SUMMARY
Skeletal muscle comprises a heterogeneous population of fibers with important physiological differences. Fast fibers are glycolytic and fatigue rapidly. Slow fibers utilize oxidative metabolism and are fatigue-resistant. Muscle diseases such as sarcopenia and atrophy selectively affect fast fibers, but the molecular mechanisms regulating fiber type-specific gene expression remain incompletely understood. Here, we show that the transcription factor NFATc1 controls fiber type composition and is required for fast-to-slow fiber type switching in response to exercise in vivo. Moreover, MyoD is a crucial transcriptional effector of the fast fiber phenotype, and we show that NFATc1 inhibits MyoD-dependent fast fiber gene promoters by physically interacting with the N-terminal activation domain of MyoD and blocking recruitment of the essential transcriptional coactivator p300. These studies establish a molecular mechanism for fiber type switching through direct inhibition of MyoD to control the opposing roles of MyoD and NFATc1 in fast versus slow fiber phenotypes.
INTRODUCTION
Skeletal muscle is composed of a heterogeneous population of muscle fibers that display several distinct properties (Schiaffino and Reggiani, 2011). Type I (slow twitch) muscle fibers utilize oxidative metabolism, are rich in mitochondria, display increased vascularization, and are fatigue-resistant. Type II (fast twitch) fibers utilize glycolytic metabolism and fatigue rapidly (Schiaffino and Reggiani, 2011). Adult skeletal muscle can undergo conversion between these fiber types in response to exercise training or the pattern of motor neuron activity (Pette, 1998).
The basic helix-loop-helix (bHLH) transcription factor MyoD possesses the remarkable ability to convert most cell types into multinucleated, contractile myotubes through forced expression in cell culture and functions as an essential regulator of myogenesis (Tapscott, 2005). MyoD is also reutilized for multiple distinct roles in postnatal muscle, including fiber type control (Legerlotz and Smith, 2008). MyoD is expressed more highly in fast fibers than in slow fibers, and Myod1-null mice show shifts in fiber type of fast muscle towards a slower phenotype (Hughes et al., 1997; Macharia et al., 2010). Conversely, over-expression of an active form of MyoD in muscle results in a slow-to-fast fiber type conversion (Ekmark et al., 2007). MyoD binds DNA as a heterodimer with members of the E-protein family of bHLH proteins through the interaction of the HLH domains (Tapscott, 2005). The N-terminus of MyoD encodes an activation domain that also contains a motif referred to as the C/H domain, which has been proposed to confer “pioneer” activity to MyoD through interactions with the histone acetyltransferase p300/CBP and other chromatin remodeling enzymes (Berkes and Tapscott, 2005; Tapscott, 2005).
Nuclear factor of activated T-cell (NFAT) proteins comprise a family of five transcription factors (NFATc1-4, NFAT5) that are regulated by the phosphatase calcineurin (Hogan et al., 2003). Calcineurin signaling plays a key role in the maintenance of the slow fiber phenotype in adult muscle (Hogan et al., 2003). NFAT proteins are known to regulate slow fiber genes in cell culture systems (Chin et al., 1998; Wu et al., 2000), but a role for NFAT proteins in control of fiber type in vivo has not yet been established. Although NFATc1 is the predominant NFAT isoform expressed in adult skeletal muscle (Rana et al., 2008), studies interrogating its role in muscle in vivo have been limited because nfatc1-null mice die at embryonic day 12.5 due to cardiac valve defects (Hogan et al., 2003).
Here, we inactivated nfatc1 exclusively in skeletal muscle. We demonstrate that NFATc1 is required in vivo for the proper balance of slow Type I and fast Type II fibers and for fast-to-slow fiber switching in response to exercise. Unexpectedly, we found that NFATc1 inhibits expression of MyoD-dependent fast fiber genes in a DNA-binding independent fashion. NFATc1 inhibits MyoD function by physically interacting with and blocking the essential interaction of the MyoD N-terminal activation domain with p300 and subsequent activation of MyoD target genes. These studies demonstrate that NFATc1 is an essential regulator of fiber type-specific gene expression and fiber type switching in response to exercise and establish NFATc1 as a novel negative regulator of MyoD.
RESULTS
MyoD function is negatively regulated by NFATc1
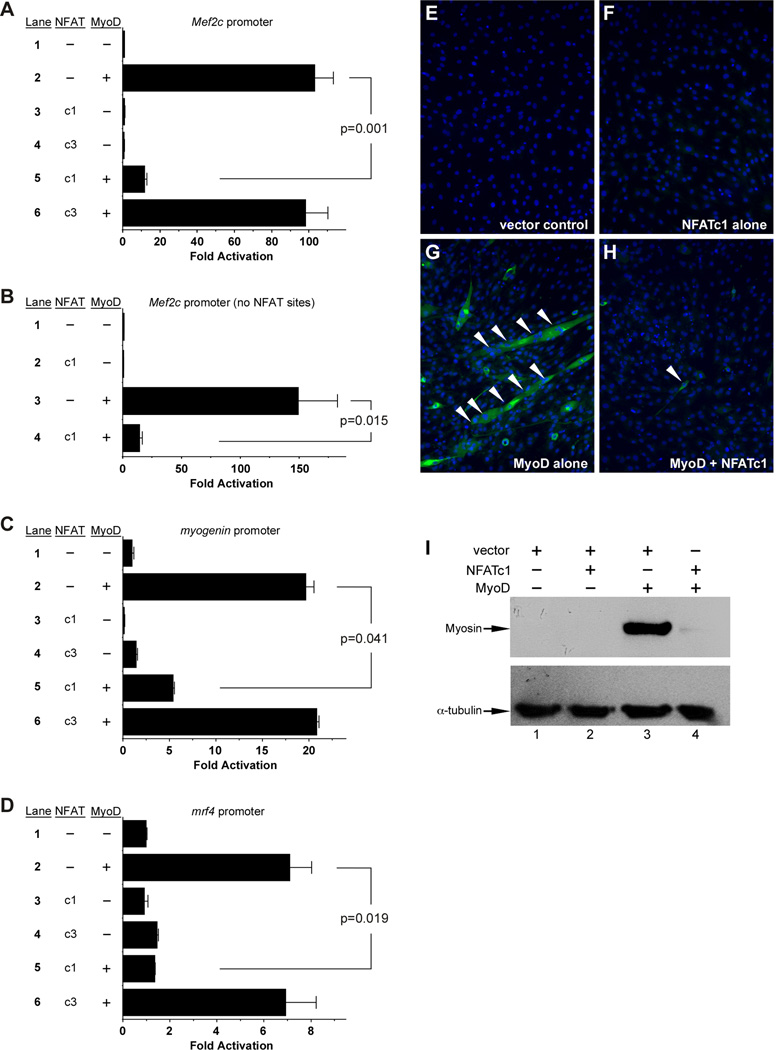
The Mef2c gene is a direct transcriptional target of MyoD via a skeletal muscle-specific promoter (Dodou et al., 2003; Wang et al., 2001). To gain additional insight into the regulation of Mef2c in skeletal muscle, we analyzed its promoter for conserved transcription factor binding sites. In addition to the previously described MyoD binding sites (E boxes), we also identified five consensus NFAT binding sites (Figure S1). Therefore, we examined the effect of several NFAT proteins on Mef2c promoter activity in the presence and absence of MyoD (Figure 1A). NFATc1, NFATc3, and NFATc4 alone had no effect on the Mef2c promoter, whereas MyoD strongly transactivated the Mef2c reporter plasmid (Figure 1A). Strikingly, MyoD-dependent transactivation was strongly repressed by NFATc1 but not by NFATc3 or NFATc4 (Figure 1A and data not shown), suggesting a role specifically for NFATc1 in the inhibition of MyoD transactivation.
Figure 1. NFATc1 inhibits MyoD activity.
(A, C, D) MyoD transactivated the Mef2c (A), myogenin (C) and mrf4 (D) promoters (lane 2); NFATc1 (c1) significantly inhibited activation (lane 5). (B) A Mef2c skeletal muscle promoter lacking NFAT sites (Figure S1) was transactivated by MyoD (lane 3), and coexpression of NFATc1 inhibited activation (lane 4). Results are reported as the mean fold activation plus SEM from 4 independent transfections. (E–I) C3H10T1/2 cells were transfected, differentiated, and assessed for fast MyHC (MY32) by immunofluorescence (E–H) and western blot (I). Cells were transfected with the pRK5 vector (E), NFATc1 alone (F), MyoD alone (G), or MyoD plus NFATc1 (H). Coexpression of NFATc1 blocked MyoD-induced MyHC expression and myotube formation (white arrowheads). Nuclei were counterstained with DAPI in all panels. (I) Western blot analysis of myosin induction in C3H10T1/2 cells transfected with vector alone (lane 1), NFATc1 alone (lane 2), MyoD alone (lane 3), and NFATc1 plus MyoD (lane 4). α-tubulin was examined as a loading control on the same cell lysates. Nearly identical results were obtained in 6 independent experiments. See also Figure S1.
To determine if NFATc1 repressed the Mef2c promoter through the NFAT sites, we generated a Mef2c reporter construct encompassing the MyoD-dependent E-boxes but lacking all of the NFATc1 binding sites (Figure S1, nucleotides shown in red). This reporter was strongly transactivated when co-transfected with MyoD, and surprisingly, MyoD-dependent activation of the reporter was still profoundly reduced when an NFATc1 expression plasmid was also co-transfected (Figure 1B). These results demonstrate that NFATc1 inhibition of MyoD activity does not require the NFAT cis-regulatory elements and suggest that it may occur through DNA-independent interaction with MyoD.
To further investigate the inhibition of MyoD-dependent transactivation by NFATc1, we examined the activation of two other established MyoD-dependent target promoters from the myogenin and mrf4 genes (Black et al., 1995; Edmondson et al., 1992) in the presence or absence of NFATc1 (Figure 1C,D). Importantly, MyoD strongly transactivated each of these other reporters, and this transactivation was repressed by NFATc1 but not by NFATc3 (Figure 1C,D).
We next examined whether NFATc1 influenced MyoD-mediated myogenic conversion (Figure 1E–I). Control transfected C3H10T1/2 cells did not express myosin heavy chain (MyHC) nor did they form multinucleated myotubes (Figure 1E). Similarly, transfection of an NFATc1 expression vector alone did not result in myotube formation or MyHC expression (Figure 1F). As expected, transfection of a MyoD expression vector resulted in robust myogenic conversion as shown by the formation of MyHC-positive myotubes (Figure 1G). Co-expression of NFATc1 with MyoD resulted in a nearly complete inhibition of myotube formation and myosin heavy chain expression (Figure 1H). Western blot analyses of fast MyHC (Figure 1I) showed nearly identical results: control and NFATc1 transfected C3H10T1/2 cells had no expression of MyHC (Figure 1I) whereas MyoD induced robust MyHC expression (Figure 1I, lane 3), and this induction was profoundly inhibited by co-expression of NFATc1 (Figure 1I, lane 4).
Generation of a skeletal muscle-specific knockout of nfatc1
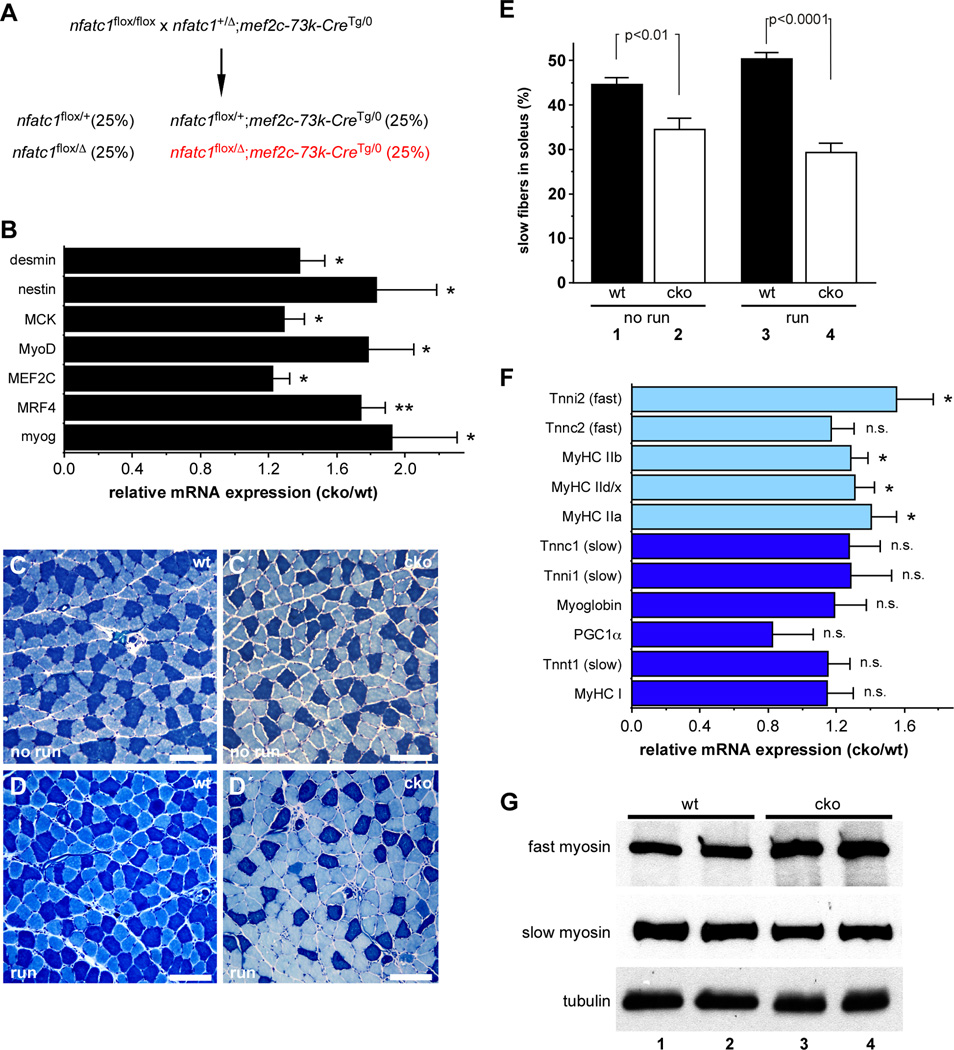
Although NFATc1 expression in skeletal muscle is well established (Hogan et al., 2003), we confirmed NFATc1 expression in the nuclei of both slow and fast myofibers, and we found that NFATc1 and MyoD were co-expressed in skeletal muscle (Figure S2). Therefore, to test the role of NFATc1 in the regulation of MyoD-dependent genes in vivo and to determine the function of NFATc1 in skeletal muscle, we generated mice with nfatc1 loss-of-function in skeletal muscle (Figure 2A). Skeletal muscle-specific nfatc1 knockout mice (nfatc1SkMKO) were born at Mendelian frequency (Figure S3A) and survived to adulthood with no overt phenotype (data not shown). We observed greater than 80% reduction in nfatc1 mRNA levels in the soleus muscles of nfatc1SkMKO mice compared to nfatc1flox/+ (control) mice by qPCR and western blot analyses (Figure S3B and S3C). We did not observe any significant differences in growth, body mass, or muscle weight between control and nfatc1SkMKO mice (Figure S3D and S3E).
Figure 2. NFATc1 is required for normal fiber type composition, gene expression, and exercise-induced fiber type switching in vivo.
(A) Strategy used to generate skeletal muscle specific knockout of nfatc1. (B) qPCR analysis of MyoD target gene expression in the soleus muscles of adult control (wt) and nfatc1SkMKO (cko) mice. *, p < 0.05; **, p < 0.01. Data shown represent the mean ratio of expression in nfatc1SkMKO compared to wild type muscle plus SEM for 6 mice of each genotype. (C–E) Metachromatic ATPase staining of soleus muscles showed a higher percentage of slow fibers (dark blue) in control (wt, panels C, D) than in nfatc1SkMKO (cko, panel C′, D′) in the absence of exercise (no run; C, C′) or following 7 days of voluntary exercise (run; D, D′). Scale bars =100µm. (E) No run wt mice had a significantly higher percentage of slow fibers than unexercised nfatc1SkMKO mice (compare lanes 1 and 2, p < 0.01). Following exercise, the percentage of slow fibers in wt soleus increased from 45% to 50% (compare lanes 1 and 3, p < 0.05); the percentage of slow fibers in the soleus of nfatc1SkMKO mice showed no statistically significant difference (compare lanes 2 and 4). Data are presented as the mean percentage of slow fibers plus SEM for 6 mice in each group. (F) qPCR analysis of fast (light blue bars) and slow (dark blue bars) fiber gene expression in the soleus muscles of unexercised control (wt) and nfatc1SkMKO (cko) male mice. Data are shown as the mean ratio of expression in nfatc1SkMKO to wild type muscle plus SEM for 6 mice of each genotype. *, p < 0.05; n.s., not significant. (G) Western blot analysis of fast and slow myosin expression from wt (lanes 1, 2) and nfatc1SkMKO (cko; lanes 3, 4) soleus muscles. α-tubulin was examined as a loading control on the same cell lysates. See also Figure S2 and Figure S3.
Expression of MyoD-dependent genes is increased in the absence of NFATc1 in vivo
The observation that NFATc1 strongly repressed MyoD-dependent activation of the Mef2c, myogenin, and mrf4 promoters (Figure 1) prompted us to examine expression of the endogenous genes in nfatc1SkMKO and control mice (Figure 2B). Importantly, expression of Mef2c, mrf4, and myogenin was significantly increased in the absence of NFATc1 in vivo (Figure 2B). Other known MyoD target genes, including desmin, nestin, muscle creatine kinase (ckm) and the Myod1 gene itself, also showed significantly increased expression in the skeletal muscle of nfatc1SkMKO mice compared to controls (Figure 2B). These data support the idea that NFATc1 represses MyoD-dependent gene activation in vivo.
NFATc1 controls skeletal muscle fiber type composition and switching in response to exercise
Given the role of calcineurin-NFAT signaling in skeletal muscle fiber type regulation based on studies performed in cell culture model systems (Chin et al., 1998; Wu et al., 2000), we reasoned that mice lacking NFATc1 in skeletal muscle might have altered fiber type composition. To test this hypothesis, we used metachromatic ATPase staining to label slow and fast fibers in the soleus and found that nfatc1SkMKO mice had a lower percentage of slow fibers than control mice (Figures 2C and 2E; 34.5% versus 44.5%, p = 0.006). These findings support a role for NFATc1 in establishing or maintaining the slow fiber phenotype and provide the first genetic evidence for NFATc1 in the regulation of fiber type composition in vivo.
The conversion of fast fibers to slow fibers can be induced in mice in response to a continuous period of voluntary exercise (Pette, 1998). The mouse soleus muscle contains a significant percentage of slow fibers under normal conditions and is extensively studied as a model for understanding signals involved in fiber type regulation. Therefore, we examined the fiber type composition of the soleus muscle following a 7-day period of voluntary exercise. Control mice showed a significant increase in the percentage of slow fibers in response to exercise (Figure 2C,2D; also Figure 2E, compare lanes 1 and 3, p=0.019). In contrast, nfatc1SkMKO mice did not undergo fiber type switching in response to exercise (Figure 2C′,2D′; also Figure 2E, compare lanes 2 and 4; p=0.137). In spite of the reduced percentage of slow fibers in the soleus muscle, we observed no differences between nfatc1SkMKO and control mice in time or distance run (Figure S3F,G). We also found a significant increase in the expression of several fast fiber genes in nfatc1SkMKO soleus muscle compared to control muscle (Figure 2F, light blue bars). Increased expression of fast myosin in the soleus muscles nfatc1SkMKO compared to controls was also observed by western blot (Figure 2G). Interestingly, we did not observe significant changes in slow fiber gene expression at the transcript level (Figure 2F, dark blue bars), even though we observed a significant reduction in the percentage of slow fibers in nfatc1SkMKO muscles (Figures 2C and 2E) and in the amount of slow myosin by western blot (Figure 2G). The observation that slow myosin protein expression decreased while mRNA was not significantly changed suggests that posttranscriptional mechanisms may be involved in maintaining the appropriate myosin expression in muscle. Our observations also suggest the possibility that NFATc1 might function in skeletal muscle fiber type control by repressing expression of MyoD-dependent fast muscle genes.
NFATc1 physically interacts with MyoD
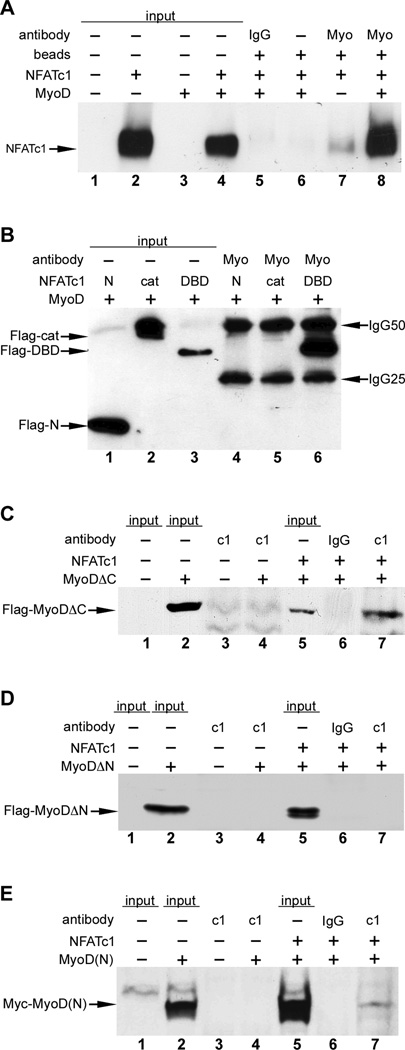
To gain further insight into the repression of MyoD activity by NFATc1, we tested whether NFATc1 physically interacts with MyoD (Figure 3). NFATc1 was efficiently co-immunoprecipitated by anti- MyoD antibody in cells expressing MyoD but not in control transfected cells (Figure 3A). We found that the DNA binding domain (DBD) of NFATc1 (REL domain) was necessary and sufficient for the physical interaction with MyoD (Figure 3B). Similarly, we examined the interaction of various domains of MyoD with NFATc1 (Figure 3C–E). MyoD truncations that contained the N-terminus and bHLH region but lacked the C terminus (ΔC) were efficiently immunoprecipitated by anti-NFATc1 antibody (Figure 3C). In contrast, MyoD truncations lacking the N-terminus (ΔN) failed to interact with NFATc1 (Figure 3D), suggesting that the MyoD N-terminus was required for interaction with NFATc1. Consistent with this notion, we found that the MyoD N-terminal activation domain alone (MyoD 1–101) was co-immunoprecipitated by anti-NFATc1 antibody (Figure 3E). These experiments demonstrate that the MyoD N-terminus is necessary and sufficient for interaction with NFATc1.
Figure 3. NFATc1 physically interacts with the N-terminus of MyoD.
C3H10T1/2 cells were transfected with the indicated plasmids, and lysates were analyzed by immunoprecipitation (IP)-western blot. Sample inputs, IgG antibody controls, and beads only controls are indicated. MyoD physically interacted with full length NFATc1 (panel A, lane 8). IP, anti-MyoD; western, anti-NFATc1. MyoD interacts with the DNA binding domain (DBD) of NFATc1 (panel B, lane 6). Expression plasmids for full length MyoD and Flag-tagged truncation fragments of NFATc1, encoding the N-terminus (N), catalytic domain (cat) or DBD were cotransfected. IP, anti-MyoD; western, anti-Flag. (C–E) Expression plasmids for full length NFATc1 and Flag-tagged MyoD lacking the C-terminus (C), Flag-tagged MyoD lacking the N-terminus (D) or a Myc-tagged N-terminal fragment of MyoD (E) were cotransfected. IP, anti-NFATc1; western, anti-Flag (panels C and D), anti-Myc (panel E). MyoDΔC was efficiently co25 immunoprecipitated by anti-NFATc1 (panel C, lane 7). The N-terminus of MyoD alone [MyoD(N)] was co-immunoprecipitated with NFATc1 (panel E, lane 7). MyoDΔN was not co-immunoprecipitated by anti-NFATc1 (panel D, lane 7).
NFATc1 inhibits MyoD transactivation
We considered several possible MyoD functions that might be inhibited by NFATc1. Notably, MyoD nuclear localization was unaffected by coexpression with either wild type or a constitutively active (cA) form of NFATc1 (Figure S4). Likewise, NFATc1 did not affect MyoD-E12 DNA binding in an in vitro electrophoretic mobility shift assay (EMSA), which also suggested that MyoD-E12 dimerization was unlikely to be affected by NFATc1 since dimerization is prerequisite to efficient DNA binding in EMSA (Figure S5).
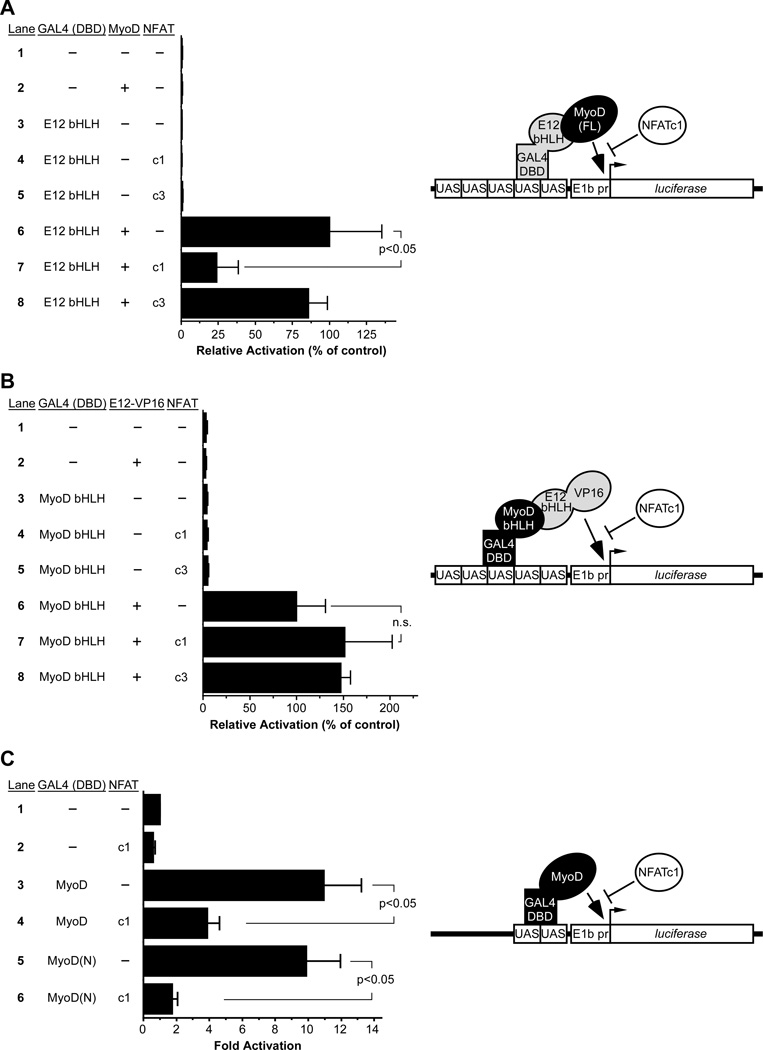
As an additional test of the effect of NFATc1 on MyoD-E protein function, we employed a modified mammalian two-hybrid assay in which the E12 bHLH domain (lacking a transactivation domain) was fused to the GAL4 DBD and was used as bait for recruitment and activation by full-length MyoD (Figure 4A, model). Co-expression of MyoD with GAL4-E12 bHLH resulted in robust activation of the GAL4-dependent UAS reporter due to MyoD-E12 bHLH dimerization and activation by MyoD (Figure 4A). NFATc1 co-expression potently inhibited the effect of MyoD on the reporter (Figure 4A, compare lanes 6 and 7), whereas NFATc3 had no effect (Figure 4A, lane 8). We also used GAL4-MyoD bHLH as bait and the E protein bHLH domain fused to the VP16 activation domain as prey (Figure 4B, model). GAL4-MyoD bHLH lacks a transactivation domain and therefore had no effect on reporter activation on its own, but co-expression of E12-VP16 resulted in potent activation (Figure 4B). Importantly, coexpression of NFATc1 had no effect on activation by E12-VP16 (Figure 4B, compare lanes 6 and 7), suggesting that NFATc1 does not inhibit MyoD-E protein dimerization. Taken together, the results in Figures 4A and 4B support the idea that NFATc1 inhibits transactivation by MyoD.
Figure 4. NFATc1 inhibits the MyoD N-terminal activation domain.
C3H10T1/2 cells were transfected with the indicated GAL4(DBD) fusion protein expression plasmids and the UAS reporter plasmid pG5E1b-luciferase. Full length MyoD, NFATc1 (c1), NFATc3 (c3), E12-VP16 or parental expression vector (dashes) were also co-transfected as indicated. (A) MyoD dimerization with GAL4- E12(bHLH) caused potent activation (lane 6), which was significantly inhibited by NFATc1 (lane 7). (B) E12-VP16-GAL4-MyoD(bHLH) dimers strongly activated the pG5E1b-luciferase reporter (lane 6) and this was not inhibited by NFATc1 (lane 7). (C) Fusion of full length MyoD or the N terminal fragment of MyoD [MyoD(N)] to the GAL4 DBD resulted in potent activation of the UAS reporter due to the strong activation domains of MyoD. Co-expression of NFATc1 significantly inhibited the MyoD activation domain (compare lanes 3, 4 and lanes 5, 6). Results are reported as the mean fold activation plus SEM from 4 independent transfections; n.s., not significant. Models depicting the mammalian two hybrid (A, B) and one-hybrid (C) assays are shown to the right of their respective graphs. See also Figure S4 and Figure S5.
As an explicit test of the effect of NFATc1 on MyoD-mediated transactivation, full-length MyoD was fused directly to the GAL4 DBD and activation of the UAS-dependent reporter was examined (Figure 4C, model). MyoD was sufficient to activate the reporter more than 10-fold, and coexpression of NFATc1 significantly inhibited activation of the reporter (Figure 4C, compare lanes 3 and 4). Importantly, the MyoD N-terminus, which encodes the major transactivation domain of MyoD (Berkes and Tapscott, 2005; Tapscott, 2005), was also sufficient to activate the reporter when fused to the GAL4 DBD, and NFATc1 co-expression significantly inhibited that activation (Figure 4C, compare lanes 5 and 6). This observation, combined with the observation that NFATc1 specifically interacted with the Nterminus of MyoD (Figure 3), suggests that a direct protein-protein inhibitory interaction accounts for the mechanism underlying NFATc1-mediated inhibition of MyoD transactivation.
NFATc1 disrupts MyoD interaction with p300
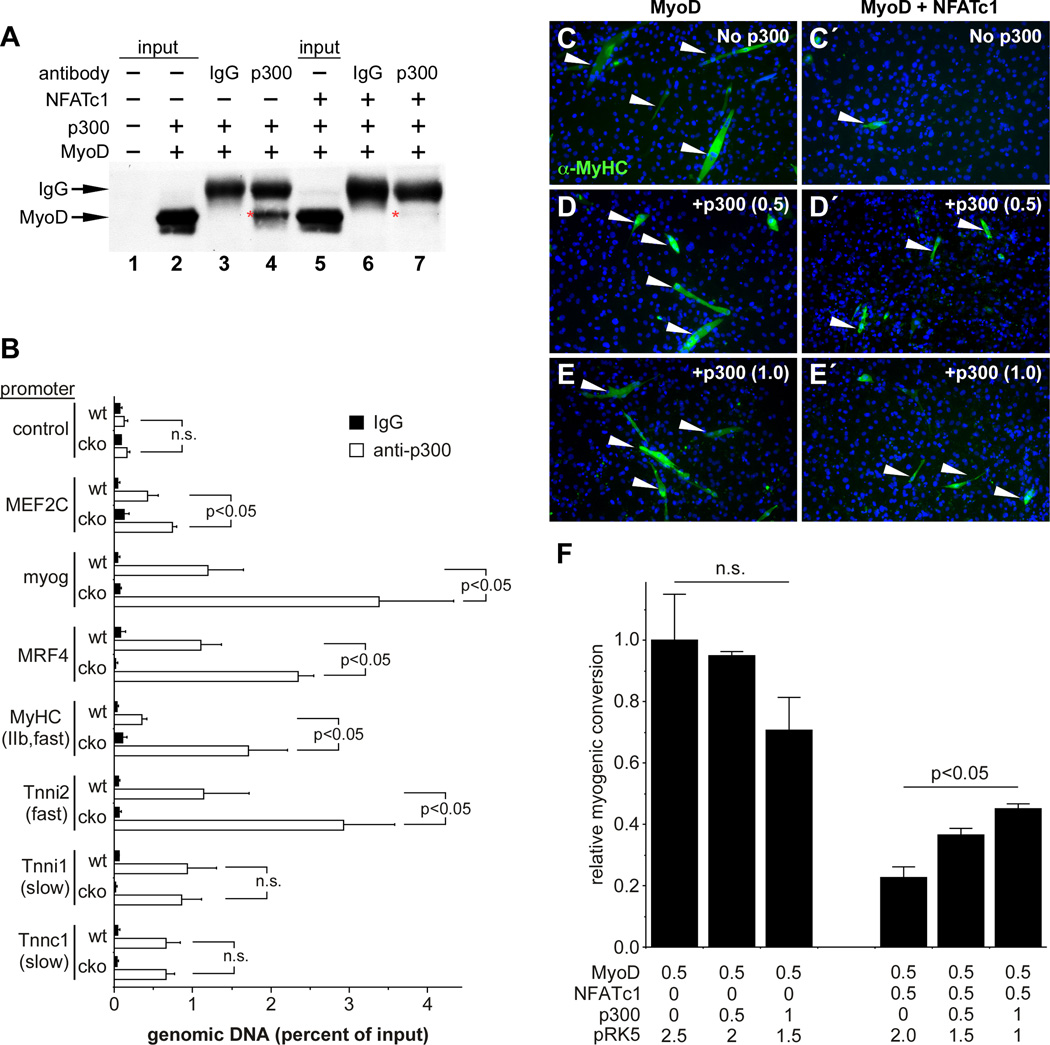
The N-terminus of MyoD interacts with the histone acetyltransferase p300, and that interaction is essential for MyoD-mediated conversion of fibroblasts into myotubes (Puri et al., 1997; Sartorelli et al., 1997). Therefore, we tested the hypothesis that NFATc1 might inhibit MyoD-p300 interaction (Figure 5A). Co-transfection of MyoD and p300 resulted in efficient co-immunoprecipitation of MyoD with anti-p300 antibody (Figure 5A, lane 4, red asterisk). Importantly, co-expression of NFATc1 inhibited p300-MyoD interaction (Figure 5A, lane 7, red asterisk).
Figure 5. NFATc1 disrupts MyoD interaction with p300.
C3H10T1/2 cells were co-transfected with MyoD and p300 plus or minus NFATc1, and lysates were analyzed by anti-p300 immunoprecipitation followed by anti-MyoD western blot (A), or cells were allowed to differentiate and were analyzed for myotube formation (C–E). (A) MyoD co-immunoprecipitates with p300 (lane 4, red asterisk); this interaction was inhibited by NFATc1 (lane 7, red asterisk). IgG isotype controls and sample inputs are indicated. (C–F) MyoD converted fibroblasts into multinucleated, MyHC+ (green staining) myotubes (white arrowheads) (C), which was potently inhibited by NFATc1 (C′). (D, E) Addition of p300 expression plasmid partially overcame the inhibitory effect of NFATc1 on MyoD (white arrowheads in D′, E′). The amount of p300 expression plasmid (µg) is indicated in parentheses. (F) Conversion to myotubes was quantified as the fraction of MY32+ cells in the MyoD alone control. MyHC+ cells were counted in 10 fields from 3 mice for each condition. Relative myogenic conversion plus standard deviation is indicated. The amount (µg) of each transfected expression plasmid is indicated. (B) ChIP-qPCR analyses for p300 occupancy of a negative control genomic region and the promoter regions of 7 genes from wt and cko soleus muscles, as indicated. Data from 3 independent experiments on different animals are shown as the percentage of input genomic DNA plus SEM. Enrichment for each promoter was calculated by the ΔCt method and normalized to input. Black bars, immunoprecipitation with isotype control IgG; white bars, immunoprecipitation with anti-p300 antibody. Data in B and F were analyzed by two-way ANOVA test with Bonferroni's multiple comparison post hoc analysis. n.s., not significant.
The inhibition of MyoD-p300 interaction by NFATc1 suggested the possibility that NFATc1 might be blocking recruitment of p300 to MyoD-dependent target genes in vivo. To test this idea, we examined p300 occupancy at MyoD-dependent targets, fast fiber genes, and slow fiber genes from wild type and nfatc1SkMKO soleus muscles (Figure 5B). Strikingly, p300 occupancy at the Mef2c, myogenin, and mrf4 promoters was higher in nfatc1SkMKO mice than in controls (Figure 5B). In addition, p300 occupancy was higher at fast fiber genes in the absence of NFATc1 but was unchanged at slow fiber genes (Figure 5B). These data support a role for NFATc1 in blocking MyoD recruitment of p300 in vivo.
Previous studies have suggested that p300 is rate limiting for MyoD-mediated conversion of fibroblasts to myotubes (Sartorelli et al., 1997). Therefore, we predicted that addition of excess p300 might be sufficient to rescue the NFATc1-mediated inhibition of MyoD. In the absence of p300, NFATc1 nearly completely inhibited MyoD-mediated conversion of C3H10T1/2 cells into myotubes (Figure 5C, C′). Remarkably, addition of p300 in the presence of MyoD and NFATc1 partially overcame the NFATc1- mediated inhibition of MyoD conversion (Figure 5D′–5E′). Quantification of the MyoD conversion in the presence and absence of excess p300 showed that statistically significant rescue of NFATc1 inhibition of MyoD occurred in a dose-dependent fashion (Figure 5F). Together, these data strongly support the notion that NFATc1 inhibits MyoD by disrupting interaction of p300 with the MyoD Nterminal transcriptional activation domain.
DISCUSSION
Regulation of skeletal muscle fiber type by NFATc1 inhibition of MyoD
NFAT proteins are major effectors of calcineurin signaling, and calcineurin-dependent transcriptional pathways are involved in the control of skeletal muscle fiber type (Chin et al., 1998; Naya et al., 2000; Parsons et al., 2004; Parsons et al., 2003). Several prior studies have supported a role for the NFAT family in the activation of slow fiber genes and concomitant repression of fast fiber genes in cell culture-based studies and through gain-of-function in vivo muscle electroporation studies (Calabria et al., 2009; McCullagh et al., 2004; Rana et al., 2008). Interestingly, delivery of a slow pattern of electrical activity mimicking endurance exercise to fast muscle fibers causes fast-to-slow fiber type switching (Pette, 1998; Schiaffino and Reggiani, 2011) and also results in the inactivation of MyoD (Ekmark et al., 2007). Given the opposing functions of MyoD and NFAT in the control of fast versus slow fiber gene expression in response to electrical stimulation (Calabria et al., 2009; Ekmark et al., 2007; McCullagh et al., 2004), and our findings that NFATc1 potently represses MyoD function, we hypothesize that NFATc1 promotes the slow fiber phenotype in part through repression of MyoD activity. Interestingly, all myogenic bHLH proteins contain an N-terminal activation domain but differ in whether or not they contain an N-terminal C/H domain (Berkes and Tapscott, 2005; Tapscott, 2005). Thus, it would be interesting to determine whether NFATc1 also interacts with other members of the MyoD family and whether these interactions influence gene expression or fiber type control.
Distinct roles for NFAT isoforms in skeletal muscle
In the studies here, we show clearly that NFATc1 blocks MyoD activity, whereas NFATc3 does not. We also show that NFATc1 physically associates with MyoD. Previous studies have suggested that NFATc3 also physically associates with MyoD and that this association promotes MyoD activity (Armand et al., 2008; Delling et al., 2000). In contrast, we did not observe cooperative activation of the myogenin promoter by NFATc3 and MyoD (Figure 1), nor did we observe augmentation of MyoD activity in the myotube conversion assay by NFATc3 (data not shown). Together, these observations suggest that the physical interaction of MyoD with NFATc1 versus NFATc3 likely results in distinct coactivator and corepressor interactions by different NFAT family members. Our studies also suggest that NFATc1 maybe the primary NFAT isoform downstream of calcineurin responsible for regulating skeletal muscle fiber type since loss-of-function studies for the other nfat genes found no defects in skeletal muscle fiber type (Graef et al., 2001; Horsley et al., 2001; Kegley et al., 2001).
Implications for NFATc1 regulation of MyoD in sarcopenia and muscle atrophy
Slow Type I fibers remain relatively constant in size with age, whereas fast Type II muscle fiber size diminishes (Porter et al., 1995). The atrophy that occurs in fast muscle fibers during sarcopenia is believed to play a major role in the pathogenesis of frailty and the functional impairment that occurs with old age or extended bed rest (Evans, 2010). Basal expression of myogenic bHLH genes increases in proportion to the degree of sarcopenia in senescent muscle (Kim et al., 2005). Our findings that NFATc1 promotes the slow fiber phenotype and that nfatc1SkMKO mice have increased expression of myogenin, mrf4, and Myod1, suggest that NFATc1 might play a role in maintaining slow fibers in aging muscle. Thus, NFATc1 and its upstream and downstream regulatory pathways, including calcineurin signaling, might represent interesting therapeutic targets for modulating the balance of slow and fast fibers in afflictions such as muscle atrophy and sarcopenia.
EXPERIMENTAL PROCEDURES
Knockout and transgenic mice
mef2c-73k-Cre and nfatc1flox/flox mice have been described (Aliprantis et al., 2008; Heidt and Black, 2005). To generate nfatc1+/Δ mice, male nfatc1flox/flox mice were crossed to female Mef2c-AHF-Cre mice (Rojas et al., 2008). When crossed from the female, Mef2c-AHF-Cre functions as a universal deleter line, resulting in Cre-dependent recombination in all cells in all offspring (B. Black, unpublished observations). Voluntary exercise assays using 42-day old male mice were conducted as described previously (Jaehnig et al., 2006). All experiments using animals were reviewed and approved by the UCSF IACUC and complied with institutional and federal guidelines.
RNA extraction, real-time quantitative PCR (qPCR), and chromatin immunoprecipitation (ChIP)
Total RNA was prepared from mouse tissues using the RNeasy Fibrous Tissue Mini Kit (Qiagen). RNA was treated for 1 h at 37°C with DNaseI, followed by cDNA synthesis using the Omniscript RT kit (Qiagen). For ChIP, soleus muscles of 42-day old control and nfatc1SkMKO run mice were frozen, minced, and fixed with 1% formaldehyde in PBS for 10 min at 37°C. ChIP was performed as previously described (Lee et al., 2006). Immunoprecipitation was performed with rabbit α-Acetyl CBP/p300 (#4771, Cell Signaling) or control IgG (ab46540 ChIP grade, Abcam). Immunoprecipitated chromatin was eluted and reverse cross-linked overnight at 65°C. qPCR was performed using the MAXIMA SYBR Green kit (Fermentas) and a 7900HT Fast Real Time PCR System. The SDS 2.4 software package was used to extract raw data. Primer sequences are available on request.
Plasmids and mutagenesis
Plasmids Mef2c-73k-lacZ (mef2c-lacZ), Mgn184-luciferase, pG5E1b-luciferase, RK5-MyoD, GAL4(DBD)-E12bHLH, E12-VP16, GAL4(DBD)-MyoD, and GAL4(DBD)-MyoD(bHLH) have been described (Black et al., 1998; Dodou et al., 2003; Heidt et al., 2007). Plasmid Mef2c[−106]-73k-lacZ (no NFAT sites) contains nucleotides −106 to +105 relative to the Mef2c skeletal muscle transcriptional start site. Plasmid mrf4-lacZ was generated by cloning a previously described 390-bp region of the mouse mrf4 promoter (Black et al., 1995) into the promoterless vector pAUG-β-gal (Rojas et al., 2008). Mouse NFATc1, NFATc3, and NFATc4 cDNAs were subcloned into pRK5 to create expression plasmids. A 316bp fragment encoding the first 101 amino acids of mouse MyoD cDNA was cloned into vector PCS2+MT to create the MyoD N-terminal expression plasmid. Truncation and deletion mutants of MyoD and NFATc1 were generated by PCR and confirmed by sequencing; amplicons were digested and subcloned into pRK5-FLAG. The p300 cDNA was obtained from Addgene and subcloned into pRK5 for expression studies.
Cell culture, transfections, and reporter assays
Transfections were performed using Lipofectamine LTX (Invitrogen) with 3 µg plasmid DNA per 35 mm dish. Cells were incubated in DMEM with 10% fetal bovine serum for 16 h post-transfection and then were switched to DMEM with 2% horse serum for 5 days for myotube conversion assays or for 36 h for all other transfection experiments. Cellular extracts were normalized and assayed for β-galactosidase or luciferase activity as previously described (Heidt et al., 2007; Rojas et al., 2008). All transfections lacking an expression plasmid contained parental vector DNA such that each transfection had an equivalent amount of plasmid DNA.
Co-immunoprecipitation and western blotting
Cells or tissues were washed in 4°C PBS and then lysed and homogenized in either cracking buffer (835mM sucrose, 1.7% SDS, 170mM Tris-HCl [pH 8.8]) or RIPA buffer (50mM Tris [pH 7.4], 150mM NaCl, 2mM EDTA, 1% NP-40, 0.1% SDS) in the presence of a protease inhibitor cocktail (Roche) for tissue culture cells or muscle tissue, respectively. Lysates were incubated at 4°C overnight with primary antibody or isotype control and then for 2 h with 100µg Protein G Sepharose beads (Sigma). Samples were then subjected to SDS-PAGE and western blot using standard procedures. The following antibodies were used: rabbit α-MyoD antibody (sc-760, Santa Cruz), mouse anti-myosin fast (MY-32, Sigma, 1:1000), mouse anti-slow myosin (M8421, Sigma, 1:1000), mouse α-NFATc1 antibody (clone 7A6, BD Pharmingen, 1:1000), rabbit α-Acetyl CBP/p300 (#4771, Cell Signaling). For Flag and Myc tagged proteins, mouse α-Flag (M2, Sigma, 1:1000) and mouse α-myc (clone 9E10, Sigma, 1:1000) were used. Mouse anti-tubulin (DSHB, 1:500) was used as a loading control. HRP-conjugated antimouse (A4416, Sigma, 1:1000) was used as the secondary antibody.
Immunohistochemistry, immunofluorescence, and metachromatic ATPase staining
Immunohistochemical staining using the Vectastain Elite ABC kit (Vector Laboratories; PK-6102) and immunofluorescence were performed as described (Rojas et al., 2008). The following antibodies were used: rabbit anti-NFATc1 (sc-13033, Santa Cruz, 1:100), goat anti-MyoD (sc-31942, Santa Cruz, 1:100), mouse anti-slow myosin (M8421, Sigma, 1:250), and mouse anti-fast myosin (MY-32, Sigma, 1:250). Biotin-conjugated goat anti-rabbit (Molecular Probes, 1:300), Alexa Fluor 594 donkey antimouse (Molecular Probes, 1:500), Alexa Fluor 488 donkey anti-rabbit (Molecular Probes, 1:500), Alexa Fluor 594 donkey anti-goat (Molecular Probes, 1:500) and Alexa Fluor 647 chicken anti-mouse (Molecular Probes, 1:500) were used as secondary antibodies. Metachromatic ATPase staining with 0.1% toluidine blue was performed as described (Ogilvie and Feeback, 1990).
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. De Val and C. Anderson for assistance with these studies and O. Weiner, S. Coughlin, and A. Chawla for helpful discussions. We are grateful to C.P. Chang and L. Glimscher for providing mice and J. Molkentin and M. Ott for providing plasmids. B. C. is supported by AHA postdoctoral fellowship 14POST18620052. This work was supported by grants HL089707, HL064658, and DE019118 from the NIH to B.L.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.L.E. and B.C. designed and conducted experiments, analyzed data, and helped write the manuscript. B.L.B. conceived the study, designed experiments, interpreted results, and wrote the manuscript.
REFERENCES
- Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, Ostrowski MC, Olsen BR, Glimcher LH. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest. 2008;118:3775–3789. doi: 10.1172/JCI35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armand AS, Bourajjaj M, Martinez-Martinez S, el Azzouzi H, da Costa Martins PA, Hatzis P, Seidler T, Redondo JM, De Windt LJ. Cooperative synergy between NFAT and MyoD regulates myogenin expression and myogenesis. J Biol Chem. 2008;283:29004–29010. doi: 10.1074/jbc.M801297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Black BL, Martin JF, Olson EN. The mouse MRF4 promoter is trans-activated directly and indirectly by muscle-specific transcription factors. J Biol Chem. 1995;270:2889–2892. doi: 10.1074/jbc.270.7.2889. [DOI] [PubMed] [Google Scholar]
- Black BL, Molkentin JD, Olson EN. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol Cell Biol. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabria E, Ciciliot S, Moretti I, Garcia M, Picard A, Dyar KA, Pallafacchina G, Tothova J, Schiaffino S, Murgia M. NFAT isoforms control activity-dependent muscle fiber type specification. Proc Natl Acad Sci U S A. 2009;106:13335–13340. doi: 10.1073/pnas.0812911106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling U, Tureckova J, Lim HW, De Windt LJ, Rotwein P, Molkentin JD. A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol Cell Biol. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Xu SM, Black BL. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech Dev. 2003;120:1021–1032. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Cheng TC, Cserjesi P, Chakraborty T, Olson EN. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol Cell Biol. 1992;12:3665–3677. doi: 10.1128/mcb.12.9.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekmark M, Rana ZA, Stewart G, Hardie DG, Gundersen K. De-phosphorylation of MyoD is linking nerve-evoked activity to fast myosin heavy chain expression in rodent adult skeletal muscle. J Physiol. 2007;584:637–650. doi: 10.1113/jphysiol.2007.141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WJ. Skeletal muscle loss: cachexia, sarcopenia, and inactivity. Am J Clin Nutr. 2010;91:1123S–1127S. doi: 10.3945/ajcn.2010.28608A. [DOI] [PubMed] [Google Scholar]
- Graef IA, Chen F, Chen L, Kuo A, Crabtree GR. Signals transduced by Ca(2+)/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell. 2001;105:863–875. doi: 10.1016/s0092-8674(01)00396-8. [DOI] [PubMed] [Google Scholar]
- Heidt AB, Black BL. Transgenic mice that express Cre recombinase under control of a skeletal muscle-specific promoter from mef2c. Genesis. 2005;42:28–32. doi: 10.1002/gene.20123. [DOI] [PubMed] [Google Scholar]
- Heidt AB, Rojas A, Harris IS, Black BL. Determinants of myogenic specificity within MyoD are required for noncanonical E box binding. Mol Cell Biol. 2007;27:5910–5920. doi: 10.1128/MCB.01700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Horsley V, Friday BB, Matteson S, Kegley KM, Gephart J, Pavlath GK. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J Cell Biol. 2001;153:329–338. doi: 10.1083/jcb.153.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SM, Koishi K, Rudnicki M, Maggs AM. MyoD protein is differentially accumulated in fast and slow skeletal muscle fibres and required for normal fibre type balance in rodents. Mech Dev. 1997;61:151–163. doi: 10.1016/s0925-4773(96)00631-4. [DOI] [PubMed] [Google Scholar]
- Jaehnig EJ, Heidt AB, Greene SB, Cornelissen I, Black BL. Increased susceptibility to isoproterenol-induced cardiac hypertrophy and impaired weight gain in mice lacking the histidine-rich calcium-binding protein. Molecular and cellular biology. 2006;26:9315–9326. doi: 10.1128/MCB.00482-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegley KM, Gephart J, Warren GL, Pavlath GK. Altered primary myogenesis in NFATC3(−/−) mice leads to decreased muscle size in the adult. Dev Biol. 2001;232:115–126. doi: 10.1006/dbio.2001.0179. [DOI] [PubMed] [Google Scholar]
- Kim JS, Kosek DJ, Petrella JK, Cross JM, Bamman MM. Resting and load-induced levels of myogenic gene transcripts differ between older adults with demonstrable sarcopenia and young men and women. J Appl Physiol. 2005;99:2149–2158. doi: 10.1152/japplphysiol.00513.2005. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerlotz K, Smith HK. Role of MyoD in denervated, disused, and exercised muscle. Muscle & nerve. 2008;38:1087–1100. doi: 10.1002/mus.21087. [DOI] [PubMed] [Google Scholar]
- Macharia R, Otto A, Valasek P, Patel K. Neuromuscular junction morphology, fiber-type proportions, and satellite-cell proliferation rates are altered in MyoD(−/−) mice. Muscle Nerve. 2010;42:38–52. doi: 10.1002/mus.21637. [DOI] [PubMed] [Google Scholar]
- McCullagh KJ, Calabria E, Pallafacchina G, Ciciliot S, Serrano AL, Argentini C, Kalhovde JM, Lomo T, Schiaffino S. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc Natl Acad Sci U S A. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J Biol Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- Ogilvie RW, Feeback DL. A metachromatic dye-ATPase method for the simultaneous identification of skeletal muscle fiber types I, IIA, IIB and IIC. Stain Technol. 1990;65:231–241. doi: 10.3109/10520299009105613. [DOI] [PubMed] [Google Scholar]
- Parsons SA, Millay DP, Wilkins BJ, Bueno OF, Tsika GL, Neilson JR, Liberatore CM, Yutzey KE, Crabtree GR, Tsika RW, et al. Genetic loss of calcineurin blocks mechanical overload-induced skeletal muscle fiber type switching but not hypertrophy. J Biol Chem. 2004;279:26192–26200. doi: 10.1074/jbc.M313800200. [DOI] [PubMed] [Google Scholar]
- Parsons SA, Wilkins BJ, Bueno OF, Molkentin JD. Altered skeletal muscle phenotypes in calcineurin Aalpha and Abeta gene-targeted mice. Mol Cell Biol. 2003;23:4331–4343. doi: 10.1128/MCB.23.12.4331-4343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pette D. Training effects on the contractile apparatus. Acta Physiol Scand. 1998;162:367–376. doi: 10.1046/j.1365-201X.1998.0296e.x. [DOI] [PubMed] [Google Scholar]
- Porter MM, Vandervoort AA, Lexell J. Aging of human muscle: structure, function and adaptability. Scand J Med Sci Sports. 1995;5:129–142. doi: 10.1111/j.1600-0838.1995.tb00026.x. [DOI] [PubMed] [Google Scholar]
- Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana ZA, Gundersen K, Buonanno A. Activity-dependent repression of muscle genes by NFAT. Proc Natl Acad Sci U S A. 2008;105:5921–5926. doi: 10.1073/pnas.0801330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Molecular and cellular biology. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Valdez MR, McAnally J, Richardson J, Olson EN. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128:4623–4633. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.