Abstract
In this study, we combine available high resolution structural information on eukaryotic ribosomes with low resolution cryo-EM data on the Hepatitis C Viral RNA (IRES) human ribosome complex. Aided further by the prediction of RNA-protein interactions and restrained docking studies, we gain insights on their interaction at the residue level. We identified the components involved at the major and minor contact regions, and propose that there are energetically favorable local interactions between 40S ribosomal proteins and IRES domains. Domain II of the IRES interacts with ribosomal proteins S5 and S25 while the pseudoknot and the downstream domain IV region bind to ribosomal proteins S26, S28 and S5. We also provide support using UV cross-linking studies to validate our proposition of interaction between the S5 and IRES domains II and IV. We found that domain IIIe makes contact with the ribosomal protein S3a (S1e). Our model also suggests that the ribosomal protein S27 interacts with domain IIIc while S7 has a weak contact with a single base RNA bulge between junction IIIabc and IIId. The interacting residues are highly conserved among mammalian homologs while IRES RNA bases involved in contact do not show strict conservation. IRES RNA binding sites for S25 and S3a show the best conservation among related viral IRESs. The new contacts identified between ribosomal proteins and RNA are consistent with previous independent studies on RNA-binding properties of ribosomal proteins reported in literature, though information at the residue level is not available in previous studies.
Keywords: RNA-Protein interactions, Cryo electron microscopy, Hepatitis C, Protein modeling
Introduction
The 5′ UTR of the Hepatitis C Viral (HCV) RNA is known to promote internal initiation of translation. The Internal Ribosome Entry Site (IRES) adopts a unique tertiary fold that binds directly to the 40S subunit of the ribosome and initiates translation in the absence of most of the canonical eukaryotic initiation factors.1-3 Thus, internal initiation by the HCV IRES is similar to the mode of translation initiation in prokaryotes where the mRNA binds to the 30S ribosomal subunit in a factor-independent manner. IRES binding induces conformational changes in the head region of the 40S subunit.4 Initiation factors eIF3 and eIF2 (as a ternary complex with tRNA and GTP) bind to the IRES-40S complex to form a 48S-like pre-initiation complex, which later associates with the 60S subunit resulting in a translation competent 80S assembly.3,5,6 In addition to the canonical eIF2-dependent mode, the HCV IRES can use an alternate pathway in which the 48S complex is formed in an eIF2-independent manner by the cooperative binding of Met-tRNAiMet and eIF5B. The switch to the eIF5B-dependent mode occurs when the availability of the ternary complex decreases due to eIF2α phosphorylation, induced under various stress conditions.7-9Factor-less translation initiation mechanisms are also used by IRES elements found in other viruses belonging to the Flaviviridae and Dicistroviridae families3,10-13.
The unique mechanism of the HCV IRES-mediated translation initiation promises the development of different inhibitor molecules that specifically target the interaction of the IRES RNA with ribosomes.14-17 It has been reported that the tertiary scaffold of the IRES RNA has a significant role in the initiation process1,18. The secondary structure of the HCV IRES RNA consists of four major stem loops named domains I-IV.19,20 Mutational analysis, cross-linking studies and RNasefootprinting experiments have implicated that domains II, IIIe+f and IIId bind to 40S subunit.14,21-28
The 40S ribosomal proteins S2, S3, S10, S15, S16/S18 and S27 have been found to crosslink with the HCV IRES in 4-thiouridine-mediated cross-linking experiments, while UV cross-linking experiments29-31 show that S5 forms a complex with the IRES. Gel digestion experiments highlight interactions of the IRES with S3, S5, S7, S18 and p40.32 The 3.93Å and 4.15Å resolution structures of small subunits of a eukaryotic ribosome are influential in getting insights into the locations and folds of ribosomal components.33,34 More recently, the structure of the human 80S ribosome was solved at 5Å resolution using cryo-electron microscopy.35
The cryo-EM structure of the IRES-bound human 80S ribosome, solved by Boehringeret al.22 at 15Å resolution provided remarkable insights into the interactions between the HCV IRES and human ribosome. Three major contact points involving interactions of domain II near the exit site of the small subunit head, domain IIId near the ribosomal platform and the region preceding IIId at the platform region have been reported. The low resolution of this cryo-EM data however restricts detailed analysis of the contact points.
Recently domain IIId has been shown to interact with the expansion segment 7 of the 18S rRNA36 and this base-pairing is reported to act as an anchor for ribosome recruitment by the HCV IRES.22 Domain IIId of the HCV IRES was shown to be essential for ribosome binding using mutation and footprinting experiments.22-24 Kikuchi et al.37 showed that RNA aptamers with a consensus loop sequence of ACCCA bound to IIId in a sequence-specific manner and inhibited the binding of 40S. The CCC triplet was found to be critical for binding to domain IIId.
Domain II binds to the ribosomal head at the exit site and interacts with the ribosomal protein S5.22,28,30 Interestingly, domain II deletion mutant-bound to 40S subunit with a nearly wild type affinity is unable to induce conformational change in the 40S, which is required for internal initiation.4 While domain IIId mainly contributes to the binding affinity of 40S, domain II has important roles in the IRES-mediated initiation process including 60S binding,5 eIF2 release,38 removal of eIF3j39 and also in inducing conformational changes involving the 18S rRNA at the decoding groove.28,36
Here we set-out to identify the 40S ribosomal components along with their residues that interact with the HCV IRES RNA and the local regions of the IRES involved in the interaction. In order to achieve this goal we integrated available experimental information mainly from cryo-electron microscopy with high resolution crystallographic structures, which became available after the cryo-EM-deduced low-resolution structure of the IRES-ribosome complex was published. A similar strategy was used recently by Filbin and Kieft40 to understand the probable interactions between domain II of the IRES and domain IV near the start codon binding site. However this study was not aimed at identifying the ribosomal components that interact with the IRES.
To locate the probable sites of contact, we also used interaction information on proteins bound to nucleic acids, that are structurally similar to human ribosomal proteins. By homology considerations we ensured the proper mapping of contact points between the RNA and ribosomal components. The distances involving probable interface residues were used to add interaction restraints for flexible RNA-protein docking. The present attempt provides a further understanding on the structural organization of the 40S subunit in complex with the HCV RNA and enabled us to identify new contact regions between the HCV RNA and ribosomal proteins.
The structure of the ribosome-bound CSFV IRES domain II deletion mutant has been solved very recently11 and the results are in agreement with our model for the HCV IRES – 40S interactions. Here we also propose interactions between domain II and IV with ribosomal proteins and provide residue-base interaction details from our docked model.
Results
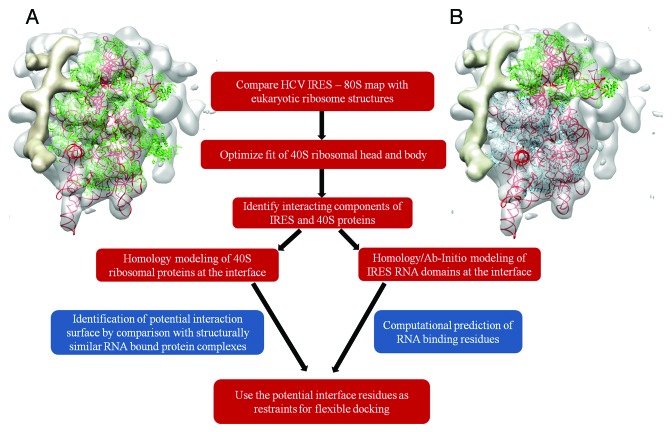
In the current work, the cryo EM-derived structure of the HCV IRES – 80S complex22 was used as a framework for identifying interactions between the 40S ribosomal proteins and the HCV IRES RNA. In addition we also used the RNA-binding modes observed in the 3-D structures of homologs of ribosomal proteins. The low resolution nature of the cryo-EM structure of the HCV IRES – ribosome complex22 limits the direct integration of atomic models of ribosomal proteins in the density map. Keeping the cryo-EM structure of the HCV IRES-ribosome complex as a framework, the identification of interacting components requires the localization of ribosomal proteins and HCV IRES RNA domains at the points of contact. Apart from providing a global view on the structure of macromolecular complexes, the cryo-EM-derived density maps also present low resolution fingerprints of the components that can be described based on the shape of density.
Though the full-length structure of the HCV IRES RNA was solved in the unbound form based on small-angle X-ray scattering data,41 the relative domain motions upon binding with the 40S subunit is best captured by fitting different RNA domain structures in the density. The initial placement of these components in the density map was based on the work of Boehringeret al.22 (PDB ID: 2AGN). The fitted components were then refined by local density optimization using CHIMERA.42,43
We compared the low resolution cryo-EM structure of the HCV IRES-80S complex with the atomic resolution structures of eukaryotic ribosomes to identify locations of ribosomal proteins in the density map. The fitted conformations of Tetrahymena thermophila33 (PDB ID: 2XZM) and human 40S structures35 (PDB IDs: 3J3A+3J3D) with the HCV IRES – 80S complex map have correlation scores of 0.70 and 0.71 respectively (Fig. 1A). The ribosomal head is known to undergo conformational changes upon IRES binding.4We manually separated the ribosomal head and body after the selection of components using Chimera. Further optimization of the human 40S head and body resulted in fits with better correlation coefficient scores of 0.69 and 0.75 respectively (Fig. 1B). The 40S head and body of the Tetrahymena ribosome fitted with lower correlation scores of 0.67 and 0.72 respectively.
Figure 1. WorkFlow used to model HCV IRES – 40S ribosomal protein interactions. (A) Comparison of the atomic model of the human 40S ribosome35 (PDB ID: 3J3A/3J3D) with the cryo-EM map of the HCV IRES – 80S complex. Proteins are shown in green and rRNA in red. (B) Optimized fits of ribosomal head (proteins in green) and body (proteins in blue). Figure rendered in CHIMERA.42
Having identified the probable ribosomal proteins at the sites of contact, the RNA-binding properties of these proteins were studied independently to assess and validate their interaction with the IRES RNA (Fig. 1). Models of human ribosomal proteins (Table S1) were superposed on the fitted human 40S ribosome structures (head and body). The ribosomal proteins that interact with the IRES RNA are expected to have two RNA-binding surfaces, one for binding rRNA and the other for contact with the IRES. The ability of these proteins to interact with nucleic acids at a site different from the rRNA-binding face, was investigated.
Protein –ligand interaction sites are observed to be conserved among similar folds44,45 and nucleic acid binding proteins often employ specialized domains or motifs for interaction.46-49 Hence the nucleic acid binding regions can be predicted by the structural analysis of homologs or analogous proteins with the same fold. This was done with the help of structure comparison techniques such as DALI.50 RNA-protein contacts were defined using an inter-atom distance cut-off of 5Å based on earlier studies.51,52 In the absence of relevant data on related protein folds with nucleic acid binding properties, tools like RNABindR53 and BindN54 were used to predict the regions involved in RNA contacts. These methods identify nucleic acid binding regions based on their sequence patterns, interface residues propensity53 and amino acid properties like pKa, hydrophobicity and molecular mass.54
The set of residues which were found at the interface of the map density fit and also predicted to bind to the IRES RNA, were used to define interaction restraints for HADDOCK55-57 to generate energetically favorable binding poses. To restrict deviations from the fitted models (based on optimization of the human 40S head and body), distance restraints were incorporated to maintain orientation during docking. The distances observed were allowed to vary up to 5Å and multiple distance restraints were applied for each docking exercise. Cases of multi-component interactions involving the S5, S25, domain II and S26, S1e and the IIIef+pseudoknot were modeled using the multi-body docking protocol in HADDOCK.56,57
While docking, 20% or 50% of the proposed interface residues and neighbors within 5Å distance were randomly excluded from the restraint set. The addition of ambiguity at the interaction site allows sampling of different poses around the observed interface. This helps account for uncertainty in the proposed set of residues. Flexibility was also added in the docking process with the use of multiple NMR ensemble models if available, allowing flexible movements in the proposed interface regions. The top 200 energetically favorable poses were clustered at 10Å RMSD cut-off and those interaction models from the three largest clusters were analyzed further. The selected poses were filtered to identify a model that gave the same or better correlation scores when compared with the model derived by optimizing ribosomal head and body.
Table 1 summarizes the new major contacts identified in this study and presents laboratory experiments supporting these interactions.
Table 1.HCV IRES domains and ribosomal components involved in major contacts. Direct experimental supports on the involvement of one or both of these interacting components are listed. The likely role of this interaction in internal initiation are also indicated.
| HCV IRES domain | Ribosomal protein (or rRNA) | Direct experimental supports | Indirect experimental supports | Probable role in internal initiation |
|---|---|---|---|---|
| II | S25 | Lytle et al., 2002 (25); Odreman-Macchioli et al., 2001(26); Landry et al., 2009 (72) |
Nishiyama et al., 2007 (70); Muhs et al., 2011(71) |
Conformational change in head region necessary for initiation (and associated functions like eif2 release mediated by II) |
| IV | S5 | Ray and Das, 2004 (14); Malygin, A.A. et al. 2013 (36) |
Schmeing et al., 2011(62); Armache et al., 2010 (13) |
Placement of start codon at P site |
| IV | S28 | Armache et al., 2010 (13); Lomakin and Steitz, 2013 (63) | Placement of start codon at P site | |
| IIIe | S3a | Laletina et al. 2006 (74); Otto et al. 2002 (31); Malygin et al. 2013 (76) |
Bhat, P et al., 2012 (78); | Stabilize pseudoknot conformation for positioning start codon at P site |
| Pseudoknot | S26 | Yu et al. 2005 (83); Sharifulin et al. 2012 (82) |
Placement of start codon at P site | |
| jIIIabc | S27 | Malygin et al. 2013 (76) | Support 40S binding |
The HCV IRES domain II – 40S Head
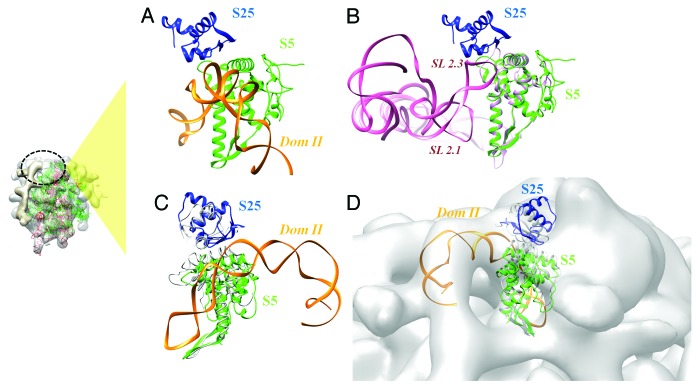
The cryo-EM structures of the HCV IRES-ribosome complex generated by Spahnet al.4 and Boehringer22 showed that domain II of the IRES RNA interacts with the head region of the 40S subunit near the exit site. In our analysis we note that at the head of 40S where domain II binds, a concave surface suitable for interaction with a helical RNA structure is present (Fig. S1Aand Figure 1A). The fitted NMR model of domain II58 has a correlation score of 0.81 with the density map. The 40S ribosomal proteins S5 and S25 form the interface at this site and positively charged patches were observed at the surface which interacts with domain II of the IRES (Figure S1B). The fitted conformation based on the human 40S model is in agreement with the independent results obtained by Filbin and Kieft.40
Analysis of related systems involving homologs and similar folds suggested two non-rRNA interactions involving S5 and S25. The CrPV IRES RNA is known to interact with S5 and S25 and a model generated based on its cryoEM structure suggests that the sites of contact overlap with the binding site of the HCV IRES domain II59 (Fig. 2A and B). The C-terminal helix and the preceding loop of S5 make multiple contacts with the CrPV IRES domain SL2.1 and SL2.3 (Fig. S1C and 2A). Mapping the interacting residues from this model to the human S5 structure suggested that most of these residues are conserved (Table 2 and Figure 2A). On the other hand, residues K66 and R76 of the human S25 are also reported to bind to the CrPV IRES domain SL2.3.60
Figure 2. Interaction between domain II and the head region of 40S. (A) Locations of domain II and ribosomal proteins S5 (green) and S25 (blue), based on density fits. (B) Interaction between the CrPV IRES RNA and 40S ribosomal protein S5 (yeast).59 (A) and (B) highlights similar interfaces used by the HCV and CrPV IRES RNAs. (C) Docked model of domain II – S5 – S25 complex. The conformations of S5 and S25 (based on ribosomal head fit) before docking are shown in gray. (D) The fit of docked model in the cryo-EM density (gray surface) corresponds to the ribosomal head. The site of interaction involving domain II with respect to the complete ribosome is indicated in the inset on the left.
Table 2. Active site residues proposed for guided docking by HADDOCK. The structurally similar RNA-bound complexes used to identify interacting residues are given. If a related fold information was not used, ab-initio RNA prediction method used to predict binding sites, are listed. The residues proposed either based on structurally similar interactions or by ab-initio predictions, are listed in parentheses. Those added additionally to the list based on the vicinity to IRES RNA, are listed outside parentheses.
| Ribosomal protein | Interface residues involved in docking restraints | Structurally similar interaction / Prediction | IRES bases |
|---|---|---|---|
| S5e | (V134,R136,R127), (T104,I178,K182,N186,Y188,K191,E195,R198,S202) |
2WDM-G, 2NOQ-F |
80–87, 67–73, 90–95, 97–101 |
| S25 | K114,(K66,R76) | Muhs et al. 2011 | 80–87, 67–73, 90–95, 97–101 |
| S26 | (R51,K66),Y62 | BindN | 305–308,329 |
| S3a (S1e) | (K199, Q202) | RNABindR | 299,300 |
| S27 | (K36,Y41,R80,Q83) | 3IZR-m | 234–237 |
The β hairpin of S5 interacts with E site tRNA (PDB ID: 2WDM)61 which reflects the possibility of similar structural contact with the apical loop of domain II (Fig. S1D and S2B). Based on these interactions with E site tRNA and CrPV IRES, we compiled a list of potential interacting residues of S5 and S25 that occur at the surface of contact with domain II (Table 2).
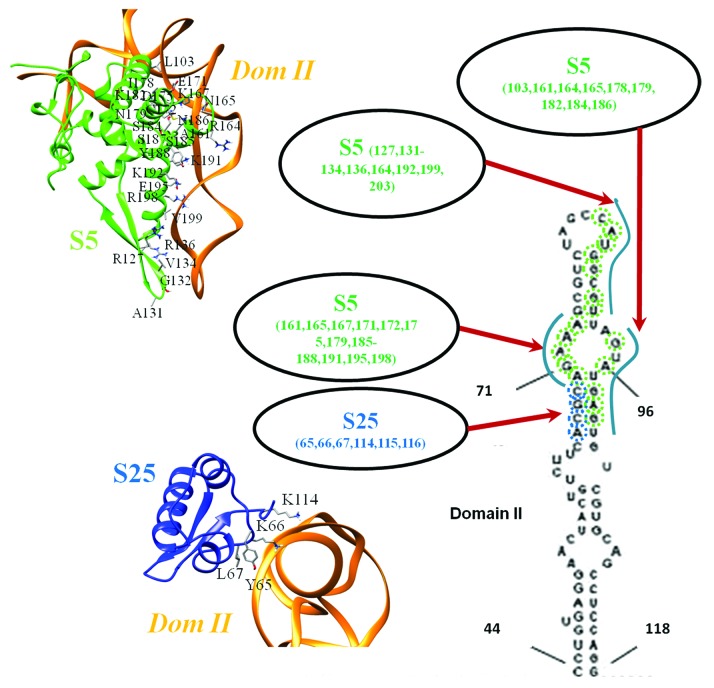
The best poses obtained by flexible docking with interaction restraints showed minimal variability and belonged to a single cluster. The selected docked model from a 20% random exclusion trial has a density correlation score of 0.83 as against a score of 0.81 with the fitted models prior to docking (Fig. 2C and D). The region involving two C-terminal helices (residues 164–204) of S5 make multiple contacts with domain II loop formed by bases 70–74; 92–97 (Fig. 3, Table 3). S25 has relatively fewer interactions involving residues in the C-terminus and a central loop (residues 65–67) (Fig. 3, Table 3). A long stretch of Lys residues forms a flexible loop at the N-terminus of S25,33 but also occurs at the interface with domain II. Hence this segment may also participate in the interaction.
Figure 3. Residue-base interactions involving domain II. The contact regions between domain II and human ribosomal proteins S5 and S25 are highlighted, based on the docked model. S5 is shown in green while S25 is in blue. The bases involved in contact are indicated on the secondary structure of domain II and the structural interfaces involving S5 and S25 are also shown.
Table 3. Pairwise interactions. Residue – base contacts (< 5Å distance) observed between 40S ribosomal proteins and the HCV IRES RNA, are listed.
| Pairwise interactions: S5 | Pairwise interactions: S25 |
|---|---|
| ALA-131 CYT-83 ALA-161 ADE-72 ALA-161 GUA-71 ALA-161 GUA-94 ARG-127 GUA-87 ARG-127 GUA-88 ARG-136 GUA-87 ARG-136 GUA-88 ARG-164 GUA-94 ARG-164 URI-91 ARG-198 ADE-74 ASN-165 ADE-70 ASN-165 GUA-71 ASN-165 GUA-94 ASN-179 ADE-70 ASN-179 GUA-98 ASN-186 ADE-70 ASN-186 ADE-98 ASN-186 GUA-71 ASN-186 GUA-98 ASN-203 GUA-87 ASN-203 URI-86 ASP-175 ADE-70 ASP-175 GUA-71 CYS-172 GUA-71 GLU-171 ADE-70 GLU-195 ADE-73 GLU-195 ADE-74 GLY-132 CYT-83 GLY-132 CYT-84 ILE-178 ADE-99 LEU-103 GUA-100 LYS-167 ADE-70 LYS-167 GUA-70 LYS-182 ADE-99 LYS-182 GUA-100 LYS-191 ADE-72 LYS-191 ADE-73 LYS-192 GUA-90 SER-184 ADE-99 SER-184 GUA-98 SER-185 ADE-72 SER-187 ADE-72 SER-187 GUA-71 THR-133 CYT-84 THR-133 URI-86 TYR-188 ADE-72 TYR-188 GUA-71 VAL-134 ADE-85 VAL-134 CYT-84 VAL-134 GUA-87 VAL-134 URI-86 VAL-199 GUA-87 |
GLY-115 GUA-68 GLY-116 GUA-68 LEU-67 CYT-69 LEU-67 GUA-68 LYS-114 ADE-66 LYS-114 CYT-67 LYS-114 GUA-68 LYS-66 CYT-69 LYS-66 GUA-68 TYR-65 CYT-69 |
| Pairwise interactions: S26 | |
| ARG-51 URI-329 TYR-62 URI-306 | |
| Pairwise interactions: S3a | |
| GLN-202 GUA-299 GLN-202 GUA-300 LYS-195 GUA-301 LYS-195 URI-302 LYS-199 GUA-299 LYS-199 GUA-300 LYS-199 GUA-301 | |
| Pairwise interactions: S27 | |
| CYS-40 URI-234 ILE-43 GUA-235 LYS-36 CYT-236 LYS-36 GUA-235 LYS-42 GUA-235 LYS-42 URI-234 SER-78 CYT-236 TYR-41 GUA-233 TYR-41 GUA-235 TYR-41 URI-234 ARG-80 CYT-151 ARG-81 GUA-150 |
The HCV IRES domain IV and 40S Exit Site
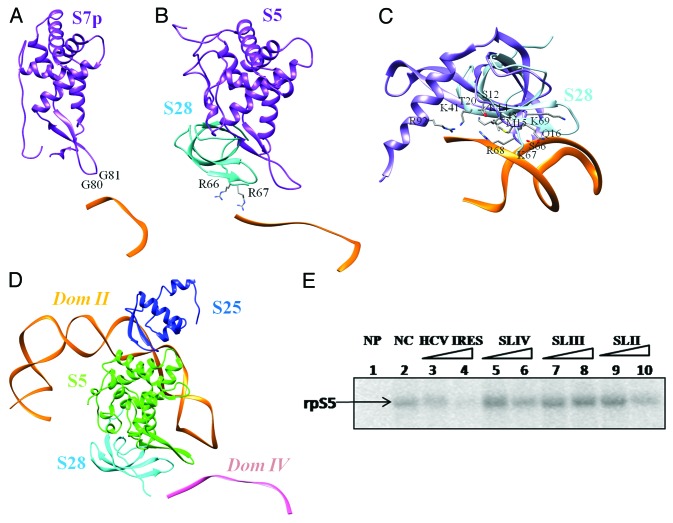
The structures of mRNA-bound ribosomes solved by X-ray crystallography61,62 (Fig. 4A) show that the β-hairpin and the C-terminal helix of S7 bind to mRNA at the exit site. Structures of eukaryotic ribosome13,63 highlight that both S28 and S5 can interact with mRNA at this site (Fig. 4B and Figure S3A). The structure of the HCV IRES SLIV was modeled using the mRNA structure bound to the Triticum aestivum ribosome13 as a template to map interactions with S5 and S28. This model reflects contacts at the stage where the start codon is placed at the P site. Based on these interactions, Ala 131 and Gly 132 of S5 are likely to interact with bases 339 and 340, just preceding the start codon (Fig. 4C).
Figure 4. SLIV interactions with S5 and S28. (A) Contacts between ribosomal protein S5p (S7) (purple) β-hairpin and mRNA at the exit site61 (PDB ID: 2WDM). (B) Interactions between mRNA and 40S ribosomal proteins S5 (purple) and S28 (cyan) highlighted in the crystal structure of Oryctolagus ribosome63 (PDB ID: 4KZZ). (C) Comparison of human ribosomal S28 model (cyan) with 30S ribosomal protein S12 (PDB ID: 3OHY, purple).64The interface residues observed in 30S protein S12 are also highlighted. (D) The docked model of domain II-S5-S25 complex relative to S28 and domain IV model. (E) S5 protein was UV cross -linked with α- 32P UTP labeled HCV IRES in the absence (Lane 2) or presence of 50 fold and 100 fold molar excess of unlabeled HCV IRES (18–383), SLIV (domain IV), SLIII (domain III) and SLII (domain II) in vitro transcribed RNAs (Lanes 3–10). NP indicates no protein control (Lane 1).
The structure of S28 is characterized by a four stranded β-sheet forming an OB fold.33 A structural relative is the 30S ribosomal protein S12 (PDB ID: 3OHY64) which also shows RNA-binding properties at this region (Fig. 4C). Though strict conservation of DNA/RNA-binding residues is not observed, residues known to have DNA-binding properties are preserved at equivalent positions (Fig. S3B and S3C). Such residues on the surface likely to bind to the bases following the pseudoknot are highlighted in the figure.
Among the different ribosomal proteins, S5 has two binding sites on the IRES RNA, namely domain II and IV (Fig. 4D). We performed UV cross-linking studies of ribosomal protein S5 (rpS5e) with α- P32 UTP labeled HCV IRES in the absence (Fig. 4E: Lane 2) and presence of 50 fold and 100 fold molar excess of either of the unlabeled HCV IRES (18–383), SLIV (domain IV), SLIII (domain III) and SLII (domain II) in vitro transcribed RNAs (Fig. 4E: Lanes 3–10). It can be seen that SLIV and SLII interferes with the HCV IRES – S5 interaction, unlike SLIII. This suggests that SLII and SLIV stem loops interact with the S5 protein, further supporting our computational modeling-based inferences.
The HCV IRES domain IIIef + Pseudoknot and the 40S platform
Cryo-EM studies on the HCV IRES–ribosome complexes4,22 show that the IRES domains between II and IIId (in the tertiary fold), interact with the platform region of the 40S. This involves the pseudoknot and domains IIIef and IV of the IRES (Fig. 5A). The stem-loop structure of domain IV that holds the start codon (Fig. 1A), is shown to unwind to a single stranded form upon 40S binding.40This helps in placing the start codon at the ribosomal P site. The IRES density observed at the 40S platform could therefore be associated to IIIef and pseudoknot. However the region involving the start codon is not distinguishable in the density map.
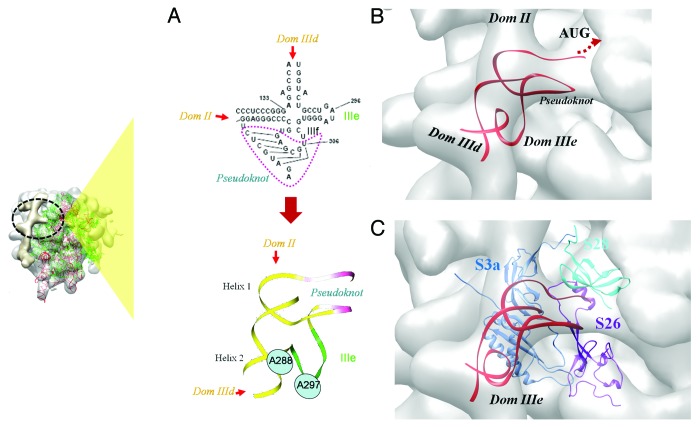
Figure 5. IRES-40S contact at the ribosomal platform region. (A) The secondary structure and 3D model of the HCV IRES comprising domains IIIef and pseudoknot. The position of two bases A288 and U297 that are reported to base-pair18,75 are highlighted. (B) Model of the IIIef+pseudoknot generated by interactive manipulation and structure refinement. The model fitted in the 40S platform density (gray) is shown. Locations of IIId, IIIe and II are also indicated (C) The docked model of IIIef+pseudoknot (red) and 40S ribosomal proteins S26 (purple), S3a (S1e) (blue), fitted in the cryoEM density (gray). The relative location of S28 (cyan) is also shown. The site of interaction involving domain IIIef+pseudoknot with respect to the complete ribosome, is indicated by the inset on the left.
Figure 5A shows the secondary structure of the region of the IRES involving IIIef and pseudoknot. It involves two RNA helices (helix I and 2), the pseudoknot and IIIe. The crystal structure of this part of the IRES has been solved at 3.55Å resolution18 and it shows interactions involving IIIe in the tertiary structure. To understand the conformational changes upon ribosome binding, we adopted an ab-initio RNA modeling approach to generate a structure based on EM density (see Methods). A coarse-grained model was generated initially by interactive manipulation and structure refinement, followed by flexible fitting to optimize the model based on density.
Helix 1 has been modeled in the A-form and it could be fitted at the IRES density leading to domain II while helix 2 has been fitted in the density arising from IIId (Fig. 5A). The NMR-derived model of IIIe24 has been used for modeling while the pseudoknot was initially modeled in the A-form. The cryo-EM density corresponding to this region has been used as a framework to find the relative orientations of these sub-components. Considering backbone continuity and distance from the decoding groove, the density at the mRNA exit channel (Fig. 5B) clearly belongs to the pseudoknot. Based on the shape of this density Boehringeret al.22 had also proposed that this region might correspond to the density of the pseudoknot.
The structure of the pseudoknot was then modified to link the backbone and fit into the density map using MANIP65 and ERNA-3D.66These methods allow interactive manipulation of RNA structure, taking into consideration stereochemistry and torsion angles. ERNA-3D permits modifications with reference to the cryo-EM density. The relative orientations of sub-domains were altered to have a maximum overlap with density. The fit with density has also been optimized and assessed after model manipulation, based on the correlation coefficient score. Subsequently, the geometry was refined and steric clashes were relieved by carrying out energy minimization. Part of domain IIIe which was modeled in continuity with pseudoknot and Helix 2, remains outside the density (Fig. 5B). To optimize the conformation and improve the fit, the model was further modified by carrying out molecular dynamics simulations using the density gradient force.67,68 The correlation score obtained with the final model was 0.78.
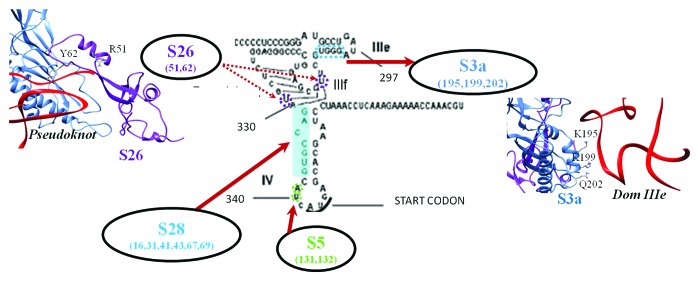
The global organization of different RNA components of the model was similar to that of the crystal structure. However structural changes in the pseudoknot and IIIe have been observed and the model has an RMSD of 2.8Å when compared with the crystal structure (Fig. S4A). Ribosomal proteins S28, S5 and S26 are found in the close proximity of the pseudoknot (Fig. 5C). Two lysine residues (Lys 195 and Lys 199) in the C-terminal helix of the ribosomal protein S3a (S1e) are close to the IIIe phosphate backbone in the fitted model (Fig. S4B). The search for related folds with RNA-binding properties did not give reliable results and the prediction of RNA-binding residues by RNABindR53 suggests possible interactions with Lys 199 and Gln 202 (Fig. S5A). These residues were considered as ambiguous interaction restraints while performing docking with the IIIef+pseudoknot.
The ribosomal protein S26 is characterized by a flexible fold13,33 dominated by loops. A segment of S26 (residues 51–66) involving a loop and short helix was found in the vicinity of the pseudoknot. Structurally similar interactions were not identified while searching for related folds. However BindN54 predicted Arg 51 and Lys 66 as RNA binding sites (Fig. S5B). These two residues along with Tyr 62 which is in close vicinity of the pseudoknot, were used as ambiguous restraints for docking.
The model of the IIIef+pseudoknot was docked with the 40S ribosomal proteins S26 and S3a. The selected pose belongs to a single largest cluster from a 50% random exclusion trial. The fit of this model has a correlation score of 0.70 as opposed to a score of 0.66 based on the model prior to docking (Fig. 5C and Figure S4C). Residues Lys 195, Lys 199 and Gln 202 of S3a and Arg 51 and Tyr 62 of S26 form the interface with IIIe and the pseudoknot respectively (Fig. 6).
Figure 6. Interactions involving IIIef and pesudoknot.Residue-base interactions involving HCV IRES IIIef+pseudoknot and the human 40S ribosomal proteins S28, S26, S5 and S3a. Residue numbers are given below the protein names and interacting bases are highlighted.
Minor contacts
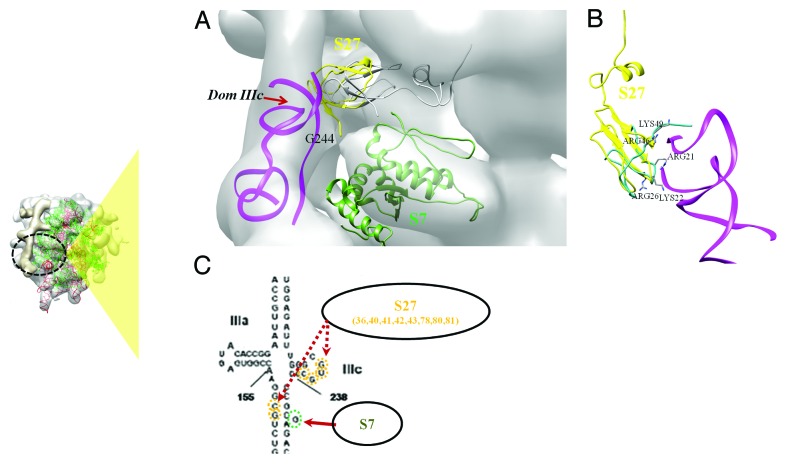
Apart from the major contacts discussed above, two regions of relatively weak density overlaps were also noted. Both these interactions correspond to the junction JIIIabc (including IIIa and IIIc) of the IRES and involve ribosomal proteins S27 and S7 as interacting partners.
The loop region of domain IIIc is in close proximity with the ribosomal protein S27 (Fig. 7A). The structure of S27 comprises of a C4 zing finger motif.33 It shares a similar fold with the eukaryotic 60S ribosomal protein 14013 (Fig. 7B and ST2). The conformation of a RNA-binding loop stabilized by Zn ion co-ordination, is highly conserved among the two proteins (Fig. S6A&S6B). It is involved in base specific interactions with the RNA loop, with a high representation of purine bases. The presence of a conserved tyrosine in the RNA binding loop of S27 also suggests a possible contact with a guanine base in the RNA loop (Fig. S6A). Four residues (Lys 36, Tyr 41, Arg 80 and Gln 83) found at equivalent positions in the alignment were used as interaction restraints (Table 2 and Figure S6B).
Figure 7.Interactions involving jIIIabc. (A) The docked model of jIIIabc (with IIIc and IIIa) (magenta) in complex with S27 (yellow) is shown fitted in the cryoEM density22 (gray). Locations of domain IIIc and bulge base G243 are also indicated. The fitted model of S7 (green) is also shown. (B) Structural alignment of human 40S S27 (yellow) and 60S ribosomal protein l4013 (green). The residues of l40 at the interface with rRNA are highlighted. (C) Residue-base interactions involving HCV IRES IIIef+pseudoknot and human 40S ribosomal proteins S27 and S7. Residue numbers are given below the protein names and interacting bases are highlighted. The site of interaction involving domain jIIIabc with respect to the complete ribosome, is indicated in the inset on the left.
The docked model of the jIIIabc-S27 complex has a correlation score of 0.71 in the density while the fitted models prior to docking had a score of 0.65. The selected cluster had only 11 poses but the interaction energy was lower when compared with the biggest cluster, and the poses fitted with a better correlation score. Residues involving Tyr 41, Lys 42, Arg 80 and Arg 81 are found at the interface with IIIc (Table 3, Figure 7C) and bases in IIIc loop and G 150, C 151 at the RNA stem between IIIc and IIId are involved in S27 binding.
A weak density overlap was observed at the interface between S7 and jIIIabc (Fig. 7A). A single base (G243) bulge was observed at this interface density. Residue Arg 81, which is predicted to have RNA-binding ability (Figure S6C), lies close to the contact density and can bind to the RNA bulge.
Discussion
Both RNA-RNA and RNA-protein contacts contribute to the interaction between the HCV IRES and ribosomal 40S subunit. Several experimental studies demonstrate that the IIId domain of the IRES is important in maintaining the affinity of interactions with the ribosome.23,24,33, 37 Base pair contact between the loop regions of IIId and ES7 can help in anchoring the IRES scaffold on the 40S subunit. Base pairing interaction between a eukaryotic mRNA and the helix 26 of 18S rRNA has also been reported during translation initiation.69
Ribosomal proteins also contribute significantly to the IRES-ribosome contact. We provide structural evidence on the interaction of the ribosomal protein S25 with domain II of the IRES (Fig. 2). Domain II is known to induce conformational changes in the head region of the ribosome but contributes minimally to binding affinity.4Hence the interactions involving ribosomal proteins S25 and S5 are essential in preparing the machinery for translation. Cryo-EM study of the CrPV IRES-bound ribosome has also shown IRES interactions with S25 near the exit site.70 Detailed studies have been performed recently in the context of S25 interaction with Dicistroviridae IRES elements.70,71 Interestingly a conserved stem loop SL2.3 of the CrPV IRES binds to S25 at a similar interface as in the case of the HCV IRES.71 Mutation experiments have shown the involvement of S25 to be essential for Dicistroviridae and HCV IRES activities.72
The tertiary structure of S25 is characterized by a winged helix DNA-binding motif that has high similarity with the FadR transcription factor. Interestingly, many residues present at the FadR-DNA interface73 are also conserved in the human S25, suggesting a similar mode of interaction with 18S rRNA (Fig. S7A-C). A significant level of conservation of the three residues involved in the base specific contact is seen among eukaryotic S25 (Fig. S2). Considering the nature of amino acid-base contacts, we point out the 18 rRNA bases involved in interaction (Fig. S2E). Interestingly, variation of the residues involved in interaction is associated with changes in rRNA bases further highlighting the contact points (Fig. S7D&E).
We note that RNA bases mainly at the loop regions of domain II are found at the interface with S5 and S25 (Fig. 3). It has been shown in a previous study that mutations on this part of domain II reduce the degree of interaction with S5.26 It is also reported that this region is protected from RNase cleavage, in the ribosome-bound form.25 A recent study demonstrates the role of domain II in promoting the switch from translation initiation to the elongation phase.28 The interaction between the apical loop of domain II and β-hairpin of S5, as observed in our model, is reported.
The results of mutational analysis and RNA footprinting1,27 indicate that the domain IIIe of the IRES is important for ribosome binding. Several experimental evidences suggested that domain IIIe has a critical role in the interaction with the ribosome14,23,25,27 and studies have been undertaken to identify the location of IIIe relative to the 40S, in the bound state.74 Our model generated based on the cryo-EM density indicates that IIIe binds the C-terminal helix of ribosomal protein S3a (Fig. 5,6). Domain IIIe also participates in tertiary RNA structure interactions that are important for positioning the start codon at the P site.18,75 A recent study76 confirms that exposed Lys residues of S27 and S3a are important for interaction with jIIIabc and domain III. In each of these binding sites, two Lys residues are observed in our docked model and Lys forms 66% and 25% of S3a and S27 interface residues, respectively. Multiple Lys mediated contacts with IRES bases are found in both the contact sites (Table 3).
The pseudoknot present upstream of the start codon has also been shown to be essential for ribosome binding and translation initiation.1,77,78 Tertiary interactions between domains IIId and region involving IIIe+f and the pseudoknot was also postulated by Kieft et al.1 The densities corresponding to the apical regions of IIId and IIIe overlap and are in continuity with ES7. The contacts connecting distant regions in the RNA identified in the current analysis help in preparing the IRES scaffold for binding the ribosome and positioning the start codon at the P site for initiation. The contacts identified between the pseudoknot (and its 3′) region and ribosomal proteins S28 and S26 can have a significant contribution to this process (Fig. 5 and 6). All the three proteins are known to interact with mRNA near the exit site.79-81The ribosomal protein S26 lacking a eubacterial counterpart, is a key component of the ribosomal binding site of the mRNA region 5′ of the codon positioned at the exit site.82
Cross-linking, gel digestion and mass spectroscopy experiments also suggest that IRES interacts with the 40S ribosomal proteins S3, S10, S14, S16, S18 and p40.29-32,83 Most of these proteins are found in the vicinity of the IRES RNA (Fig. S8). S0A has a long C-terminal loop that lies close to the pseudoknot but the main fold of the protein is too distant for any direct contacts. Interactions between IIIe and proteins S5, S16, S3a and S0A, were recognized using UV cross-linking.84 Locations of S5 and S16 are clearly far from IIIe for any direct contact while S0A loops close to the pseudoknot and S3A binds directly to IIIe. The possibility of IRES interacting with S18 was also reported.32 S18 is located close to the domain II binding site at the 40S – 60S interface but a direct contact could not be deciphered and the shortest distance was more than 20Å.
Another study proposed interactions of domain IIId with S3a, S14 and S16.85IIId is known to interact with the expansion segment 7 (ES7) and S3a binds to the apical region of ES7 (Figure S9A). Hence S3a is in close vicinity of IIId loop, while S16 and S14 are too far for any direct contacts. Similarly, domain II apical region crosslinks with S14 and S16.85The C-terminal loop of S16 is close to the decoding site and at about 20Å from the apical region of domain II (Figure S9B), while S14 is closer with a shortest distance of about 10Å from II. Many of the ribosomal proteins have long-terminal loops that coil around rRNA and may traverse a long distance.
Several eukaryote-specific ribosomal proteins like S25, S26, S28, S3a, S27 and S7 participate in making contact with the ribosome (Fig. 3, 6 and 7 and Figure S10). These proteins adopt characteristic folds specialized for single or double stranded DNA/RNA-binding. With the help of structural information on homologous/analogous folds and RNA-binding residue predictions, we compiled a probable set of interface residues. These were further used as ambiguous interaction restraints to obtain energetically favorable interactions based on flexible docking. Most of these proteins bind to the irregular RNA structures forming the loop regions of IRES domains (Fig. 3,6,7 and Figure S10). Direct experimental supports for interactions based on our model are reported in Table 1 and the likely roles of these interactions in internal initiation are also indicated.
The residues involved in IRES interactions are conserved among mammalian ribosomal proteins (Figs. S11–13). Viral IRES sequences from porcine teschovirus (PTV), canine herpesvirus (CHV), rodent herpesvirus (RodHV), hepatitis GBVB (HGBVB), classical swine fever virus (CSFV) and bovine viral diarrhea virus (BVDV) were aligned with the HCV IRES sequence to check conservation of the 40S binding bases. Interaction sites of S25 and S3a are highly conserved (Fig. S14A), while the S5 binding surface is partially conserved and less conserved in the rest of the surface (Fig. S14, S15). Hence different amino acid – base interactions may be involved and could be characteristic of the IRES function. Many of these interactions may also be auxiliary or non-specific interactions.
The coordinates for the model of the HCV IRES RNA and 40S ribosomal protein interactions are available at http://nslab.mbu.iisc.ernet.in/supplementary.html. The distance restrains files used for docking are also available at this site. All the structure-based sequence alignments generated in this study are also available as supplementary data files. (data S1).
Conclusions
The ability to initiate translation in the absence of many of the canonical initiation factors reflects the significance of the IRES tertiary fold in the initiation process. The distinctive mode of translation initiation by the HCV IRES makes it a putative target for anti-viral therapy. A number of anti-viral therapeutic strategies involving the introduction of foreign nucleic acids into cells have been developed in the past. These nucleic acids are either antisense RNA or ribozymes that specifically target HCV IRES and block internal initiation.16,86 A clear understanding of the mode of interaction between the HCV IRES and the ribosome is essential to make these approaches effective.
By integrating the available information on the 3D structure and RNA-binding properties of the 40S ribosomal proteins with low resolution cryo-EM data on the IRES-human ribosome complex, we propose the probable interacting components and the sites of contact. The HCV IRES is observed to make multiple contacts with the ribosome, involving both RNA-RNA and RNA-protein interactions. Eukaryotic specific ribosomal components (S7, S3a (S1e), S26, S27, S28 and rRNA expansion segment 7) play a major role in binding the IRES. The results of this study also provide various possibilities for precise experimental studies on these interactions and further potential use for drug design.
Methods
The cryo-electron microscopic density maps of Hepatitis C Viral IRES RNA bound to the human 80S ribosome complex22 have been obtained from the EMDB database (http://www.ebi.ac.uk/pdbe-srv/emsearch/, EMDB IDs: emd-1138). MAPMAN87 was used to interconvert various map formats and also to alter density values. Various programs implemented in SITUS package88 have been used to visualize voxel histograms, to generate density for an atomic model at a given resolution, and to perform a six-dimensional search of an atomic model in the electron density map. Colores was used to compare eukaryotic 40S ribosome structures (2XZM,33 3J3A/3J3D35) with the HCV IRES- 80S map. The top ranking poses were optimized further to fit in the density using the Chimera local optimization tool, scored based on the correlation coefficient value calculated independent of the mean density (about zero). Chimera’s interactive atom selection utility was used for selecting the 40S ribosomal head and body and they were separately optimized to fit with the map (Fig. 1).
Modeling 40S ribosomal proteins
Sequences of the human 40S ribosomal proteins were obtained from SWISS-PROT (http://www.expasy.ch/sprot/). Models of the human 40S ribosomal proteins observed at the interface with IRES were generated using the crystal structure of Tetrahymena ribosome as a template. Modeler v9.289 was used for generating homology models based on target-template alignment. Twenty models were generated and the one with the best DOPE score90 was chosen for further analysis (Table S1). Models of human ribosomal proteins (at the interface) were superimposed on the fitted human 40S ribosome structures (head and body) using Chimera MatchMaker.42
Fitting IRES domains
NMR models of domain II of IRES were placed on the reported density at the exit site4,22 and locally optimized to select the model with the best correlation score and minimum steric clash with the fitted ribosome components.
RNA modeling
The model of the IIIef+pseudoknot was generated in two steps
Coarse-grain modeling and structure refinement
MANIP65 and ERNA -3d66 were used to model RNA structures by assembling and modifying helices. MANIP presents a platform for interactive modeling of RNA structures, taking into consideration standard stereochemistry. RNA stem regions (Fig. 5) were modeled as A-form helices while the pseudoknot was initially modeled in the A-form and then modified to fit in the density. The region of the cryo-EM map assigned to this RNA component forms a skeleton upon which the model can be built. Different possibilities of placing the RNA helices were assessed, maintaining the continuity of the backbone and constraining the distance to the decoding groove. To release major steric clashes, energy minimization was performed using AMBER ff99 force field,91 available as a module in the NAB RNA modeling package.92
Flexible fitting in density
To optimize the density fit allowing flexible movements, molecular dynamic simulation was performed using MDFF67,68 for 500 ps and gscale value of 0.1. The backbone torsion angles and base pairs were constrained during the course of the simulation.
The coordinates for the stem region between the junction and IIId are not available from the crystal structure of jIIIabc.93This stem structure with the G243 bulge was modeled using the McFold/McSym94 based on the assigned secondary structure. ModeRNA95 was used to generate a model of jIIIabc along with this stem region, using the coordinates of the crystal structure of jIIIabc and the model of the stem joining IIId as templates. To optimize the backbone conformation and base pairs, energy minimization was performed using AMBER ff99 force field.91
RNA structure comparison was performed using R3D96 and Chimera.42 Multiple RNA sequence alignments were generated using LocaRNA,97 while protein sequences were aligned using ClustalW.98
Identification of potential RNA binding residues
The ribosomal proteins that interact with the IRES RNA are expected to have two RNA binding surfaces, one for binding rRNA and other for IRES contact. DALI server50 was used to identify structurally similar proteins for a given model. The models of the human 40S ribosomal proteins were used to search for similar structures available in the PDB and only the RNA-bound structures were selected for further analysis. RNA-protein contacts were defined using inter-atom distance cut-off of 5Å based on earlier work.51,52 Prediction of residues of the proteins potentially involved in RNA contacts was performed using RNABindR53 and BindN.54
Experimental validation of S5-IRES interaction
Plasmid Constructs
Plasmid pET28a‐S5 (a gift from Dr S. Fukushi, Biomedical Laboratories, Japan) was used for expressing the poly (His) ‐tagged human ribosomal protein S5.
Purification of ribosomal protein S5
Human ribosomal protein S5 was expressed in bacteria (BL21) and purified as mentioned earlier78. Culture was induced with 0.4 mM IPTG at 0.4 O.D 600 and purified using Ni2+–nitrilotriacetic acid–agarose (Qiagen, Hilden, Germany) under non‐denaturing conditions. Protein was eluted with 250mM and 500mM imidazole, dialyzed and stored in 20% glycerol.
UV Cross-linking experiment
UV cross-linking was performed as mentioned earlier78. α32P UTP-labeled HCV IRES RNA (100fmol) was incubated with the purified S5 protein (15 pmol) in presence or absence of unlabeled domain II, domain III and domain IV RNAs at 30°C for 15 min in an RNA binding buffer and then irradiated with a hand-held UV lamp for 20 min on ice. The mixture was treated with 30 µg of RNase A (Sigma) at 37 °C for 45 min. The protein-nucleotidyl complexes were separated on SDS-10% PAGE and analyzed by phosphoimaging.
Availability of data
The model coordinates, the entire supplementary information which includes sequence alignments used in this work and the distance restraint files used in docking are available in http://nslab.mbu.iisc.ernet.in/supplementary.html
Supplementary Material
Acknowledgment
We are grateful to the two anonymous reviewers of our manuscript for critical comments and for excellent suggestions. This project was funded by Department of Biotechnology, New Delhi, India.
References
- 1.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J MolBiol. 1999;292:513–29. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 2.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–62. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 5.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–80. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 6.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 7.Galmozzi E, Aghemo A, Colombo M. Eukaryotic initiation factor 5B: a new player for the anti-hepatitis C virus effect of ribavirin? Med Hypotheses. 2012;79:471–3. doi: 10.1016/j.mehy.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat StructMolBiol. 2008;15:836–41. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 9.Thakor N, Holcik M. IRES-mediated translation of cellular messenger RNA operates in eIF2α- independent manner during stress. Nucleic Acids Res. 2012;40:541–52. doi: 10.1093/nar/gkr701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–13. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539–43. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarnow P. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J Virol. 2003;77:2801–6. doi: 10.1128/JVI.77.5.2801-2806.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. ProcNatlAcadSci U S A. 2010;107:19754–9. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray PS, Das S. Inhibition of hepatitis C virus IRES-mediated translation by small RNAs analogous to stem-loop structures of the 5′-untranslated region. Nucleic Acids Res. 2004;32:1678–87. doi: 10.1093/nar/gkh328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das S, Ott M, Yamane A, Tsai W, Gromeier M, Lahser F, Gupta S, Dasgupta A. A small yeast RNA blocks hepatitis C virus internal ribosome entry site (HCV IRES)-mediated translation and inhibits replication of a chimeric poliovirus under translational control of the HCV IRES element. J Virol. 1998;72:5638–47. doi: 10.1128/jvi.72.7.5638-5647.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian N, Mani P, Roy S, Gnanasundram SV, Sarkar DP, Das S. Targeted delivery of hepatitis C virus-specific short hairpin RNA in mouse liver using Sendai virosomes. J Gen Virol. 2009;90:1812–9. doi: 10.1099/vir.0.010579-0. [DOI] [PubMed] [Google Scholar]
- 17.Welch PJ, Tritz R, Yei S, Leavitt M, Yu M, Barber J. A potential therapeutic application of hairpin ribozymes: in vitro and in vivo studies of gene therapy for hepatitis C virus infection. Gene Ther. 1996;3:994–1001. [PubMed] [Google Scholar]
- 18.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–66. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown EA, Zhang H, Ping LH, Lemon SM. Secondary structure of the 5′nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–68. [PMC free article] [PubMed] [Google Scholar]
- 21.Chandramouli P, Topf M, Ménétret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–48. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/S1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat StructBiol. 2000;7:1105–10. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 25.Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8:1045–55. doi: 10.1017/S1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odreman-Macchioli F, Baralle FE, Buratti E. Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J BiolChem. 2001;276:41648–55. doi: 10.1074/jbc.M104128200. [DOI] [PubMed] [Google Scholar]
- 27.Psaridi L, Georgopoulou U, Varaklioti A, Mavromara P. Mutational analysis of a conserved tetraloop in the 5′ untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. FEBS Lett. 1999;453:49–53. doi: 10.1016/S0014-5793(99)00662-6. [DOI] [PubMed] [Google Scholar]
- 28.Filbin ME, Vollmar BS, Shi D, Gonen T, Kieft JS. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat StructMolBiol. 2013;20:150–8. doi: 10.1038/nsmb.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukushi S, Okada M, Kageyama T, Hoshino FB, Katayama K. Specific interaction of a 25-kilodalton cellular protein, a 40S ribosomal subunit protein, with the internal ribosome entry site of hepatitis C virus genome. Virus Genes. 1999;19:153–61. doi: 10.1023/A:1008131325056. [DOI] [PubMed] [Google Scholar]
- 30.Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J BiolChem. 2001;276:20824–6. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 31.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–23. doi: 10.1017/S1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu H, Li W, Noble WS, Payan D, Anderson DC. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J Proteome Res. 2004;3:949–57. doi: 10.1021/pr0499592. [DOI] [PubMed] [Google Scholar]
- 33.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–6. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 34.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–9. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 35.Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–5. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 36.Malygin AA, Kossinova OA, Shatsky IN, Karpova GG. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013;41:8706–14. doi: 10.1093/nar/gkt632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kikuchi K, Umehara T, Fukuda K, Hwang J, Kuno A, Hasegawa T, Nishikawa S. Structure-inhibition analysis of RNA aptamers that bind to HCV IRES. Nucleic Acids Res Suppl. 2003;3:291–2. doi: 10.1093/nass/3.1.291. [DOI] [PubMed] [Google Scholar]
- 38.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraser CS, Hershey JW, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat StructMolBiol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit’s decoding groove. RNA. 2011;17:1258–73. doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérard J, Leyrat C, Baudin F, Drouet E, Jamin M. Structure of the full-length HCV IRES in solution. Nat Commun. 2013;4:1612. doi: 10.1038/ncomms2611. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J ComputChem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.PintilieGD, ZhangJ, GoddardTD, ChiuW, GossardDC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J StructBiol2010; 170:427-38. [DOI] [PMC free article] [PubMed]
- 44.Russell RB, Sasieni PD, Sternberg MJ. Supersites within superfolds.Binding site similarity in the absence of homology. J MolBiol. 1998;282:903–18. doi: 10.1006/jmbi.1998.2043. [DOI] [PubMed] [Google Scholar]
- 45.Panchenko AR, Kondrashov F, Bryant S. Prediction of functional sites by analysis of sequence and structure conservation. Protein Sci. 2004;13:884–92. doi: 10.1110/ps.03465504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen YC, Sargsyan K, Wright JD, Huang YS, Lim C. Identifying RNA-binding residues based on evolutionary conserved structural and energetic features. Nucleic Acids Res. 2014;42:e15. doi: 10.1093/nar/gkt1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Draper DE, Reynaldo LP. RNA binding strategies of ribosomal proteins. Nucleic Acids Res. 1999;27:381–8. doi: 10.1093/nar/27.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–90. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parca L, Gherardini PF, Truglio M, Mangone I, Ferrè F, Helmer-Citterich M, Ausiello G. Identification of nucleotide-binding sites in protein structures: a novel approach based on nucleotide modularity. PLoS One. 2012;7:e50240. doi: 10.1371/journal.pone.0050240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–1. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis BA, Walia RR, Terribilini M, Ferguson J, Zheng C, Honavar V, Dobbs D. PRIDB: a Protein-RNA interface database. Nucleic Acids Res. 2011;39:D277–82. doi: 10.1093/nar/gkq1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Q, Ren S, Lu M, Zhang Y, Zhu D, Zhang X, Li T. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genomics. 2013;14:651. doi: 10.1186/1471-2164-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, Honavar V, Dobbs D. RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res. 2007;35:W578-84. doi: 10.1093/nar/gkm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Huang C, Yang MQ, Yang JY. BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC SystBiol. 2010;4(Suppl 1):S3. doi: 10.1186/1752-0509-4-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am ChemSoc. 2003;125:1731–7. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 56.Karaca E, Melquiond AS, de Vries SJ, Kastritis PL, Bonvin AM. Building macromolecular assemblies by information-driven docking: introducing the HADDOCK multibody docking server. Mol Cell Proteomics. 2010;9:1784–94. doi: 10.1074/mcp.M000051-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Dijk M, Bonvin AM. A protein-DNA docking benchmark. Nucleic Acids Res. 2008;36:e88. doi: 10.1093/nar/gkn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat StructBiol. 2003;10:1033–8. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 59.Schüler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat StructMolBiol. 2006;13:1092–6. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 60.Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, et al. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 2011;39:5264–75. doi: 10.1093/nar/gkr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyltransferase center of the intact 70S ribosome. Nat StructMolBiol. 2009;16:528–33. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmeing TM, Voorhees RM, Kelley AC, Ramakrishnan V. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat StructMolBiol. 2011;18:432–6. doi: 10.1038/nsmb.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500:307–11. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. ProcNatlAcadSci U S A. 2010;107:17158–63. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Massire C, Westhof E. MANIP: an interactive tool for modelling RNA. J Mol Graph Model. 1998;16:197–205, 255-7. doi: 10.1016/S1093-3263(98)80004-1. [DOI] [PubMed] [Google Scholar]
- 66.Mueller F, Brimacombe R. A new model for the three-dimensional folding of Escherichia coli 16 S ribosomal RNA. I. Fitting the RNA to a 3D electron microscopic map at 20 A. J MolBiol. 1997;271:524–44. doi: 10.1006/jmbi.1997.1210. [DOI] [PubMed] [Google Scholar]
- 67.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–83. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–80. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dresios J, Chappell SA, Zhou W, Mauro VP. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat StructMolBiol. 2006;13:30–4. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- 70.Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviralintergenic internal ribosome entry site. Nucleic Acids Res. 2007;35:1514–21. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, et al. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 2011;39:5264–75. doi: 10.1093/nar/gkr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–64. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Aalten DM, DiRusso CC, Knudsen J. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 2001;20:2041–50. doi: 10.1093/emboj/20.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laletina ES, Graĭfer DM, Malygin AA, Shatskiĭ IN, Karpova GG. [Molecular environment of the subdomain IIIe loop of the RNA IRES element of hepatitis C virus on the human 40S ribosomal subunit] BioorgKhim. 2006;32:311–9. doi: 10.1134/s1068162006030101. [DOI] [PubMed] [Google Scholar]
- 75.Easton LE, Locker N, Lukavsky PJ. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009;37:5537–49. doi: 10.1093/nar/gkp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malygin AA, Shatsky IN, Karpova GG. Proteins of the human 40S ribosomal subunit involved in hepatitis C IRES binding as revealed from fluorescent labeling. Biochemistry (Mosc) 2013;78:53–9. doi: 10.1134/S0006297913010069. [DOI] [PubMed] [Google Scholar]
- 77.Wang C, Le SY, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–37. [PMC free article] [PubMed] [Google Scholar]
- 78.Bhat P, Gnanasundram SV, Mani P, Ray PS, Sarkar DP, Das S. Targeting ribosome assembly on the HCV RNA using a small RNA molecule. RNA Biol. 2012;9:1110–9. doi: 10.4161/rna.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Demeshkina N, Laletina E, Meschaninova M, Ven’yaminova A, Graifer D, Karpova G. Positioning of mRNA codons with respect to 18S rRNA at the P and E sites of human ribosome. BiochimBiophysActa. 2003;1627:39–46. doi: 10.1016/S0167-4781(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 80.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–21. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi Y, Hirayama S, Odani S. Ribosomal proteins cross-linked to the initiator AUG codon of a mRNA in the translation initiation complex by UV-irradiation. J Biochem. 2005;138:41–6. doi: 10.1093/jb/mvi096. [DOI] [PubMed] [Google Scholar]
- 82.Sharifulin D, Khairulina Y, Ivanov A, Meschaninova M, Ven’yaminova A, Graifer D, Karpova G. A central fragment of ribosomal protein S26 containing the eukaryote-specific motif YxxPKxYxK is a key component of the ribosomal binding site of mRNA region 5′ of the E site codon. Nucleic Acids Res. 2012;40:3056–65. doi: 10.1093/nar/gkr1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu Y, Ji H, Doudna JA, Leary JA. Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci. 2005;14:1438–46. doi: 10.1110/ps.041293005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laletina E, Graifer D, Malygin A, Ivanov A, Shatsky I, Karpova G. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006;34:2027–36. doi: 10.1093/nar/gkl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009;37:1141–51. doi: 10.1093/nar/gkn1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Romero-López C, Díaz-González R, Barroso-delJesus A, Berzal-Herranz A. Inhibition of hepatitis C virus replication and internal ribosome entry site-dependent translation by an RNA molecule. J Gen Virol. 2009;90:1659–69. doi: 10.1099/vir.0.008821-0. [DOI] [PubMed] [Google Scholar]
- 87.Kleywegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN - programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. ActaCrystallogr D BiolCrystallogr. 1996;52:826–8. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 88.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J StructBiol. 1999;125:185–95. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 89.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J MolBiol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 90.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–24. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sorin EJ, Pande VS. Empirical force-field assessment: The interplay between backbone torsions and noncovalent term scaling. J ComputChem. 2005;26:682–90. doi: 10.1002/jcc.20208. [DOI] [PubMed] [Google Scholar]
- 92.MackeT, CaseDA. Modeling unusual nucleic acid structures. In: Leontes NB, SantaLucia J, eds. In Molecular Modeling of Nucleic Acids ACS Symposium Series, 1998:379-93. [Google Scholar]
- 93.Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat StructBiol. 2002;9:370–4. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 94.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–5. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 95.Rother M, Rother K, Puton T, Bujnicki JM. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39:4007–22. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rahrig RR, Leontis NB, Zirbel CL. R3D Align: global pairwise alignment of RNA 3D structures using local superpositions. Bioinformatics. 2010;26:2689–97. doi: 10.1093/bioinformatics/btq506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38:W373-7. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.