Abstract
The PsbQ-like protein, termed CyanoQ, found in the cyanobacterium Synechocystis sp. PCC 6803 is thought to bind to the lumenal surface of photosystem II (PSII), helping to shield the Mn4CaO5 oxygen-evolving cluster. CyanoQ is, however, absent from the crystal structures of PSII isolated from thermophilic cyanobacteria raising the possibility that the association of CyanoQ with PSII might not be a conserved feature. Here, we show that CyanoQ (encoded by tll2057) is indeed expressed in the thermophilic cyanobacterium Thermosynechococcus elongatus and provide evidence in support of its assignment as a lipoprotein. Using an immunochemical approach, we show that CyanoQ co-purifies with PSII and is actually present in highly pure PSII samples used to generate PSII crystals. The absence of CyanoQ in the final crystal structure is possibly due to detachment of CyanoQ during crystallisation or its presence in sub-stoichiometric amounts. In contrast, the PsbP homologue, CyanoP, is severely depleted in isolated PSII complexes. We have also determined the crystal structure of CyanoQ from T. elongatus to a resolution of 1.6 Å. It lacks bound metal ions and contains a four-helix up-down bundle similar to the ones found in Synechocystis CyanoQ and spinach PsbQ. However, the N-terminal region and extensive lysine patch that are thought to be important for binding of PsbQ to PSII are not conserved in T. elongatus CyanoQ.
Electronic supplementary material
The online version of this article (doi:10.1007/s11120-014-0010-z) contains supplementary material, which is available to authorized users.
Keywords: Photosynthetic oxygen evolution, PsbQ, PsbQ’, CyanoP, Psb27, Peroxiredoxin
Introduction
The water oxidation reaction of oxygenic photosynthesis is catalysed by the photosystem II (PSII) complex located in the thylakoid membranes of chloroplasts and cyanobacteria. Crystal structures of monomeric and dimeric oxygen-evolving PSII complexes isolated from the thermophilic cyanobacteria Thermosynechococcus vulcanus and Thermosynechococcus elongatus have been determined (Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005; Guskov et al. 2009; Broser et al. 2010; Umena et al. 2011). Each PSII monomer contains about 20 subunits, depending on the preparation, most of which are integral to the membrane (reviewed by Müh et al. 2008). In the case of cyanobacteria three extrinsic proteins (PsbO, PsbU and PsbV) are attached to the lumenal surface of the crystallised complex where in vivo they help to shield the Mn4CaO5 oxygen-evolving complex from aberrant reduction (Shen et al. 1998). A different set of proteins (PsbO, PsbP, PsbQ and PsbR) is associated with PSII in green algae and higher plant chloroplasts, but their binding sites remain unclear (reviewed by Bricker et al. 2012). For red algae and diatoms, an intermediate situation exists in which a PsbQ-like subunit (termed PsbQ’) is present in addition to the PsbO, PsbU and PsbV subunits, while a fifth subunit, Psb31, is also found in diatoms (reviewed by Enami et al. 2008). PsbP-like and PsbQ-like proteins are also expressed in higher plant chloroplasts, but they have roles outside PSII. For instance, two PsbQ-like proteins are components of the thylakoid NADH dehydrogenase-like (NDH) complex in Arabidopsis (Yabuta et al. 2010).
Homologues of PsbP and PsbQ are also found in cyanobacteria (Thornton et al. 2004). The function of these two proteins, designated here as CyanoP and CyanoQ, respectively, is still obscure (reviewed by Fagerlund and Eaton-Rye 2011), particularly as they are not present in the published crystal structures of PSII (Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005; Guskov et al. 2009; Umena et al. 2011). Most work on the structure and function of CyanoQ has come from studies of the mesophilic cyanobacterium Synechocystis sp. PCC 6803, hereafter Synechocystis, where it is known to be a subunit of oxygen-evolving PSII complexes (Roose et al. 2007). Synechocystis cells lacking CyanoQ grow photoautotrophically as well as WT under optimal growth conditions but do show some growth inhibition when exposed to nutrient stress such as by depleting the medium of calcium and chloride (Thornton et al. 2004) and iron (Summerfield et al. 2005). Analysis of isolated PSII complexes lacking CyanoQ from Synechocystis suggests that CyanoQ stabilises binding of PsbV and helps protect the oxygen-evolving Mn4CaO5 complex from reduction in the dark (Kashino et al. 2006).
The crystal structure of Escherichia coli-expressed Synechocystis CyanoQ, determined to a resolution of 1.8 Å, is similar to that of PsbQ from spinach with a root mean square deviation (RMSD) for the Cα atoms of 1.4 Å despite only 17 % identity in primary structure (Jackson et al. 2010). Both crystallised proteins consist of a four-helix bundle and contain bound Zn2+, although a metal-free structure has also been determined for Synechocystis CyanoQ (Jackson et al. 2010); the physiological relevance of these metal-binding sites is currently unknown. In contrast, much less is known about CyanoQ in the thermophilic cyanobacteria used for structural studies of PSII. Indeed the association of CyanoQ with PSII in either T. elongatus or T. vulcanus has yet to be demonstrated. Here, we describe the crystal structure of E. coli-expressed CyanoQ from T. elongatus and provide evidence that CyanoQ co-purifies with isolated PSII and strikingly is still present in samples used to generate PSII crystals lacking CyanoQ.
Materials and methods
Thermosynechococcus elongatus BP1 strains
A His-tagged CP43 strain (CP43-His) of Thermosynechococcus elongatus (Sugiura and Inoue 1999) was kindly provided by Dr Miwa Sugiura, and a His6-tagged derivative of CP47 (CP47-His) by Dr Diana Kirilovsky. The WT strain was the same as that used by Ferreira et al. (2004).
Construction of plasmid for over-expression of CyanoQ
The DNA sequence corresponding to the CyanoQ homologue of T. elongatus (tll2057) without the sequence encoding the predicted signal peptide and lipid-binding Cys24 residue was cloned into a pRSET-A vector modified as described in Bialek et al. (2013). The corresponding PCR fragment was amplified from T. elongatus genomic DNA using Phusion polymerase (NEB, UK) and primers CyanoQ-XhoI-F (5′-TATATACTCGAGGGCGGCCCCAGTGCCACCACTCCACCCCCACCCACCTA-3′) and CyanoQ-EcoRI-R (5′-TATATAGAATTCTTACTAGGACAACTCAGGCAAGCTGTTGAGAT-3′) introducing underlined restriction sites, double digested with XhoI and EcoRI and ligated (Quick Ligation Kit, NEB, UK) into the modified and XhoI/EcoRI linearised pRSET-A. The vector was then transformed into KRX E. coli cells (Promega, UK).
Expression, purification and crystallisation of CyanoQ
Expression of His6-tagged CyanoQ was induced by the addition of 2 g/L of rhamnose, and cells were grown at 18 °C overnight. Cells were lysed with a sonicator (Sonics and Materials, CT, USA) in lysis buffer (50 mM Tris–HCl pH 7.9, 500 mM NaCl, 1 mM MgCl2) supplemented with one Complete Protease Inhibitor Cocktail-EDTA Tablet (Roche, UK) per 50 ml lysis buffer. Broken cells were spun down for 10 min at 4 °C at 18,000×g, and the supernatant was mixed with a Ni-iminodiacetic acid resin (Generon, UK). Non-specifically bound proteins were removed by washing 3 times with wash buffer (20 mM Tris–HCl pH 7.9, 500 mM NaCl, 60 mM imidazole), and His6-CyanoQ was eluted with elution buffer (20 mM Tris–HCl pH 7.9, 500 mM NaCl, 1 M imidazole). Purified His6-CyanoQ was dialysed overnight against 20 mM Tris–HCl pH 7.9, 200 mM NaCl at 4 °C. The His-tag was removed by thrombin (GE Healthcare, UK) digestion at a ratio of 1 unit of thrombin per 100 µg of purified CyanoQ. Proteolysis was performed overnight at 4 °C and the digested sample was reloaded onto a nickel-iminodiacetic acid column. The flow-through containing CyanoQ without the His-tag was concentrated at 4 °C to around 10 mg/ml with a centrifugal concentrator device with a molecular weight cut off (MWCO) of 3500 (Sartorius, Germany). Crystals appeared in hanging drop vapour diffusion, above 1.8 M ammonium sulphate, with drops of protein solution and an equal volume of mother liquor. Crystals were cryoprotected in the mother-liquor solution with 30 % (v/v) glycerol, then flash-cooled in liquid nitrogen.
Protein structure determination
Data were integrated and scaled with MOSFLM (Leslie and Powell 2007) and programmes of the CCP4 suite (Winn et al. 2011). 5 % of reflections were set aside as the Free set for cross-validation. The structure was solved by molecular replacement using the CyanoQ structure from Synechocystis (Jackson et al. 2010). The model was truncated using Chainsaw (Stein 2008) mode, and used as a model in PHASER (McCoy et al. 2007). The structure was refined in REFMAC (Murshudov et al. 2011) with cycles of manual model-building in COOT (Emsley and Cowtan 2004). Validation was performed using the MolProbity server (Davis et al. 2007). The atomic model and structure factors have been deposited in the PDB under accession number 3ZSU.
Sequence alignment and structural conservation
The full protein sequence of CyanoQ (Tll2057) from T. elongatus was searched against cyanobacterial genomes using BLAST (Altschul et al. 1990) having gapless chromosome assembly level on NCBI. Sequences were aligned in ClustalW2 and analysed by Prosite (De Castro et al. 2006).
Isolation of PSII complexes from T. elongatus
Thermosynechococcus elongatus cultures were grown in DTN medium (Mühlenhoff and Chauvat 1996) at a constant temperature of 45 °C under continuous illumination (~60 μmol photons·m−2·s−1) by an Innova 44 shaker (New Brunswick Scientific) at 120 rpm. Typically 5-L Erlenmeyer flasks were used to grow five 3.5-l cultures to give a total culture volume of about 17.5 l. Cells were harvested at an optical density of about 1 at 750 nm using a Sartocon cross flow filtration system (Sartorius) followed by centrifugation at 10,000 rpm (JA14 rotor, Beckman Coulter Ltd.) for 5 min at room temperature. The cell pellet was re-suspended in RSB buffer (40 mM MES–NaOH pH 6.5, 15 mM MgCl2, 15 mM CaCl2, 1.2 M betaine and 10 % (v/v) glycerol) to a volume of 50–75 ml and disrupted by 2 passes at 25,000 psi using a T5 cell disruptor set to 4 °C (Constant Systems Ltd). Unbroken cells were removed by centrifugation at 1,000×g (JA14 rotor, Beckman Coulter Ltd.) for 5 min at 4 °C, and membranes were pelleted and washed three times with the same buffer by centrifugation at 184,000×g (Ti45 rotor, Beckman Coulter Ltd.) for 20 min at 4 °C. Membranes were then resuspended in 20 mM MES–NaOH pH 6.5, 10 mM MgCl2, 20 mM CaCl2, 25 % (v/v) glycerol and stored at −0 °C. These membranes were then used to isolate PSII oxygen-evolving complexes from WT T. elongatus using the two-step anion-exchange chromatography procedure described by Kern et al. (2005).
Dimeric His-tagged oxygen-evolving complexes were isolated from a His-tagged CP47 strain of T. elongatus by Ni-affinity purification followed by anion-exchange chromatography as described by Nowaczyk et al. (2006) except for the following modifications: freshly grown cells were broken in 20 mM MES–NaOH pH 6.5, 2.5 mM CaCl2, 2.5 mM MgCl2, 10 % (v/v) glycerol and 1.2 M betaine, and unbroken cells were removed by centrifuging at 1,000 g (JA14 rotor, Beckman Coulter Ltd.) for 5 min at 4 °C; the resulting supernatant was diluted to a Chl concentration of 1 mg/ml and the thylakoid membranes were solubilised for 10 min at 4 °C with 1 % (w/v) n-dodecyl-β-D-maltoside (β-DDM) at a detergent to Chl ratio of 18:1 followed by a 30-min spin at 4 °C and 184,000 g (Ti70 rotor, Beckman Coulter Ltd.); the extract was incubated for 45 min with Ni-affinity resin (Probond Resin, Invitrogen) equilibrated in buffer E (20 mM MES–NaOH pH 6.5, 2.5 mM CaCl2, 2.5 mM MgCl2, 0.5 M D-mannitol and 0.03 % (w/v) β-DDM); after loading, the Ni-affinity column was washed with 6 column volumes of buffer E + 5-mM histidine; His-tagged PSII complexes were eluted by application of a 100-mM histidine isocratic step gradient in buffer E and loaded directly onto a Bio-Rad UNO Q-12 column using a AKTA Purifier 10 system (GE Healthcare Life Sciences); PSII complexes were eluted through the application of a 5–200-mM MgSO4 gradient in buffer E (at 2 mM/min and 4 ml/min). The third peak containing active PSII dimeric complexes (Nowaczyk et al. 2006) was concentrated using Vivaspin centrifugal concentrators (100,000 MWCO) before storing at −80 °C.
Preparation of crude thylakoid membranes and pull-down experiments
Pull-down experiments were done using the His-tagged CP43 strain of T. elongatus and cobalt resin prepared by charging chelating Sepharose fast flow resin according to the manufacturer’s instructions (GE Healthcare Life Sciences). Crude thylakoid membranes were prepared from T. elongatus by glass bead breakage and differential centrifugation as described by Boehm et al. (2009) and re-suspended in buffer A (50 mM MES–NaOH pH 6.0, 10 mM MgCl2, 5 mM CaCl2, 10 % (w/v) glycerol) as used by Kashino et al. (2002). Thylakoids were solubilised with 1 % (w/v) β-DDM at a Chl concentration of 0.2 mg/ml for 10 min on ice in a final volume of 0.5 ml. After pelleting insoluble material by centrifuging in a microfuge, 0.45 ml of the supernatant was removed and diluted by addition of 0.45 ml of buffer A to which was added 0.1 ml of cobalt resin (50 µl of resin resuspended to final volume of 100 µl by addition of buffer A). Samples were then incubated on a rotating wheel at 4 °C for 2 h. After removal of the membrane extract, the cobalt resin was washed four times with 500 µl of buffer A, with the final wash kept for analysis. Bound proteins were eluted with 100 µl of buffer A containing 100-mM imidazole followed by 100 µl of 1× SDS sample buffer used for electrophoresis. Chelating Sepharose lacking bound metal ions was used as a control.
Salt washes of purified PSII complexes and thylakoid membranes
PSII complexes in buffer A2 (20 mM MES–NaOH pH 6.5, 1 mM MgCl2, 1 mM CaCl2, 10 % (w/v) glycerol, 0.03 % (w/v) β-DDM) purified either by two-step anion-exchange or by nickel-affinity chromatography were incubated with buffer A2 supplemented with 1 M CaCl2 on ice for 30 min in the dark. Immediately after incubation samples were concentrated on 100,000 MWCO Vivaspin 500 centrifugal concentrators (Sartorius AG). Green retentate and flow-through containing removed extrinsic proteins were desalted by two buffer exchanges using Vivaspin 500 centrifugal concentrators, with MWCO of 100,000 and 3,000, respectively. Chlorophyll concentration was adjusted to 1 mg/ml and the volume of the filtrate was adjusted to match the volume of the green retentate. In the case of thylakoid membranes, proteins were extracted by high salt or high pH using the Freeze–Thaw approach described by Boehm et al. (2009).
Protein analysis, isolation of protein and immunoblotting
Thermosynechococcus elongatus CyanoP and Psb27 were over-expressed in E. coli and purified as described previously (Michoux et al. 2010, 2012). These proteins plus CyanoQ isolated here were used to raise antibodies in rabbit. Protein samples were separated on 18 % (w/v) polyacrylamide gels containing 6 M urea as described by Boehm et al. (2009). Immunoblotting analyses were performed as described by Boehm et al. (2009) using the following antibodies and dilutions: αD1 (1:5000), αPsbO (1:1000), αCyanoP (1:2500), αCyanoQ (1:5000) and αPsb27 (1:2500). Chlorophyll a (Chl a) content and protein concentrations (using BSA as standard) were determined as described by Boehm et al. (2009). For densitometry gels were analysed by Image Studio Lite (LI-COR, Inc).
Results and discussion
CyanoQ associates with PSII complexes isolated from T. elongatus
The CyanoP and CyanoQ orthologues in T. elongatus are encoded by tlr2075 (Michoux et al. 2010) and tll2057, respectively. Despite detailed analysis of the subunit composition of His-tagged PSII complexes isolated from T. elongatus by mass spectrometry (Sugiura et al. 2010), neither CyanoQ nor CyanoP has been detected. To investigate whether CyanoQ or CyanoP are able to associate with PSII isolated from T. elongatus, we first performed pull-down experiments by binding solubilised membrane extracts obtained from a His-tagged CP43 strain of T. elongatus (CP43-His) to a cobalt resin and analysing bound proteins released by 100-mM imidazole. Immunoblotting experiments revealed that a significant proportion of CyanoQ co-purified with CP43-His (Fig. 1). By contrast, no detectable CyanoQ bound to the cobalt resin when a non-tagged WT sample was tested. As expected, the D1 and PsbO subunits of PSII co-purified with His-tagged CP43, as did significant amounts of Psb27, which is known to be a component of non-oxygen-evolving PSII complexes (Nowaczyk et al. 2006; Grasse et al. 2011). In contrast only trace amounts of CyanoP co-purified with CP47-His under the experimental conditions used.
Fig. 1.
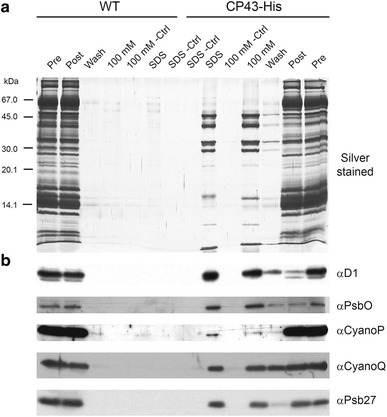
Association of CyanoQ with His-tagged CP43. Detergent solubilised membrane extracts from either WT or His-tagged CP43 strains of T. elongatus (CP43-His) were mixed with cobalt resin and the bound proteins eluted by 100-mM imidazole (100 mM) followed by SDS solubilising buffer (SDS) for analysis by a SDS-PAGE and silver staining and b immunoblotting. Pre solubilised extract added to resin; Post solubilised extract after incubation with cobalt resin; Wash last wash before elution; Ctrl control in which resin lacking Co was used
A commonly used method to isolate highly active oxygen-evolving dimeric PSII complexes from T. elongatus for structural studies involves a two-step anion-exchange chromatography protocol (Kern et al. 2005). This type of preparation has been successfully used to generate high-quality PSII crystals yielding diffraction data of up to 3 Å resolution (Loll et al. 2005; Murray et al. 2008a, b). The PSII preparation analysed here (which produced 400-µm-long PSII crystals) also contained detectable levels of the alpha subunit of the ATPase (Tlr0435) and, interestingly, a predicted thioredoxin peroxidase/peroxiredoxin (Tll1454), which is homologous to a peroxiredoxin (2-CysPrx) thought to interact with PSII in chloroplasts (Muthuramalingam et al. 2009) (Fig. 2). Immunoblotting of the PSII complex revealed that CyanoQ was indeed present and had been purified to about the same degree as the D1 subunit (approximate 10-fold enrichment on chlorophyll basis compared with thylakoid membranes). The presence of CyanoQ, which comigrates with PsbV upon SDS-PAGE, was confirmed by mass spectrometry (data not shown). In contrast less than 5 % of CyanoP and Psb27 originally found in the membrane were retained in the PSII fraction. More detailed analysis of individual fractions by immunoblotting confirmed that CyanoP and Psb27 had been removed during purification of dimeric PSII whereas CyanoQ co-purified (Figs. S1, S2). Loss of CyanoP on purification of PSII is in line with earlier studies on Synechocystis (Ishikawa et al. 2005).
Fig. 2.
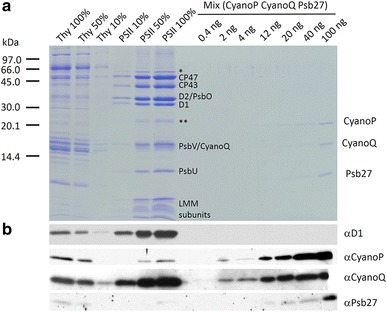
a SDS-PAGE analysis of serial dilutions of solubilised thylakoids membranes and T. elongatus PSII complexes isolated using the two-step anion-exchange chromatography method and known amounts of a mix of recombinant non-tagged CyanoP, CyanoQ and Psb27 proteins. 100 % level corresponds to 1 µg of Chl and amount in mix refers to amount of each of the proteins. Protein detected by Coomassie Blue staining. Single asterisk indicates migration of AtpA and double asterisk the migration of thioredoxin peroxidase as determined by mass spectrometry. Assignment of PSII subunits was determined through immunoblotting and mass spectrometry. LMM low molecular mass subunits of PSII. b Semi-quantitative immunoblotting analysis to determine CyanoP, CyanoQ and Psb27 levels in thylakoids and PSII
We attempted to estimate the stoichiometry of CyanoQ present in the isolated PSII complex using a semi-quantitative immunoblotting approach (Fig. 2). A number of assumptions are made in this method including equal cross-reactivity of the native protein and E. coli-expressed version and the use of a protein assay to determine the amount of the standard; however this method has been applied previously to estimate levels of CyanoP and CyanoQ in Synechocystis (Thornton et al. 2004). Using the recombinant protein standards, we tentatively estimate that 20 ng of CyanoQ is present in PSII protein complexes containing 0.1 μg of Chl a. Assuming 35 Chl a are bound per PSII monomer and a molecular mass of 14,329 Da for CyanoQ, this would mean a CyanoQ:PSII monomer ratio of 0.4:1. In the case of Synechocystis, estimates range from 1.2 CyanoQ per 1 CP47 in membranes, determined by immunoblotting (Thornton et al. 2004), to approximately 0.25–0.30 CyanoQ per PSII based on the yield of His-tagged CyanoQ-containing PSII complexes (Roose et al. 2007). For CyanoP (molecular mass of 18,031 Da), assuming 1.3 ng of protein is present in PSII complexes containing 0.5 μg of Chl a (Fig. 2), the same calculation suggests that less than 1 % of PSII complexes in our preparation contain CyanoP.
Overall these data suggest that CyanoQ in T. elongatus co-purifies with dimeric PSII when isolated by anion-exchange chromatography. Absence of CyanoQ in PSII crystals obtained from this type of preparation could be due to detachment during crystallisation, such as by high salt (Fig. S3), or the fact that only PSII complexes lacking CyanoQ crystallised under the conditions tested. Importantly, we also found that the amount of CyanoQ associated with isolated PSII is variable with much less CyanoQ present in His-tagged PSII complexes isolated by the immobilised metal affinity chromatography and anion-exchange chromatography method described by Nowaczyk et al. 2006 (Fig. S3), which might explain why CyanoQ had not until now been detected in isolated His-tagged PSII complexes. In contrast, we have so far been unable to find conditions where CyanoP remains fully attached to PSII complexes.
CyanoQ is a likely lipoprotein in T. elongatus
Like the situation in Synechocystis (Ujihara et al. 2008), both CyanoP and CyanoQ from T. elongatus contain a characteristic lipobox sequence, as detected by Prosite (De Castro et al. 2006), suggesting that they might be processed at the N-terminus and anchored to the membrane via lipidation of a cysteine residue (Fig. S4). Previous membrane washing experiments using either a high-salt treatment (2 M NaCl or 1 M CaCl2) or an alkaline treatment (pH 12.0), coupled with immunochemical detection, have shown that CyanoP is tightly bound to the membrane consistent with its assignment as a lipoprotein, whereas the non-lipidated extrinsic PsbO subunit is more easily removed (Michoux et al. 2010). Analysis of the same samples revealed that CyanoQ behaved like CyanoP and the lipidated Psb27 subunit of PSII (Nowaczyk et al. 2006) and was more resistant to extraction than PsbO (Fig. S5).
Expression and crystallisation of the CyanoQ protein from T. elongatus
CyanoQ in Synechocystis and T. elongatus are relatively divergent with only 31 % sequence identity (Fig. 3 and Fig. S4). To gain insights into the structure of CyanoQ from T. elongatus, a cleavable N-terminal His6-tagged derivative lacking the predicted lipidated Cys24 (Fig. 3) residue was over-expressed in E. coli and the protein purified by immobilised nickel-affinity chromatography to near homogeneity (Fig. S6a). The His-tag was removed by thrombin cleavage and CyanoQ was re-purified and concentrated to 10 mg/ml (Fig. S6b). The predicted product contains residues 25–152 of CyanoQ plus 5 additional residues (GSELE) at the N-terminus. Crystallisation screens, performed using hanging drop plates, resulted in the formation of crystals, which were further optimised to grow in 1.8 M ammonium sulphate (Fig. S6c).
Fig. 3.
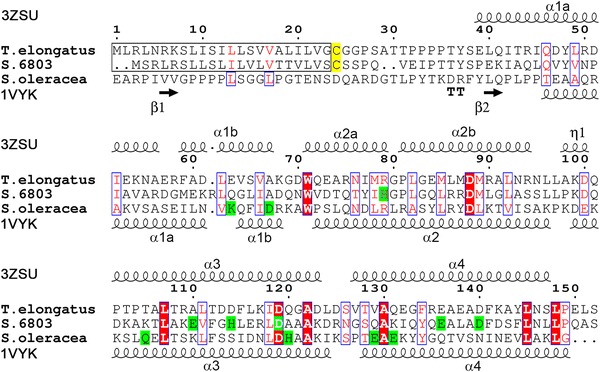
Sequence alignment of CyanoQ from T. elongatus, Synechocystis and PsbQ from spinach. Secondary structures are shown for CyanoQ from T. elongatus (3ZSU) and PsbQ from spinach (1VYK). Zinc-binding sites and lipidated cysteine residues are highlighted in green and yellow, respectively. Predicted signal peptides for CyanoQ are boxed in black. Numbering according to CyanoQ sequence from T. elongatus. Absolutely conserved and similar residues are shown as white letters on red background and red letters on white background, respectively, as calculated by ESPript (Gouet et al. 2003)
Structural features and comparison with other CyanoQ proteins
A native data set was obtained by X-ray crystallography at a resolution of 1.6 Å and the structure solved by molecular replacement using the crystal structure of CyanoQ from Synechocystis (PDB:3LS0, for details see Table 1). The refined co-ordinates of the 3D model of CyanoQ from T. elongatus have been deposited at the Protein Data Bank using the accession code 3ZSU. The first nine N-terminal residues as well as the last C-terminal residue of CyanoQ could not be detected in the electron density map so only residues 34–151 were fitted. Topologically the protein belongs to four-helix bundle superfamily and its fold is classified as mainly alpha up-down bundle (CATH 1.20.120.290) with four α-helices, of which the first two are broken, and one 310 helix (Fig. 4a). The three-dimensional structure of CyanoQ from thermophilic T. elongatus showed a high level of similarity with the two structures of CyanoQ (with and without bound zinc) from the mesophilic Synechocystis (Jackson et al. 2010) with a RMSD of 1.6 Å for the C α atoms (Table 2 and Fig. S7).
Table 1.
Data collection and refinement statistics for the CyanoQ crystal structure
| CyanoQ data | |
|---|---|
| X-ray source | Diamond I03 |
| Data processing | Mosflm/Scala |
| Space group | P 21 21 21 |
| Unit-cell parameters | a = 47.165 Å, b = 47.165 Å, c = 106.700 Å, α = β = 90°, γ = 120° |
| Wavelength (Å) | 1.0722 |
| Resolution (Å) | 53.4–1.6 (1.69–1.60) |
| Measured reflections | 130,767 (19,307) |
| Unique reflections | 18,728 (2707) |
| Mn (I/sd) | 10.8 (3.7) |
| Completeness (%) | 99.38 (100.0) |
| Multiplicity | 6.98 (7.13) |
| R meas (%) | 0.11 (0.62) |
| Solvent content (%) | 48.6 |
| R work/R free (%) | 16.7/19.0 |
| Protein atoms | 974 |
| Solvent atoms | 79 |
| RMSD from ideal | |
| Bond lengths (Å) | 0.022 |
| Bond angles (°) | 1.982 |
| Average B factor (Å2) | 18.2 |
| Ramachandran favoured region (%) | 100 |
| Ramachandran allowed region (%) | 0 |
Fig. 4.
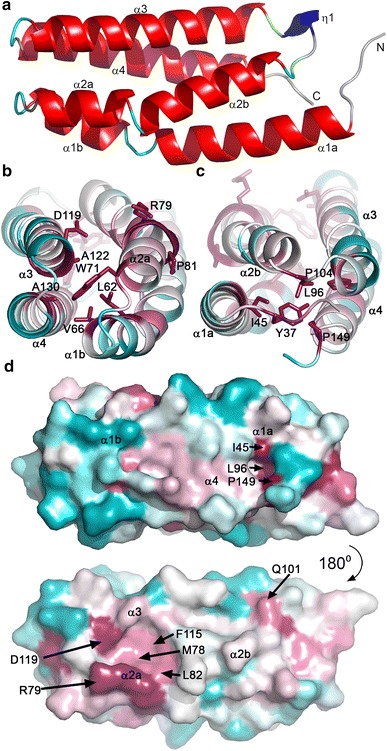
a Overall structure of CyanoQ from T. elongatus coloured according to DSSP (Kabsch and Sander 1983): α-helices (α1-α4, red), 310 helix (blue, η1), hydrogen-bonded turns (cyan) and bends (green). b top and c bottom view of the protein coloured according to sequence conservation in cyanobacteria with most conserved residues shown as sticks. Bottom view in c corresponds to the end of CyanoQ containing the N- and C-termini. d Consurf (Ashkenazy et al. 2010) analysis of two conserved cavities (H4-H1 in upper view and H2–H3 in lower view; see text for details) with most conserved residues shown in dark pink and magenta. The most divergent regions are coloured in cyan
Table 2.
Comparison of sequence identities and similarities (%, top) and structural RMSD (bottom) of CyanoQ from T. elongatus (3ZSU), Synechocystis with and without zinc (3LS1 and 3LS0) and PsbQ from spinach (1VYK and 1NZE)
| 3ZSU | 3LS0 | 3LS1 | 1VYK | 1NZE | |
|---|---|---|---|---|---|
| T. elongatus | Synechocystis | S. oleracea | |||
| 3ZSU | 31/50 | 31/50 | 14/24 | 14/24 | |
| 3LS0 | 1.6 Å | 100/100 | 17/33 | 17/33 | |
| 3LS1 | 2.0 Å | 0.7 Å | 17/33 | 17/33 | |
| 1VYK | 1.6 Å | 1.4 Å | 1.6 Å | 100/100 | |
| 1NZE | 1.5 Å | 1.4 Å | 1.6 Å | 0.5 Å | |
Although CyanoQ is likely to be lipidated in vivo in both Synechocystis and T. elongatus, this is not a universal feature of CyanoQ as the lipobox sequence and Cys residue needed for lipidation are absent in a number of other cyanobacteria (Fig. S4). These include Acaryochloris marina, a chlorophyll d-containing cyanobacterium and the siderophilic (having an affinity for iron) cyanobacterium JSC-12, whereas no protein homologous to CyanoQ could be detected in the Prochlorococcus spp., the two thermophilic species Synechococcus sp. JA-3-3Ab and Synechococcus sp. JA-2-3B’a(2-13) and the thylakoid-less Gloeobacter violaceus (De Las and Roman 2005; Fagerlund and Eaton-Rye 2011).
According to our sequence alignment, there are only two regions with absolutely conserved amino-acid residues across the cyanobacterial lineage. These regions flank helix 2a, the shortest one out of six found in this protein. The first amino-acid residue of helix 2a, Trp71, is absolutely conserved in the analysed CyanoQ sequences (Fig. S4). The indole nitrogen is exposed towards the solvent, and in this structure a 2.8 Å hydrogen bond is created between Trp71Nε1 and Asp125Oδ1. A typical Ncap motif (Richardson and Richardson 1988) is observed for helix 2a where a main-chain carbonyl oxygen of Asp70 creates an hydrogen bond with the backbone amide nitrogen of Glu73. The other absolutely conserved residues are found right after the C-terminus of helix 2a and consist of a Gly80Pro81 motif that is immediately preceded by a positively charged amino acid, either arginine as in T. elongatus or in most cases histidine. Both glycine and proline are well known as the most efficient ‘helix breakers’ and in fact they separate helix 2a from helix 2b in CyanoQ (Fig. 4a).
Strongly conserved residues are found at both the apex and the base of the protein (Fig. 4b, c). Interestingly, these residues seem to shield the interior from the solvent by capping both ends of the protein. In agreement with the Synechocystis structures, we also observe two cavities, termed the H4-H1 and H2-H3 cavities by Jackson et al. (2010), composed of well-conserved residues (Fig. 4d). The smaller H4-H1 cavity is formed by Ile45, Leu96 and Pro149. In the case of T. elongatus the larger H2-H3 cavity is composed of a cluster of Met78, Arg79, Leu82, Phe115 and Asp119 surrounding the Gly80Pro81 motif. In the vicinity of this cavity, but absent in our structure, is found one of the Zn2+ ions in Synechocystis CyanoQ (Jackson et al. 2010).
Comparison of CyanoQ and PsbQ
Currently there are two available structures of PsbQ from higher plants, both from spinach. The earlier structure (Calderone et al. 2003) lacks the first 37 residues whereas the later structure (Balsera et al. 2005) contains thirteen of these residues. Despite the low sequence similarity to spinach PsbQ, both CyanoQ and PsbQ are structurally similar (Table 2). One notable difference is the absence of the GlyPro helix breaking motif in algal or plant PsbQ sequences. As a consequence, the spinach structure shows a single 25 residue-long helix rather than the two helices (2a and 2b) observed in CyanoQ. In addition, PsbQ contains a much longer N-terminal sequence, which might be important for binding to PSII (Kuwabara et al. 1986).
All three crystallised proteins differ in their isoelectric points as calculated by Protparam (Gasteiger et al. 2005) with pI values of 4.5 for T. elongatus CyanoQ, 5.6 for Synechocystis CyanoQ and 9.25 for spinach PsbQ. This is reflected in their surface charge distribution (Fig. 5). Both CyanoQ proteins show only a small patch of positively charged surface around T. elongatus Arg109, whereas the equivalent region of the PsbQ protein contains a large patch of lysine residues thought to be involved in binding to PSII (Meades et al. 2005) (Fig. 5, top).
Fig. 5.
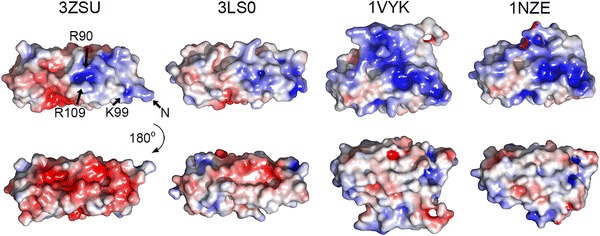
Solvent accessible surface charges of CyanoQ from T. elongatus (3ZSU), Synechocystis (3LS0) and spinach PsbQ (1VYK and 1NZE). Colour range spans from -5 (red) to 5 (blue) kT/e. Differences between the two spinach structures result from the fact that fewer residues could be fitted in 1NZE. Arrows point at Cα of selected residues. Arg109 is resolved in dual conformation
Significant differences in surface charge are also observed on the opposite faces of PsbQ and CyanoQ (Fig. 5): PsbQ is relatively uncharged whereas CyanoQ is negatively charged (Fig. 5, bottom row). Given the differences in composition of the extrinsic PSII subunits in cyanobacteria and plants, this face of the protein may be involved in interactions with these subunits or with assembly factors or possibly other protein components in the thylakoid membrane.
Comparison of zinc-binding sites
Zinc ions have been shown to bind to plant PsbQ (Calderone et al. 2003; Balsera et al. 2005) and CyanoQ from Synechocystis (Jackson et al. 2010), although the binding sites are not conserved (Fig. S7). Zinc has also been shown to bind to plant PsbP (Kopecky et al. 2012) and CyanoP from T. elongatus (Michoux et al. 2010) and Synechocystis (Jackson et al. 2012). The physiological relevance of these metal binding sites is currently unknown. In Synechocystis CyanoQ two zinc ions are coordinated by six amino-acid residues (Fig. 3 and Fig. S7). Despite the fact that five out of the six corresponding positions are occupied by potential metal ligands in T. elongatus CyanoQ, no zinc cations are present in the crystal structure. Unlike Synechocystis CyanoQ, where it was possible to obtain both zinc-bound and metal-free structures, our attempts to crystallise T. elongatus CyanoQ with zinc failed. Although there were no bound Zn2+ ions in our structure, we were able to fit a sulphate ion into the electron density. This anion is coordinated by three consecutive residues, Ser126ValThr128, found at the beginning of helix 4, at the apex of the protein.
Possible binding sites for CyanoQ in PSII
Very recent chemical cross-linking experiments have suggested that Synechocystis CyanoQ might interact with both PsbO and CP47, at the interface of the two monomeric PSII complexes (Liu et al. 2014). The cross-linking data indicate that Asp440 of CP47 (numbering according to Liu et al. 2014) is in van der Waal’s contact with Lys102 of Synechocystis CyanoQ, and that Lys120 of Synechocystis CyanoQ is within 12 Å of both Lys59 and Lys180 of PsbO. Although Asp440 of CP47 is conserved in both Synechocystis and T. elongatus, Lys102 and Lys120 of Synechocystis CyanoQ are replaced by Thr105 and Asp123, respectively, in T. elongatus CyanoQ (3ZSU numbering) (Fig. S8). These cross-linked residues in CyanoQ are found in a region containing helices α2a, α2b and α3 and the H2-H3 cavity (Jackson et al. 2010) (Fig. 4). Highly conserved residues Arg79 and Asp119 found in the H2–H3 cavity highlighted in Fig. 4d are therefore good candidates for interacting with PsbO, whereas residue Gln101 might interact with CP47 (Fig. S8).
In contrast, a recent structural analysis of the isolated PSII complex from the red alga Cyanidioschyzon merolae suggests that PsbQ’ binds near to CP43 (Krupnik et al. 2013) rather than CP47. Given the significant structural differences between PsbQ and CyanoQ with regard the N-terminus and surface charge, we do not yet exclude the possibility that PsbQ and CyanoQ bind at different locations in PSII.
Summary
We have provided evidence that CyanoQ binds to PSII complexes isolated from the thermophilic cyanobacterium T. elongatus, although the degree of association is dependent on the purification method. The crystal structures of CyanoQ and spinach PsbQ are very similar despite limited sequence identity with a four-helix bundle the common structural feature. This robust fold is likely to be conserved in the other members of the PsbQ family. Changes in the surface properties through mutation would explain how binding specificity could be altered to allow PsbQ-like proteins to bind outside PSII.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the staff of Diamond Light Source for their assistance, and the BBSRC (BB/E006388/1 and BB/I00937X/1) and EPSRC (EP/F00270X/1) for financial support. We are grateful to Dr Miwa Sugiura for providing the His-tagged CP43 strain of T. elongatus, and Dr Diana Kirilovsky for sending the His-tagged CP47 strain. Special thanks to Dr Michael Hippler for mass spectrometry analysis.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsera M, Arellano JB, Revuelta JL, de las Rivas J, Hermoso JA. The 1.49 Å resolution crystal structure of PsbQ from photosystem II of Spinacia oleracea reveals a PPII structure in the N-terminal region. J Mol Biol. 2005;350:1051–1060. doi: 10.1016/j.jmb.2005.05.044. [DOI] [PubMed] [Google Scholar]
- Bialek W, Wen S, Michoux F, Beckova M, Komenda J, Murray JW, Nixon PJ. Crystal structure of the Psb28 accessory factor of Thermosynechococcus elongatus photosystem II at 2.3 Å. Photosynth Res. 2013;117:375–383. doi: 10.1007/s11120-013-9939-6. [DOI] [PubMed] [Google Scholar]
- Boehm M, Nield J, Zhang P, Aro EM, Komenda J, Nixon PJ. Structural and mutational analysis of band 7 proteins in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2009;191:6425–6435. doi: 10.1128/JB.00644-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker TM, Roose JL, Fagerlund RD, Frankel LK, Eaton-Rye JJ. The extrinsic proteins of Photosystem II. Biochim Biophys Acta. 2012;1817:121–142. doi: 10.1016/j.bbabio.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Broser M, Gabdulkhakov A, Kern J, Guskov A, Muh F, Saenger W, Zouni A. Crystal structure of monomeric photosystem II from Thermosynechococcus elongatus at 3.6 Å resolution. J Biol Chem. 2010;285:26255–26262. doi: 10.1074/jbc.M110.127589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone V, Trabucco M, Vujicic A, Battistutta R, Giacometti GM, Andreucci F, Barbato R, Zanotti G. Crystal structure of the PsbQ protein of photosystem II from higher plants. EMBO Rep. 2003;4:900–905. doi: 10.1038/sj.embor.embor923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Rivas J, Roman A. Structure and evolution of the extrinsic proteins that stabilize the oxygen-evolving engine. Photoch Photobio Sci. 2005;4:1003–1010. doi: 10.1039/b506874f. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Enami I, Okumura A, Nagao R, Suzuki T, Iwai M, Shen JR. Structures and functions of the extrinsic proteins of photosystem II from different species. Photosynth Res. 2008;98:349–363. doi: 10.1007/s11120-008-9343-9. [DOI] [PubMed] [Google Scholar]
- Fagerlund RD, Eaton-Rye JJ. The lipoproteins of cyanobacterial photosystem II. J Photochem Photobiol B. 2011;104:191–203. doi: 10.1016/j.jphotobiol.2011.01.022. [DOI] [PubMed] [Google Scholar]
- Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Totowa: Humana Press; 2005. pp. 571–607. [Google Scholar]
- Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucl Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasse N, Mamedov F, Becker K, Styring S, Rogner M, Nowaczyk MM. Role of novel dimeric photosystem II (PSII)-Psb27 protein complex in PSII repair. J Biol Chem. 2011;286:29548–29555. doi: 10.1074/jbc.M111.238394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2.9 Å resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Schroder WP, Funk C. Functional analysis of the PsbP-like protein (sll1418) in Synechocystis sp. PCC 6803. Photosynth Res. 2005;84:257–262. doi: 10.1007/s11120-005-0477-8. [DOI] [PubMed] [Google Scholar]
- Jackson SA, Fagerlund RD, Wilbanks SM, Eaton-Rye JJ. Crystal structure of PsbQ from Synechocystis sp. PCC 6803 at 1.8 Å: implications for binding and function in cyanobacterial photosystem II. Biochemistry. 2010;49:2765–2767. doi: 10.1021/bi100217h. [DOI] [PubMed] [Google Scholar]
- Jackson SA, Hinds MG, Eaton-Rye JJ. Solution structure of CyanoP from Synechocystis sp. PCC 6803: new insights on the structural basis for functional specialization amongst PsbP family proteins. Biochim Biophy Acta. 2012;1817:1331–1338. doi: 10.1016/j.bbabio.2012.02.032. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kamiya N, Shen JR. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7 Å resolution. Proc Natl Acad Sci USA. 2003;100:98–103. doi: 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashino Y, Lauber WM, Carroll JA, Wang Q, Whitmarsh J, Satoh K, Pakrasi HB. Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry. 2002;41:8004–8012. doi: 10.1021/bi026012+. [DOI] [PubMed] [Google Scholar]
- Kashino Y, Inoue-Kashino N, Roose JL, Pakrasi HB. Absence of the PsbQ protein results in destabilization of the PsbV protein and decreased oxygen evolution activity in cyanobacterial photosystem II. J Biol Chem. 2006;281:20834–20841. doi: 10.1074/jbc.M603188200. [DOI] [PubMed] [Google Scholar]
- Kern J, Loll B, Luneberg C, DiFiore D, Biesiadka J, Irrgang KD, Zouni A. Purification, characterisation and crystallisation of photosystem II from Thermosynechococcus elongatus cultivated in a new type of photobioreactor. Biochim Biophy Acta. 2005;1706:147–157. doi: 10.1016/j.bbabio.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Kopecky V, Jr, Kohoutova J, Lapkouski M, Hofbauerova K, Sovova Z, Ettrichova O, Gonzalez-Perez S, Dulebo A, Kaftan D, Smatanova IK, Revuelta JL, Arellano JB, Carey J, Ettrich R. Raman spectroscopy adds complementary detail to the high-resolution X-ray crystal structure of photosynthetic PsbP from Spinacia oleracea. PLoS One. 2012;7:e46694. doi: 10.1371/journal.pone.0046694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupnik T, Kotabova E, van Bezouwen LS, Mazur R, Garstka M, Nixon PJ, Barber J, Kana R, Boekema EJ, Kargul J. A reaction center-dependent photoprotection mechanism in a highly robust photosystem II from an extremophilic red alga, Cyanidioschyzon merolae. J Biol Chem. 2013;288:23529–23542. doi: 10.1074/jbc.M113.484659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Murata T, Miyao M, Murata N. Partial degradation of the 18-kDa protein of the photosynthetic oxygen-evolving complex-a study of a binding-site. Biochim Biophys Acta. 1986;850:146–155. doi: 10.1016/0005-2728(86)90019-8. [DOI] [Google Scholar]
- Leslie AGW, Powell HR. Processing diffraction data with mosflm. In: Read RJ, Sussman JL, editors. Evolving Methods for Macromolecular Crystallography. Dordrecht: Springer; 2007. pp. 41–51. [Google Scholar]
- Liu H, Zhang H, Weisz DA, Vidavsky I, Gross ML, Pakrasi HB. MS-based cross-linking analysis reveals the location of the PsbQ protein in cyanobacterial photosystem II. Proc Natl Acad Sci USA. 2014;111(12):4638–4643. doi: 10.1073/pnas.1323063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meades GD, Jr, McLachlan A, Sallans L, Limbach PA, Frankel LK, Bricker TM. Association of the 17-kDa extrinsic protein with photosystem II in higher plants. Biochemistry. 2005;44:15216–15221. doi: 10.1021/bi051704u. [DOI] [PubMed] [Google Scholar]
- Michoux F, Takasaka K, Boehm M, Nixon PJ, Murray JW. Structure of CyanoP at 2.8 Å: implications for the evolution and function of the PsbP subunit of photosystem II. Biochemistry. 2010;49:7411–7413. doi: 10.1021/bi1011145. [DOI] [PubMed] [Google Scholar]
- Michoux F, Takasaka K, Boehm M, Komenda J, Nixon PJ, Murray JW. Crystal structure of the Psb27 assembly factor at 1.6 Å: implications for binding to photosystem II. Photosynth Res. 2012;110:169–175. doi: 10.1007/s11120-011-9712-7. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Chauvat F. Gene transfer and manipulation in the thermophilic cyanobacterium Synechococcus elongatus. Mol Gen Genet. 1996;252:93–100. doi: 10.1007/BF02173209. [DOI] [PubMed] [Google Scholar]
- Murray JW, Maghlaoui K, Kargul J, Sugiura M, Barber J. Analysis of xenon binding to photosystem II by X-ray crystallography. Photosynth Res. 2008;98:523–527. doi: 10.1007/s11120-008-9366-2. [DOI] [PubMed] [Google Scholar]
- Murray JW, Maghlaoui K, Kargul J, Ishida N, Lai T-L, Rutherford AW, Sugiura M, Boussac A, Barber J. X-ray crystallography identifies two chloride binding sites in the oxygen evolving centre of Photosystem II. Energ Environ Sci. 2008;1:161–166. doi: 10.1039/b810067p. [DOI] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczyk MM, Hebeler R, Schlodder E, Meyer HE, Warscheid B, Rogner M. Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell. 2006;18:3121–3131. doi: 10.1105/tpc.106.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Roose JL, Kashino Y, Pakrasi HB. The PsbQ protein defines cyanobacterial Photosystem II complexes with highest activity and stability. P Natl Acad Sci USA. 2007;104:2548–2553. doi: 10.1073/pnas.0609337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JR, Qian M, Inoue Y, Burnap RL. Functional characterization of Synechocystis sp. PCC 6803 delta psbU and delta psbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry. 1998;37:1551–1558. doi: 10.1021/bi971676i. [DOI] [PubMed] [Google Scholar]
- Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J Appl Cryst. 2008;41:641–643. doi: 10.1107/S0021889808006985. [DOI] [Google Scholar]
- Sugiura M, Inoue Y. Highly purified thermo-stable oxygen-evolving photosystem II core complex from the thermophilic cyanobacterium Synechococcus elongatus having His-tagged CP43. Plant Cell Physiol. 1999;40:1219–1231. doi: 10.1093/oxfordjournals.pcp.a029510. [DOI] [PubMed] [Google Scholar]
- Sugiura M, Iwai E, Hayashi H, Boussac A. Differences in the Interactions between the subunits of photosystem II dependent on D1 protein variants in the thermophilic cyanobacterium Thermosynechococcus elongatus. J Biol Chem. 2010;285:30008–30018. doi: 10.1074/jbc.M110.136945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield TC, Shand JA, Bentley FK, Eaton-Rye JJ. PsbQ (Sll1638) in Synechocystis sp. PCC 6803 is required for photosystem II activity in specific mutants and in nutrient-limiting conditions. Biochemistry. 2005;44:805–815. doi: 10.1021/bi048394k. [DOI] [PubMed] [Google Scholar]
- Thornton LE, Ohkawa H, Roose JL, Kashino Y, Keren N, Pakrasi HB. Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechocystis 6803. Plant cell. 2004;16:2164–2175. doi: 10.1105/tpc.104.023515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujihara T, Sakurai I, Mizusawa N, Wada H. A method for analyzing lipid-modified proteins with mass spectrometry. Anal Biochem. 2008;374:429–431. doi: 10.1016/j.ab.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta crystallogr D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta S, Ifuku K, Takabayashi A, Ishihara S, Ido K, Ishikawa N, Endo T, Sato F. Three PsbQ-like proteins are required for the function of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 2010;51:866–876. doi: 10.1093/pcp/pcq060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


