Abstract
Exosomes, membranous nanovesicles, naturally carry bio-macromolecules and play pivotal roles in both physiological intercellular crosstalk and disease pathogenesis. Here, we showed that B cell-derived exosomes can function as vehicles to deliver exogenous miRNA-155 mimic or inhibitor into hepatocytes or macrophages, respectively. Stimulation of B cells significantly increased exosome production. Unlike in parental cells, baseline level of miRNA-155 was very low in exosomes derived from stimulated B cells. Exosomes loaded with a miRNA-155 mimic significantly increased miRNA-155 levels in primary mouse hepatocytes and the liver of miRNA-155 knockout mice. Treatment of RAW macrophages with miRNA-155 inhibitor loaded exosomes resulted in statistically significant reduction in LPS-induced TNFα production and partially prevented LPS-induced decrease in SOCS1 mRNA levels. Furthermore, exosome-mediated miRNA-155 inhibitor delivery resulted in functionally more efficient inhibition and less cellular toxicity compared to conventional transfection methods. Similar approaches could be useful in modification of target biomolecules in vitro and in vivo.
Keywords: RNAs delivery, Exosomes, Macrophages, miRNA-155, TNFα
BACKGROUND
Extracellular vesicles (EVs) are heterogeneous membranous vesicles (40–5000nm diameter) that originate from different cells and are released into biofluids and the intercellular microenvironment. The smallest subpopulation (40–150nm), termed exosomes, derives from inward budding of the inner endosomal membrane followed by fusion of multivesicular bodies (MVBs) with the plasma membrane.1–3 Exosomes are enriched in proteins of the tetraspanin family such as CD63, CD9 and CD81,3 and play pivotal roles in both physiological crosstalk between cells and disease pathogenesis. Exosomes are subject of intensive research and exert their biological or pathophysiological functions by delivering diverse bio-macromolecules including different types of RNAs (mRNAs and noncoding regulatory RNAs), cytoplasmic proteins, membranous proteins, and lipids.4–6 Exosomes are involved in various pathologic conditions including tumorigenesis, establishing pre-metastatic tumor niche, stimulating pathologic angiogenesis, promoting tumor immune escape by modulating T cell activities, and facilitating spread of HIV.7–11
Exosomes as natural nano-sized membranous vesicles are one of the new frontiers of nanomedical research. Given their nanoscale size, potential ability to express targeting ligands and deliver bio-macromolecules, and their excellent biocompatibility, exosomes are emerging as attractive biological nanovesicle platforms for loading and carrying bio-macromolecules for nanomedicine applications.12–13 Exosomes are naturally adapted for transport and intercellular delivery of proteins and nucleic acids and can also deliver pharmaceutical proteins and nucleic acids such as mRNA, miRNA and siRNA.14–16 Exosomes express several characteristics of suitable nano-vehicles as they are small, relatively homogenous, and stable.12–14 Moreover, exosomes provide benefit of mediating gene delivery without inducing adverse immune reactions and pro-inflammatory responses,14 which is in contrast with many frequently used gene therapy methods including viral vectors, liposomes, and lipid nanoparticles that activate the host immune system, induce toxicity, and trigger inflammatory responses.14,17–19
Monocytes, macrophages, and dendritic cells represent pivotal cell types in innate immunity and participate in disease pathology. The inflammatory process is a key factor in progression and pathogenesis of different diseases including liver disease,20 rheumatoid arthritis,21 atherosclerosis,22 cancer,23 and in infections including tuberculosis,24 human immunodeficiency virus (HIV), and leishmaniasis.25,26 Inflammation produces cytokines such as tumor necrosis factor (TNFα), which are the key drivers of both disease progression and pathogenesis. Due to the mechanistic role of monocytes and macrophages in inflammation, targeted drug delivery to these cells to modulate their pro-inflammatory activation has been an active line of research in recent years. However, these cells revealed to be difficult targets,27, 28 particularly where intracellular delivery of an active macromolecule was necessary for gene therapy.27, 29 Thus, introducing new delivery systems for targeting macrophages is of great interest and could potentially introduce new treatment paradigms for a range of diseases.
Consistent with the role of macrophages in inflammation, our group and others previously showed that miRNA-155 exerts a positive regulation on the release of TNFα through enhancing its translation upon lipopolysaccharide (LPS) stimulation.20 Given the potential role of miRNA-155 in LPS-induced TNFα production and the importance of macrophage inflammatory activation in different diseases including alcoholic liver disease, multiple sclerosis, inflammatory bowel disease, we hypothesized that it might be useful to harness exosomes as vehicles to deliver a miRNA-155 inhibitor.
In this study, we evaluated whether exosomes (murine B cell (M12.4) derived) could deliver exogenous miRNA-155 inhibitor or miRNA-155 mimic. Here, we optimized loading and isolation conditions for B cell-derived exosomes to deliver miRNA-155 mimic and miRNA-155 inhibitor to primary mouse hepatocytes and RAW 264.7 macrophages, respectively. Our results suggest that exosomes derived from B cells can be harnessed in gene therapy to introduce exogenous miRNA-155 inhibitor to RAW 264.7 cells and functionally decrease TNFα production. In vivo, miRNA-155 loaded exosomes successfully delivered exogenous miRNA-155 mimic to the liver and isolated hepatocytes in miRNA-155 knockout mice.
METHODS
Cell culture and exosome isolation
Murine B cells (M12.4) were cultured in RPMI medium plus 10% exosome-depleted FBS (Exo-FBS™) (Mountain View, CA, USA), and 1% penicillin/streptomycin (Gibco®, NY, USA). After 12 hours, the cells were exposed to CD40 (5 μg/ml) (PeproTech. Rocky Hill, New Jersey, USA) and IL-4 (50 ng/ml) (PeproTech. Rocky Hill, New Jersey, USA). Three days later, the culture media was harvested and exosomes were isolated. RAW 264.7 macrophages were cultured in Dulbecco’s modified medium (Invitrogen) containing 10% FBS at 37 °C in a 5% CO2 atmosphere and used for exosome production and co-culture experiments.
For exosome isolation from different sources (non stimulated B cells, stimulated B cells and RAW macrophages), supernatants were centrifuged at 1500g for 5 minutes to remove cells and 10000 × g for 20 minutes to deplete residual cellular debris. Afterward, samples were serially filtered through 0.8μm, 0.44μm and 0.2μm. The filtered supernatant was used to precipitate exosomes with Exoquick-TC™ (according to the manufacturer’s guidelines) or immunomagnetic isolation for exosomal marker, CD63. For CD63 isolation, supernatants were condensed using Amicon® Ultra-15 Centrifugal Filter Devices and exosomes were positively selected using anti-CD63 immunomagnetic capturing with a primary anti-CD63 (BioLegend, Tokyo, Japan) followed by corresponding secondary antibody coupled to magnetic beads (Miltenyi Biotec). The MidiMACS™ Separator was used with LD columns. After isolation, exosomes were re-suspended in PBS. Immunomagnetic beads selection method was used for characterization and optimization of the procedure. For cell culture studies, Exoquick-TC™ isolation method was used.
Characterization of exosomes
The size, concentration, morphology, and surface marker (CD63) of isolated exosomes were identified by Nanoparticle Tracking Analysis (NTA), transmission electron microscopy (TEM) and western blot as described in supplementary method section.
RNA Isolation, mRNA and miRNA analysis
Cells were lysed in QIAzol Lysis reagent (Qiagen, Maryland, USA) and total RNA was extracted using Direct-zol™ RNA MiniPrep isolation kit (Zymo Research Corp, Irvine, CA). SOCS1 and 18S mRNA levels were analyzed using real-time quantitative PCR (qPCR). We used TaqMan miRNA Assays (Applied Biosystems, Foster City, CA) for detection of miRNA-155 expression according to manufacturer’s protocol, as described previously.20 Detailed protocols are described in supplementary method section.
Optimizing loading conditions of exosomes with miRNA-155 mimic
To standardize loading conditions of exosomes to achieve successful outcome and reproducible results, we optimized loading conditions for B cell derived exosomes.15 Re-suspended exosomes were diluted in Gene Pulser®electroporation buffer (Bio-Rad Laboratories, Berkeley, CA) in 1:1 ratio. miRNA-155 mimic or negative control 1 for miRNA mimic (Ambion, Grand Island, NY) at final amount of 150 pmol were added to 0.25μg/μl, 0.5μg/μl, 1μg/μl, and 1.5μg/μl of exosome sample. The mixtures were transferred into cold 0.2 cm electroporation cuvettes and electroporated at various voltages (0.130kV to 0.200kV) at 100 μF. After optimization of the voltage, effect of different capacitance was assessed. A Gene pulser II System (Bio-Rad Laboratories, Berkeley, CA) was used for electroporation. The exosomes were treated with one unit of RNase H to eliminate free floating miRNA-155 mimic outside the exosomes and re-isolated using Exoquick-TC™. The relative amount of encapsulated miRNA-155 was determined using TaqMan miRNA Assays as described previously.
Efficiency of different isolation methods in recovery of loaded exosomes
To identify the most efficient isolation method to re-isolate exosomes after electroporation procedure, we compared CD63 immunomagnetic isolation, ultracentrifugation, and Exoquick-TC™. First, exosome were isolated using CD63 immunomagnetic beads as described previously. Then, miRNA-155 mimic (150 pmol) was electroporated into 0.5μg/μl of exosomes. The RNase H treatment was done for 1 hour to eliminate free floating unloaded miRNA-155 mimic and exosomes re-isolated with different isolation methods including immunomagnetic isolation, Exoquick-TC™, and ultracentrifugation. Immunomagnetic isolation and Exoquick-TC™ methods are performed as described previously. Ultracentrifugation was done at 100,000 × g for 90 minutes using fixed angle 75 Ti rotor (Beckman Coulter, Miami, FL, USA), at 4°C. The experiments were done in triplicate and the amount of recovered miRNA-155 was quantified by qPCR.
Assessing efficiency of miRNA-155 mimic encapsulation into the exosomes using an exogenous control
In order to evaluate the fraction of loaded exosomes with miRNA-155 mimic, B cell derived exosomes were isolated and loaded using the optimal conditions (like voltage and concentrations), as determined in the previous section. After loading, exosomes were re-pelleted using Exoquick-TC™ reagent. To eliminate the presence of miRNA-155 mimic aggregates outside of the exosomes, the exosome pellet was treated with one unit of RNase H. Afterward, the total RNA was extracted and 2μl of exogenous cel-miRNA-39 was spiked to the samples as an exogenous control to calculate loading efficiency. The fraction of miRNA-155 mimic that was loaded into the exosomes was calculated using the following formula:
Where f (miRNA-LE (loading efficiency)) is fraction of miRNA which was loaded into the exosomes; Vc is volume of added exogenous control; Cc is concentration of exogenous control; MWc is molecular weight of exogenous control; Vs is volume of electroporated miRNA sample; Cs is concentration of sample; MWs is molecular weight of sample (loaded miRNA-155 mimic); REs is relative expression of sample (miRNA-155 mimic) and REc is relative expression of control (cel- miRNA-39). Using this formula, our “optimal” loading conditions resulted in efficient loading of 55.06% of miRNA-155 mimic into the exosomes.
Enzyme-linked Immunosorbent Assay (ELISA) and Lactate Dehydrogenase (LDH) cytotoxicity assay
The amount of TNFα in cell-free supernatants of RAW 264.7 cells was measured by Mouse TNFα ELISA kit (BD Biosciences, San Diego, CA, USA). The LDH cytotoxicity assay was done based on the manufacturer’s recommendation. Detailed procedures are described in supplementary method section.
Co-culture experiments and transfections
The exosomes were loaded with miRNA-155 mimic or negative control 1 for miRNA mimic (Ambion, Grand Island, NY) and co-cultured with primary mouse hepatocytes for 6 hours followed by washing and media replacement. The amount of miRNA-155 was assessed after 24 hours. snoRNA202 was used as internal control for qPCR analysis.
For miRNA-155 inhibition experiments, RAW macrophages were seeded 1 day before treatment and different treatment conditions and controls including miRNA-155 inhibitor (Ambion, Grand Island, NY) loaded exosomes, control inhibitor (Ambion, Grand Island, NY) loaded exosomes, miRNA-155 inhibitor with FuGENE® HD (Promega, Madison, WI), and miRNA-155 inhibitor with HiPerFect (Qiagen, Hilden, Germany), were applied for 24 hours. The same starting amount of miRNA-155 inhibitor (300 pmol) used for loading the exosomes and chemical transfection methods. FuGENE® HD and HiPerFect were applied following manufacturer’s recommendations (detailed procedure is available at supplementary method section). Afterward, cells were washed and treated with 100ng/ml LPS for 6 hours. Relative expression of miRNA-155 and SOCS1 expression were measured by qPCR and TNFα protein levels were measured in supernatants by ELISA as described in the previous sections.
In vivo delivery of miRNA-155 mimic loaded exosomes to miRNA-155 knockout mice
Detailed procedure of animal studies and in vivo delivery of miRNA-155 mimic loaded exosomes to miRNA-155 knockout mice are described in supplementary method section.
Statistical Analysis
Based on data distribution, Kruskal-Wallis nonparametric test or one-way analyses of variance (ANOVA) were performed to make comparisons between different groups. Student’s t test or Mann-Whitney U test were done for comparing two groups. Data are presented as mean ± standard deviation (SD). P values less than 0.05 was considered as significant.
RESULTS
Characterization of B cell derived exosomes
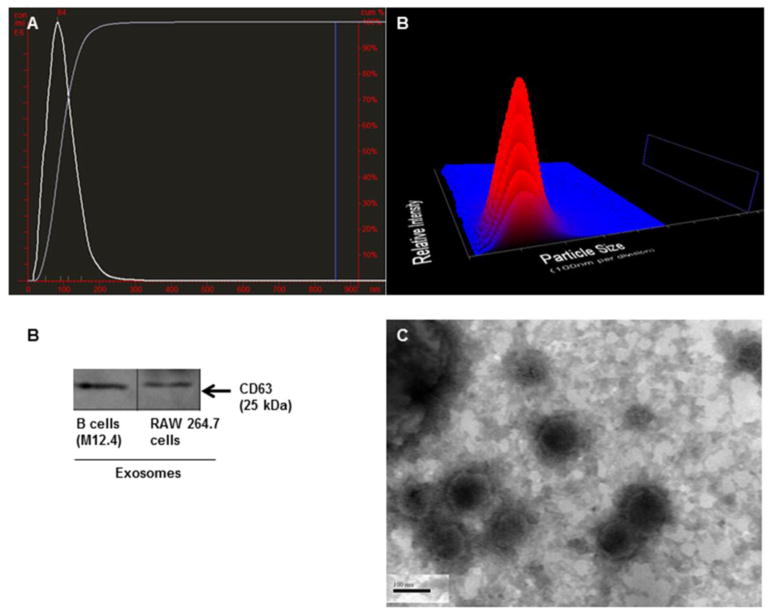
Exosomes derived from B (M12.4) and macrophages (RAW 264.7) cell lines were isolated as described in the method section. The average size of exosomes for stimulated B cell line (with IL-4/CD40), non-stimulated B cell line, and RAW macrophages were 98 nm, 108 nm and 102 nm, respectively, as measured by Nanoparticle Tracking Analysis (NTA)(NanoSight), which were all in the range of exosome size described earlier.1 Figure 1A shows the size versus concentration (particles/ml) of exosomes derived from stimulated B cells. The identity of isolated exosomes was confirmed by the presence of exosomal marker, CD63 (Figure 1B). We further examined the B cell exosome with electron microscopy and confirmed the size range of less than 150 nm (Figure 1C). As previously described by Saunderson et al., activation of B cells with IL-4/CD40 enhances the release of exosomes.30 Consistent with this report, NTA (Nanosight) showed that after stimulation of B cells with IL-4/CD40, the number of exosomes increased more than 200 fold compared to non-stimulated B cells (Figure 2A).
Figure 1. Charachterization of exosomes.
(A) Average size of stimulated B cell-derived exosomes was 98nm with the mode of 84nm. 3D graph represents particle size versus intensity versus concentration (particles/ml) of stimulated B cell-derived exosomes. (B) Isolated exosomes from murine B cells (M12.4) and RAW 264.7 cells expressed exosomal marker, CD63. (C) TEM image of B cell exosomes showed the diameter of less than 150 nm.
Figure 2. Stimulation of B cell exosomes and characterization of miRNA-155 profile.
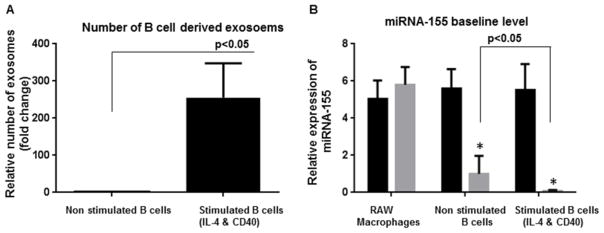
(A) The exosome production was increased in B cells upon stimulation with IL-4 and CD40. The number of exosomes was quantified using NanoSight. (B) Baseline levels of miRNA-155 in different groups including RAW macrophages, non-stimulated B cells, stimulated B cells with IL-4 &CD 40, and pertinent exosomes are shown. Although a considerable amount of miRNA-155 was present in the parental B cells, they were not sorted into the exosomes (p<0.05). Exosomes derived from stimulated B cells showed statistically significant lower levels of miRNA-155 compared to other experimental groups (p<0.001). The results represent three independent experiments. (*indicates p < 0.05 versus parental cells)
An ideal vehicle for miRNA-155 inhibitor/mimic delivery should have minimal miRNA-155 baseline level; thus, we compared miRNA-155 baseline expression in B cells and B cell-derived exosomes. Exosomes derived from naive (non-stimulated B cells) had significantly lower miRNA-155 levels compared to the parental cells (Figure 2B). We found that while stimulated B cells intracellularly expressed high levels of miRNA-155, exosomes derived from stimulated B cells had the lowest miRNA-155 level compared to stimulated B cells (p<0.001), non-stimulated B cells (p<0.001), and exosomes derived from non-stimulated B cells (p<0.05) (Figure 2B). The relative expression of miRNA-155 was similar between RAW cells and RAW cell-derived exosomes (Figure 2B). These observations, first, suggested that miRNA levels can be different between the parent cells and their exosomes depending on the cell type and, second, that stimulated B cell-derived exosomes have very low levels of miRNA-155.
Optimization of B cell exosome loading using miRNA-155 mimic model
Since utilizing exosomes as delivery vehicles should be tailored for specific exosome, specific cargo, and cell type,15 we optimized the loading efficiency for murine B cell (M12.4) derived exosomes to use them as miRNA-155 mimic or miRNA-155 inhibitor delivery vesicles. Our results showed that stimulated B cell-derived exosomes were almost devoid of endogenous miRNA-155 and they can be used as vehicles for delivery of exogenous miRNA-155 nucleotides.
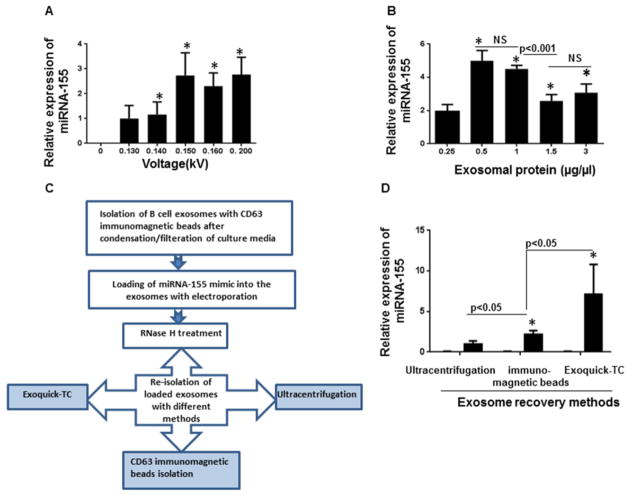
To optimize electroporation loading, B cell-derived exosomes were isolated with CD63 immunomagnetic beads and electroporated in the presence of miRNA-155 mimics (150 pmol) and 1mM EDTA at various voltages (0.130 to 0.200kV) (Figure 3A). We used EDTA in the process because EDTA prevents interactions of aluminium ions with buffer components and macromolecules in the electroporation cuvette.31 Next, exosomes were treated with RNase H to eliminate unloaded miRNA-155 mimic outside the exosomes and re-pelleted using Exoquick™. The amount of miRNA-155 packaged into the exosomes was quantified by TaqMan qPCR. Five different voltages with constant capacitance (100μf) were evaluated for delivery of miRNA-155 mimic. Voltages between 0.14 kV to 0.200 kV showed significantly more efficiency compared to a lower voltage (0.130) (p<0.05) (Figure 3A). In contrast, variation of capacitance (0–400 μF) did not affect the efficiency of electroporation (data not shown).
Figure 3. Optimization of loading of miRNA-155 mimic to the exosomes.
(A) miRNA-155 mimic was introduced to the exosomes using various voltages (0.130 to 0.200kV). Exosomes+ miRNA-155 mimic without exertion of electric pulse served as the negative control. After electroporation, the exosomes were re-pelleted using Exoquick-TC™ and the effect of voltage variation on electroporation efficiency was determined by detection of relative amount miRNA-155 mimic encapsulated into the exosomes using qPCR. Voltages between 0.14 kV to 0.200 kV showed significantly more efficiency in comparison with a lower voltage (0.130kV) (p<0.05). (*indicates p < 0.05 versus the lowest voltage, 0.13kV). (B) Different amount of exosomes containing 0.25μg/μl, 0.5μg/μl, 1μg/μl, 1.5μg/μl, and 3 μg/μl exosomal proteins were electroporated in similar conditions. The exosomal protein concentrations of 0.5 μg/μl to 1μg/μl were the most efficient concentrations for loading. The efficiency of loading significantly decreased when the concentrations of exosomal protein were more than 1μg/μl (p<0.001). (*indicates p < 0.05 versus the lowest concentration, 0.25μg/μl). (C) Schematic experimental design for evaluating effect of different isolation method in exosome recovery. (D) The same amount of miRNA-155 mimic was loaded into the exosoms via electroporation and re-isolated with different methods (Ultracentrifugation, Exoquick-TC™ and CD63 immunomagnetic beads). Exoquick-TC™ showed more efficiency compared to ultracentrifugation and CD63 immunomagnetic isolation (p<0.05). CD63 immunomagnetic isolation method was significantly more efficient compared to ultracentrifugation (p<0.05). The results represent three independent experiments. (*indicates p < 0.05 versus ultracentrifugation)
To identify the most efficient exosome-to-miRNA ratio for loading of B cell-derived exosomes, we used different exosome concentrations (0.25 to 3μg/ml), based on exosomal protein concentration, and electroporated them with the same amount of miRNA-155 mimic. We found that the exosomal density of 0.5μg/ml to 1 μg/ml resulted in efficient encapsulation of miRNA-155 mimic. With an increase in exosome concentration to the 3μg/ml, the loading efficiency of miRNA-155 mimic into the exosomes was decreased significantly (p<0.001) (Figure 3B).
Recovery of exosomes after miRNA-155 loading
Equal number of isolated exosomes were electroporated with the same amount of miRNA-155 mimic and efficiency of three different methods for re-isolation of the exosomes including ultracentrifugation, immunomagnetic isolation method and Exoquick™ were compared (Figure 3C&3D). We found that Exoquick™ had the best isolation efficiency compared to CD63 immunomagnetic isolation method (p<0.05) and ultracentrifugation (p<0.05). The CD63 immunomagnetic selection method was superior compared to ultracentrifugation for isolation of exosomes (p<0.05) (Figure 3D). With the most optimized loading conditions, about 55 % of the initial miRNA-155 mimic was loaded into the exosomes.
Efficient delivery of exogenous miRNA-155 mimic to primary mouse hepatocytes
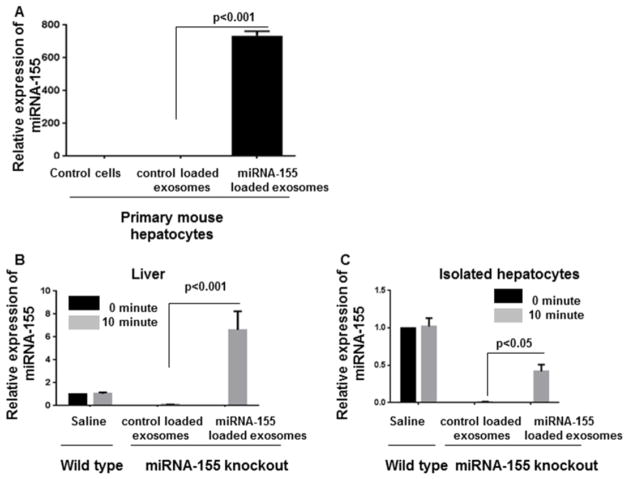
To assess the functional ability of exosomal delivery of miRNA-155 mimic, we loaded B cell-derived exosomes with miRNA-155 mimic using optimal loading conditions. Then exosomes were co-cultured with primary mouse hepatocytes for 6 hours, followed by washing off extra exosomes and replacement of medium. The readout after 24 hours showed that exosomes were able to successfully deliver miRNA-155 mimic to primary mouse hepatocytes, indicated by the approximately 700 fold increase in intracellular miRNA-155 levels (Figure 4A).
Figure 4. Delivery of miRNA-155 mimic to the primary mouse hepatocytes and liver of miRNA-155 knockout mice.
B cell-derived exosomes were loaded with miRNA-155 using optimal loading conditions. (A)The exosomes were co-cultured with primary mouse hepatocytes for 6 hours followed by washing and media replacement. The amount of miRNA-155 was assessed after 24 hours. Exosomes were able to successfully deliver miRNA-155 mimic to primary mouse hepatocytes (p<0.001). (B&C) miRNA-155 mimic or control mimic loaded exosomes were injected intravenously to the miRNA-155 knockout mice (n=4). The levels of miRNA-155 in the liver and hepatocytes were significantly increased in the miRNA-155 loaded exosomes and not in the control mimic loaded exosomes (p<0.05), 10 minutes after injection. snoRNA202 was used as internal control for qPCR analysis.
In vivo delivery of exogenous miRNA-155 mimic to miRNA-155 knockout mice
Next, we tested ability of exosomes to mediate miRNA-155 mimic delivery in vivo. Intravenous administration (IV) of miRNA-155 loaded exosomes into miRNA-155 knockout mice resulted in successful delivery of miRNA-155 mimic to the liver (Figure 4B). Furthermore, a statistically significant increase in the levels of miRNA-155 was detected in the isolated hepatocytes of miRNA-155 knockout mice, 10 minutes after IV injection with exosomes containing miRNA-155 mimic and not control mimic (p<0.05) (Figure 4C). These results suggest in vivo delivery of miRNA-155 by exosomes to the liver.
Efficient delivery of exogenous miRNA-155 inhibitor to RAW macrophages via exosomes
Although nucleic acid-based treatment, especially siRNA treatment, has introduced a promising therapeutic method, only few studies evaluated exosome-mediated delivery of siRNA and miRNA inhibitors/mimics to the cells. miRNA inhibitors are synthetic miRNA target analogs, which are fully complementary, chemically modified oligonucleotides, and are capable of inhibiting miRNAs function.32 The main problem with delivery of RNA inhibitors is their negative charge which jeopardizes their penetration to the hydrophobic cellular membrane even in the presence of a transfection reagent.33 Our group previously reported the LPS-induced miRNA-155 overexpression in RAW 264.7 macrophages, resulted in TNFα production.20
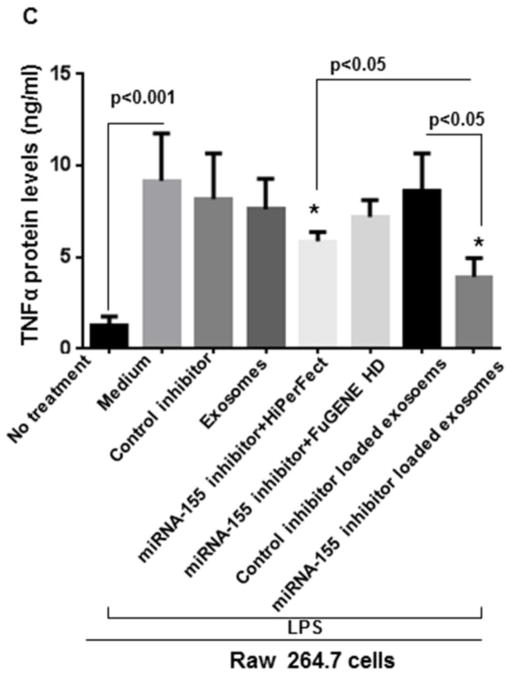
Here, using the optimized conditions of exosome loading, we investigated whether B cells-derived exosomes can act as carriers to deliver a miRNA-155 inhibitor to RAW 264.7 macrophages and exert functional modulation of miRNA-155 target genes. Thus, the miRNA-155 inhibitor was electroporated into B cell-derived exosomes using the optimized loading conditions. We used various control groups including non-specific control inhibitor (electroporated and non electroporated), miRNA-155 inhibitor with and without transfection reagents (FuGENE® HD and HiPerFect), and control inhibitor loaded exosomes (Figure 5A). B cell-derived exosomes showed functional delivery of the miRNA-155 inhibitor indicated by a statistically significant decrease in LPS-induced miRNA-155 levels and subsequent statistically significant increase in SOCS1 levels in cells treated with exosomes loaded with the miRNA-155 inhibitor and not with the control miRNA inhibitor (p<0.05) (Figure 5A & 5B). Additionally, exosome-mediated miRNA-155 inhibition resulted in a significant decrease in LPS-induced TNFα protein levels compared to transfection reagents (FuGENE® HD and HiPerFect) and control inhibitor (p<0.05) (Figure 5C).
Figure 5. Delivery of miRNA-155 inhibitor to RAW 264.7 cells via exosomes.
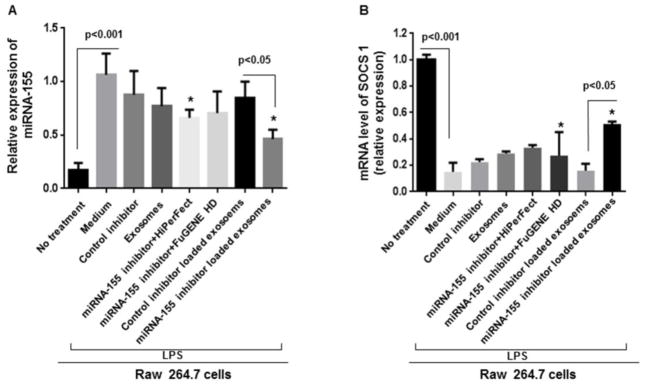
Cells were seeded 1 day before treatment and different treatment conditions and controls were applied for 24 hours. Afterward, cells were treated with 100ng/ml LPS for 6 hours. Relative expression of miRNA-155 and SOCS1 expression were measured by qPCR. TNFα protein levels were measured in supernatants by ELISA. (A) Relative expression of miRNA-155 is shown in different conditions versus LPS stimulated RAW 264.7 cells. miRNA-155 inhibitor loaded exosomes effectively inhibited miRNA-155 expression compared to control inhibitor loaded exosomes (p<0.05). (B) SOCS1 mRNA levels were significantly increased after exosome-mediated inhibition of miRNA-155 (p<0.05) (C) miRNA-155 loaded exosomes significantly diminished TNFα protein levels compared to control inhibitor loaded exosomes and transfection reagents (FuGENE® HD and HiPerFect), after LPS treatment. The results represent three independent experiments. (*indicates p<0.05 versus medium+LPS treated RAW 264.7 cells).
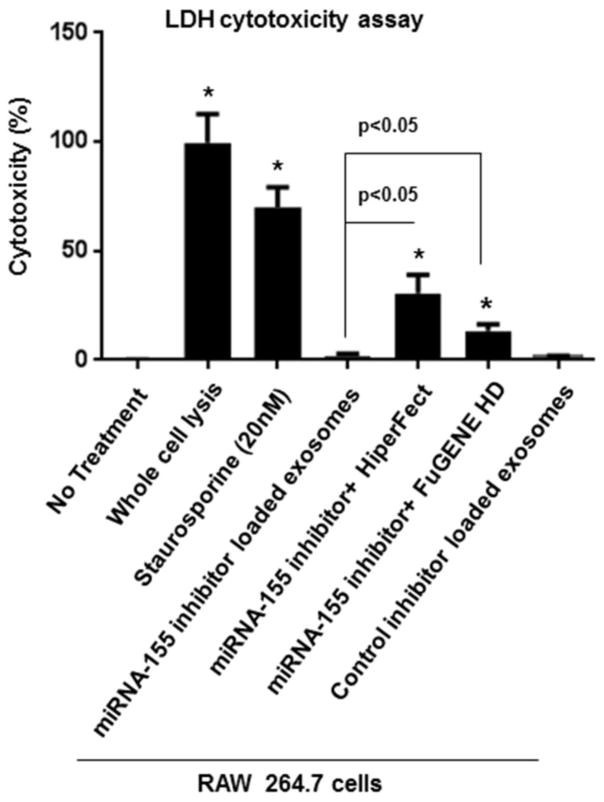
Exosomes-based delivery has minimal cytotoxicity
The level of dead cells did not increase with exosome-mediated inhibition of miRNA-155, while by using traditional transfection reagents (FuGENE® HD and HiPerFect) the lactate dehydrogenase (LDH) levels and the percentage of cytotoxicity were significantly increased, 36 hours after co-culture (p<0.05) (Figure 6).
Figure 6. Evaluating cytotoxicty of exosome-based miRNA-155 inhibitor delivery and transfection reagants.
After 36 hours of co-culture, miRNA-155 loaded exosomes showed minimal toxicity, while miRNA-155 inhibitor + FuGENE HD, and miRNA-155 inhibitor + HiPerFect showed significant cytotoxicity (p<0.05). Results are representative of three independent experiments. (*indicates p < 0.05 versus non treated RAW 264.7 macrophages).
DISCUSSION
In this report, exogenous miRNA-155 mimic and miRNA-155 inhibitor were successfully introduced into murine B cell-derived exosomes (membranous nanovesicles) and was shown that loaded exosomes can deliver miRNA-155 mimic and miRNA-155 inhibitor to primary mouse hepatocytes and RAW 264.7 macrophages, respectively. B cell exosomes effectively delivered miRNA-155 inhibitor into the RAW 264.7 cells, causing inhibition of miRNA-155, and functionally led to a statistically significant decrease of TNFα protein levels. SOCS1 is reported to be an evolutionary conserved target of miRNA-155 and its expression is inversely correlated with miRNA-155 level. 34, 35 Consistently, after successful delivery of miRNA-155 inhibitor to RAW 264.7 cells, the mRNA level of SOCS1 was increased. In comparison with transfection reagents (FuGENE® HD and HiPerFect), using the same amount of miRNA-155 inhibitor, miRNA-155 loaded exosomes mediated more efficient inhibition of miRNA-155, reflected by a statistically significant decrease in TNFα protein levels.
It was previously shown that exosomes can naturally transfer a variety of biologically active cargos such as mRNA, miRNA, mitochondrial DNA, genomic DNA, and proteins. 36–39 However, the efficiency of exosome mediated delivery has not yet been systemically assessed, and it may vary based on the recipient cell type, special cargo and exosome source.19 Especially, recent evidence showed that different exosomes have different ability to naturally pack and deliver special subsets of cellular RNA and proteins which might be non-identical to the parental cells.36,37 Thus, we first characterized and optimized loading conditions for B cell-derived exosomes to use them as vehicles to deliver miRNA-155 inhibitor. The experimental design of exosome-based miRNA inhibitor/mimic delivery is shown in Supplementary Figure 1.
B cells differentiation results in production of exosomes that are enriched in CD63, CD9, CD19, CD22, Tsg101, and MHC-II.30,40–43 CD40 and IL-4 synergistically stimulate proliferation and exosome release in B cells.30,43,44 It has been shown that B cells stimulated with CD40 and IL-4 maintained their B cell associated phenotypic traits such as maintaining the expression of CD40 and MHC-II and remaining responsive to IL-4 stimulation.30,41,43 Our result showed that B cell-derived exosomes are ideal candidates to deliver miRNA-155 inhibitor, since upon stimulation with IL-4 and CD40 large quantities of exosomes were generated and they were almost devoid of miRNA-155. Moreover, B cell-derived exosomes appeared to be more efficient vehicles for miRNA-155 inhibitor delivery compared to the conventional transfection reagents. Additional advantage of the exosome-based delivery is lack of cytotoxicity and cell death compared to the transfection reagents.
Regarding the prominent role of miRNA-155 in the signaling pathways of monocytes and macrophages,20,45 miRNA-155 mimic and miRNA-155 inhibitor were chosen as the exogenous bio-macromolecules in this study. miRNA inhibitors are synthetic miRNA analogs, which are fully complementary, chemically modified oligonucleotides, and are capable of inhibiting target miRNAs functions.32 The functional model of inhibiting miRNA-155 that we introduced in this study could be applied to other disease models such as cancers, neurological disorders, multiple sclerosis, and autoimmune disorders where miRNA-155 over expression induces pathological conditions.20,46
We optimized electroporation conditions in terms of voltage, capacitance, exosome concentration, and re-isolation method for effective miRNA-155 loading and recovery of B cell exosomes. Using optimal conditions, 55% of miRNA-155 mimic were loaded successfully into the exosomes. To make sure that this amount of miRNA is indeed the fraction of miRNA which is loaded inside the exosomes and not the outside miRNA-155 aggregates, we treated the suspension of exosomes after loading with RNase H to eliminate free floating miRNA-155 mimic. Moreover, we applied EDTA to the electroporation buffer to prevent precipitation of miRNA-155 mimic in the electroporation procedure.32 As low efficiency of exosome isolation methods is one of the major limiting factors in fostering exosome-mediated drug delivery;47 we compared the efficiency of three different isolation methods in terms of recovery of miRNA-155 loaded exosomes. The result of our study showed that both immunomagnetic selection method and polymer based isolation method (Exoquick™) have better efficiency in recovery of exosomes compared to ultracentrifugation. Exoquick™ precipitation method resulted in more recovery of exosomes compared to the other methods.
The promising efficiency of exosome loading was confirmed by co-culturing miRNA-155 mimic loaded exosomes with primary mouse hepatocytes. The level of miRNA-155 was increased more than 700 fold in the primary mouse hepatocytes co-cultured with miRNA-155 loaded exosome compared to the control mimic. Furthermore, our in vivo study using miRNA-155 knockout mice showed that exosomes mediated successful delivery of miRNA-155 to the liver and to primary mouse hepatocytes at 10 minutes after IV injection. Altogether, these results provide convincing evidence that the exogenous miRNA-155 mimic was effectively introduced into the exosomes and exosomes can mediate miRNA-155 delivery to primary mouse hepatocytes in vivo and in vitro.
Recent advances in understanding biological processes have broadened the scope of therapeutic options to RNA-based therapeutics. These therapeutic approaches range from introducing specific RNA, using RNA interfaces (RNAi) such as siRNA to silence gene expression, or to introduce miRNA inhibitors and miRNA mimics. However, most forms of RNA are inherently unstable and prone to RNase effect, potentially immunogenic, and negatively charged. This negative charge jeopardizes their effective delivery to the hydrophobic cellular membrane even in the presence of transfection reagents.33,48 Viral and non-viral vectors are already broadly used for delivery of RNA/DNA molecules into target cells both in vitro and in vivo. However, the present vectors have not been completely successful in clinical trials, as they induce immunogenicity and show cytotoxicity, particularly in repeated administrations.19 Our results indicate that co-culturing of exosomes (alone) with RAW macrophages did not induce TNFα production. Moreover, in contrast with chemical transfection methods, exosome-based delivery showed no cytotoxicity.
Exosome-based drug delivery confers a number of advantages over other vector or liposome-based delivery methods. First, exosomes act as natural membranous nanocarriers of bio-macromolecules between cells and they can easily transpass the plasma membrane and mediate effective RNA delivery. Second, exosomes can be isolated from the recipient’s own body fluids or cell culture and be used for loading the biological drug; indeed, in our experiment, B cells seemed to be a generous viable source of exosomes for this purpose. Autologous exosomes, which are non-immunogenic, would be a perfect source of personalized drug vehicles. Third, non-toxicity of exosomes compared to transfection reagents is another key advantage.
In summary, our results provide evidence for exosome-based delivery of RNA-based therapeutics in vitro and in vivo. Interestingly, in our study, the B cell-derived exosomes mediated effective delivery of miRNA-155 inhibitor and caused post-transcriptional gene knockdown (TNFα) in the recipient cells. miRNA-155 mimic loaded exosomes significantly increased miRNA-155 levels in primary mouse hepatocytes and the liver of miRNA-155 knockout mice. Although in this study we focused on the exosome-mediated delivery of miRNA-155 mimic to the liver and primary hepatocytes, it would be interesting to assess the bio-distribution of exosome-mediated delivery in other organs. In vivo findings of this study mechanistically showed that exosomes can mediate successful delivery of miRNA-155 mimic to the liver and hepatocytes as early as 10 minutes after IV injection. The in vivo study was performed at a short time point based on the reported peak of bio-distribution of exosomes, miRNAs, and RNA interfaces in pharmacokinetics studies.49, 50 Further investigations at various time points to elucidate kinetics of exosome-based delivery in vivo could be subject of future studies. Our results mechanistically demonstrated that B cell-derived exosomes could be used as efficient vehicles in RNA-based therapeutic strategies and provide proof of concept for using exosomes as efficient delivery nano-vehicles with minimal cytotoxicity. This identifies exosomes as novel frontiers in expanding nanomedicine applications.
Supplementary Material
Acknowledgments
This work was supported by NIH PHS grant AA020744 to G.S.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Momen-Heravi F, Balaj L, Alian S, Tigges J, Toxavidis V, Ericsson M, et al. Alternative methods for characterization of extracellular vesicles. Front Physiol. 2012;3:354. doi: 10.3389/fphys.2012.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–57. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 3.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson RJ, Mathivanan S. Extracellular microvesicles: the need for internationally recognised nomenclature and stringent purification criteria. J Proteomics Bioinform. 2012;5:ii–ii. [Google Scholar]
- 5.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, et al. Mechanism of transfer of functional micrornas between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Momen-Heravi F, Balaj L, Alian S, Trachtenberg AJ, Hochberg FH, Skog J, et al. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front Physiol. 2012;3:162. doi: 10.3389/fphys.2012.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–91. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiteside TL, Mandapathil M, Szczepanski M, Szajnik M. Mechanisms of tumor escape from the immune system: adenosine-producing Treg, exosomes and tumor-associated TLRs. Bull Cancer. 2011;98:E25–31. doi: 10.1684/bdc.2010.1294. [DOI] [PubMed] [Google Scholar]
- 9.Mu W, Rana S, Zöller M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia. 2013;15:875–87. doi: 10.1593/neo.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7. 5 cells. Proc Natl Acad Sci U S A. 2013;110:13109–13. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izquierdo-Useros N, Naranjo-Gómez M, Erkizia I, Puertas MC, Borràs FE, Blanco J, et al. HIV and mature dendritic cells: Trojan exosomes riding the Trojan horse? PLoS Pathog. 2010;6:e1000740. doi: 10.1371/journal.ppat.1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kooijmans SA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomedicine. 2012;7:1525–41. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hood JL, Wickline SA. A systematic approach to exosome-based translational nanomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4:458–67. doi: 10.1002/wnan.1174. [DOI] [PubMed] [Google Scholar]
- 14.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 15.Wahlgren J, De L Karlson T, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suntres ZE, Smith MG, Momen-Heravi F, Hu Jie, Zhang Xin, Wu Ying, et al. Therapeutic uses of exosomes. Exosomes Microvesicles. 2013;1:1. [Google Scholar]
- 17.Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol Ther. 2011;19:1769–79. doi: 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seow Y, Wood MJ. Biological gene delivery vehicles: Beyond viral vectors. Mol Ther. 2009;17:767–777. doi: 10.1038/mt.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcus ME, Leonard JN. FedExosomes: Engineering Therapeutic Biological Nanoparticles that Truly Deliver. Pharmaceuticals(Basel) 2013;6:659–680. doi: 10.3390/ph6050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286:1436–44. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y, Park HJ, Kang MI, Lee HS, Lee SW, Lee SK, et al. Adipokines, inflammation, insulin resistance, and carotid atherosclerosis in patients with rheumatoid arthritis. Arthritis Res Ther. 2013;15:R194. doi: 10.1186/ar4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren H, Zhou X, Luan Z, Luo X, Han S, Cai Q, et al. The Relationship between Carotid Atherosclerosis, Inflammatory Cytokines, and Oxidative Stress in Middle-Aged and Elderly Hemodialysis Patients. Int J Nephrol. 2013;2013:835465. doi: 10.1155/2013/835465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonioli L, Blandizzi C, Pacher P, Haskó G. Immunity, inflammation and cancer: a leading role for adenosine. Nat Rev Cancer. 2013;13:842–57. doi: 10.1038/nrc3613. [DOI] [PubMed] [Google Scholar]
- 24.Chono S, Tanino T, Seki T, Morimoto K. Efficient drug targeting to rat alveolar macrophages by pulmonary administration of ciprofloxacin incorporated into mannosylated liposomes for treatment of respiratory intracellular parasitic infections. J Control Release. 2008;127:50–8. doi: 10.1016/j.jconrel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Kole L, Das L, Das PK. Synergistic effect of interferon-gamma and mannosylated liposome-incorporated doxorubicin in the therapy of experimental visceral leishmaniasis. J Infect Dis. 1999;180:811–20. doi: 10.1086/314929. [DOI] [PubMed] [Google Scholar]
- 26.Eue L. Growth inhibition of human mammary carcinoma by liposomal hexadecylphosphocholine: participation of activated macrophages in the antitumor mechanism. Int J Cancer. 2001;92:426–33. doi: 10.1002/ijc.1201. [DOI] [PubMed] [Google Scholar]
- 27.Kelly C, Jefferies C, Cryan SA. Targeted liposomal drug delivery to monocytes and macrophages. J Drug Deliv. 2011;2011:727241. doi: 10.1155/2011/727241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Rooijen N, Sanders A. The macrophage as target or obstacle in liposome-based targeting strategies. Int J Pharm. 1998;162:45–50. [Google Scholar]
- 29.Burke B, Sumner S, Maitland N, Lewis CE. Macrophages in gene therapy: cellular delivery vehicles and in vivo targets. J Leukoc Biol. 2002;72:417–28. [PubMed] [Google Scholar]
- 30.Saunderson SC, Schuberth PC, Dunn AC, Miller L, Hock BD, MacKay PA, et al. Induction of exosome release in primary B cells stimulated via CD40 and the IL-4 receptor. J Immunol. 2008;180:8146–52. doi: 10.4049/jimmunol.180.12.8146. [DOI] [PubMed] [Google Scholar]
- 31.Kooijmans SA, Stremersch S, Braeckmans K, de Smedt SC, Hendrix A, Wood MJ, et al. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J Control Release. 2013;172:229–38. doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Robertson B, Dalby AB, Karpilow J, Khvorova A, Leake D, Vermeulen A. Specificity and functionality of microRNA inhibitors. Silence. 2010;1:10. doi: 10.1186/1758-907X-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang S, Zhang HW, Lu MH, Lu MH, He XH, Li Y, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–27. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 35.Davey GM, Heath WR, Starr R. SOCS1: a potent and multifaceted regulator of cytokines and cell-mediated inflammation. Tissue Antigens. 2006;67:1–9. doi: 10.1111/j.1399-0039.2005.00532.x. [DOI] [PubMed] [Google Scholar]
- 36.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 37.Crescitelli R, Lässer C, Szabó TG, Kittel A, Eldh M, Dianzani I, et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013;12:2. doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, et al. Mechanism of transfer of functional micrornas between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seow Y, Wood MJ. Biological gene delivery vehicles: Beyond viral vectors. Mol Ther. 2009;17:767–777. doi: 10.1038/mt.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arita S, Baba E, Shibata Y, Niiro H, Shimoda S, Isobe T, et al. B cell activation regulates exosomal HLA production. Eur J Immunol. 2008;38:1423–34. doi: 10.1002/eji.200737694. [DOI] [PubMed] [Google Scholar]
- 41.McLellan AD. Exosome release by primary B cells. Crit Rev Immunol. 2009;29:203–17. doi: 10.1615/critrevimmunol.v29.i3.20. [DOI] [PubMed] [Google Scholar]
- 42.Muntasell A, Berger AC, Roche PA. T cell-induced secretion of MHC class II-peptide complexes on B cell exosomes. EMBOJ. 2007;26:4263–72. doi: 10.1038/sj.emboj.7601842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rousset F, Peyrol S, Garcia E, Vezzio N, Andujar M, Grimaud JA, et al. Long-term cultured CD40-activated B lymphocytes differentiate into plasma cells in response to IL-10 but not IL-4. Int Immunol. 1995;7:1243–53. doi: 10.1093/intimm/7.8.1243. [DOI] [PubMed] [Google Scholar]
- 44.Schultze JL, Michalak S, Seamon MJ, Dranoff G, Jung K, Daley J, et al. CD40-activated human B cells: an alternative source of highly efficient antigen presenting cells to generate autologous antigen-specific T cells for adoptive immunotherapy. J Clin Invest. 1997;100:2757–65. doi: 10.1172/JCI119822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subedi A, Park PH. Autocrine and paracrine modulation of microRNA-155 expression by globular adiponectin in RAW 264. 7 macrophages: Involvement of MAPK/NF-κB pathway. Cytokine. 2013;64:638–41. doi: 10.1016/j.cyto.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Escobar T, Yu CR, Muljo SA, Egwuagu CE. STAT3 activates miR-155 in Th17 cells and acts in concert to promote experimental autoimmune uveitis. Invest Ophthalmol Vis Sci. 2013;54:4017–25. doi: 10.1167/iovs.13-11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Momen-Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, et al. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013;394:1253–62. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Chiu M, Xie Z, Chiu M, Liu Z, Chen P, et al. Synthetic microRNA cassette dosing: pharmacokinetics, tissue distribution and bioactivity. Mol Pharm. 2012;9:1638–44. doi: 10.1021/mp2006483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi Y, Nishikawa M, Shinotsuka H, Matsui Y, Ohara S, Imai T, et al. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J Biotechnol. 2013;165:77–84. doi: 10.1016/j.jbiotec.2013.03.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.