Abstract
Hereditary motor and sensory neuropathy associated with agenesis of the corpus callosum (HMSN/ACC or ACCPN) is an autosomal recessive disease caused by the disruption of the SLC12A6 gene, which encodes the K-Cl cotransporter-3 (KCC3). A ubiquitous deletion of KCC3 in mice leads to severe locomotor deficits similar to ACCPN patients. However, the underlying pathological mechanism leading to the disease remains unclear. Even though a recent study suggests that the neuropathic features of ACCPN are mostly due to neuronal loss of KCC3, the specific cell type responsible for the disease is still unknown. Here we established 4 tissue specific KCC3 knockout mouse lines to explore the cell population origin of ACCPN. Our results showed that the loss of KCC3 in parvalbumin-positive neurons led to significant locomotor deficit, suggesting a crucial role of these neurons in the development of the locomotor deficit. Interestingly, mice in which KCC3 deletion was driven by the neuron-specific enolase (NSE) did not develop any phenotype. Furthermore, we demonstrated that nociceptive neurons targeted with Nav1.8-driven CRE and Schwann cells targeted with a desert hedgehog-driven CRE were not involved in the development of ACCPN. Together, these results establish that the parvalbumin-positive neuronal population is an important player in the pathogenic development of ACCPN.
Keywords: K-Cl cotransport, sensory neurons, neuropathy, parvalbumin, locomotion, behavior
INTRODUCTION
Peripheral neuropathy associated with agenesis of the corpus callosum (ACCPN) is an autosomal recessive disease characterized by progressive sensorimotor neuropathy, mental retardation, dysmorphic features (high arched palate, hypertelorism and syndactyly) and complete or partial agenesis of the corpus callosum [2,16]. Genetic linkage in fourteen families mapped the autosomal recessive disorder to chromosome 15q [7] and several mutations in SLC12A6, the human gene which encodes the K-Cl cotransporter-3 (KCC3), were later identified in HMSN/ACC patients [5,13,23,28]. In a mouse model where the KCC3 gene was disrupted to produce a global knockout, an early onset and severe locomotor deficit similar to the crippling human peripheral neuropathy disorder was observed [13]. The mice also developed high blood pressure [1,5], age-related deafness [5], and renal dysfunction [30]. At the ultra-structural level, KCC3-deficient mice exhibited axonal and peri-axonal swelling, suggesting both neuronal and Schwann cell defects [6].
In a recent study, Shekarabi et al used a synapsin 1 promoter-CRE mouse to drive deletion of KCC3 in neurons, and demonstrated that loss of neuronal KCC3 reproduced the neuropathy phenotype observed in the KCC3 knockout mouse [25]. However, since synapsin 1 promoter is present in all neurons, it remains to be determined which specific neuronal cell type play underlies the development of the peripheral neuropathy, and whether KCC3 in Schwann cells are also involved in the ontogeny of the disease.
In the present study, we created several novel mouse models to target deletion of KCC3 in specific cell types by using specific transgenic CRE lines. CRE recombinase is an enzyme derived from the P1 Bacteriophage which catalyzes the specific recombination between two 34 bp DNA recognition sites (loxP sites). The artificial loxP sites are introduced in the target gene, Slc12a6 in this case, and a mutant mouse is produced. The recombinase which is under a tissue-specific promoter is expressed from DNA (transgene) inserted in the genome of a separate mouse. The Sc12a6 gene with loxP sites flanking an exon of interest and the recombinase transgene are brough together by crossing the two lines of mice. Deletion of KCC3 in small sensory neurons, driven by the Nav1.8 Na+ channel promoter, resulted in mice that exhibited no motor coordination phenotype. In contrast, deletion of KCC3 in larger type-Ia proprioceptive afferent neurons driven by parvalbumin resulted in a significant loss of locomotion. In these mice, there was also trend towards a hyperactivity phenotype. Surprisingly, deletion of KCC3 driven by enolase-2, which is supposedly expressed in most mature neurons, failed to recapitulate this phenotype. The KCC3 deletion in the Schwann cells driven by desert hedgehog also failed to induce a locomotor phenotype. Histology analysis showed that parvalbumin-positive neurons and enolase2 positive neurons in dorsal root ganglion had different expression profiles, consistent with the distinct performances in locomotor tests. Finally, histological analysis revealed a significant pathology associated with cells that were immunoreactive to parvalbumin. Therefore, our study demonstrates that parvalbumin-positive neurons played a key role in producing the ACCPN-like locomotor phenotype.
METHODS
Generation of tissue-specific KCC3 knockout mice
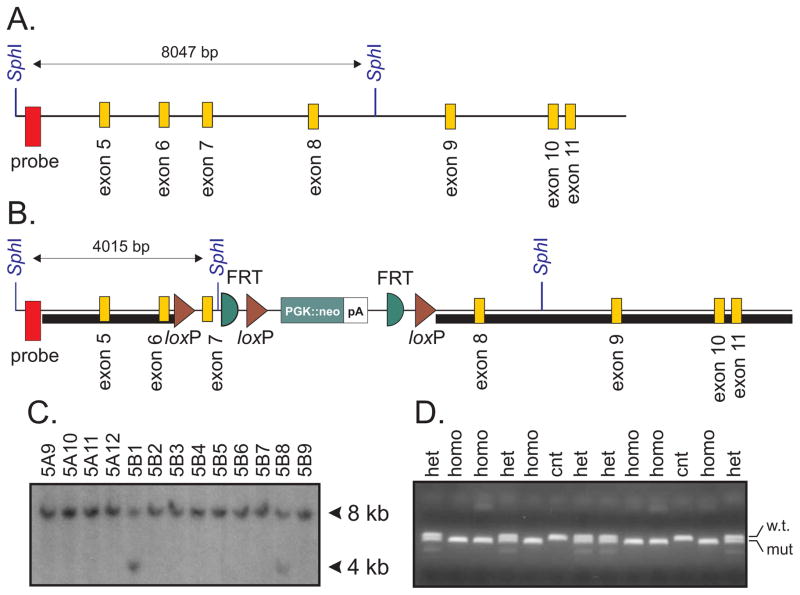
All procedures performed with mice were approved by the Vanderbilt University Institutional Animal Care and Use Committee. We disrupted the mouse Slc12a6 gene by inserting loxP sites around exon 7 (131 bp), followed by a neomycin resistance gene cassette flanked by FRT (Flippase Recombination Target) sites. The targeting vector was constructed using recombineering techniques to drop the short 5′ end and long 3′ end arms of recombination from Bacterial Artificial Chromosome (BAC) clone bMQ-302F12 (Geneservice Ltd, Cambridge, UK) into a vector containing 3 loxP sites, 2 FRT sites, and a PGK-driven neomycin resistance gene cassette. Briefly, a 2526 bp fragment (short arm) was dropped from the BAC clone between unique sites located upstream of the first loxP site. A small PCR fragment consisting of exon 7 surrounded by short intronic sequences was then ligated downstream of the first loxP site. In a final step, larger 7.5 kb fragment (large arm) was dropped from the BAC clone downstream of the last loxP site. The construct, verified by map digest and partial sequencing, was linearized using NotI. To target the gene for homologous recombination, TL-1 ES cells were electroporated with the linearized construct and grown on fibroblast feeder cells in Dulbecco’s Modified Eagle Medium (D-MEM) medium supplemented with 15% fetal bovine serum, 50 mg/ml gentamicin, 1000U/ml LIF, 90 mM β-mercaptoethanol, and 0.2 mg/ml G418. Three hundred and fifty independent neomycin-resistant colonies were picked and grown in 96-well plates on feeder layer, expanded, and analyzed for the presence of the mutant gene by performing Southern blot analysis using genomic DNA digested with SphI and hybridized with a 32P-labeled probe consisting of 347 bp upstream of the left arm of recombination. Eight positive clones were identified and one clone (5B1) was injected into C57BL/6J blastocysts. Two chimeric males (> 90% brown fur) were mated with C57BL/6J females and germline transmission was obtained. Since these mice carried one allele containing the neomycin-resistance gene cassette (3 loxP), they were mated with Flp recombinase (FLPeR) mice (obtained from Susan Dymicki, Harvard and backcrossed in our laboratory for > 10 generations in C57BL/6J background) to eliminate the neomycin-resistance gene cassette.
Heterozygous KCC3flox mice were first crossed to obtain viable homozygous mice. The mice were genotyped using oligonucleotide primers flanking the loxP site located upstream of exon 7: forward primer: 5′ TGTGACAGACACTTCCTACAAGCC 3′ and reverse primer: 5′ TCAGACTTTGGGAAATTGAACGTAAC 3′. The PCR amplification was done using an annealing temperature of 60°C and yielded a 294 bp fragment from the mutant allele and a smaller 254 bp fragment from the wild-type allele. The mice were then crossed with the different transgenic CRE mice to obtain in a first step mice carrying the CRE transgenes with one copy of the KCC3floxed allele and in a second step mice carrying or not the CRE transgenes with two copies of the KCC3floxed allele, i.e. tissue-specific KCC3 knockout mice. Behavioral tests were performed with age of mice ranging from 90 – 130 days.
Accelerated Rotarod
Neuromotor coordination task was performed using an accelerating rotating cylinder (model 47600: Ugo Basile, S.R. Biological Research Apparatus, Comerio, Italy). The cylinder was 3 cm in diameter and was covered with scored plastic. Mice were confined to a 4 cm long section of the cylinder by gray Plexiglas dividers. Three to five mice were placed on the cylinder at once. The rotation rate of the cylinder increased over a 5 min period from 4 to 40 rpm. The latency of each mouse to fall off the rotating cylinder was automatically recorded by the device. Mice that remained on the rotarod during the 300 s trial period were removed and given a score of 300 s. The test was performed for three trials a day for 3 consecutive days.
Open field
Exploratory and locomotor activities were tested using an open-field activity chamber surrounded by a Plexglas enclosure within sound-attenuating cubicles (ENV-022MD-027; Med Associates, Inc., St. Albans, VT). The mice were placed in the center of the open field and their activity was monitored for 60 min. The test was performed at ambient RT (~25°C), moderate light (60 lx), and background noise (80 dB). Distance traveled and time spend in the periphery (thigmotaxis) of the chamber were quantified by the number of beam crossings.
Hot-plate assay in Nav1.8-CRE x KCC3f/f mice
The hotplate assay was performed by placing the mice individually on a platform maintained at 52°C to 55°C (Hot-Plate Analgesia Meter; Columbus Instruments, Columbus, OH). A plastic cylinder measuring 15 cm (diameter) and 20 cm (height) confined the mouse to the surface of the hotplate. The time necessary for the mouse to respond to the thermal stimulus (hindpaw fluttering, licking, or withdrawal) was measured using a stopwatch. After the initial response or the maximal cutoff time of 15 s, the mouse was removed from the hotplate and returned to its home cage. A minimal recovery period of 1 day was implemented between assays at the two temperatures.
Dorsal root ganglia fixation and immunofluorescence staining
Dorsal root ganglia were dissected from the lower thoracic to mid-lumbar regions of the vertebral column and were placed in a 4% (w/v) solution of PFA (Sigma Aldrich) in 0.1 M PBS (4°C, pH 7.4) overnight. Following fixation, DRG neurons were transferred to a 30% (w/v) sucrose solution in 0.1 M PBS for 24 hours~48 hours before embedding into O.C.T. compound (Sakura Tissue-Tek, Thermo Fisher Scientific) blocks. The O.C.T. blocks were stored at −20°C before being transferred for cryostat sectioning. Sections were cut 10 μm thick in the cryostat microtome at −20°C and were immediately transferred to a RT microscope slide by gentle touching. The slides were stored at −20°C for further histology study. Routine H&E staining was performed for general histology. For immunostaining, the slides were defrosted at RT for 10 min., and then permeabilized with 0.1% Triton X-100 for 5 min. After 2 washes in PBS for 5 min. each, the slides were blocked for 30 min with 1% BSA + 1% goat serum in PBS. Primary antibodies (anti-PDI antibody (Abcam, Ab27043 1:200), anti-NSE antibody (Abcam, ab53025, 1:250), anti-parvalbumin antibody (Abcam, MAB1572, 1:500), anti-Cre antibody (Abcam, ab24607, 1:500)) were applied in 1%BSA with PBS at 4°C, overnight. On the second day, the slides were washed twice separately with high salt PBS and regular PBS, then blocked with 1% BSA + 4% goat serum in PBS for 30 min. Secondary antibodies (100 ul, Cy3 conjugated anti-rabbit antibody (1:1000, Jackson Immunochemicals) or Alexa Fluor anti-mouse IgG (H+L) (1:400, Invitrogen) were then applied and incubated for 60 min at RT. After final washes with high salt PBS and regular PBS, coverslips were mounted on slides using VectaShield (Vector Laboratories, Burlingame, CA) and sealed with nail polish. Fluorescence signal was visualized using a Carl Zeiss LMS 510 META confocal microscope.
Spinal cord fixation and immunohistochemistry
Mice were euthanized and lumbar vertebra were collected and submerged in 10% neutral buffered formalin for overnight fixation. Following overnight fixation, vertebrae were decalcified in 23% formic acid for 48 hours. The vertebrae were cut in cross sections, placed in tissue cassettes, processed routinely, embedded in paraffin, cut in 4μm sections, mounted on slides and stained with hematoxylin and eosin for identification of dorsal root ganglia. Once the sections containing DRGs were identified, unstained slides were cut for immunohistochemical labeling with anti-mouse parvalbumin (Cat# p3088, Sigma) at a 1:200 dilution on a Bond Max Autostainer (Leica), using EDTA antigen retrieval for 20 min, and Ultravision™ Quanto Mouse-on-mouse blocking kit (Thermo Fisher Scientific).
Western blot analysis
After euthanasia, brains from Eno2-CRE x KCC3f/f and KCC3f/f control mice were promptly removed and flash frozen in liquid Nitrogen. Tissue was then homogenized using a dounce/Teflon pestle in 0.32M sucrose buffer (0.32 M sucrose, 5 mM Tris-Cl pH 7.5, 2 mM EDTA, 2.5 mM β-mercaptoethanol and protease inhibitors), spun at 4,000 rpm for 10 min, followed by 9,000 rpm for 20 min, and 100,000 rpm for 1 h. Pellets (microsomal fractions) were resuspended in sucrose buffer and the protein concentration was measured using standard Bradford Assay. Equal amount of protein (200 ug) was loaded on a 9% SDS-polyacrylamide gel, separated, and electrotransferred onto 0.45 μm polyvinylidine fluoride membranes (ThermoFisher Scientific). The membrane was incubated for 2 h at RT in blocking solution (5% nonfat milk in TBST, i.e. 150 mM NaCl, 10 mM Tris, 0.5% Tween 20), and incubated with rabbit polyclonal anti-KCC3 antibody (1:1000) overnight at 4°C. The membrane was thoroughly washed in TBST, incubated with horseradish peroxidase-conjugated secondary antibody in blocking solution (1:5000) for 1 h at RT, and washed again for 2 h in TBST. Protein bands were visualized by chemiluminescence (ECL Plus, Amersham Biosciences, Piscataway, NJ).
Statistical analysis
For rotarod experiment, statistical analyses were performed using SAS software (version 9.3, SAS Institute, Inc., Cary, NC). To account for the effects of animal group, days, and trials on rotorod times, a three-way repeated measures ANOVA test was utilized. The error terms of the different animal groups was modeled as independent variables while both the error terms of days and trials were correlated variables. Numerous covariate matrices were used to model both days and trials with the matrix that best fit the data ultimately being used (autoregressive model of order 1). To analyze the trends of rotorod times based on days and trials, Bonferroni-corrected linear and quadratic contrasts were constructed for both of these variables. P < 0.05 was considered statistically significant. For other analyses, two-tailed paired t-tests were performed using GraphPad Prism (version 3.0, GraphPad Sotwares, Inc., La Jolla, CA). Data were noted to be significant at P < 0.05 and highly significant at P < 0.001.
RESULTS
To determine the cellular origin of HSMN/ACC, we created several conditional KCC3 knockout mouse lines by using CRE mediated recombination under the control of tissue-specific promoters. First, we created a mouse in which exon 7 of the Slc12a6 gene was flanked by loxP sites. A construct targeting exon 7 was engineered (Figure 1) and electroporated in mouse embryonic stem cells. After germline transmission and elimination of the neomycin-resistance gene cassette, a 3 kb fragment was PCR-amplified to demonstrate recombination in the proper locus. Sequencing of the fragment revealed correct 5′ sequence and presence of the exon and of loxP sites at the 3′ end, demonstrating recombination in the proper KCC3 locus.
Figure 1. Generation of the KCC3flox/flox mouse.
A. Schematic representation of the Slc12a6 (KCC3) gene around exon 5–11. Highlighted are two SphI restriction sites separated by 8 kb genomic sequence. Location of a 347 bp PCR fragment/32P-labelled probe (red box) is also indicated. B. Construct containing a 2950 bp genomic DNA sequence representing the 5′ arm of recombination (short black box), loxP sites flanking exon 7, followed by a PGK-driven neomycin resistance gene cassette flanked by FRT sites, and a 7500 bp 3′ arm of recombination (long black box). A SphI site was inserted downstream of exon 7, reducing the size of the SphI fragment recognized by the 5′ probe from 8 to 4 kb. C. Portion of the ES cell screening Southern blot (SphI digest) showing 13 clones with 8 kb genomic fragment (wild-type allele), including 2 clones with an additional 4 kb fragment (mutant allele). D. Genotyping of 2 litters of pups from heterozygous breeding showing PCR fragments from wild-type and mutant alleles.
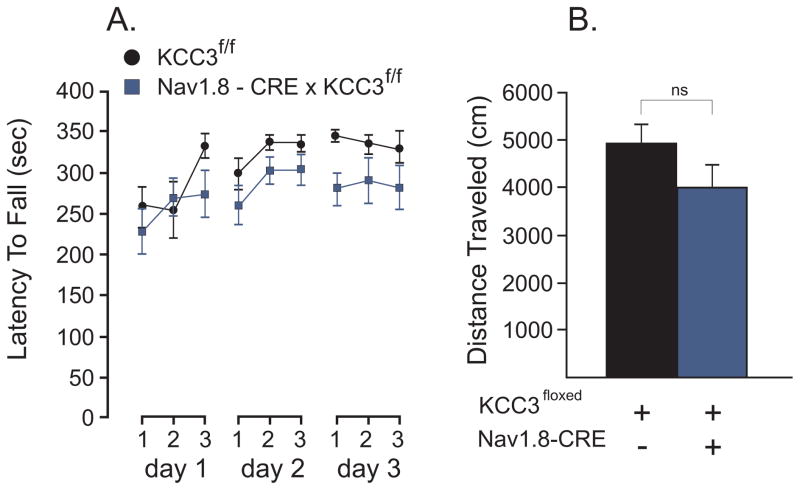
As previous reports have shown that KCC3 KO mice display severe motor and locomotor abnormalities [5,13], we subjected all our conditional KCC3 knockout mice to a standard protocol of accelerating rotarod: 3 trials a day for 3 days. The test assesses fore- and hindlimb balance coordination as well as a learning component for this motor task [14]. Locomotor activity was also assessed in an open-field chamber equipped with infrared beams. Each mouse was separately monitored for distance traveled during a specific time period. For our first tissue-specific knockout mouse line, we crossed KCC3flox/flox mice with animals that express the Cre recombinase under the Nav1.8 promoter [26]. Nav 1.8 is a voltage-gated sodium channel which is expressed in a subset of sensory neurons, 85% or more of which are nociceptors. Thus, we generated a mouse where KCC3 deletion is targeted in nociceptive neurons. As shown in Figure 2, we observed no difference between wild-type and Nav1.8-driven KCC3 knockout mice (F(1, 15) = 2.76; P = 0.1173, 8–9 mice per group). When we performed statistical analysis of trials or days, we observed a very significant difference: F(6, 90) = 4.4, P = 0.0006 for trials and F(2, 30) = 7.88, P = 0.0018 for days, indicating that the mice are learning the task or performing better with each trial and each day of trials. The relationship between trials was linear: F(2, 90) = 5.84, P = 0.002, instead of quadratic: F(1,90) = 0.57, P = 0.05778. There was also no difference in total distance travelled in the open field test, as the two tailed t-test demonstrated a P value of 0.3696 (t = 0.9270, df = 14, n = 8–9 mice per group). As Nav1.8 is expressed in nociceptive neurons, we also tested the mice for a nociception phenotype. There was no statistical differences between the two groups of mice in the response latency (withdrawal times) to heat-evoked nociceptive stimuli at 52°C (t = 0.8307, df = 15, P = 0.4192), or 55°C (t = 0.6465, df = 15, P = 0.5277).
Figure 2. Absence of locomotor phenotype in Nav1.8-driven KCC3 knockout mice.
A. Accelerated rotorod test (4 to 40 rpm, 5 min) performed in control (KCC3f/f) and tissue-specific knockout (Nav1.8-CRE x KCC3f/f) mice. Test was given three times a day for 3 consecutive days. The latency to fall from the rotating rod to the platform was measured in seconds. B. Locomotor activity as measured by the distance traveled (cm) in the open filed chamber over a period of 60 min. Statistical analyses showed no significant differences between groups (see text). N = 8–9 mice per group.
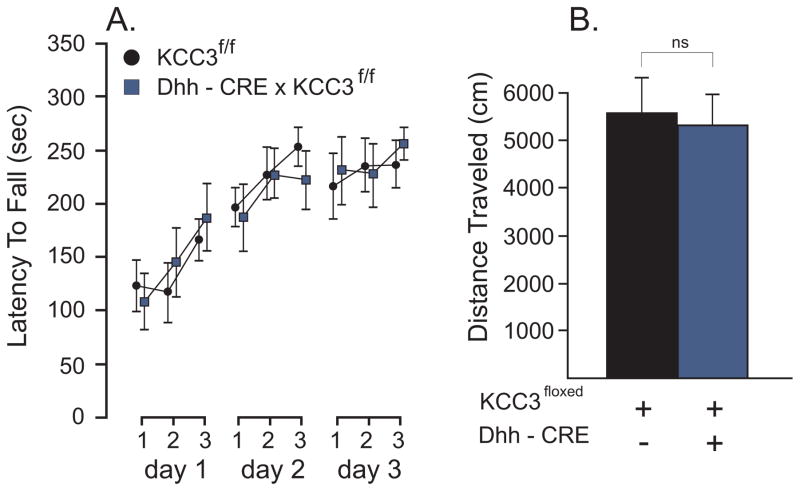
To explore the possibility that the locomotor deficit is caused by KCC3 deletion in Schwann cells, we utilized a transgenic mouse expressing the CRE recombinase under the desert hedgehog (Dhh) promoter, desert hedgehog being a protein expressed exclusively in Schwann cells [21] and Sertoli cells in testis [4]. Figure 3 presents the data of the accelerated rotorod and open field tests performed in control (KCC3f/f) and Schwann cell knockout (Dhh-CRE x KCC3f/f) sibling mice. No differences were observed between genotypes in the rotorod test (F(1, 14) = 0; P = 0.9741, 8 mice per group). Again, all mice learned the task as the differences between trials and days were highly significant: F(6, 84) = 4.82, P = 0.0003 and F(2, 28) = 18.99, P < 0.,0001), respectively. Similarly, we observed no phenotype in the open field test (t = 0.2828, df = 15, P = 0.7812).
Figure 3. Absence of locomotor phenotype in Dhh-driven KCC3 knockout mice.
A. Accelerated rotorod test (4 to 40 rpm, 5 min) performed in control (KCC3f/f) and tissue-specific knockout (Dhh-CRE x KCC3f/f) mice. Test was given three times a day for 3 consecutive days. The latency to fall from the rotating rod to the platform was measured in seconds. B. Locomotor activity as measured by the distance traveled (cm) in the open filed chamber over a period of 60 min. Statistical analyses showed no significant differences between groups (see text). N = 10 mice per group.
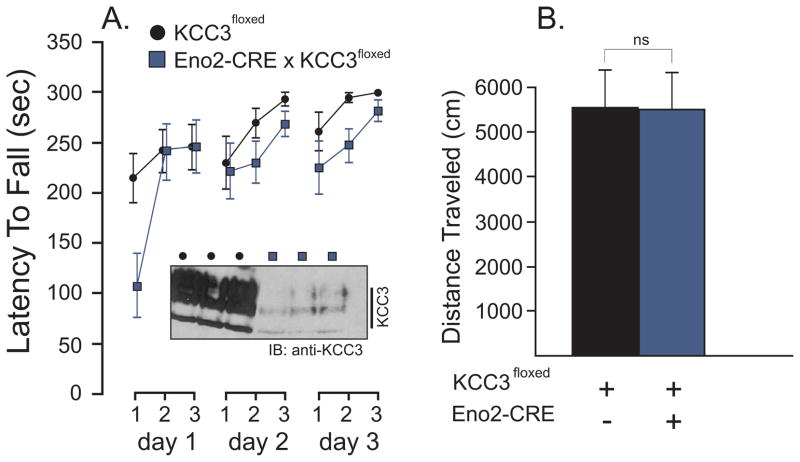
To confirm that neurons were involved in the development of the phenotype, we used enolase-2-CRE (Eno2-CRE) mice to drive deletion of KCC3 in neurons. Surprisingly, we observed no locomotor deficit in these mice (Figure 4). There was no difference between genotypes in the latency to fall the accelerating rod: F(1,10) = 3.37, P = 0.0962 nor in the ability to learn the task, as there was highly significant difference between trials (F(6, 60) = 8.44, P < 0.0001) or days (F(2, 20) = 8.38, P = 0.0023). There was also no difference in the distance traveled in the open-field test (two-tailed t test with t = 0.03740, df = 10, P = 0.9709). To determine the extent of KCC3 deletion in central neurons, we performed a Western blot analysis of KCC3 from brains obtained from control (KCC3f/f) and tissue-specific knockout (Eno2-CRE x KCC3f/f) mice. As seen in the inset of Figure 4, there was significant decrease in KCC3 expression in the brain of Eno2-CRE x KCC3f/f mice, indicating that this promoter efficiently drove deletion of the cotransporter in neuronal populations in the brain. Expression of the CRE recombinase in sensory DRG neurons was examined by immunofluorescence in a later Figure.
Figure 4. Absence of locomotor phenotype in Eno2-driven KCC3 knockout mice.
A. Accelerated rotorod test (4 to 40 rpm, 5 min) performed in control (KCC3f/f) and tissue-specific knockout (Eno2-CRE x KCC3f/f) mice. Test was given three times a day for 3 consecutive days. The latency to fall from the rotating rod to the platform was measured in seconds. Inset: Western blot analysis of KCC3 expression in brain of 3 controls and 3 Eno2-CRE driven knockout mice. B. Locomotor activity as measured by the distance traveled (cm) in the open filed chamber over a period of 60 min. Statistical analyses showed no significant differences between groups (see text). N = 6 mice per group.
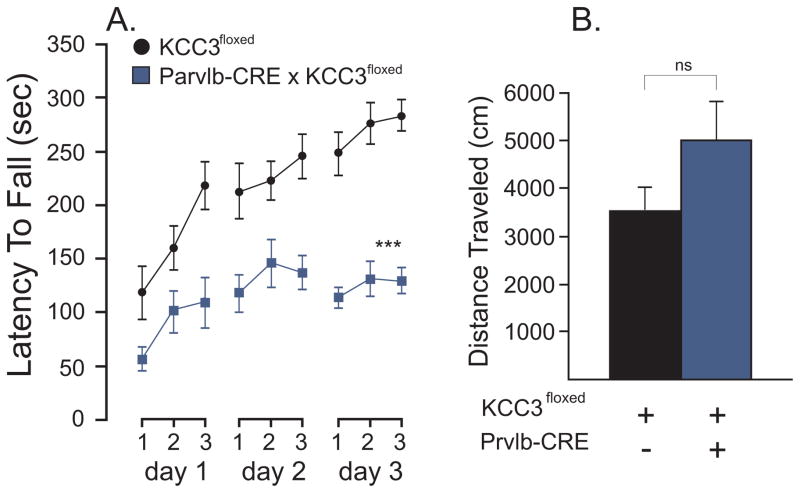
Next, we targeted deletion of KCC3 in proprioceptive neurons [3] by using a parvalbumin-CRE (Prvlb-CRE) transgenic mouse [12]. When locomotion and balance coordination was measured in Prvlb-CRE x KCC3f/f mice compared to their control littermate (Fig. 5), we observed a phenotype with knockout mice falling significantly faster: < 150 sec versus >250 sec: F(1,17) = 42, P < 0.0001. The knockout mice, however, were able to improve their performance as the difference between trials was highly significant (F(6,102) = 10.38, P < 0.0001), as was the difference between days (F(2,34) = 20.76, P < 0.001). The differences in the average rotorod times between the two animal groups were similar for each of the three days (F(2,34) = 3.04, P = 0.0592) and each of the three trials within each day (F(6,102) = 1.89, P = 0.0897), indicating absence of group day interaction or group trial interaction. When the locomotor activity was assessed through the open field, we observed a trend towards hyperactivity, although it did not reach statistical significance.
Figure 5. A locomotor phenotype is observed in parvalbumin-driven KCC3 knockout mice.
A. Accelerated rotorod test (4 to 40 rpm, 5 min) performed in control (KCC3f/f) and tissue-specific knockout (Eno2-CRE x KCC3f/f) mice. Test was given three times a day for 3 consecutive days. The latency to fall from the rotating rod to the platform was measured in seconds. B. Locomotor activity as measured by the distance traveled (cm) in the open filed chamber over a period of 60 min. Statistical analyses showed highly significant difference between groups in the rotorod test (see text), but no difference in the activity/open field test. N = 8–11 mice per group. *** P < 0.001, highly significant.
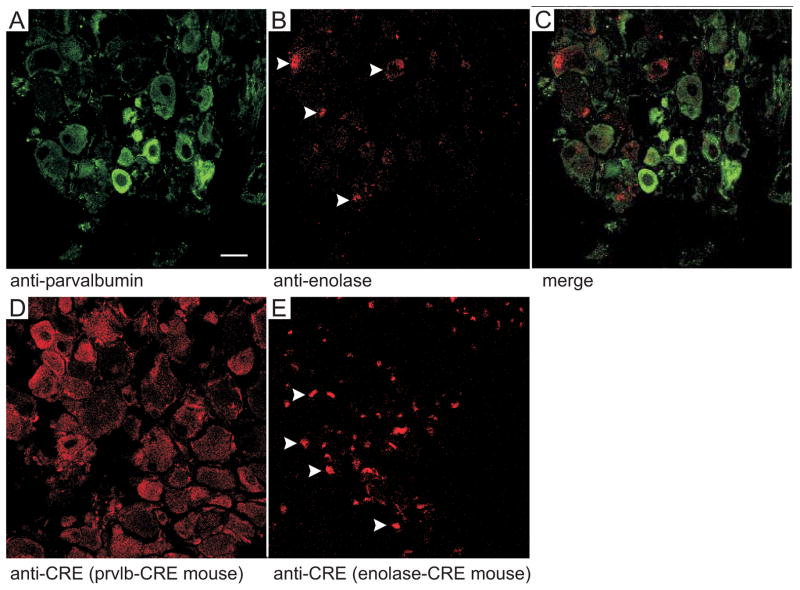
Using immunofluorescence, we examined the expression of parvalbumin and enolase-2 in isolated dorsal root ganglion from wild-type mice, and expression of CRE in DRGs isolated from Prvlb-CRE x KCC3f/f and Eno2-CRE x KCC3f/f mice. As described in the method section, the ganglia were promptly removed from the spinal cord, fixed, cryoprotected, embedded, and cryosectioned. As seen in Figure 6A, strong parvalbumin staining was observed in a subset of large cells, whereas other cells showed weak or no staining. In contrast, enolase staining was punctated in dorsal root ganglia (Fig. 6B), demonstrating a very different pattern of expression (merge, Fig. 6C). Consistent with this staining pattern in wild-type mice, CRE expression was observed in large neurons in the Prvlb-CRE mouse, while punctated staining was observed in the Eno2-CRE mouse.
Figure 6. Parvalbumin and enolase 2 were expressed in different structures in DRGs.
A–C. Longitudinal sections of DRG neurons from wild-type (n=3) mice were analyzed using immunofluorescence antibody against parvalbumin (A, green) and enolase 2 (B, red). Panel C represents the summation of both signals. D–E. Longitudinal sections of DRG neurons from parvalbumin-CRE x KCC3f/f mice and Eno2-CRE x KCC3f/f mice were immunostained with CRE antibody. CRE expression was observed in both mice and mirrored the signal seen with the two protein-specific antibodies.
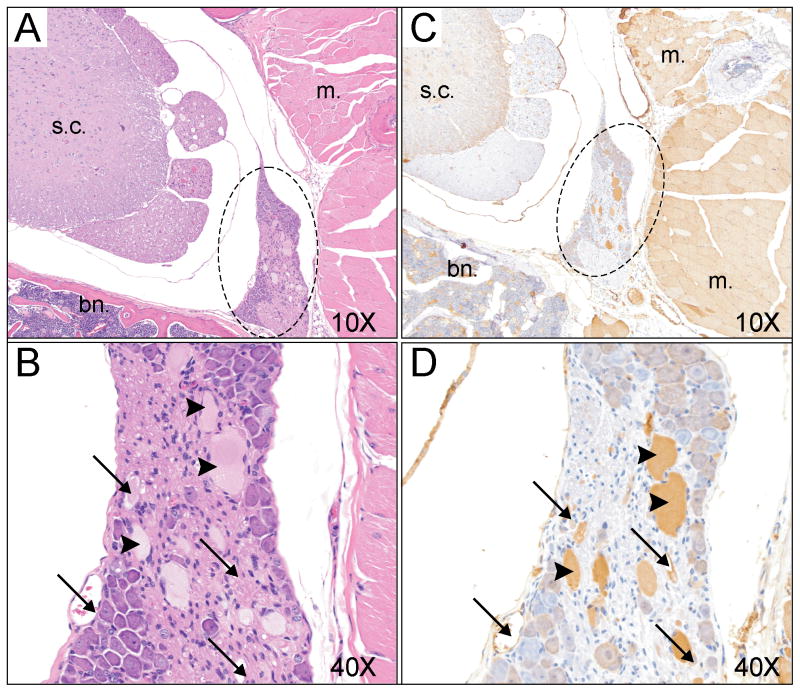
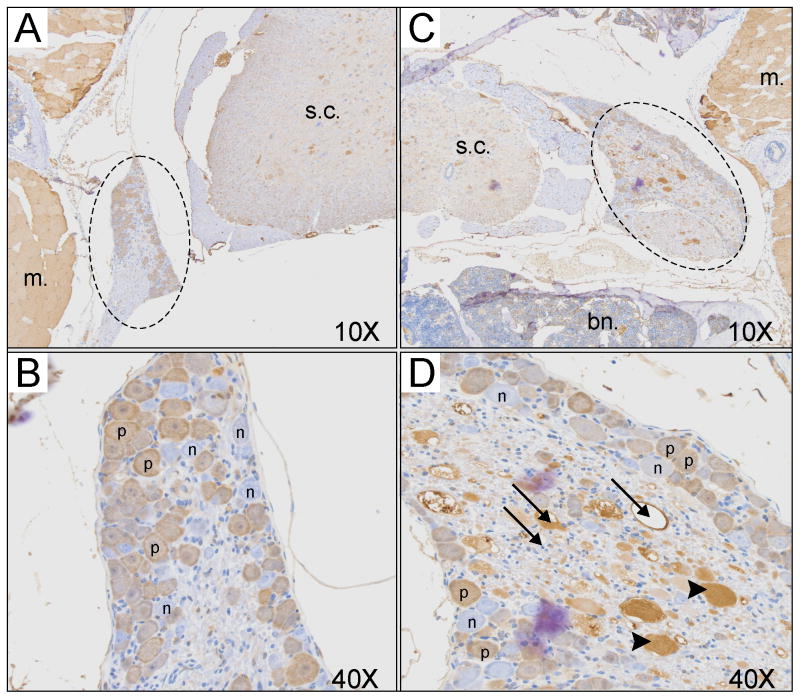
Further analyses of dorsal root ganglia were performed by immunocytochemistry. Entire spinal cord sections were H&E stained and when dorsal root ganglia were observed, the next contiguous section was stained with anti-parvalbumin antibody. Large vacuoles, reminiscent of what was observed in brain and spinal cord in the global KCC3 knockout [5], were observed in dorsal roots of Prvlb-CRE x KCC3 knockout mice (Figure 7, arrows). In addition, many structures containing dense material (arrow heads) were also observed in the DRG of these mice. The vacuoles and structures were immuno-reactive with the anti-parvalbumin antibody (Figure 7C, 7D). To better understand the histopathlogy of the dorsal root ganglia in these tissue-specific knockout mice, we isolated spinal cords from wild-type mice and KCC3 global knockout mice. The tissues were subjected to the exact same protocol, from animal perfusion to parvalbumin antibody detection. While no vacuoles or dense materials were observed in DRGs from wild-type mice (Figure 8A, B), pathology similar to that observed in parvalbumin-driven KCC3 knockout was also observed in the DRGs of global knowckout mice (Figure 8C, D).
Figure 7. Histological analysis of dorsal root ganglia in Prvlb-CRE x KCC3f/f mice.
A. Partial view of spinal cord with dorsal root with H&E stain at a magnification of 10x. B. Higher magnification (40x) of the dorsal root of Panel A. C. Contiguous section showing spinal cord stained with anti-parvalbumin antibody. D. Higher magnification (40x) of the dorsal root showing parvalbumin staining of structures surrounding vacuoles (arrows) as well as some dense material which is parvalbumin immunoreactive. The following structures are labeled: s.c. = spinal cord; m. = muscle; bn. = bone, dashed circle = dorsal root ganglion.
Figure 8. Histological analysis of dorsal root ganglia from wild-type and global KCC3 knockout mice.
A. Partial view of spinal cord with dorsal root and parvalbumin staining in wild-type mouse at a magnification of 10x. B. Hi gher magnification (40x) of the dorsal root of Panel A, showing absence of pathology. C. Partial view of spinal cord with dorsal root and parvalbumin staining in global KCC3 knockout mouse at a magnification of 10x. B. Higher magnification (40x) of the dorsal root of Panel C, showing pathology similar to that observed in Figure 7. The following structures are labeled: s.c. = spinal cord; m. = muscle; bn. = bone, dashed circle = dorsal root ganglion, p = positive neuron, n = negative neuron.
DISCUSSION
To address the cellular origin of the locomotor deficit associated with the disruption of KCC3 in mice and the development of the early onset peripheral neuropathy disorder observed in HSN/ACC patients [5,13], we created several tissue-specific knockout lines. Only when KCC3 was targeted in parvalbumin-positive neurons did we detect a locomotor deficit similar to the one observed in the global knockout animals. Parvalbumin is a member of the large family of EF-hand calcium-binding proteins, which comprises more than 200 members in human [22]. It is essential for Ca2+ homeostasis and the modulation of intracellular Ca2+ concentration in specific populations of neurons [8]. In the brain, parvalbumin is almost exclusively expressed in subpopulations of inhibitory fast-spiking GABAergic interneurons located in many different brain regions such as cortex, cerebellum, hippocampus and the reticular nucleus of the thalamus [8,19,20]. Deficits in parvalbumin-expressing interneurons have been linked to cognitive impairments in animal models of Alzheimer disease [29]. In the peripheral nervous system, parvalbumin is expressed in proprioceptive type Ia fibers (Figure 9). The locomotor deficit observed in the Prvlb-CRE x KCC3f/f mice is unlikely related to a disruption of parvalbumin in these mice. Indeed, the transgenic mouse was created by inserting the CRE recombinase in the parvalbumin gene locus using an internal ribosomal entry site (IRES) at the end of the exons encoding the calcium binding protein. Thus, both parvalbumin and CRE transcripts are expressed under the parvalbumin promoter. Parvalbumnin expression was verified by our immunocytochemistry studies of dorsal root ganglia which showed parvalbumin expression in the tissue-specific knockout mice (Figures 7). Furthermore, it was shown that disruption of parvalbumin in mice had no phenotype in the rotarod assay [9]. Therefore, the observed phenotype likely originates from disruption of KCC3 in proprioceptive DRG neurons. In fact, we demonstrated that the abnormal structures in the dorsal root ganglia of either tissue-specific or global KCC3 knockout, i.e. vacuoles and dense material, had parvalbumin immunoreactivity (Figures 7 and 8). Thus, the pathology clearly involves parvalbumin fibers. This observation makes the involvement of CNS interneurons (e.g. in cerebellum) in the locomotor phenotype less likely. On the other hand, decreased KCC3 expression in CNS interneurons might explain the hyperactivity observed in the neuronal-specific KCC3 knockout mouse [25], and the hyperactivity trend observed in our parvalbumin-driven KCC3 knockout mouse. Decreased activity of parvalbumin-positive interneurons would result in an increased excitatory network drive, leading to hyperactivity. Interestingly, network hyperexcitability and increased susceptibility to epileptic seizures were also demonstrated in parvalbumin null mice [24]. Although not tested in this study, global KCC3 knockout mice also demonstrate a reduced threshold to develop seizures [5].
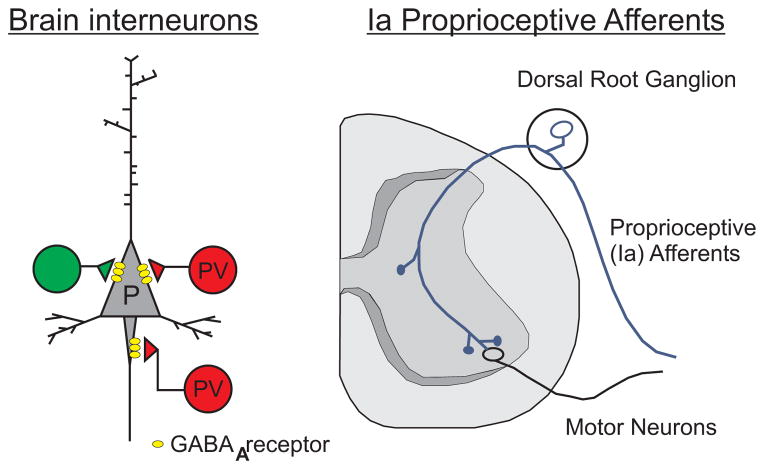
Figure 9. Schematic representation of parvalbumin expression in neurons.
In brain, parvalbumin is expressed in fast-spiking GABAergic interneurons (PV) which provide inhibitory input to pyramidal neurons (P). Another type of interneuron is depicted green. In the periphery, parvalbumin is expressed in type Ia large diameter proprioceptive afferents neurons which project to both middle spinal cord interneurons and to ventral motor neurons.
None of the other 3 tissue specific knockout mice showed the same locomotor deficit as the Parvlb-CRE x KCC3f/f mice. Because global KCC3 knockout mice demonstrated hypomyelination and periaxonal swelling [6], we postulated that Schwann cells might contribute to the phenotype, and more specifically KCC3 as a mechanism which drive salts and water out of the cell might be involved in the process of myelin compaction. We therefore targeted KCC3 in Schwann cells by crossing our floxed mouse with Desert hedgehog (Dhh)-driven CRE mice. These mice have been used to successfully delete Pten in Schwann cells [15]. Dhh is a member of the Hedgehogs family of intercellular signaling proteins that are best known for controlling tissue patterning during development [10]. Dhh is specifically expressed in Schwann cells of the nervous system and Sertoli cells in the testis in mammals. Mutations in human DHH have been identified resulting in gonade dysgenesis associated with polyneuropathy [27]. In addition, Dhh knockout (KO) mice have abnormal perineurial sheath formation, suggesting a key role of controlling the connective tissue sheaths formation around peripheral nerves [4,21]. Our results showed no locomotor phenotype in these mice, indicating no participation of Schwann cell KCC3 in the development of ACCPN. Note that this observation does not rule out the participation of Schwann cells themselves in ACCPN, as these cells clearly interact with both normal and degenerating neurons in the nerves of KCC3 knockout mice or ACCPN patients. The lack of phenotype also indicates that the Schwann cell deficit observed in the global KCC3 knockout [6] and in patients [16] is likely to be secondary to the neuronal degeneration.
To test whether the locomotor and balance deficit is related to an inability to sense mechanical pressure, we crossed the KCC3f/f line with Nav1.8-driven CRE mice [26]. Indeed, Nav1.8 is a voltage-gated sodium channel alpha subunit which is found within small-diameter sensory DRG neurons and is involved in temperature and mechanical pressure perception as well as nociception [17]. As no locomotor deficit was observed in these mice, we can conclude that the accelerated rotarod phenotype observed in the global KCC3 knockout mice is not related to a role of the cotransporter in these small sensory fibers. As we had the Nav1.8-CRE x KCC3f/f mouse line, we also tested the mice for a heat-evoked nociception phenotype at two noxious temperatures (52°C and 55°C). There was no difference observed between genotypes in the latency to respond to the thermal stimuli. Our data therefore indicate that KCC3 is unlikely to play a key role in the physiology of these neurons.
In our studies, we included a CRE line, Eno2-CRE, that we thought would produce knockout of KCC3 in all neurons. This line was our version of the synapsin 1-CRE line used by Shekarabi and coworkers [25]. Neuron specific enolase (Eno2, γ-enolase) is a glycolytic enzyme that catalyzes the conversion of phosphoglycerate to phosphoenol pyruvate in neurons [18]. Enolase 2 expression started shortly after synaptogenesis and is shown to be expressed in most mature neurons and neuroendocrine cells, thus making it a useful mature neuron specific marker [24, 25]. We were surprised by the absence of a locomotor phenotype in Eno2-CRE x KCC3f/f mice (Figure 4). To generate the transgenic Eno2-CRE mice, a 1.8-kb DNA promoter element which is located upstream of the rat NSE gene was inserted upstream of the CRE recombinase [11]. As demonstrated by our Western blot analysis of KCC3 expression, recombination was extremely efficient in the brain of Eno2-CRE positive mice. These data suggest that expression of the recombinase is optimal in central but not peripheral neurons. This could be due to either poor enolase-2 expression or weak promoter activity of the transgene in sensory neurons. Our immunofluorescence data (Figure 6) showed punctated CRE expression signal in DRG, different from the large cell bodies staining we observed in Parvlb-CRE mice.
In conclusion, our study establishes a key role for parvalbumin-expressing neurons in the development of the locomotor phenotype associated with HSN/ACC. The precise function of KCC3 in these cells is still unknown. Our study also indicates that the lack of function of KCC3 in Schwann cells nor in neurons sensing mechanical signals does not contribute to the locomotor phenotype observed in the HSN/ACC disorder.
Acknowledgments
We thank Thomas M. Austin for the statistical analysis of the rotarod data. This work was supported by NIH grants NS036758 and GM074771 to ED. The Translational Pathology Shared Resource supported by NCI/NIH Cancer Center Support Grant 2P30 CA068485-14 and the Vanderbilt Mouse Metabolic Phenotyping Center Grant 5U24DK059637-13.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adragna NC, Chen Y, Delpire E, Lauf PK, Morris M. Hypertension in K-Cl cotransporter-3 knockout mice. Adv Exp Med Biol. 2004;559:379–385. doi: 10.1007/0-387-23752-6_35. [DOI] [PubMed] [Google Scholar]
- 2.Andermann F, Andermann E, Joubert M, Karpati G, Carpenter S, Melancon D. Familial agenesis of the corpus callosum with anterior horn cell disease: a syndrome of mental retardation, areflexia and paraparesis. Trans Amer Neurol Ass. 1972;97:242–244. [Google Scholar]
- 3.Arber S, Ladle DR, Lin JH, Jessell FE. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000:101. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- 4.Bitgood MJ, Shen L, McMahon AP. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr Biol. 1996;6:298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 5.Boettger T, Rust MB, Maier H, Seidenbecher T, Schweizer M, Keating DJ, et al. Loss of K-Cl co-transporter KCC3 causes deafness, neurodegeneration and reduced seizure threshold. EMBO J. 2003;22:5422–5434. doi: 10.1093/emboj/cdg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byun N, Delpire E. Axonal and periaxonal swelling precede peripheral neurodegeneration in KCC3 knockout mice. Neurobiol Dis. 2007;28:39–51. doi: 10.1016/j.nbd.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casaubon LK, Melanson M, Lopes-Cendes I, Marineau C, Andermann E, Andermann F, et al. The gene responsible for a severe form of peripheral neuropathy and agenesis of the corpus callosum maps to chromosome 15q. Am J Hum Genet. 1996;58:28–34. [PMC free article] [PubMed] [Google Scholar]
- 8.Celio MR. Calbindin D-28k and parvalbumin in the rat nervous system. Neuroscience. 1990;35:375–475. doi: 10.1016/0306-4522(90)90091-h. [DOI] [PubMed] [Google Scholar]
- 9.Farré-Castany MA, Schwaller B, Gregory P, Barski J, Mariethoz C, Eriksson JL, et al. Differences in locomotor behavior revealed in mice deficient for the calcium-binding proteins parvalbumin, calbindin D-28k or both. Behav Brain Res. 2007;178:250–261. doi: 10.1016/j.bbr.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Ferent J, Traiffort E. Hedgehog: Multiple Paths for Multiple Roles in Shaping the Brain and Spinal Cord Neuroscientist. 2014. [DOI] [PubMed] [Google Scholar]
- 11.Frugier T, Tiziano FD, Cifuentes-Diaz C, Miniou P, Roblot N, Dierich A, et al. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2000;9:849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- 12.Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard HC, Mount DB, Rochefort D, Byun N, Dupré N, Lu J, et al. Mutations in the K-Cl cotransporter KCC3 cause a severe peripheral neuropathy associated with agenesis of the corpus callosum. Nat Genet. 2002;32:384–392. doi: 10.1038/ng1002. [DOI] [PubMed] [Google Scholar]
- 14.Karl T, Pabst R, von Horsten S. Behavioral phenotyping of mice in pharmacological and toxicological research. Exp Toxicol Pathol. 2003;55:69–83. doi: 10.1078/0940-2993-00301. [DOI] [PubMed] [Google Scholar]
- 15.Keng VW, Watson AL, Rahrmann EP, Li H, Tschida BR, Moriarity BS, et al. Conditional Inactivation of Pten with EGFR Overexpression in Schwann. Cells Models Sporadic MPNST Sarcoma. 2012;2012:620834. doi: 10.1155/2012/620834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labrisseau A, Vanasse M, Brochu P, Jasmin G. The andermann syndrome: agenesis of the corpus callosum associated with mental retardation and progressive sesorimotor neuronopathy. Can J Neurol Sci. 1984;11:257–261. doi: 10.1017/s0317167100045509. [DOI] [PubMed] [Google Scholar]
- 17.Liu M, Wood JN. The roles of sodium channels in nociception: implications for mechanisms of neuropathic pain. Pain Med. 2011;12:S93–S99. doi: 10.1111/j.1526-4637.2011.01158.x. [DOI] [PubMed] [Google Scholar]
- 18.Marangos PJ, Schmechel DE. Neuron specific enolase, a clinically useful marker for neurons and neuroendocrine cells. Annu Rev Neurosci. 1987;10:269–295. doi: 10.1146/annurev.ne.10.030187.001413. [DOI] [PubMed] [Google Scholar]
- 19.Münkle MC, Waldvogel HJ, Faull RL. The distribution of calbindin, calretinin and parvalbumin immunoreactivity in the human thalamus. J Chem Neuroanat. 2000;19:155–173. doi: 10.1016/s0891-0618(00)00060-0. [DOI] [PubMed] [Google Scholar]
- 20.Ohshima T, Endo T, Onaya T. Distribution of parvalbumin immunoreactivity in the human brain. J Neurol. 1991;238:320–322. doi: 10.1007/BF00315329. [DOI] [PubMed] [Google Scholar]
- 21.Parmantier E, Lynn B, Lawson D, Turmaine M, Namini SS, Chakrabarti L, et al. Schwann cell-derived Desert hedgehog controls the development of peripheral nerve sheaths. Neuron. 1999;23:713–724. doi: 10.1016/s0896-6273(01)80030-1. [DOI] [PubMed] [Google Scholar]
- 22.Popelár J, Rybalko N, Burianová J, Schwaller B, Syka J. The effect of parvalbumin deficiency on the acoustic startle response and prepulse inhibition in mice. Neurosci Lett. 2013;553:216–220. doi: 10.1016/j.neulet.2013.08.042. [DOI] [PubMed] [Google Scholar]
- 23.Rudnik-Schöneborn S, Hehr U, von Kalle T, Bornemann A, Winkler J, Zerres K. Andermann syndrome can be a phenocopy of hereditary motor and sensory neuropathy--report of a discordant sibship with a compound heterozygous mutation of the KCC3 gene. Neuropediatrics. 2009;40:129–133. doi: 10.1055/s-0029-1234084. [DOI] [PubMed] [Google Scholar]
- 24.Schwaller B, Tetko IV, Tandon P, Silveira DC, Vreugdenhil M, Henzi T, et al. Parvalbumin deficiency affects network properties resulting in increased susceptibility to epileptic seizures. Mol Cell Neurosci. 2004;25:650–653. doi: 10.1016/j.mcn.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Shekarabi M, Moldrich RX, Rasheed S, Salin-Cantegrel A, Laganière J, Rochefort D, et al. Loss of Neuronal Potassium/Chloride Cotransporter 3 (KCC3) Is Responsible for the Degenerative Phenotype in a Conditional Mouse Model of Hereditary Motor and Sensory Neuropathy Associated with Agenesis of the Corpus Callosum. J Neurosci. 2012;32:3865–3876. doi: 10.1523/JNEUROSCI.3679-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stirling LC, Forlani G, Baker MD, Wood JN, Matthews EA, Dickenson AH, et al. Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. Pain. 2005;113:27–36. doi: 10.1016/j.pain.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Umehara F, Tate G, Itoh K, Yamaguchi N, Douchi T, Mitsuya T, et al. A novel mutation of desert hedgehog in a patient with 46, XY partial gonadal dysgenesis accompanied by minifascicular neuropathy. Am J Hum Genet. 2000;67:1302–1305. doi: 10.1016/s0002-9297(07)62958-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uyanik G, Elcioglu N, Penzien J, Gross C, Yilmaz Y, Olmez A, et al. Novel truncating and missense mutations of the KCC3 gene associated with Andermann syndrome. Neurology. 2006;66:1044–1048. doi: 10.1212/01.wnl.0000204181.31175.8b. [DOI] [PubMed] [Google Scholar]
- 29.Verret L, Mann EO, Hang GB, Barth AM, Cobos I, Ho K, et al. Inhibitory interneuron deficit links altered network activity and cognitive dysfunction in Alzheimer model. Cell. 2012;149:708–721. doi: 10.1016/j.cell.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T, Delpire E, Giebisch G, Hebert SC, Mount DB. Impaired fluid and bicarbonate absorption in proximal tubules (PT) of KCC3 knockout mice. FASEB J. 2003;17:A464. [Google Scholar]