Abstract
FST is one of the most frequently-used indices of genetic differentiation among groups. Though FST takes values between 0 and 1, authors going back to Wright have noted that under many circumstances, FST is constrained to be less than 1. Recently, we showed that at a genetic locus with an unspecified number of alleles, FST for two subpopulations is strictly bounded from above by functions of both the frequency of the most frequent allele (M) and the homozygosity of the total population (HT). In the two-subpopulation case, FST can equal one only when the frequency of the most frequent allele and the total homozygosity are 1/2. Here, we extend this work by deriving strict bounds on FST for two subpopulations when the number of alleles at the locus is specified to be I. We show that restricting to I alleles produces the same upper bound on FST over much of the allowable domain for M and HT, and we derive more restrictive bounds in the windows M ∈ [1/I, 1/(I − 1)) and HT ∈ [1/I, I/(I2 − 1)). These results extend our understanding of the behavior of FST in relation to other population-genetic statistics.
1. Introduction
Genetic differentiation among groups is a phenomenon of central importance in population genetics, informing inferences about selection, migration, and demography. FST, one of Wright’s (Wright, 1951) fixation indices, is perhaps the most frequently used measurement of genetic differentiation among groups. One reason for the popularity of FST is its theoretical richness. For example, FST can be interpreted as an index of the reduction in heterozygosity that accompanies population structure (Nei, 1987), as a proportion of variance in allelic types accounted for by population structure (Holsinger and Weir, 2009), or as an index comparing mean coalescence times within subpopulations to mean coalescence times within the whole population (Slatkin, 1991).
Though FST has interpretations in terms of several major frameworks in population genetics, there has been a strong temptation to view FST as a simple measurement of the degree of genetic differentiation among groups, with increasing values indicating increased differentiation. Indeed, Wright himself provided heuristic guidelines as to what ranges of FST values may be considered as representing “moderately great” or “very great” differentiation (Wright, 1978, p. 85), lending credence to the idea that FST can be interpreted without reference to allelic diversity at the locus or other properties of the allele frequencies used in its computation.
However, as many investigators have noted—with Wright first among them (Wright, 1978, p. 82)—FST measures a very specific form of genetic differentiation. Namely, FST measures the extent to which different subpopulations have progressed toward fixation on different alleles. When there are exactly two subpopulations and exactly two alleles with positive frequency, FST is maximized when the two subpopulations have fixed on different alleles and, as a result, share no alleles in common.
One of the challenges of interpreting FST is that FST is dependent on the within-subpopulation diversity and other properties of the allele frequencies at the loci for which it is calculated (Charlesworth, 1998; Nagylaki, 1998; Hedrick, 1999, 2005; Long and Kittles, 2003; Ryman and Leimar, 2008; Jost, 2008; Long, 2009; Meirmans and Hedrick, 2011; Maruki et al., 2012). Recently, we considered the relationship of FST to both the frequency of the most frequent allele, M, and the homozygosity of the total population, HT (Jakobsson et al., 2013). These two statistics capture important aspects of the allele frequencies and diversity of a locus, and their relationship to each other is well understood (Rosenberg and Jakobsson, 2008; Reddy and Rosenberg, 2012). We calculated the upper bound on FST as a function of M and as a function of HT when the number of alleles is left unspecified.
Here, we extend these results by deriving and reporting bounds on FST when the number of alleles is specified to be a fixed value I. The extension reported here parallels the specified-I extension by Reddy and Rosenberg (2012) to the unspecified-I work of Rosenberg and Jakobsson (2008) on the relationship between homozygosity and the frequency of the most frequent allele.
We begin by describing the framework we adopt for conceptualizing FST. Next, we derive strict bounds when the number of alleles is specified, first as a function of the frequency of the most frequent allele and then as a function of total homozygosity.
2. Model
Consider a polymorphic locus with up to I alleles (I ≥ 2) in a population with K subpopulations of equal size. The frequency of allele i in subpopulation k is pki. All allele frequencies are non-negative, and within each subpopulation, the allele frequencies sum to 1. That is, pki ≥ 0 for all k and i, and for each k, . The mean allele frequency across subpopulations for allele i is . We assume that the allele frequencies are the parametric values for the subpopulations under study. We do not consider estimation of the allele frequencies from samples, nor do we consider the evolutionary sources of the allele frequencies in each subpopulation.
We define the frequency M of the most frequent allele as the highest mean allele frequency across subpopulations. That is, M = max {p̅1, p̅2, …, p̅I}. It is possible that more than one allele has mean frequency M.
The homozygosity within subpopulation k is the sum of the squares of the allele frequencies within subpopulation k, . The mean homozygosity across subpopulations is
In contrast, the total homozygosity is the sum of the squares of the mean allele frequencies across subpopulations,
With I alleles, both HS and HT lie in [1/I, 1]. Note that the homozygosities within each subpopopulation are expectations for the proportion of homozygotes in the subpopulation under Hardy-Weinberg equilibrium, and HT is the expected fraction of homozygotes in the whole population if the total population were at Hardy-Weinberg equilibrium with no structure.
Nei (1973) considered a version of Wright’s FST termed GST. From here forward, we work with this formulation, calling it F,
| (1) |
We restrict our attention to the case of K = 2. Table 1 presents a summary of the notation used for the two-subpopulation case.
Table 1.
Notation for the two-subpopulation case.
| Allele | ||||||
|---|---|---|---|---|---|---|
| Subpopulation | 1 | 2 | … | I | Sum | Sum of Squares |
| 1 | p11 | p12 | … | p1I | 1 | H1 |
| 2 | p21 | p22 | … | p2I | 1 | H2 |
| Mean | p̅1 | p̅2 | … | p̅I | 1 | HT |
3. Bounds on F as a function of M
Our goal is to identify bounds on F in terms of the frequency of the most frequent allele M and the homozygosity of the total population HT when the number of alleles I is specified. When I is specified, we do not require that all I alleles have positive frequency in the total population; we merely forbid the presence of more than I alleles with positive frequency. For both M and HT, we first identify circumstances in which the bounds obtained by Jakobsson et al. (2013) for unspecified I hold strictly and circumstances in which new strict bounds are required.
3.1. Bounds on F in terms of M when I is left unspecified
We previously found that when there are two subpopulations of equal size and an unspecified number of alleles at the locus, F can only reach values near 1 when the frequency of the most frequent allele and total homozygosity are near 1/2 (Jakobsson et al., 2013). Specifically, in terms of the frequency of the most frequent allele, M, we have
| (2) |
| (3) |
| (4) |
3.2. Circumstances in which the unspecified-I bounds for F in terms of M apply strictly
When the number of alleles is unspecified—and therefore permitted to be arbitrarily large—F is bounded by the functions of M given in Eq. (2). Under what conditions do these bounds apply when the number of alleles is specified?
First, we note that the domain of M is restricted by I; M ∈ [1/I, 1]. Because the sum of the allele frequencies is 1 and M is the largest of these frequencies, M must be at least as great as the mean of the I frequencies, or 1/I.
Second, for any M allowed given the number of alleles I, the lower bound on F is always 0. To see this, pick a set of allele frequencies with a desired largest allele frequency M. Set the allele frequencies in both subpopulations to be equal to these values. In this case, HS = HT, and Eq. (1) shows that F = 0.
Third, we previously showed that for M ∈ [1/2, 1], it is possible to achieve the upper bound on F given in Eq. (4) with I = 2 alleles (Jakobsson et al., 2013, Eq. 7). Because our framework allows us to set some of the I allele frequencies to be 0 in both subpopulations, we can achieve the previously obtained upper bound on F with I > 2 alleles by setting I − 2 of the allele frequencies to zero in both subpopulations and then following the procedure of Jakobsson et al. (2013) for the remaining two alleles. That is, we set the allele frequencies of the two subpopulations to differ as much as possible, choosing either (p11, p21) = (1, 2 M − 1) or (p11, p21) = (2 M − 1, 1).
Similarly, when I > 2 and M ∈ [1/I, 1/2), we previously showed that the upper bound on F given in Eq. (3) can be achieved when for each subpopulation, there are exactly ⌈(2 M)−1⌉ alleles that have positive frequency in the subpopulation, all of which have frequencies of 0 in the other subpopulation (Jakobsson et al., 2013, Eq. 9). When there are two subpopulations, it is possible to have ⌈(2 M)−1⌉ distinct alleles in each subpopulation if
| (5) |
Because M ≥ 1/I, the maximum value that ⌈(2 M)−1⌉ can take with I alleles is ⌈I/2⌉. When I is even, ⌈I/2⌉ = I/2, implying that the condition in Eq. (5) is met. Thus, when the number of alleles I is even, the upper bound on F from Eq. (3) applies for M ∈ [1/I, 1/2). However, when I is odd, 2 ⌈I/2⌉ = I + 1 > I, and the condition is not always met. Indeed, the condition in Eq. (5) is only met when M ≥ 1/(I − 1), and it is not met when M ∈ [1/I, 1/(I − 1)).
Combining these conclusions, we can state that for even I, the bounds on F given in Eq. (2) apply strictly for all allowed values of M ∈ [1/I, 1]. When I is odd, the bounds on F from Eq. (2) apply strictly for M ∈ [1/(I − 1), 1]. For odd I and M ∈ [1/I, 1/(I − 1)), the lower bound on F is 0, and the upper bound on F from Eq. (3) cannot be achieved; this upper bound can therefore be tightened.
3.3. Bounds on F in terms of M when I is specified
We begin by stating our main results for the bounds on F in terms of M ∈ (0, 1) when the number of alleles, I, is specified to be an integer greater than or equal to 2. We then complete the proof, leaving many of the details for the appendices.
Theorem 1. Suppose that F is defined as in Eq. (1), M is the frequency of the most frequent allele at a locus, and I is the number of alleles at the locus. I is an integer, and I ≥ 2. If I is even, then
| (6) |
and if I is odd, then
| (7) |
where
| (8) |
| (9) |
Proof. We have already argued that the bounds on F in terms of M are the same as in the case of unspecified I when I is even or when I is odd and M ≥ 1/(I −1). It remains to prove that if I is odd and M ∈ [1/I, 1/(I − 1)), then F ≤ M/(2 − I M). The proof has four steps.
We show that for M ∈ [1/I, 1/(I − 1)), when F is at its maximum in terms of M, no more than one allele has positive frequency in both subpopulations (Appendix A).
We show that when M ∈ [1/I, 1/(I − 1)), each subpopulation has positive frequency for at least (I +1)/2 alleles. In conjunction with the result of step (A) and the fact that I is odd, this result implies that when F is maximized, each subpopulation has positive frequency for exactly (I +1)/2 alleles and exactly one allele has positive frequency in both subpopulations. We also show that the allele with positive frequency in both subpopulations is not the most frequent allele unless all alleles have the same frequency (Appendix B). Steps (A) and (B) allow us to write the allele frequencies in each subpopulation as shown in Table 2.
(A) and (B) reduce the I = 3 case to a single-variable optimization problem, which we solve directly to find that for I = 3 and M ∈ [1/3, 1/2), the maximum value of F is M/(2 − 3 M) (Appendix C).
For odd I ≥ 5, we show that when F is maximized, at least (I − 3)/2 alleles have frequency 2 M in subpopulation 1 and frequency 0 in subpopulation 2. Similarly, at least (I − 3)/2 of the remaining alleles have frequency 0 in subpopulation 1 and frequency 2 M in subpopulation 2. We then obtain the arrangement of allele frequencies shown in Table 3, from which we can directly solve the case of I ≥ 5 as a two-variable optimization problem in p1I and p2I. Doing so reveals that setting p1I = p2I = 1 − M (I − 1) and setting other allele frequencies as shown in Table 3 maximizes F as a function of M. For odd I ≥ 5 and M ∈ [1/I, 1/(I − 1)), the maximum value of F that results is M/(2 − IM) (Appendix D). This completes the proof.
Table 2.
Allele frequencies in each subpopulation when no more than one allele has positive frequency simultaneously in both subpopulations. (I − 1)/2 alleles have positive frequencies in subpopulation 1 but frequency zero in subpopulation 2, and another (I − 1)/2 alleles have positive frequencies in subpopulation
| Allele | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subpopulation | 1 | 2 | … | (I − 1)/2 | (I + 1)/2 | … | I − 1 | I | ||
| 1 | 2M | p12 | … | 0 | … | 0 | p1I | |||
| 2 | 0 | 0 | … | 0 | … | p2(I−1) | p2I | |||
| Mean | p̅1 | p̅2 | … | … | p̅(I−1) | p̅I | ||||
Table 3.
Allele frequencies in each subpopulation for maximizing F in terms of M. Using this arrangement, we can maximize F directly in terms of p1I and p2I. Exactly (I − 3)/2 columns have allele frequencies as in column 1, and another (I − 3)/2 columns have allele frequencies as in column (I + 1)/2.
| Allele | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subpopulation | 1 | … | (I − 1)/2 | (I + 1)/2 | … | I − 1 | I | ||
| 1 | 2M | … | 0 | … | 0 | p1I | |||
| 2 | 0 | … | 0 | 2M | … | p2I | |||
| Mean | M | … | M | … | (p1I + p2I)/2 | ||||
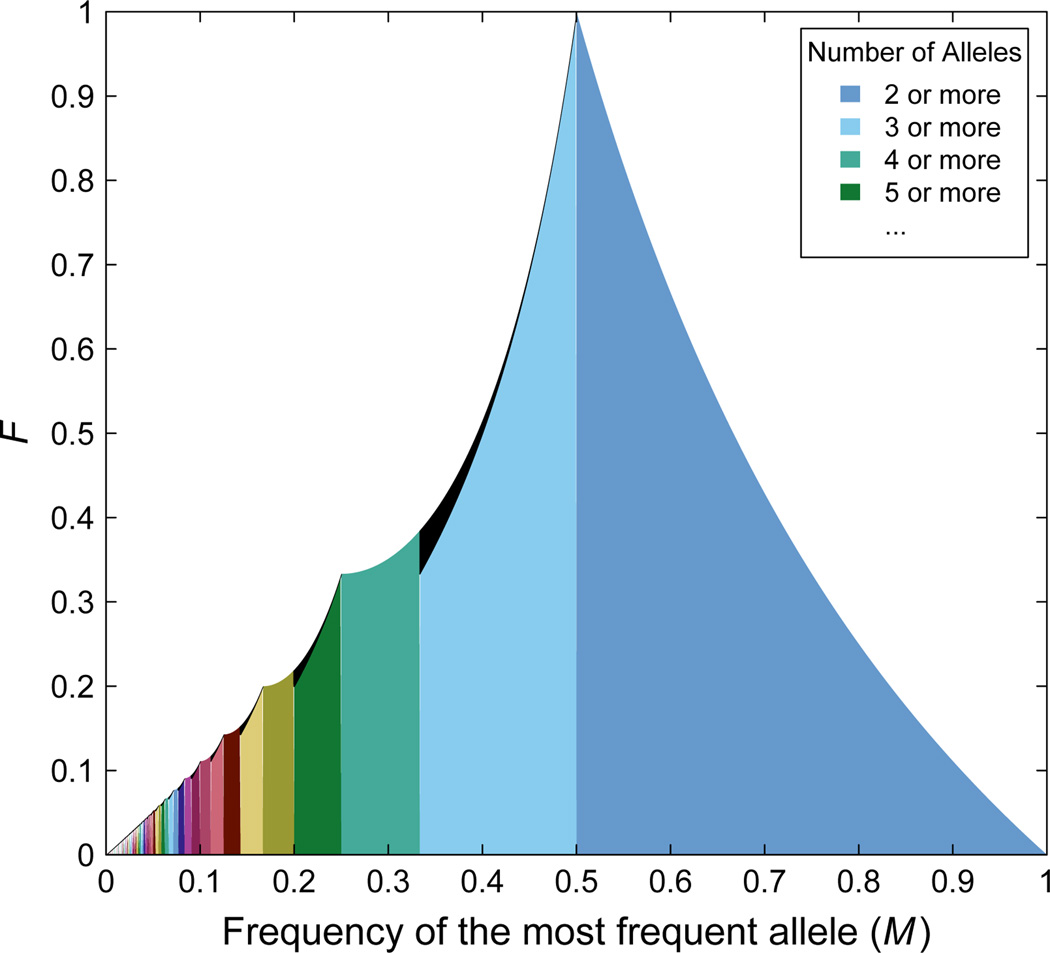
Figure 1 shows the upper bound on F as a function of M for specified I. The figure shows that limiting to a specified number of alleles I has important effects on the allowable domain of M. In addition, when I is odd, the maximum value of F for M ∈ [1/I, 1/(I − 1)) is lower than when I is unspecified, particularly when I is small. If I is odd and M = 1/I, then F ≤ 1/I. Thus, the bottom-left extrema of the black regions fall on the line F = M. The total area of the black regions in Figure 1 between the arbitary-I and fixed-I upper bounds, representing parts of the space accessible when I is unspecified but no longer accessible when I is specified, is approximately 0.002971 (Appendix E). The total area of all shaded regions, representing the mean maximal value of F over the unit interval for M in the unspecified-I case, is approximately 0.358538 (Jakobsson et al., 2013).
Figure 1.
The upper bound on F as a function of the frequency of the most frequent allele M. The differently colored vertical bands represent (M, F) pairs that become possible as the number of alleles at the locus increases; the vertical bands stretch horizontally from M = 1/I to M = 1/(I − 1) for I ∈ {2, 3, 4, …}. The regions colored in black that stretch horizontally from M = 1/I to M = 1/(I − 1) represent (M, F) pairs that are not allowed when the number of alleles is I but are achievable when the number of alleles increases. In other words, when the number of alleles is I, the colored regions from M = 1/I to M = 1 represent allowed (M, F) pairs, as do any black regions to the right of M = 1/(I − 1). For M ∈ [1/I, 1/(I − 1)) and I even, the upper bound is computed from Eq. (6). For M ∈ [1/I, 1/(I − 1)) and I odd, the black region stretches from the curve given in Eq. (7) to the curve given in Eq. (6). The lower bound on F is 0 for all values of M.
4. Bounds on F as a function of HT
To find bounds on F in terms of HT when I is specified, we follow an argument that is similar in structure to the one we used to find bounds on F in terms of M. We begin by identifying the cases in which the arbitrary-I bounds are not strict when I is specified. Once these cases are identified, we make arguments to reduce the number of variables before proceeding to direct optimization.
4.1. Bounds on F in terms of HT when I is left unspecified
We previously showed that when there are two subpopulations of the same size and an unspecified number of alleles at the locus, F is constrained by the homozygosity of the total population at the locus (Jakobsson et al., 2013). Specifically, in terms of the homozygosity of the total population, HT, where
| (10) |
| (11) |
| (12) |
4.2. Circumstances in which the unspecified-I bounds for F in terms of HT apply strictly
Just as with M, the domain of HT is restricted by the number of alleles, HT ∈ [1/I, 1] (Reddy and Rosenberg, 2012, Lemma 4). As stated above, the lower bound on F is 0 for any choice of allele frequencies for the total population and thus for any HT.
For HT ≥ 1/2, we have shown elsewhere that the upper bound on F given in Eq. (12) can be achieved with I ≥ 2 by setting and p1i = p2i = 0 for all i > 2 (Jakobsson et al., 2013).
For HT < 1/2, comparison of Eq. (A.3) and Eq. (11) shows that F achieves its upper bound in terms of HT when . For even I, we can achieve the upper bound on F when HT = 1/I by setting I/2 alleles to have frequency 2/I in subpopulation 1 and setting the other I/2 alleles to have frequency 2/I in subpopulation 2. In this case, HT = 1/I, , and F = 1/(I − 1) = HT/(1 − HT), which is the arbitrary-I upper bound for HT ∈ (0, 1/2). Further, Theorem 1ii of Rosenberg and Jakobsson (2008) guarantees that we can specify a set of ⌈H−1⌉ alleles to have homozygosity H. Because , setting I/2 alleles to give H1 = 2 HT in subpopulation 1, setting I/2 alleles to have homozygosity H2 = 2 HT in subpopulation 2, and setting no alleles to have positive frequency in both subpopulations simultaneously will achieve the upper bound on F from Eq. (11) for all HT ∈ [1/I, 1/2).
For odd I, the upper bound on F from Eq. (11) can be achieved when HT = I/(I2 − 1) by setting (I+1)/2 alleles to have frequency 2/(I + 1) in one subpopulation and setting the other (I − 1)/2 alleles to have frequency 2/(I − 1) in the other subpopulation. In this case, HT = I/(I2 − 1), , and F = I/(I2 − I − 1) = HT /(1 − HT), which is the upper bound from Eq. (11). Further, the upper bound on F can be achieved for HT ∈ [I/(I2−1), 1/(I − 1)) by setting H1 = 2/(I − 1) using (I − 1)/2 alleles and setting H2 = 4 HT − 2/(I − 1) using (I + 1)/2 alleles, with no alleles simultaneously having positive frequency in both subpopulations. For HT ∈ [I/(I2 − 1), 1/(I − 1)), H2 ∈ [2/(I + 1), 2/(I − 1)). This range of H2 values requires alleles, which is exactly the number of alleles we can set to have positive values in subpopulation 2.
For odd I and HT ∈ [1/(I − 1), 1/2), we can use only I − 1 of the alleles and the approach outlined above for even numbers of alleles to achieve the upper bound in Eq. (11). That is, for odd I and HT ∈ [1/(I − 1), 1/2), we can obtain H1 = 2 HT using (I − 1)/2 alleles and H2 = 2 HT using (I − 1)/2 other alleles, so that only I − 1 of the I available alleles have nonzero frequency (each in exactly one subpopulation).
Combining these results, we can confirm that for HT ∈ [1/I, 1), the bounds on F in terms of HT from Eq. (10) apply strictly when I is specified except when I is odd and HT ∈ [1/I, I/(I2 − 1)), in which case the strict upper bound on F remains to be determined. To find the upper bound on F in this region, we follow an argument similar to the one we used for M, reducing the number of variables as much as possible before attempting the optimization.
4.3. Bounds on F in terms of HT when I is specified
We state our main results for the bounds on F in terms of HT when the number of alleles, I, is specified to be an integer greater than or equal to 2. We then outline the proof, again leaving many of the details to the appendices.
Theorem 2. Suppose that F is defined as in Eq. (1), HT is the homozygosity of the total population at a locus, and I is the number of alleles at the locus. I is an integer, and I ≥ 2. If I is even, then
| (13) |
and if I is odd, then
| (14) |
where
| (15) |
| (16) |
| (17) |
| (18) |
Proof. We have already shown that the bounds on F in terms of HT are the same in the specified-I case as in the unspecified-I case of Jakobsson et al. (2013) when I is even or when I is odd and HT ≥ I/(I2 − 1). It remains to show that when I is odd and HT ∈ [1/I, I/(I2 − 1)), the upper bound on F is as shown in Theorem 2, Eqs. (15) and (16). The proof has four steps.
We proved in Appendix A that for all possible sets of population-level allele frequencies with M ≤ 1/2, the maximum F is achieved when no more than one allele has positive frequency in both subpopulations. If HT ∈ [1/I, I/(I2 − 1)) for I ≥ 3, then M < 1/2, so we can again exclude possible solutions in which more than one allele has positive frequency in both subpopulations.
We prove in Appendix F that when HT ∈ [1/I, I/(I2 − 1)) and F is maximized in terms of HT, each subpopulation must have positive frequency for exactly (I + 1)/2 alleles, counting the allele for which both subpopulations are allowed to have positive frequency, which we label allele I. This gives us the arrangement in Table 2, but because we are not currently considering M, we replace the 2 M in the first row and column with p11.
We show that the arrangement of allele frequencies can be updated to the one in Table 4. That is, we show that if F is maximized in terms of HT, I is odd, and HT ∈ [1/I, I/(I2 − 1)), then (I − 1)/2 alleles have a shared positive frequency in subpopulation 1 and frequency 0 in subpopulation 2 and another (I − 1)/2 alleles have a (possibly distinct) shared positive frequency in subpopulation 2 and frequency 0 in subpopulation 1. We write these shared frequencies in terms of the frequencies of allele I in the two subpopulations, where allele I is the allele that has positive frequency in both subpopulations. The subpopulation allele frequencies of allele I are p1I and p2I. We further show that the value of p2I that maximizes F while keeping HT fixed can be written as a function of p1I. We call this maximizing value . Using and the arrangement in Table 4, . Thus, maximizing F in terms of HT is equivalent to minimizing the product , a function of a single variable, p1I (Appendix G).
- We give the details of the minimization of in Appendix H. Completing the optimization reveals that the range with which we are concerned, HT ∈ [1/I, I/(I2 − 1)), must be split into two segments, [1/I, (I2 + I − 1)/(I3 + I2 − I − 1)) and [(I2 + I − 1)/(I3 + I2 − I − 1), I/(I2 − 1)). For HT ∈ [1/I, (I2 + I − 1)/(I3 + I2 − I − 1)), the maximum F is achieved by setting
This gives the inequality F ≤ U (HT), with U (HT) as in Eq. (15).(19)
Table 4.
Allele frequencies for maximizing F in terms of HT. Exactly (I − 1)/2 columns in the table have frequencies identical to those shown in column 1, and exactly (I − 1)/2 columns have frequencies identical to those shown in column (I + 1)/2.
| Allele | |||||||
|---|---|---|---|---|---|---|---|
| Subpopulation | 1 | … | (I + 1)/2 | … | I | ||
| 1 | … | 0 | … | p1I | |||
| 2 | 0 | … | … | ||||
| Mean | p̅1 | … | p̅(I + 1)/2 | … | p̅I | ||
For HT ∈ [(I2 + I − 1)/(I3 + I2 − I − 1), I/(I2 − 1)), the maximum F is achieved by setting
| (20) |
and
| (21) |
or by switching these assignments and setting p1I to equal the expression on the right side of Eq. (21) and setting p2I to equal the expression on the right side of Eq. (20). This gives the inequality F ≤ u(HT), with u(HT) as in Eq. (16). This completes the proof of Theorem 2.
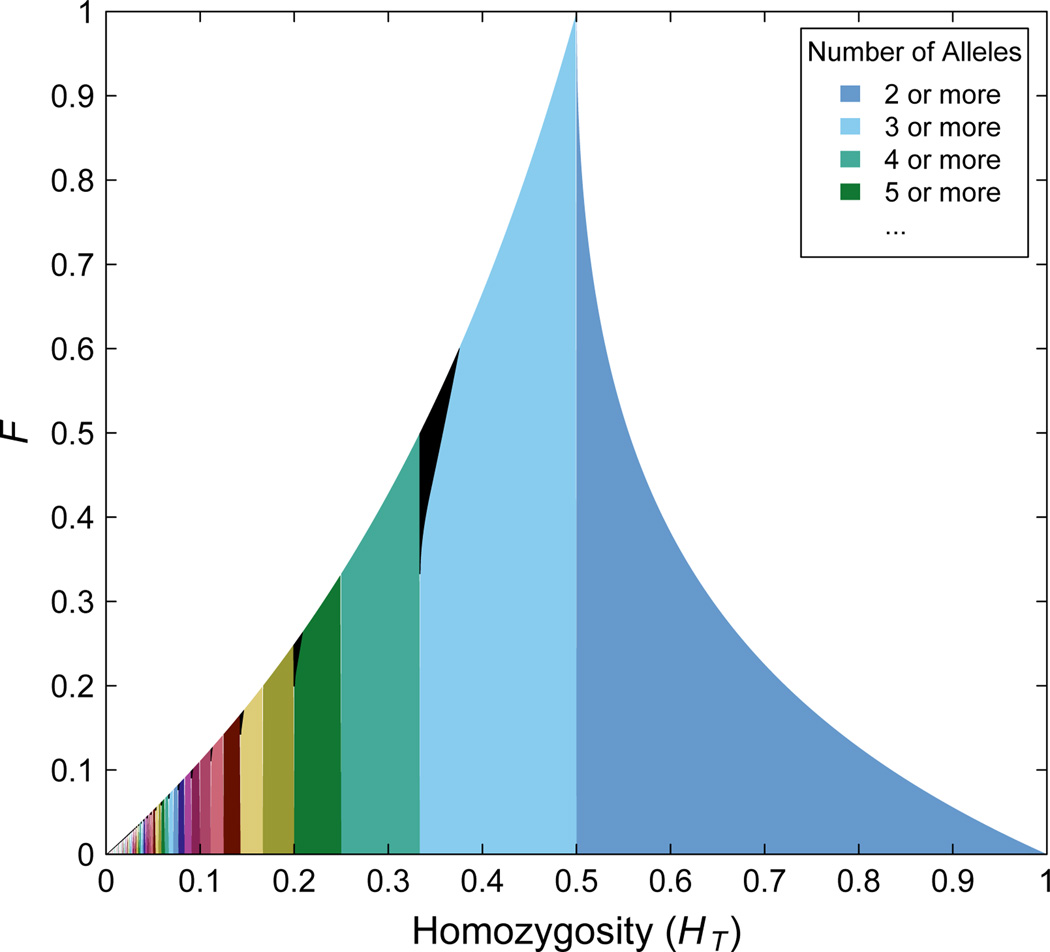
Figure 2 shows the upper bound on F as a function of HT for specified I. As in the case of M, limiting to a specified number of alleles I has important effects on the domain of HT. When I is odd, the maximum value of F for HT ∈ [1/I, I/(I2 − 1)) is lower than when I is unspecified. Analogously to the case of M, if I is odd and HT = 1/I, then F ≤ 1/I, which implies that the bottom-left extrema of the black regions in Figure 2 fall on the line F = HT. However, unlike in the case of M, in which a single function describes the upper bound on F in the interval M ∈ [1/I, 1/(I −1)), we can see that for odd I, the interval HT ∈ [1/I, 1/(I − 1)) is split into three components, one where U (HT) is the upper bound, a second where u(HT) is the upper bound, and a third where R(HT) is the upper bound.
Figure 2.
The upper bound on F as a function of the homozygosity of the total population HT. The differently colored vertical bands represent (HT, F) pairs that become possible as the number of alleles at the locus increases; the vertical bands stretch horizontally from HT = 1/I to HT = 1/(I − 1) for I ∈ {2, 3, 4, …}. The regions colored in black that stretch horizontally from HT = 1/I to HT = I/(I2 − 1) represent (HT, F) pairs that are not allowed when the number of alleles is I but are achievable when the number of alleles is larger than I. In other words, when the number of alleles is I, the colored regions from HT = 1/I to HT = 1 represent allowed (HT, F) pairs, as do any black regions where HT ≥ I/(I2 − 1). For HT ∈ [1/I, I/(I2 − 1)) and I even, the upper bound is computed from Eq. (13). For M ∈ [1/I, 1/(I − 1)) and odd I, the black region stretches from the curves given in Eq. (15) and Eq. (16) to the curve given in Eq. (17). Numerical integration reveals that the total area of the black regions between the arbitary-I and fixed-I upper bounds is ≈ 0.002955. The total area of the shaded regions is 1 − ln 2 ≈ 0.306853 (Jakobsson et al., 2013). The lower bound on F is 0 for all values of HT.
5. Discussion
We have extended the work of Jakobsson et al. (2013) by finding strict bounds on FST in terms of the frequency of the most frequent allele M and the homozygosity of the total population HT when the number of alleles I is specified. Specifying the number of alleles I restricts the domain of both the frequency of the most frequent allele and the homozygosity of the total population to the interval [1/I, 1) rather than the whole unit interval. In addition to this domain restriction, the upper bound on FST changes when the number of alleles is odd in a portion of the interval near its left endpoint. In particular, compared with the unspecified-I case, the upper bound on FST in terms of M changes for odd I and M ∈ [1/I, 1/(I − 1)), and the upper bound on FST in terms of HT changes for odd I and HT ∈ [1/I, I/(I2 − 1)). In the case of M, the width of the interval in which the upper bound changes is given by 1/[I(I − 1)], and the proportion of the domain on M for which the bound changes is 1/(I −1)2. In the case of HT, the upper bound changes for an interval of width 1/(I3 − I), which is 1/[(I − 1)2(I + 1)] as a proportion of the domain on HT. Thus, for M and especially for HT, the proportion of the space for which the upper bound on F changes when the number of alleles is specified becomes smaller as the number of alleles grows.
Our extension to the work of Jakobsson et al. (2013) is analogous to the extension of the results of Rosenberg and Jakobsson (2008) by Reddy and Rosenberg (2012). Rosenberg and Jakobsson (2008) determined the bounds on homozygosity in terms of the frequency of the most frequent allele when the number of alleles is left unspecified. Reddy and Rosenberg (2012) found that the bounds on the frequency of the most frequent allele in terms of the homozygosity of a single population are more constrained when the number of alleles is specified than when the number of alleles is left unspecified, especially for small numbers of alleles. Similarly, we find that the extent to which the bounds on FST in terms of the frequency of the most frequent allele and the homozygosity of the total population change decreases when the number of alleles increases. However, in contrast to Reddy and Rosenberg’s (2012) results, we find that the bounds on FST in terms of the frequency of the most frequent allele and the homozygosity of the total population only change shape relative to the case of an unspecified number of alleles when the number of alleles at the locus is odd.
One feature of the approach we have taken here and in other contexts (Rosenberg and Jakobsson, 2008; VanLiere and Rosenberg, 2008; Reddy and Rosenberg, 2012; Jakobsson et al., 2013) is that we have worked with parametric allele frequencies, considering population-genetic statistics as functions of sets of non-negative numbers constrained to sum to one rather than as outcomes of evolutionary processes. It has been pointed out that ultimately, the performance of population-genetic statistics in contexts of biological interest is what determines their usefulness. In particular, Rousset (2013) notes that “model-free” approaches like ours fail to identify the biological conditions under which FST calculations will produce biased results with respect to biological goals such as, for example, examining differences in coalescence times for different sets of lineages. We agree that studying the performance of FST and other proposed measures of population differentiation (Hedrick, 2005; Jost, 2008) under specific evolutionary models is necessary for fully articulating the effects of the mathematical properties of population-genetic statistics that we identify (Whitlock, 2011; Alcala et al., 2014). We would add another potential concern: we discuss the dependence of the parameter FST on properties of the allele frequencies, but estimators of FST also have properties that depend on locus allele frequencies, as demonstrated, for example, by Bhatia et al. (2013), who discussed the behavior of various estimators of FST in the presence of rare variants. At the same time, we hasten to note that the benefit of our parametric mathematical approach is that the results we identify hold under all possible population models that employ the statistics we study and define them in the same way. As such, our results are a starting point for studying the properties of population-genetic statistics in interesting biological scenarios and can help in the identification of biological contexts in which the mathematical properties we identify may be important. Further, they are available as a guide even when data analysts use FST to comment on applications and theoretical possibilities that fall outside the rich set of theoretically-motivated interpretations of FST.
Acknowledgments
We thank Lars Andersen and Ethan Jewett for discussions of this work. Financial support was provided by NIH R01HG005855 and a Stanford Graduate Fellowship.
Appendix A At maximum F, no more than one allele has positive frequency in both subpopulations
As a first step in finding the upper bound on F for odd I and M ∈ [1/I, 1/(I − 1)), we prove that for any set of population-level allele frequencies with M ≤ 1/2, the maximum value of F is achieved when no more than one allele simultaneously has positive frequency in both subpopulations.
Assume that there exist two alleles that both have positive frequency in both subpopulations. Call the alleles 1 and 2, and call the frequencies of alleles 1 and 2 in subpopulation 1 a and b. Call the frequencies of alleles 1 and 2 in subpopulation 2 c and d, as shown in Table A.5. Note that a + b ≤ 1 and c + d ≤ 1. Without loss of generality, assume that
| (A.1) |
| (A.2) |
That is, assume that we have labeled the alleles and subpopulations such that allele 1 has a mean frequency at least as great as allele 2 and such that allele 1 has frequency in subpopulation 1 at least as great as its frequency in subpopulation 2. The sums a + c and b + d are guaranteed to be less than or equal to 1 because M ≤ 1/2.
To maximize F, we use an expression from Jakobsson et al. (2013, Eq. 30). Noting that in the case of two subpopulations, , we can write
| (A.3) |
Because HT is a function of the mean (or total population) allele frequencies at the locus, an arrangement of the allele frequencies that keeps the mean allele frequencies the same for every allele but decreases will increase F. We will show that whenever there are two alleles with positive frequency in both subpopulations and mean allele frequencies less than or equal to 1/2, we can reduce but keep HT (and M) the same by replacing the allele frequencies at alleles 1 and 2 so that no more than one allele has positive frequency in both subpopulations.
To prove this claim, consider two cases. First, if b ≥ c, we rearrange frequencies in the way shown in the left side of Table A.6. We add c to a and d and subtract it from b. We are allowed to add c to a while still producing valid allele frequencies because a + c ≤ 1. Similarly, we can add c to d because c + d ≤ 1, and we can subtract c from b because b ≥ c, so b − c ≥ 0. Making these changes does not change the mean allele frequency for any allele, so HT does not change, nor does M. Thus, if ac + bd > (b − c)(d+c), then decreases as a result of the rearrangement, and F will increase. The inequality ac + bd > (b − c)(d + c) is equivalent to the inequality a + c > b −d. This inequality is guaranteed to be true because we assumed in Eq. (A.1) that a + c ≥ b + d and because d is positive. Thus, when b ≥ c, rearranging as in the left side of Table A.6 increases F.
Taking the second case of b < c, we rearrange in the way shown in the right side of Table A.6, adding b to a and d and subtracting it from c. Following reasoning similar to that used in the case of b ≥ c, we find that F increases if ac + bd > (a + b)(c − b). This inequality is equivalent to d + b > c − a. We assumed in Eq. (A.2) that a ≥ c, so because d + b > 0 and c − a ≤ 0, d + b > c − a. Thus, combining with the b ≥ c case, whenever M ≤ 1/2 and the two subpopulations have positive allele frequencies for more than one allele, F can be increased without changing M or HT by rearranging the subpopulation allele frequencies so that no more than one allele has positive frequency in both subpopulations.
This result allows us to eliminate candidates for maximum F in terms of M or HT in which more than one allele simultaneously has positive frequency in both subpopulations.
Table A.5.
Notation for a case with two or more shared alleles.
| Allele | |||||
|---|---|---|---|---|---|
| Subpopulation | 1 | 2 | … | ||
| 1 | a | b | … | ||
| 2 | c | d | … | ||
| Mean | … | ||||
| Product | ac | bd | … | ||
Table A.6.
A scheme for rearranging two shared alleles to get one shared allele and larger F.
| b ≥ c | b < c | |||||||
|---|---|---|---|---|---|---|---|---|
| Subpopulation | Allele 1 | Allele 2 | Allele 1 | Allele 2 | ||||
| 1 | a + c | b − c | a + b | 0 | ||||
| 2 | 0 | d + c | c − b | d + b | ||||
| Mean | ||||||||
| Product | 0 | (b − c)(d + c) | (a + b)(c − b) | 0 | ||||
Appendix B. At maximum F in terms of M, exactly one allele has positive frequency in both subpopulations, and it is not the most frequent
Assume that the single shared allele that is allowed to have positive frequency in both subpopulations is allele I. We can deduce three important facts from the results of Appendix A.
First, when I is odd and M ∈ [1/I, 1/(I − 1)), both subpopulations must have positive frequency for allele I. To prove this, assume without loss of generality that the number of alleles with positive frequency in subpopulation 2 is less than or equal to the number of alleles with positive frequency in subpopulation 1. If one subpopulation has an allele frequency of 0 for allele I, then subpopulation 2 can have positive frequency for at most (I − 1)/2 alleles. The most frequent allele in subpopulation 2 must then have an allele frequency of at least 2/(I − 1), which implies that the mean allele frequency for that allele must be at least 1/(I − 1). This means that M ≥ 1/(I − 1), which is outside the range with which we are concerned.
Second, taking the shared allele into account, it follows that each subpopulation must have positive frequency for exactly (I + 1)/2 alleles.
Third, if allele I is the allele for which both subpopulations are allowed to have positive frequency, then allele I is not the most frequent allele unless all alleles have the same frequency and M = 1/I. We prove this claim using a rearrangement strategy similar to the one we used in Appendix A. Label two alleles allele 1 and allele 2. Call the frequency of allele 1 in subpopulation 1 a, the frequency of allele 2 in subpopulation 1 b, the frequency of allele 1 in subpopulation 2 c, and let the frequency of allele 2 in subpopulation 2 be 0, as shown in the left side of Table B.7. Assume that b < a + c ≤ 2/(I − 1), with a + c ≤ 2/(I − 1) because M ≤ 1/(I − 1). Also, assume that excluding a from consideration, b is the largest allele frequency in subpopulation 1. Excluding allele 1, frequency equal to 1 − a must be spread over (I − 1)/2 alleles, so b ≥ 2(1 − a)/(I − 1). At the same time, c ≤ 2/(I − 1) − a. This guarantees that b ≥ c for the cases we are considering, because 2(1 − a)/(I − 1) ≥ 2/(I − 1) − a whenever I ≥ 3.
Because b ≥ c, we can rearrange the allele frequencies as shown in the right side of Table B.7, adding c to a, subtracting c from b, and switching the two alleles’ frequencies in subpopulation 2. This rearrangement does not change any of the mean allele frequencies and thus does not change M. The rearrangement will increase F if ac > (b − c)c. But this inequality is equivalent to b < a + c, which is what we assumed initially, so F does increase. Thus, as long as the mean allele frequencies are not the same for every allele, the most frequent allele will have positive frequency in only one subpopulation when F is maximized conditional on M. (If the mean frequencies are the same for every allele, then every mean allele frequency is equal to M, including the mean frequency of the shared allele.)
Thus, we can update the arrangement shown in Table 1 to the one shown in Table 2. For the remainder of the proof of Theorem 1, we assume that the shared allele that is allowed to have positive frequency in both subpopulations is allele I, and we assume without loss of generality that the most frequent allele is allele 1, which has positive frequency in subpopulation 1. We have reduced the number of variables from 2I − 3 to I − 2.
Table B.7.
The allele for which both subpopulations have positive frequency is not the most frequent allele unless all mean allele frequencies are equal.
| Start | Rearrangement | |||||||
|---|---|---|---|---|---|---|---|---|
| Subpopulation | Allele 1 | Allele 2 | Allele 1 | Allele 2 | ||||
| 1 | a | b | a + c | b − c | ||||
| 2 | c | 0 | 0 | c | ||||
| Mean | ||||||||
| Product | ac | 0 | 0 | (b − c)c | ||||
Appendix C. Upper bound on F in terms of M for I = 3 and M ∈ [1/3, 1/2)
The results of Appendix A and Appendix B allow us to solve directly the I = 3 case in terms of M as a single-variable optimization problem. When considering the I = 3 case, the structure specified in Table 2 gives the layout shown in Table C.8. Because M is fixed, only one allele frequency in subpopulation 2 is free to vary. Plugging the allele frequencies shown in Table C.8 into Eq. (A.3) gives
| (C.1) |
Obtaining the upper bound
Because M is the largest mean allele frequency allowed, p23 ∈ [1 − 2M, 4M − 1]. The constraint p23 ≥ 1 − 2M is found by noting that the mean frequency of allele 1, or M, must be greater than or equal to the mean frequency of allele 2, or (1 − p23)/2. The constraint p23 ≤ 4M − 1 arises from a similar argument comparing the frequencies of alleles 1 and 3.
To maximize F, we must consider p23 = 1 − 2M, p23 = 4M − 1, and any maxima of Eq. (C.1) with respect to p23 as candidate values for p23. Taking the derivative of Eq. (C.1) with respect to p23 and simplifying gives
| (C.2) |
The denominator of ∂F/∂p23 is non-negative and in fact is strictly positive for the values we consider, as it can only equal 0 when , a condition that generates values of p23 outside of [0, 1] when M ∈ [1/3, 1/2). The numerator is a concave-down quadratic function in p23. Thus, if the roots are real, then ∂F/p23 is positive between its roots. ∂F/p23 equals zero when
| (C.3) |
The larger of these two solutions is always greater than 1 because M ∈ [1/3, 1/2). Because ∂F/p23 is positive between its roots, the smaller solution represents a local minimum of F. As we seek to maximize F for p23 ∈ [1 − 2M, 4M − 1], we can ignore both of these solutions as candidates. The maximum value of F will occur when p23 is either as large or as small as possible; that is, when either p23 = 1 − 2M or p23 = 4M − 1.
When p23 = 1 − 2M,
| (C.4) |
and when p23 = 4M − 1,
| (C.5) |
Subtracting the right side of Eq. (C.5) from the right side of Eq. (C.4) gives
| (C.6) |
Table C.8.
Maximizing F when I = 3 and M ∈ .
| Allele | |||||
|---|---|---|---|---|---|
| Subpopulation | 1 | 2 | 3 | ||
| 1 | 2M | 0 | 1 − 2M | ||
| 2 | 0 | 1 − p23 | p23 | ||
| Mean | M | ||||
When the right side of Eq. (C.6) is non-negative, choosing p23 = 1 − 2M maximizes F. Both the numerator and denominator of the right side of Eq. (C.6) are concave-down quadratics in M and take positive values between their roots. The denominator is positive for M ∈ (0, 2/3), and the numerator is non-negative for M ∈ [1/3, 1/2]. Thus, for M ∈ [1/3, 1/2], the right side of Eq. (C.6) is non-negative, and setting p23 = 1 − 2M maximizes F. We can now state strict bounds on F in terms of M when I = 3:
| (C.7) |
The bound for 1/2 ≤ M < 1 comes from Eq. (4).
Appendix D. Upper bound on F in terms of M for odd I ≥ 5 and M ∈ [1/I, 1/(I − 1))
To maximize F for odd I ≥ 5 and M ∈ [1/I, 1/(I − 1)), we return to the situation of I − 2 variables described in Table 2. We will reduce the number of variables to 2 and then solve the optimization problem directly.
To reduce the number of variables, we make use of an expression for F from Jakobsson et al. (2013, Eq. 8),
| (D.1) |
Obtaining the upper bound
We assume that the allele for which both subpopulations are allowed to have positive frequency is allele I. Plugging in the allele frequency structure from Table 2 and defining and lets us write
| (D.2) |
Eq. (D.2) makes clear that conditional on p1I and p2I, F is maximized when and are maximized. and are sums of squares of non-negative numbers that add up to a fixed sum and that are each bounded above by a constant—2M in this case. Lemma 3 of Rosenberg and Jakobsson (2008) guarantees that such sums of squares are maximized by setting as many of the numbers as possible to be equal to the upper bound. In this case, that means setting as many alleles as possible to have frequency 2M. Within each subpopulation, when M ∈ [1/I, 1/(I − 1)), at least (I − 3)/2 alleles can be set to have frequency 2M. To see this, note that the allele frequencies in a subpopulation must sum to 1, so the number of alleles that can be set to frequency 2M is given by, in the case of subpopulation 1, ⌊(1 − p1I)/(2M)⌋. It follows that
| (D.3) |
The first step is true because p1I ≤ 2M, the second step because (1 − 2M)/(2M) is decreasing in M for M < 1/2 (and thus for M < 1/(I − 1) when I ≥ 3), and the third step because I is an odd integer, so (I − 3)/2 is an integer.
When we set (I − 3)/2 alleles in each subpopulation to have frequency 2M, we can update the arrangement in Table 2 to the one in Table 3. Plugging these allele frequencies into Eq. (D.1) gives a new expression for F,
| (D.4) |
where 2Hs is given by
| (D.5) |
With M fixed, all that remains is to pick p1I and p2I to maximize F. As in the three-allele case, we search for the largest values of F produced by choosing p1I and p2I to either be their maximum or minimum values or to be any local maxima occurring within their allowed ranges. We consider p1I first.
Taking the derivative of F with respect to p1I and simplifying gives
| (D.6) |
where
| (D.7) |
| (D.8) |
S(p1I, p2I, M) is a concave-down quadratic function in p1I, and s(p1I, p2I, M) is non-negative. Consequently, the equation ∂F/p1I = 0 has at most two real solutions. If ∂F/p1I = 0 has two real solutions, then ∂F/p1I will take positive values only in the interval between those solutions. Therefore, the larger solution will be a value of p1I at which F is locally maximized and the smaller solution will be a value of p1I at which F is locally minimized. (The roots of S, the numerator of ∂F/p1I, might not be roots of ∂F/p1I because s, the denominator, could equal zero at the same point. However, we show below that we can exclude the roots of S as candidate maxima of F for our purposes, regardless of the value of s.) The values of p1I that solve ∂F/p1I = 0 are
| (D.9) |
where
| (D.10) |
The larger of these two solutions for p1I is greater than 1—and therefore outside our allowed range for p1I—because p2I ∈ (0, 1). The smaller solution gives a local minimum, and we seek to maximize F. We can therefore ignore both solutions and simply compare the values of F given by the minimum and maximum allowed values of p1I.
The allele frequencies in subpopulation 1 must sum to one, and besides p1I, (I − 1)/2 alleles can have positive frequency of up to 2M each. Therefore, p1I ≥ 1 − M(I − 1). Because allele I cannot have mean frequency greater than M, p1I ≤ 2M − p2I.
Setting p1I = 1 − M(I − 1) in Eq. (D.4) gives
| (D.11) |
Similarly, setting p1I = 2M − p2I in Eq. (D.4) gives
| (D.12) |
Taking Fmin(p1I) − Fmax(p1I) and simplifying gives
| (D.13) |
Whenever the right side of Eq. (D.13) is non-negative, choosing p1I = 1 − M(I − 1) maximizes F. The numerator of the right side of Eq. (D.13) is a concave-down quadratic function in p2I with roots at p2I = 0 and p2I = (I + 1)M − 1. The denominator is a concave-down quadratic in p2I with roots at . The minimum value that p2I can take for M ∈ [1/I, 1/(I − 1)) is 1 − max(M)(I − 1) = 1 − (I − 1)/(I − 1) = 0. The maximum value that p2I can take for any allowed p1I is 2M − min(p1I) = 2M − [1 − (I − 1)M] = (I + 1)M − 1. Thus, for all allowed values of p2I, the numerator of the right side of Eq. (D.13) is non-negative. If the denominator is positive for allowed values of p2I, then choosing p1I = 1 − M(I − 1) maximizes F. The denominator is positive between its roots. Thus, choosing p2I = 1 − M (I − 1) maximizes F if (i) and (ii)
Condition (i) is true if:
| (D.14) |
If this interval contains the values of M for which we seek to maximize F, [1/I, 1/(I − 1)), then condition (i) holds. For I > 1, the lower bound of the interval specified by condition (i) is less than 0, as . Because 0 < 1/I for positive I, condition (i) holds if
| (D.15) |
This inequality is true when , which is true for all I > 2. Because we are only considering odd I ≥ 5, condition (i) is true.
Moreover, for M ∈ [1/I, 1/(I − 1)), the truth of condition (i) implies the truth of condition (ii). Condition (i) can be restated as , and condition (ii) can be restated as . Because M ≥ IM − 1 when M ≤ 1/(I − 1), condition (ii) is guaranteed to hold when condition (i) holds and M ≤ 1/(I − 1).
Thus, for odd I and M ∈ [1/I, 1/(I − 1)), choosing p1I = 1 − M (I − 1) and other subpopulation 1 allele frequencies as shown in Table 3 maximizes F as a function of p1I. Further, Eq. (D.4) is symmetric in (p1I, p2I), so analogous steps for p2I identify p2I = 1 − M (I − 1) as the choice that maximizes F as a function of p2I. Plugging 1 − M (I − 1) in for both p1I and p2I in Eq. (D.4) and simplifying gives the upper bound on F for odd I ≥ 5 and M ∈ [1/I, 1/(I − 1)),
| (D.16) |
Appendix E. The reduction in area under the upper bound on F in terms of M
To calculate the total area of the black regions in the Figure 1 representing parts of the space accessible when I is unspecified but not accessible when I is specified, we calculate the integral from 0 to 1/2 of the arbitrary-I upper bound on F minus the upper bound on F when I is specified. The integral of the arbitrary-I upper bound from 0 to 1/2 is
| (E.1) |
This expression comes from Jakobsson et al. 2013, Eq. 18, with the multiplication by 1/2 coming from the fact that Jakobsson et al. integrated a function of σ1 = 2M from 0 to 1 rather than integrating a function of M from 0 to 1/2. To calculate the integral of the specified-I upper bound on F, we start by summing the areas under the parts of the unspecified-I bounds that apply for even I from 4 to ∞. Modifying a result of Jakobsson et al. (2013, Eq. A1) and letting k = I/2 gives
| (E.2) |
To get this expression, we change the bounds of integration for the integral in Jakobsson et al. (2013, Eq. A1) such that we integrate over the regions corresponding to M ∈ [1/I, 1/(I − 1)) for even I ≥ 4. Because Jakobsson et al. integrated a function of σ = 2M, we multiply by 1/2 to get the corresponding integral for M. The first sum simplifies to 1 − 2 ln 2, and the second sum is evaluated numerically.
To complete the integral of the specified-I upper bound on F, we integrate M/(2 − IM), summing the definite integrals that result when integrating from 1/I to 1/(I − 1) and odd I ≥ 3:
| (E.3) |
Notice that the second term can be evaluated exactly, as
| (E.4) |
Numerically evaluating the expression that results when the expressions in Eq. (E.2) and Eq. (E.3) are subtracted from the expression in Eq. (E.1) reveals that the total area of the black regions between the arbitary-I and fixed I upper bounds is approximately 0.002971. The total area of all shaded regions is approximately 0.358538 (Jakobsson et al., 2013).
Appendix F. Exactly (I + 1)/2 alleles have positive frequency in each subpopulation when F is maximized in terms of HT
In this appendix, we are in the setting of odd I and HT ∈ [1/I, I/ (I2 − 1)). In Appendix A, we showed that when F is maximized in terms of HT, no more than one allele simultaneously has positive frequency in both subpopulations. Here, we prove that when there is no more than one allele for which both subpopulations have positive frequency, both subpopulations must have exactly (I + 1)/2 alleles with positive frequency.
Consider the situation depicted in Table F.9, which is modified from Table 4. We seek to prove that when only one allele is allowed to have positive frequency in both subpopulations and I is odd, then unless each subpopulation has positive frequency for exactly (I + 1)/2 alleles, HT ≥ I/(I2 − 1), which places HT outside the set of possibilities we are considering. We handle the I = 3 and I ≥ 5 cases separately. After dispensing with the I = 3 case directly, we prove our claim for I ≥ 5 by first minimizing HT and showing that if each subpopulation has positive frequency for exactly (I + 1)/2 alleles, then the minimum achievable value of HT is 1/I. Next, we show that when it is not the case that each subpopulation has positive frequency for exactly (I + 1)/2 alleles, the minimum achievable HT given that p1I and p2I are in the interval [0, 1] is I/(I2 − 1).
We designate the number of alleles that have positive frequency in subpopulation 1 but do not appear in subpopulation 2 by ℓ. We have arranged the allele frequencies in Table F.9 to minimize HT conditional on p1I, p2I, and ℓ, distributing the mass that remains in each subpopulation after accounting for allele I evenly over the alleles that remain accessible to that subpopulation (Reddy and Rosenberg, 2012).
Because the problem is symmetric in p1I and p2I, we can, without loss of generality, consider only values of ℓ ∈ {0, 1, …, (I − 1)/2}. Note that the number of alleles with positive frequency in subpopulation 1 is ℓ + 1 and that the number of alleles with positive frequency in subpopulation 2 is I − ℓ. Therefore, if among the candidate values of ℓ ∈ {0, 1, …, (I − 1)/2}, ℓ ≤ (I − 3)/2 implies HT ≥ I2/(I − 1), then each subpopulation must have positive frequency for exactly (I + 1)/2 alleles in order to achieve the HT values in [1/I, I/(I2 − 1)) that we consider for maximizing F.
When I = 3, HT ∈ [1/3, 3/8) only if ℓ = 1. To see this, note that if ℓ = 0, then p13 = 1, which implies p̅3 ≥ 1/2. M must be at least as large as p̅3, and when I = 3, M ≥ 1/2 implies HT ≥ 3/8 (Reddy and Rosenberg, 2012, Theorem 2). Symmetrically, if ℓ = 2, then p23 = 1, which again implies p̅3 ≥ 1/2 and HT ≥ 3/8. We cannot choose ℓ = 3
Table F.9.
Allele frequencies for minimizing HT conditional on p1I, p2I, and ℓ, where ℓ is the number of alleles that have positive frequency in subpopulation 1 but frequency 0 in subpopulation 2.
| Allele | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subpopulation | 1 | … | ℓ | ℓ + 1 | … | I − 1 | I | ||||
| 1 | … | 0 | … | 0 | p1I | ||||||
| 2 | 0 | … | 0 | … | p2I | ||||||
| Mean | … | … | p̄I | ||||||||
because at least one allele must have positive frequency in subpopulation 2. The only remaining choice is ℓ = 1, and indeed, choosing ℓ = 1, p11 = p22 = 2/3, p12 = p21 = 0, and p13 = p23 = 1/3 gives the minimum possible HT of 1/3. Thus, when I = 3, HT ∈ [1/I, I/(I2 − 1)) implies ℓ = (I − 1)/2.
We proceed to the case of I ≥ 5. The arrangement in Table F.9 gives
| (F.1) |
This function is a concave-up quadratic in p1I and p2I. As such, it will have exactly one critical point, and that point will be the global minimum.
The derivative of HT with respect to p1I is
| (F.2) |
Setting the derivative to zero gives p1I = (1 − ℓp2I)/(ℓ + 1), which minimizes HT with respect to p1I.
The derivative with respect to p2I is
| (F.3) |
Setting this derivative to zero gives p2I = [1 − (I − ℓ − 1)p1I]/(I − ℓ), which minimizes HT with respect to p2I.
Solving the system
| (F.4) |
| (F.5) |
for p1I and p2I gives
| (F.6) |
| (F.7) |
Because we consider p1I and p2I as allele frequencies, we can only achieve the global minimum when the expressions in Eq. (F.6) and Eq. (F.7) are in the interval [0, 1]. The expression in Eq. (F.6) is in [0, 1] only if ℓ ∈ [0, I/2], and the expression in Eq. (F.7) is in [0, 1] only if ℓ ∈ [(I − 2)/2, I − 1]. These conditions are both met when ℓ ∈ [(I − 2)/2, I/2]. When I is odd, the only integer in this range is (I − 1)/2. When ℓ = (I − 1)/2, the minimum HT achievable by the arrangement in Table F.9 is 1/I, which occurs when p1I = p2I = 1/I. We note that 1/I is also the minimum possible HT for any arrangement of I alleles.
Thus, setting the number of alleles with positive frequency in each subpopulation to ℓ + 1 = (I + 1)/2 allows the minimum value of HT to be achieved. It remains to show that if this is not the case—that is, if ℓ < (I − 1)/2—then HT ≥ I/(I2 − 1).
When ℓ < (I − 1)/2, we must check the minimum values of HT available on the endpoints of the allowed intervals for p1I and p2I, because the global minimum is not available. Because p1I and p2I are allele frequencies, they take values in [0, 1]. Thus, we consider three possibilities in turn: p1I = 1 or p2I = 1 (these two possibilities can be handled in one step), p1I = 0, and p2I = 0.
When p1I = 1 or p2I = 1, we can use an argument similar to the one we used for the I = 3 case. That is, setting either p1I = 1 or p2I = 1 implies p̅I ≥ 1/2. However, because HT is the sum of squares of the mean allele frequencies, . When I ≥ 5, I/(I2 − 1) < 1/4, so setting either p1I = 1 or p2I = 1 implies that HT > I/(I2 − 1). It remains to check the minimum possible values of HT when p1I = 0 or p2I = 0.
If p1I = 0, then HT is minimized by setting p2I = 1/(I − ℓ). Plugging these values into Eq. (F.1) and simplifying gives HT = I/[4ℓ(I − ℓ)]. For ℓ ∈ [0, (I − 3)/2], this function is decreasing in ℓ, so the smallest HT possible is at ℓ = (I − 3)/2. Plugging in ℓ = (I − 3)/2 gives HT = I/(I2 − 9) > I/(I2 − 1).
When p2I = 0, we minimize HT by setting p1I = 1/(ℓ + 1), and the minimum value of HT is I/[4(ℓ + 1)(I − ℓ − 1)]. For ℓ ∈ [0, (I − 3)/2], this function is decreasing in ℓ, so HT is minimized when ℓ = (I − 3)/2 and HT = I/(I2 − 1).
Combining these results shows that when ℓ ≤ (I − 3)/2, the minimum possible value of HT is I/(I2 − 1). Because we are concerned with HT ∈ [1/I, I/(I2 − 1)), we conclude that ℓ = (I − 1)/2. Setting ℓ = (I − 1)/2 implies that each subpopulation has positive frequency for exactly (I + 1)/2 alleles because the number of positive alleles in subpopulation 1 is ℓ + 1 and the number of positive alleles in subpopulation 2 is I − ℓ. This is what we sought to prove.
Appendix G. Reducing the maximization of F in terms of HT to a single-variable optimization
In this appendix, we are in the setting of odd I, HT ∈ [1/I, I/(I2 − 1)), and only one allele for which both subpopulations simultaneously have positive frequency. Our goal is to reduce the maximization of F in terms of HT to a single-variable maximization problem. When allele I is the only allele that has positive frequency in both subpopulations, maximizing F with respect to HT is equivalent to minimizing the product p1Ip2I while keeping HT fixed (Eq. A.3). With the allele frequencies arranged as specified in Table 2, replacing 2M with p1I,
| (G.1) |
Conditional on p1I and p2I and the allele-frequency arrangement specified, H1 is minimized by spreading the available mass in subpopulation 1, given by 1 − p1I, evenly over the remaining (I − 1)/2 alleles that are allowed to be positive (Reddy and Rosenberg, 2012, Lemma 3). Applying the same reasoning to H2 and plugging into Eq. (G.1) gives the inequality
| (G.2) |
Conditional on p1I and HT, equality is achieved when
| (G.3) |
Because the right side of the inequality in (G.2) is a concave-up quadratic in p2I, conditional on HT and p1I, p2I falls in the closed interval bounded by the two values on the right side of Eq. (G.3). Because we seek to minimize p1Ip2I with both p1I and p2I non-negative, we need to choose p2I to be the smallest allowed value given p1I and HT, which is either the smaller value on the right side of Eq. (G.3) or 0. However, by symmetry, choosing p2I = 0 implies
| (G.4) |
The bounds of this interval are only real when HT ≥ I/(I2 − 1), which is outside the range we are considering. As a result, we can choose p2I to be
| (G.5) |
in order to maximize F. We label the value of p2I that maximizes F as . The arrangement of allele frequencies in this scheme appears in Table 4.
Thus, for odd I and HT ∈ [1/I, I/(I2 − 1)), maximizing F is equivalent to minimizing
| (G.6) |
where is the function of p1I defined in Eq. (G.5).
Appendix H. Obtaining the upper bound on F in terms of HT by minimizing
In Appendix G, we showed that for HT ∈ [1/I, I/(I2 − 1)) and odd I, maximizing F in terms of HT is equivalent to minimizing a quantity that we label A. , where is given in Eq. (G.5). Here, we minimize A.
Appendix H.1. A geometric view
We consider a geometric approach to the problem in order to build intuition. Let us revisit some material covered differently in Appendix G.
Assume that we start with the arrangement of allele frequencies shown in Table 4 but that we have not yet defined , so where appears in Table 4, we have the variable p2I. Given an odd number of alleles I and a homozygosity HT ∈ [1/I, I/(I2 −1)), p1I and p2I can only take certain values. The values that p2I can take are in the closed interval bounded by the two expressions on the right side of Eq. (G.3), as argued in Appendix G. That is,
| (H.1) |
At the same time, p1I can only take values that lead to real-valued bounds on p2I. That is, we must choose p1I such that . Choosing
| (H.2) |
satisfies this inequality.
Figure H.3 shows (p1I, p2I) values allowed for I = 5 and four specific values of HT ∈ [1/I, I/(I2 − 1)). For any odd I and HT ∈ [1/I, I/(I2 − 1)), the region of allowed (p1I, p2I) values is symmetric around the p1I = p2I line. Given the allele-frequency arrangement in Table 4, the problem of maximizing F given H ∈ [1/I, I/(I2 −1)) is solved when the product p1Ip2I is minimized. This product can be visualized as the area of a rectangle with one vertex at the origin, two sides that stretch along the axes, and an upper-right vertex required to be in the allowed region of (p1I, p2I).
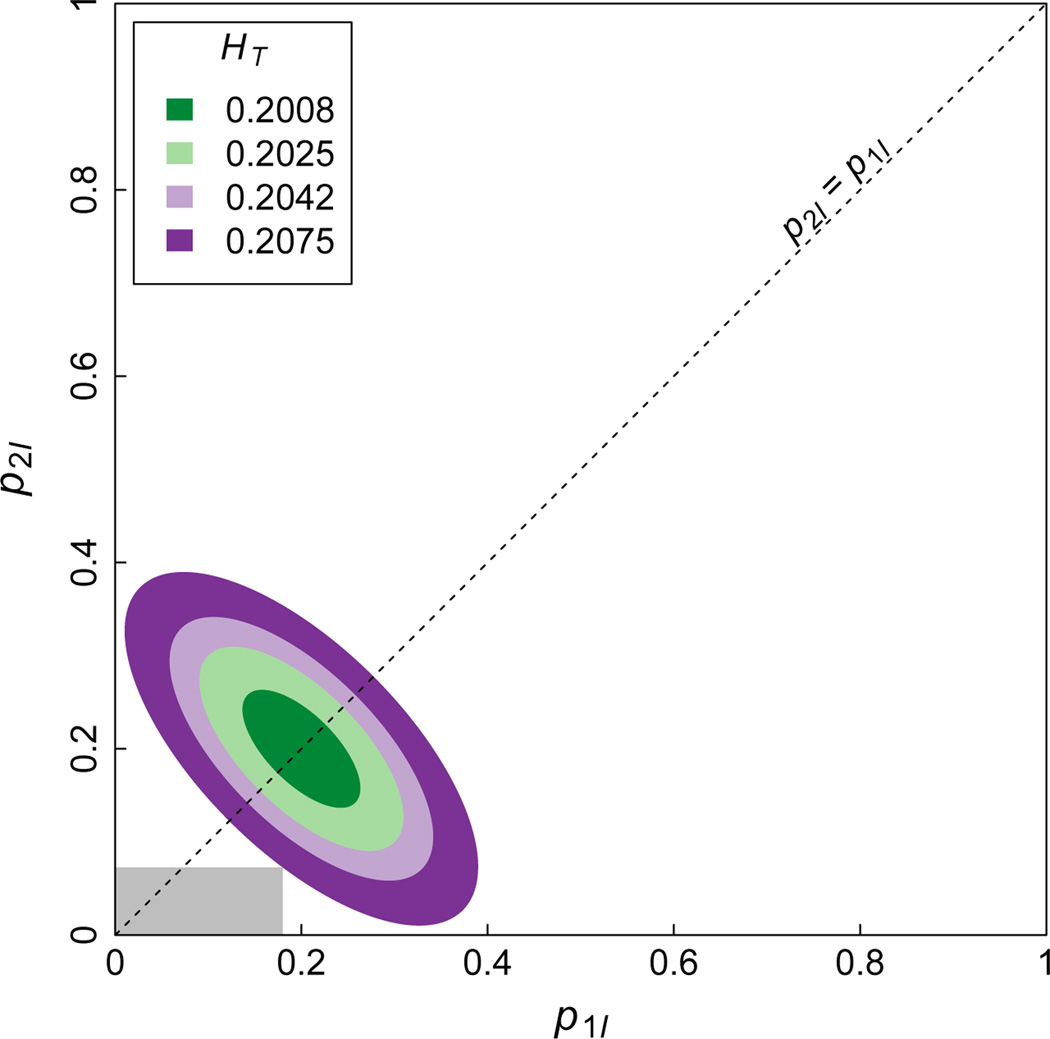
Figure H. 3.
The values of p1I and p2I that are possible when there are I = 5 alleles and HT is equal to the specific values in [1/I, I/(I2 − 1)) shown in the legend. If a pair of values is possible for (p1I, p2I) at a given HT ∈ [1/I, I/(I2 − 1)), then it is also allowed for larger HT ∈ [1/I, I/(I2 − 1)). Thus, the larger regions on the outside encompass the smaller interior regions. When HT increases to I/(I2 − 1), it is possible to set either p1I or p2I to 0. For a given HT in the relevant range, the region of allowed (p1I, p2I) values is symmetric around p1I = p2I, shown as a black dashed line on the plot. Because the problem of maximizing F given HT is solved when the product p1Ip2I is minimized, this visualization allows one to view the problem as that of finding the smallest rectangle that has its bottom-left vertex at the origin, two sides running along the axes, and its top-right vertex in the region of allowed (p1I, p2I) values allowed given HT. An example rectangle—not the one that maximizes FST—is shown in grey for HT = 0.2075.
Examination of the figure provides an intuition for the claim, proven in Appendix G, that the product of p1I and p2I is minimized when , where is the function of p1I shown in Eq. (G.5). To see this, note that this function traces the lower boundary of allowed p2I values shown in Figure H.3.
We can use Figure H.3 to make some informal predictions, proof of which will appear in the next section. First, consider a rectangle with a vertex at the origin, two sides that run along the axes, and another vertex on the curve that traces the lower bound on allowed values of p2I. Now, imagine another rectangle with an upper-right vertex that is reflected across the line p1I = p2I. It is clear that these two rectangles must have the same area, and thus that is symmetric around the value of p1I that solves . Therefore, setting must produce either a local minimum or a local maximum of A.
Second, notice that when HT is set to its smallest possible value, 1/I, the allowed region for (p1I, p2I) shrinks to the single point p1I = p2I = 1/I. Thus, at this value, F will be maximized when p1I = p2I. However, as HT approaches I/(I2 − 1), it becomes possible to set p2I to be arbitrarily close to 0 and to set p1I to be some larger number (or vice versa). Figure H.3 suggests that for some sufficiently large HT, setting p2I (or p1I) to be small and setting p1I (or p2I) to be larger will produce smaller values of A (and thus larger values of F) than setting p1I = p2I. Thus, the geometric approach suggests that for at least some values of HT (possibly just HT = 1/I), setting will maximize F, but for at least some larger values of HT, F will be maximized by setting p1I and to be different values.
Appendix H.2. Completing the minimization
We proceed with the minimization of A, which is equivalent to maximizing F. We start by finding candidate local optima for A and by ruling out the possibility that A is minimized when p1I is equal to its maximum or minimum allowed value. Next, we use properties of A and of ∂A/∂p1I to deduce some facts about the critical points of A. Finally, we use these facts to find the values of p1I that maximize F for two different ranges of HT values in [1/I, I/(I2 − 1)).
Appendix H.2.1. Identifying candidate minima
The derivative of A with respect to p1I is
| (H.3) |
Setting ∂A/∂p1I = 0 and rearranging gives
| (H.4) |
Squaring both sides and collecting terms gives a quartic equation in p1I. Dividing out (I + 1) gives
| (H.5) |
Eq. (H.5) has four solutions:
| (H.6) |
| (H.7) |
| (H.8) |
| (H.9) |
Because we squared both sides of Eq. (H.4), not all of the four solutions in Eq. (H.6–H.9) are guaranteed to be solutions of ∂A/∂p1I = 0, but all solutions of ∂A/∂p1I = 0 will be included among Eq. (H.6–H.9).
Next, we show that we need not consider the bounds of p1I when seeking to minimize A and that therefore, the only candidates for values of p1I that maximize F are the expressions in Eq. (H.6–H.9). The bounds on p1I are given in Eq. (H.2). The product rule for derivatives lets us rewrite Eq. (H.3) as
| (H.10) |
where
This expression makes clear that in the limit as p1I approaches its upper and lower bounds, the approach of
to 0 causes to approach either +∞ or −∞, depending on whether 2Ip1I −2 is positive or negative. As such, whenever p1I > 0, which is true for HT ∈ [1/I, I/(I2 − 1)) (see Section 4.2), ∂A/∂p1I also approaches +∞ or −∞ when p1I approaches its bounds in Eq. (H.2). Moreover, 2Ip1I − 2 > 0 when p1I > 1/I, so ∂A/∂p1I approaches +∞ when p1I approaches its upper bound, and ∂A/∂p1I approaches −∞ when p1I approaches its lower bound. This means that at the upper bound of p1I, A is increasing with p1I, and at the lower bound of p1I, A is decreasing with p1I, so the minimum of A for will occur in the open interval . Consequently, the minimum of A will occur when p1I is equal to one (or more) of the expressions in Eq. (H.6–H.9).
Appendix H.2.2. Properties of the critical points of
Before considering the candidates listed in Eq. (H.6–H.9), we note the following properties of A and ∂A/∂p1I, which will allow us to deduce some helpful facts:
∂A/∂p1I is negative when p1I is at its minimum and positive when p1I is at its maximum. This result is shown in the final paragraph of Appendix H.2.1.
Eq. (G.2) is symmetric in p1I and p2I.
∂A/∂p1I has no more than four critical points, where a saddle point counts for two critical points. This result holds because Eq. (H.5) is quartic.
Using (i–iii), we can deduce the following:
A must have at least one minimum for . This follows from (i). Thus, if ∂A/∂p1I = 0 has only one solution, then that solution is guaranteed to correspond to a minimum of A, which, by (ii), will occur where .
There cannot be exactly two solutions to ∂A/∂p1I = 0. If there were exactly two solutions of different types (for example, a maximum of A and a minimum of A), then the symmetry in (ii) would be violated. There cannot be two minima of A without a maximum of A or two maxima of A without a minimum of A. If there were two saddle points, then (i) would be contradicted.
If there are exactly three solutions to ∂A/∂p1I = 0, then there must be a maximum where is flanked by two equal minima that are reflections across p1I = p2I. To see this, note that if there are three solutions, then (ii) requires that one of them have p1I = p2I and that it be surrounded by two optima of the same type, one on each side. The middle solution cannot be a saddle point because symmetry would be violated. It cannot be a minimum flanked by maxima because (i) would be violated, and it cannot be a minimum flanked by saddle points because (iii) would be violated. Thus, it must be a maximum. Because it is a maximum, (i) requires that the solutions surrounding it are minima, and (ii) requires that the minima are equal.
There cannot be four or more solutions to ∂A/∂p1I = 0. If there are four solutions, then none can be saddle points of A by (iii). If none are saddle points, then there must be two maxima of A and two minima of A, but this violates (i). There cannot be more than four solutions by (iii).
Combining (A–D), the expressions in Eq. (H.6–H.9) must represent either one minimum of A or a maximum surrounded by two equal minima of A.
Appendix H.2.3. Maximizing F for odd I and HT ∈ [1/I, (I2 + I − 1)/(I3 + I2 − I − 1))
The expressions in Eq. (H.8) and Eq. (H.9) are only real when 1−I(I +1)+HT (I −1)(I +1)2 ≥ 0, which is only true when
For I > 1,
so for part of the range of HT values we consider, the expressions in Eq. (H.8) and Eq. (H.9) are real, but for part of the range, they are not. We thus must consider HT ∈ [1/I, (I2+I −1)/(I3+I2−I −1)) and HT ∈ [(I2+I −1)/(I3+ I2 − I − 1), I/(I2 − 1)) separately.
For HT ∈ [1/I, (I2 + I − 1)/(I3 + I2 − I − 1)), only the expressions in Eq. (H.6) and Eq. (H.7) are possible solutions to ∂A/∂p1I = 0, because the expressions in Eq. (H.8) and Eq. (H.9) are not real in this range of HT values. Invoking A–D lets us conclude that because there are not three solutions, there must be exactly one solution, it must have , and it must be a minimum of A.
Eq. (H.6) gives the solution to . As such, it is the sole solution of ∂A/∂p1I = 0 when HT ∈ [1/I, (I2 + I − 1)/(I3 + I2 − I − 1)), and for these values of HT, F is maximized by setting . These values of p1I and p2I can then be plugged into a special case of Eq. (A.3), modified to reflect the allele frequency arrangement in Table 4:
| (H.11) |
When this is done, the maximum F attained is
Note that setting p1I to equal the expression in Eq. (H.7) does not produce an optimum of A, as it is a fictitious root of Eq. (H.3). We can therefore exclude it as a candidate when we seek to minimize A in the next range of HT values we consider.
Appendix H.2.4. Maximizing F for odd I and HT ∈ [(I2 + I − 1)/(I3 + I2 − I − 1), I/(I2 − 1))
For the second range of HT values we must consider, HT ∈ [(I2 + I − 1)/(I3 + I2 − I − 1), I/(I2 − 1)), either A has its minimum when p1I equals the expression in Eq. (H.6), or it has a local maximum when p1I equals the expression in Eq. (H.6) and minima when p1I equals either the expression in Eq. (H.8) or the expression in Eq. (H.9). This statement follows from points (I–IV) in subsection Appendix H.2.2, along with the fact that setting p1I to equal the expression in Eq. (H.3) solves the equation .
Because these are the only two possibilities, we can distinguish them simply by comparing the value of A produced when p1I is set to equal the expression in Eq. (H.6) against the value of A produced when p1I equals either of the expressions in Eq. (H.8) or Eq. (H.9). That is, if it can be shown that the value of A produced by choosing p1I to be equal to the expression in Eq. (H.8) is smaller than the value of A produced by choosing p1I to be equal to the expression in Eq. (H.6), then A will be minimized (and F will be maximized) by setting p1I to be equal to the expression in either Eq. (H.8) or Eq. (H.9).
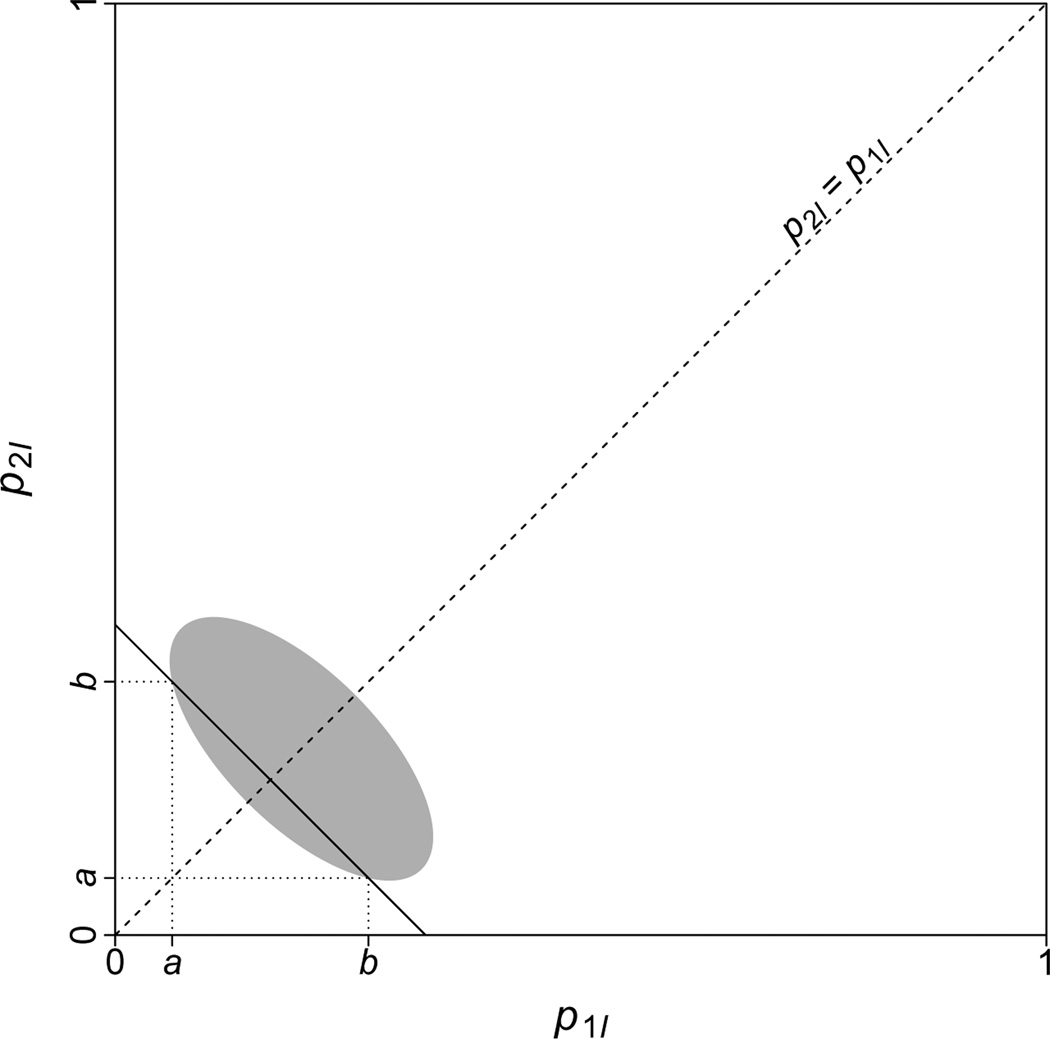
The first step is to find the value of when p1I is as in Eq. (H.8). Plugging this value of p1I directly into Eq. (G.5) to find produces an unwieldy expression. Rather than simplifying it, we can find in the alternative manner suggested in Figure H.4. To use this method, we need the equation for the line of slope −1 that intersects the curve when p1I is as in Eq. (H.8). As shown in Figure H.4, the intercept of this line is equal to the sum of a and b, where a is the p1I value for which we seek to find the associated value of , which we call b.
Figure H. 4.
An argument for identifying the value of that corresponds to a value of p1I denoted by a. To find b, we take advantage of symmetry around the p1I = p2I line. Suppose we find the line of slope −1 that intersects the curve at p1I = a (solid line in the Figure). As can be seen, this line is the line with slope −1 and intercept equal to a + b. If the same line intersects in another location, then the value of p1I at the second intersection is equal to b.
On the basis of the symmetry of the problem, we conjecture that if a is the expression in Eq. (H.8), then b must be the expression in Eq. (H.9). We verify our conjecture by checking that the line with slope −1 and intercept equal to the sum of the expressions in Eq. (H.8) and Eq. (H.9), or 2/(I + 1), intersects twice, where p1I is equal to the expressions in Eq. (H.8) and Eq. (H.9). The equation we need to solve is
| (H.12) |
One solution has p1I as in Eq. (H.8), and the other solution has p1I as in Eq. (H.9). Thus, when p1I is as in Eq. (H.8), is equal to the expression in Eq. (H.9), and when p1I is as in Eq. (H.9), is equal to the expression in Eq. (H.8).
It remains to compare the values of A generated when p1I is as in Eq. (H.6) and when p1I is as in Eq. (H.8). When p1I is as in Eq. (H.6),
| (H.13) |
In contrast, when p1I is as in Eq. (H.8),
| (H.14) |
Setting the right sides of Eq. (H.13) and Eq. (H.14) to be equal to each other gives
| (H.15) |
Squaring both sides of Eq. (H.15), rearranging, and simplifying gives a quadratic in HT:
| (H.16) |
Eq. (H.16) has only one solution, and thus, values of A produced when p1I is as in Eq. (H.6) and as in Eq. (H.8) are equal only when
| (H.17) |
This solution is the lower boundary of the interval over which we seek to minimize A. Because the expressions in Eq. (H.13) and Eq. (H.14) are only equal at one point, the expression in Eq. (H.14) is less than the expression in Eq. (H.13) for all HT > (I2 + I − 1)/(I3 + I2 − I − 1) if it is less for any HT > (I2 + I − 1)/(I3 + I2 − I − 1). For all I > 2, 1 > (I2 + I − 1)/(I3 + I2 − I − 1). When HT = 1, which is biologically impossible in our setting but mathematically valid, the expression in Eq. (H.14) is less than the expression in Eq. (H.13) when
| (H.18) |
If I > 2, then the expression on the left side of Eq. (H.18) is positive and the expression on the right is negative, so the inequality holds for all I > 2. Therefore, for HT ∈ [(I2 + I − 1)/(I3 + I2 − I − 1), I/(I2 − 1)) and all I > 2, the expression in Eq. (H.14) is less than the expression in Eq. (H.13), and choosing p1I as in Eq. (H.8) or Eq. (H.9) minimizes A. Minimizing A maximizes F with respect to HT. The upper bound on F is attained using Eq. (H.11), setting p1I to the expression in Eq. (H.8) and setting p2I to the expression in Eq. (H.9), or vice versa. The upper bound is
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcala N, Goudet J, Vuilleumier S. On the transition of genetic differentiation from isolation to panmixia: What we can learn from GST and D. Theor. Pop. Biol. 2014;93:75–84. doi: 10.1016/j.tpb.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: The impact of rare variants. Genome Research. 2013;23:1514–1521. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 1998;15:538–543. doi: 10.1093/oxfordjournals.molbev.a025953. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nature Rev. Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Edge MD, Rosenberg NA. The relationship between FST and the frequency of the most frequent allele. Genetics. 2013;193:515–528. doi: 10.1534/genetics.112.144758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Mol. Ecol. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Long JC. Update to Long and Kittles’s “Human genetic diversity and the nonexistence of biological races (2003): fixation on an index”. Hum. Biol. 2009;81:799–803. doi: 10.3378/027.081.0622. [DOI] [PubMed] [Google Scholar]
- Long JC, Kittles RA. Human genetic diversity and the nonexistence of biological races. Hum. Biol. 2003;75:449–471. doi: 10.1353/hub.2003.0058. [DOI] [PubMed] [Google Scholar]
- Maruki T, Kumar S, Kim Y. Purifying selection modulates the estimates of population differentiation and confounds genome-wide comparisons across single-nucleotide polymorphisms. Mol. Biol. Evol. 2012;29:3617–3623. doi: 10.1093/molbev/mss187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirmans PG, Hedrick PW. Assessing population structure: FST and related measures. Mol. Ecol. Resources. 2011;11:5–18. doi: 10.1111/j.1755-0998.2010.02927.x. [DOI] [PubMed] [Google Scholar]
- Nagylaki T. Fixation indices in subdivided populations. Genetics. 1998;148:1325–1332. doi: 10.1093/genetics/148.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Reddy SB, Rosenberg NA. Refining the relationship between homozygosity and the frequency of the most frequent allele. J. Math. Biol. 2012;64:87–108. doi: 10.1007/s00285-011-0406-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Jakobsson M. The relationship between homozygosity and the frequency of the most frequent allele. Genetics. 2008;179:2027–2036. doi: 10.1534/genetics.107.084772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Exegeses on maximum genetic differentiation. Genetics. 2013;194:557–559. doi: 10.1534/genetics.113.152132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman N, Leimar O. Effect of mutation on genetic differentiation among nonequilibrium populations. Evolution. 2008;62:2250–2259. doi: 10.1111/j.1558-5646.2008.00453.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Inbreeding coefficients and coalescence times. Genet. Res. 1991;58:167–175. doi: 10.1017/s0016672300029827. [DOI] [PubMed] [Google Scholar]
- VanLiere JM, Rosenberg NA. Mathematical properties of the r2 measure of linkage disequilibrium. Theor. Pop. Biol. 2008;74:130–137. doi: 10.1016/j.tpb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC. and D do not replace FST. Mol. Ecol. 2011;20:1083–1091. doi: 10.1111/j.1365-294X.2010.04996.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Ann. Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution and the Genetics of Populations Volume 4: Variability within and among natural populations. Chicago: University of Chicago Press; 1978. [Google Scholar]