central role in nuclear RNA surveillance that is known to confer non-coding RNA degradation and mRNA quality control, including exosomal RNA decay and transcript retention triggered by defective RNA processing.24,25
While Rrp6-dependent RNA decay has been shown to eliminate numerous non-coding RNAs that mostly appear to be noisy byproducts of pervasive transcription,26-29 AtRRP6L1 displays a positive regulation of Pol V-dependent lncRNAs and Pol IV-dependent siRNAs.23 Mutations in AtRRP6L1 reduce the levels of scaffold lncRNAs and siRNAs in the RdDM pathway, leading to DNA hypomethylation and release of transcriptional silencing.23 Therefore, the positive regulation of functional non-coding RNAs by AtRRP6L1 is coherent with a role of Rrp6 proteins in eliminating useless non-coding RNAs, because such positive regulation ensures transcriptional silencing via RdDM that prevents production of unwanted non-coding RNAs. AtRRP6L1 associates with Pol V-dependent lncRNAs and maintains the levels of chromatin-associated lncRNAs, i.e., scaffold lncRNAs, but not those lncRNAs that have been released from chromatin,23 suggesting that Pol V-dependent lncRNAs are subjected to AtRRP6L1-mediated transcript retention in the chromatin for a scaffold function. Pol V is present in the nucleoplasm and in perinucleolar foci, where AGO4 and the de novo DNA methyltransferase DRM2 are also localized and are therefore known as RdDM foci.30,31 AtRRP6L1 colocalizes with Pol V not only in the nucleoplasm but also in perinucleolar RdDM foci,23 supporting that the retention of Pol V lncRNAs is integral to the RdDM process.
Pol IV-dependent lncRNAs are also expected to be retained in the chromatin to facilitate RDR2-dependent double-stranded RNA synthesis, because RDR2 physically interacts with Pol IV and is inactive without Pol IV in an in vitro assay.11,32 Therefore, the impaired accumulation of Pol IV-dependent siRNAs in atrrp6l1 mutants may be explained by the function of AtRRP6L1 in mediating lncRNA retention, although Pol IV-dependent lncRNAs have yet to be detected. In contrast to Pol IV-dependent siRNAs, neither tasiRNAs nor siRNAs that are derived from inverted repeats (IR) are decreased by AtRRP6L1 dysfunction.23 Biogenesis of IR siRNAs does not require transcript retention for RNA-directed RNA polymerase action, since the Pol II-generated IR lncRNAs directly form double-stranded hairpin structures that are cleaved by DCL2 and DCL3 to generate 22nt and 24nt siRNAs, respectively.15 Meanwhile, production of tasiRNAs would not require retention of precursor lncRNAs in the nucleus, because tasiRNA precursors are produced by Pol II and cleaved by miRNA-guided AGO1, and the resultant single-stranded RNA fragments require cytoplasmically localized RDR6 and SGS3 to generate double-stranded RNAs for subsequent tasiRNA production.33,34 The differential regulation of IR siRNAs, tasiRNAs, and Pol IV-dependent siRNAs by AtRRP6L1 suggest that AtRRP6L1 is not a general regulator of siRNA biogenesis, and are consistent with AtRRP6L1 mediating lncRNA retention in chromatin. These results, together with AtRRP6L1 regulation of Pol V-dependent lncRNAs, indicate that RNA retention may be specific to those lncRNAs that function in cis.
Splicing Factors Control Accumulation of lncRNAs and siRNAs
The levels of Pol V-dependent lncRNAs were shown to be reduced in the Arabidopsis mutants defective in RDM16 and STA1, both of which are pre-mRNA splicing factors (Fig. 1).35,36 The rdm16 and sta1 mutants do not display altered mRNA accumulation or pre-mRNA splicing of known RdDM factors.35,36 Thus the effects of RDM16 and STA1 on Pol V lncRNA homeostasis are possibly direct, although the underlying mechanisms remain unclear.
STA1 is a protein that associates with the U5 snRNP complex, while RDM16 encodes a component of the U4/U6 snRNP complex.35,36 Nuclear immunostaining studies revealed that RDM16 is distributed throughout the nucleoplasm while STA1 is mainly present in nucleolus-associated Cajal bodies.35,36 Protein localization in Cajal bodies has been observed for some siRNA-producing RdDM factors, including RDR2, DCL3, and AGO4.31,37 Consistent with their nuclear localization patterns, STA1 but not RDM16 was found to be required for siRNA accumulation. Pol V reinforces production of some Pol IV-dependent siRNAs, in addition to transcribing scaffold lncRNAs.9 While it is unclear how Pol V lncRNAs may be involved in the reinforcement of siRNA production, STA1 appears to be a connection between Pol V-dependent lncRNAs and siRNAs, because STA1-dependent siRNAs are almost exclusively Pol V-dependent.36
Pol IV-dependent siRNA accumulation was also impaired in Arabidopsis mutants defective in several other RNA splicing factors, including ZOP1, SR45, MAC3A, MAC3B, MOS4, MOS12, and MOS14.38,39 ZOP1 physically associates with STA1 and, similar to STA1, is present in Cajal bodies where AGO4 is also localized.38 Because AGO4 is a downstream factor in siRNA production and because Pol IV is not localized in Cajal bodies,37 the presence of ZOP1 and STA1 in Cajal bodies suggests that these two splicing factors are involved in the downstream steps during Pol IV-dependent siRNA biogenesis. However, a role of ZOP1 in affecting Pol IV-dependent lncRNAs cannot be ruled out, since ZOP1 shows partial colocalization with Pol IV in the nucleoplasm.38 In Arabidopsis, small RNA densities in gene regions are adversely correlated with the presence of introns.40 This appears to argue against a positive role of RNA splicing in siRNA production. Meanwhile, the majority of RdDM targets are intronless TEs and intergenic regions. Therefore, the roles of RNA splicing factors in siRNA production during RdDM are likely decoupled from their functions in pre-mRNA splicing.
Conclusions
Functional lncRNAs are key regulators of many biological processes. In the canonical RdDM pathway, production of functional lncRNAs requires not only RNA polymerase transcription but also the activities of some RNA processing factors. Compared with mRNAs, lncRNAs exhibit both similar and dissimilar characteristics. Our recent results demonstrated the importance of AtRRP6L1-dependent lncRNA retention in mediating DNA methylation and transcriptional silencing. In addition, our results showed that pre-mRNA splicing factors affect accumulation of lncRNAs and siRNAs in the RdDM pathway. The importance of RNA processing to lncRNAs is being increasingly recognized; meanwhile, new questions have arisen. For instance, the mechanism underlying Rrp6-dependent RNA retention is unclear; it remains to be elucidated how RNA splicing factors are involved in lncRNA and siRNA homeostasis; and the potential roles of other RNA processing factors in regulating lncRNAs remain to be explored.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We apologize to colleagues whose work we were unable to include because of space limitations. The work in J.-K.Z.’s lab is supported by grants from National Institutes of Health, USA and by the Chinese Academy of Sciences, China.
References
- 1.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14:142–7. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pikaard CS, Haag JR, Pontes OM, Blevins T, Cocklin R. A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harb Symp Quant Biol. 2012;77:205–12. doi: 10.1101/sqb.2013.77.014803. [DOI] [PubMed] [Google Scholar]
- 4.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–52. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–24. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A. 2007;104:4536–41. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:3145–50. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–21. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AM, Strahl BD, Patel DJ, Jacobsen SE. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–9. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Ma ZY, Zeng L, Tanaka K, Zhang CJ, Ma J, Bai G, Wang P, Zhang SW, Liu ZW, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci U S A. 2013;110:8290–5. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blevins T, Pontvianne F, Cocklin R, Podicheti R, Chandrasekhara C, Yerneni S, Braun C, Lee B, Rusch D, Mockaitis K, et al. A two-step process for epigenetic inheritance in Arabidopsis. Mol Cell. 2014;54:30–42. doi: 10.1016/j.molcel.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunoyer P, Brosnan CA, Schott G, Wang Y, Jay F, Alioua A, Himber C, Voinnet O. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010;29:1699–712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–48. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–4. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wierzbicki AT. The role of long non-coding RNA in transcriptional gene silencing. Curr Opin Plant Biol. 2012;15:517–22. doi: 10.1016/j.pbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–6. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu ZW, Shao CR, Zhang CJ, Zhou JX, Zhang SW, Li L, Chen S, Huang HW, Cai T, He XJ. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 2014;10:e1003948. doi: 10.1371/journal.pgen.1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507:124–8. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aufsatz W, Mette MF, Matzke AJ, Matzke M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol Biol. 2004;54:793–804. doi: 10.1007/s11103-004-0179-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Tang K, Qian W, Duan CG, Wang B, Zhang H, Wang P, Zhu X, Lang Z, Yang Y, et al. An Rrp6-like protein positively regulates noncoding RNA levels and DNA methylation in Arabidopsis. Mol Cell. 2014;54:418–30. doi: 10.1016/j.molcel.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma. 2008;117:419–29. doi: 10.1007/s00412-008-0166-4. [DOI] [PubMed] [Google Scholar]
- 25.Butler JS, Mitchell P. Rrp6, rrp47 and cofactors of the nuclear exosome. Adv Exp Med Biol. 2011;702:91–104. doi: 10.1007/978-1-4419-7841-7_8. [DOI] [PubMed] [Google Scholar]
- 26.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–53. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 27.Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–42. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 28.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–4. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 29.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–7. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li CF, Henderson IR, Song L, Fedoroff N, Lagrange T, Jacobsen SE. Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana. PLoS Genet. 2008;4:e27. doi: 10.1371/journal.pgen.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 32.Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell. 2012;48:811–8. doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009;583:1261–6. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 34.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang CF, Miki D, Tang K, Zhou HR, Zheng Z, Chen W, Ma ZY, Yang L, Zhang H, Liu R, et al. A Pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 2013;9:e1003779. doi: 10.1371/journal.pgen.1003779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dou K, Huang CF, Ma ZY, Zhang CJ, Zhou JX, Huang HW, Cai T, Tang K, Zhu JK, He XJ. The PRP6-like splicing factor STA1 is involved in RNA-directed DNA methylation by facilitating the production of Pol V-dependent scaffold RNAs. Nucleic Acids Res. 2013;41:8489–502. doi: 10.1093/nar/gkt639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 38.Zhang CJ, Zhou JX, Liu J, Ma ZY, Zhang SW, Dou K, Huang HW, Cai T, Liu R, Zhu JK, et al. The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis. EMBO J. 2013;32:1128–40. doi: 10.1038/emboj.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ausin I, Greenberg MV, Li CF, Jacobsen SE. The splicing factor SR45 affects the RNA-directed DNA methylation pathway in Arabidopsis. Epigenetics. 2012;7:29–33. doi: 10.4161/epi.7.1.18782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christie M, Croft LJ, Carroll BJ. Intron splicing suppresses RNA silencing in Arabidopsis. Plant J. 2011;68:159–67. doi: 10.1111/j.1365-313X.2011.04676.x. [DOI] [PubMed] [Google Scholar]