Abstract
Prospective studies focusing on EGFR inhibitors in African Americans with NSCLC have not been previously performed. In this phase II randomized study, 55 African Americans with NSCLC received erlotinib 150mg/day or a body weight adjusted dose with subsequent escalations to the maximum allowable, 200mg/day, to achieve rash. Erlotinib and OSI-420 exposures were lower compared to previous reports, consistent with CYP3A pharmacogenetics implying higher metabolic activity. Tumor genetics revealed only two EGFR mutations, EGFR amplification in 17/47 samples, 8 KRAS mutations and 5 EML4-ALK translocations. Although absence of rash was associated with shorter time to progression (TTP), disease control rate, TTP, and 1-year survival were not different between the two dose groups, indicating the dose-to-rash strategy failed to increase clinical benefit. Observed low incidence of toxicity and low erlotinib exposure suggest standardized and maximum allowable dosing may be suboptimal in African Americans.
Keywords: EGFR, Erlotinib, African American, Pharmacokinetics, Pharmacogenetics
INTRODUCTION
African Americans have a higher incidence and overall mortality of lung cancer compared to Caucasians.1 The reasons for these disparities have been debated extensively, with arguments including increased susceptibility and more aggressive biologic features for lung cancer in African Americans, increased stage of disease at diagnosis, and less aggressive treatment being offered for early-stage disease.2–6 Survival analyses of patients participating in cooperative groups clinical trials, which provide for uniform extent of disease and therapy, showed statistically significantly worse survival at one year for African American patients.7
This disparity suggests an even more pressing need to develop new anticancer agents to target lung cancer in this patient population, and the low rate of enrollment of African Americans in cancer clinical trials is distressing. Moreover, the development of most novel cancer therapeutic agents occurs exclusive of African Americans. Potential metabolism/drug-disposition, tolerance, and drug target frequency issues for this ethnicity, which may be relevant to the safety and/or efficacy of these agents, are often ignored until the drugs are widely available to the general population.
Interest in molecular targeted therapy for lung cancer has been stimulated by the success of tyrosine kinase inhibitors (TKis) (i.e., gefitinib, erlotinib) in the setting of NSCLC tumors harboring epidermal growth factor receptor (EGFR) mutations (approximately 10% of NSCLC tumors).8–12 Common EGFR activating mutations in NSCLC that are sensitive to TKI therapies include amino acid deletions in exon 19 and the L858R point mutation at exon 21.8,9 However, erlotinib has been shown to be effective at delaying progression and improving overall survival (OS) as second- and third-line treatment of NSCLC, even in unselected patients, thus its current labeling.12,13 Clinical benefit from this agent may therefore be mediated by additional factors, including tumor mutations outside EGFR mutation hotspots, pharmacokinetics, pharmacogenetics, gender and race.13,14 Furthermore, the primary toxicity of this agent, skin rash, has been correlated with response and survival.15
Recent data suggests pharmacogenetic factors may influence disposition of TKIs. For example, erlotinib is inactivated through CYP3A4, CYP3A5, CYP1A1 and CYP1A2 metabolism.16,17 Ethnic diversity in cytochrome P450 polymorphisms and enzyme activity has been well described, and lower erlotinib exposure in African Americans may be expected due to metabolism from higher CYP3A4/5 and CYP1A1/2 expression compared to Caucasians.18–20 Furthermore, previous reports evaluating genetic diversity in CYP1A1/2 suggest genotypes less prevalent in African Americans may confer protective effects in never smokers and increased risk of lung cancer in ever smokers.21,22
In this first of its kind trial, we aimed to prospectively explore the roles of dosing strategy, smoking, pharmacokinetics, pharmacogenetics, and tumor genetics on outcomes following erlotinib treatment in African Americans with NSCLC.
RESULTS
Patient Characteristics
Fifty-eight patients were consented between April 2006 and July 2011. Prior to receiving treatment, one patient withdrew consent and two patients were removed from study due to deteriorating health status. Pretreatment characteristics for the 55 patients treated on study are presented in Table 1. Prior to the 3-month period for response assessment one patient was removed from the study for evaluation of pulmonary toxicity (day 42). Two additional patients died prior to response assessment (sudden death due to likely pulmonary embolism in one patient and clinical deterioration in the other). The remaining 52 patients were evaluable for the primary endpoint of DCR.
Table 1.
Patient characteristics
| Number of patients (%) |
|||
|---|---|---|---|
| Arm A (fixed dose) |
Arm B (dose adjusted) |
All (A + B) |
|
| Number of enrolled patients | 28 | 27 | 55 |
| Men | 17 (60.7) | 14 (51.9) | 31 (56.4) |
| Age (years) | |||
| Median | 63 | 59 | 62 |
| Range | 29–83 | 37–96 | 29–96 |
| Weight (Kg) | |||
| Mean | 75.8 | 72.5 | 74.2 |
| Standard Deviation | 15.2 | 13.7 | 14.4 |
| Performance Status | |||
| 0 | 7 (25.0) | 6 (22.2) | 13 (23.6) |
| 1 | 14 (50.0) | 17 (63.0) | 31 (56.4) |
| 2 | 7 (25.0) | 4 (14.8) | 11 (20.0) |
| Histologic cell type | |||
| Adenocarcinoma | 15 (53.6) | 17 (63.0) | 32 (58.2) |
| Squamous | 7 (25.0) | 5 (18.5) | 12 (21.8) |
| NSC not otherwise classified | 5 (17.9) | 2 (7.4) | 7 (12.7) |
| Adenosquamous | 0 | 1 | 1 |
| Large cell neuroendocrine | 1 | 1 | 2 |
| Sarcomatoid | 0 | 1 | 1 |
| Prior Chemotherapy | |||
| No | 4 (14.3) | 4 (14.8) | 8 (14.5) |
| 1 regimen | 19 (67.9) | 15 (55.6) | 34 (61.8) |
| 2 regimens | 5 (17.9) | 8 (29.6) | 13 (23.6) |
| Previous radiation therapy | 11 (39.3) | 11 (40.7) | 22 (40.0) |
| Smoking Status | |||
| Current | 8 (28.6) | 13 (48.1) | 21 (38.2) |
| Former | 15 (53.6) | 14 (51.9) | 29 (52.7) |
| Never | 5 (17.9) | 0 (0) | 5 (9.1) |
Treatment Toxicity
Fifty-five enrolled patients received therapy. All but 8 patients received 150 mg as starting dose; 6 and 2 patients received 175 and 200 mg starting doses, respectively. Of the 9 patients in Arm B who did not experience rash during the first cycle, 1 started at the maximum dose (200 mg), and 5 others did not receive treatment in cycle 2. All 3 patients in Arm B who were dose escalated started at 150 mg and received maximum escalated doses of 175 mg (n=1) or 200 mg (n=2). Missed doses were minimal overall, especially in cycles 1 and 2, according to patient diaries. Adverse events and treatment-related toxicities experienced during the trial are summarized in the Table 2. The predominant toxicity was rash (69% of patients), followed by anorexia and diarrhea (29% each), and nausea (15%). Overall, toxicities were mild, the majority being of grade 1–2, and only 11 patients (20%) experienced any grade 3 toxicity. Severe events included a grade 5 gastrointestinal hemorrhage from a gastric ulcer one month after beginning therapy, a grade 3 interstitial pneumonitis, a grade 3 dehydration, and a grade 2 bowel obstruction. The patient with interstitial pneumonitis was removed from study; there were no other withdrawals due to drug-related adverse events, and no dose reductions for toxicity.
Table 2.
Treatment Related Toxicities
| Toxicity, no. of pts (%) | Arm A (n=28) | Arm B (n=27) | All (n=55) |
|---|---|---|---|
| Any grade 3+ toxicity | 5 (18) | 6 (22) | 11 (20) |
| Any grade 4/5 toxicity | 1 (4) | 0 | 1 (2) |
| Rash, maximum grade | |||
| Any grade | 19 (68) | 19 (70) | 38 (69) |
| Grade 1 | 12 (43) | 10 (37) | 22 (40) |
| Grade 2 | 7 (25) | 5 (19) | 12 (22) |
| Grade 3 | 0 | 4 (15) | 4 (7) |
| Dermatologic, other than rash* | |||
| Any, grade 1–3 | 7 (25) | 9 (33) | 16 (29) |
| Diarrhea | |||
| Any grade | 9 (32) | 7 (26) | 16 (29) |
| Grade 1 | 7 (25) | 7 (26) | 14 (25) |
| Grade 2 | 2 (7) | 0 | 2 (4) |
| Grade 3+ | 0 | 0 | 0 |
| Other gastrointestinal | |||
| Anorexia, grade 1–2 | 5 (18) | 9 (33) | 14 (25) |
| Anorexia, grade 3 | 2 (7) | 1 (4) | 3 (5) |
| AST elevation | 4 (14) | 0 | 4 (7) |
| Dehydration, grade 1–2 | 0 | 1 (4) | 1 (2) |
| Dehydration, grade 3 | 1 (4) | 1 (4) | 2 (4) |
| Hyperbilirubinemia | 0 | 1 (4) | 1 (2) |
| Nausea, grade 1–3** | 2 (7) | 6 (22) | 8 (15) |
| Xerostomia | 0 | 2 (7) | 2 (4) |
| Other | |||
| Pneumonitis, grade 3 | 1 (4) | 0 | 1 (2) |
erythema, hyperpigmentation, pruritus, xerodermia.
only one grade three nausea was recorded
Treatment Efficacy
A total of 12 patients had their disease controlled at 3 months; six for each arm. In arm A DCR, TTP, and 1-year OS were 23% (95% CI, 9%–44%), 2.8 months (95% CI, 2.5–3.1), and 30% (95% CI, 13–47%), respectively (Table 3). In arm B DCR, TTP, and 1-year OS were 23% (95% CI, 9%–44%), 2.4 months (95% CI, 1.5–2.7) and 26% (95% CI, 9–42%). When patients were stratified by rash, median TTP for patients with rash (n=38) compared to patients not experiencing rash (n=17) was significantly different (TTP 2.8 vs. 1.8 months, p=0.036). However, median OS was not significantly different (5.2 vs. 5.3 months, p=0.30). Severity of rash (grades 2–3 vs. grade 1) was not associated with differences in outcomes.
Table 3.
Efficacy
| Number of patients (%) | |||
|---|---|---|---|
| Arm A (n=26) | Arm B (n=26) | All (n=52)* | |
| DCR at 3 months | 6 (23) | 6 (23) | 12 (24) |
| PR | 1 (4) | 0 | 1 (2) |
| SD | 5 (19) | 6 (23) | 11 (22) |
| PD | 20 (77) | 20 (77) | 40 (78) |
| Arm A (n=28) | Arm B (n=27) | All (n=55)** | |
| TTP, months | |||
| Median (95% CI) | 2.8 (2.5–3.1) | 2.4 (1.5–2.7) | 2.6 (1.9– 2.8) |
| Range | 0.6–14.4 | 0.3–20.2 | 0.3–20.2 |
| 1 year progression free rate (%) | 10 | 4 | 7 |
| Survival, months | |||
| Median | 5.3 | 5.2 | 5.3 |
| Range | 0.9–37.4 | 1.3–42.8 | 0.9–42.8 |
| 1 year survival rate (%) | 30 | 26 | 28 |
responses based on evaluable patients
based on all 55 treated patients
Non-compartmental pharmacokinetics
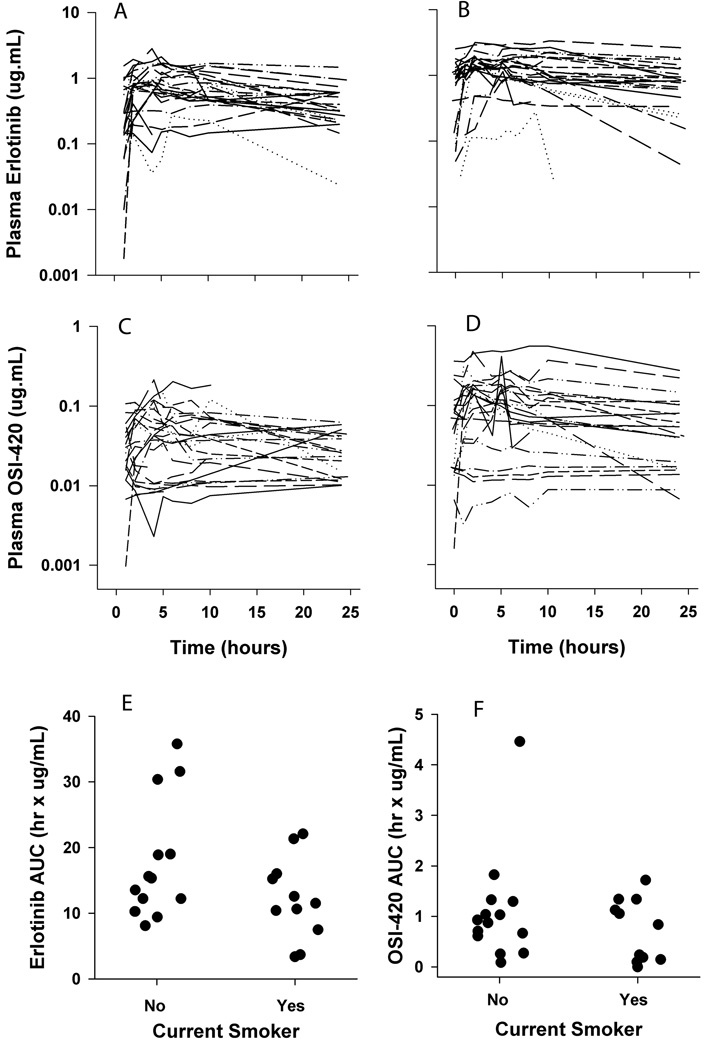
Pharmacokinetic data was obtained from 26 patients who received either 150 mg (n=20) or 175 mg erlotinib (n=5) during cycle 1. Figure 1 displays observed concentration-time profiles for erlotinib and OSI-420 for all patients on cycles 1 and 2. Non-compartmental parameter estimates for both agents are displayed in Table 4 (Table S1 also displays the data separated by dose level). No associations were observed between rash and erlotinib pharmacokinetics. However, lower mean OSI-420 Cmax was observed in patients with no or mild rash (grades 0,1; 108 ± 87 ng/mL) compared to patients with more severe rash (grades 2,3; 252 ± 182 ng/mL, p-value = .023, 2-tailed t-test). Furthermore, trends were observed where current smokers had overall decreased erlotinib AUC0–24 on day 1 of cycle 1 (12.2 ± 6.2 mg/L*hrs, n=11) compared to former/never smokers (17.4 ± 8.9 mg/L*hrs, n=14, Student’s 2-tailed t-test P = 0.12; see Figure 1 and Supplemental Figure S3). This trend was not apparent beyond cycle 1, but OSI-420 concentrations did trend higher in smokers compared to non-smokers in cycle 2 (see Supplemental Figure S4). No statistically significant associations were observed between erlotinib pharmacokinetics and outcomes, including progression-free and overall survival.
Figure 1. Pharmacokinetics and Impact of Smoking for Erlotinib (OSI-774) and OSI-420.
Semi-log concentration vs. time plots of erlotinib (panels A and B) and OSI-420 (panels C and D). Each compound was simultaneously measured with a validated LC/MS/MS assay in patient plasma samples up to 24 hours after the first dose in cycle 1 (panels A and C) and cycle 2 (panels B and D). Dot plots showing smoking status vs. (E) erlotinib (OSI-774) and (F) OSI-420 AUC during cycle 1.
Table 4.
Noncompartmental Pharmacokinetic Parameters for Erlotinib and OSI-420 in Cycles 1 and 2
| Cycle 1 |
Cycle 2 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter* (units) |
Arm A | n¥ | Arm B | n | Total | n | Arm A | n | Arm B | n | Total | n |
| Erlotinib (OSI-774) | ||||||||||||
| AUC0-24** (mg/L*hr) |
15.3 (7.5–35.8) |
13 | 12.4 (3.4–31.6) |
12 | 12.6 (3.4–35.8) |
25 | 26.3 (8.9–51.0) |
14 | 21.4 (1.4–71.5) |
10 | 22.9 (1.4–71.5) |
24 |
| Cmax£ (mg/L) |
0.91 (0.32–2.1) |
13 | 0.79 (0.16–1.9) |
12 | 0.88 (0.16–2.1) |
25 | 1.7 (0.48–3.2) |
14 | 1.6 (0.28–3.1) |
10 | 1.6 (0.28–3.2) |
24 |
| CL/F§ (L/hr) |
9.8 (4.2–20.1) |
13 | 13.5 (5.5–44.5) |
12 | 12.3 (4.2–44.5) |
25 | 5.7 (2.9–16.8) |
14 | 7.6 (2.1–110) |
10 | 6.6 (2.1–110) |
24 |
| Tmax† (hrs) |
5 (1–24) |
13 | 4 (1–8) |
12 | 4 (1–24) |
25 | 2 (1–6) |
14 | 2.5 (0–10) |
10 | 2 (0–10) |
24 |
| T1/2‡ (hrs) |
11.6 (2.1–116) |
11 | 11.2 (4.7–29.9) |
9 | 11.6 (2.1–116) |
20 | 28.4 (5.7–68.5) |
14 | 19.7 (1.7–45.6) |
7 | 22.5 (1.7–68.5) |
21 |
| OSI-420ƛ | ||||||||||||
| AUC0-24 (mg/L*hr) |
0.93 (0.088–4.5) |
13 | 0.69 (0–1.7) |
11 | 0.87 (0–4.5) |
24 | 1.4 (0.19–10.7) |
14 | 1.7 (0–6.9) |
9 | 1.5 (0–10.7) |
23 |
| AUC Ratio€ 420/774 |
0.068 (0.004–0.129) |
13 | 0.056 (0–0.109) |
11 | 0.066 (0–0.127) |
24 | 0.102 (0.011–0.209) |
14 | 0.094 (0–0.149) |
9 | 0.096 (0–0.209) |
23 |
| Cmax (mg/L) |
0.070 (0.010–0.203) |
13 | 0.048 (0–0.119) |
11 | 0.054 (0–0.203) |
24 | 0.100 (0.009–0.558) |
14 | 0.178 (0–0.412) |
9 | 0.153 (0–0.558) |
23 |
| Tmax (hrs) |
5 (1–24) |
13 | 2 (1–11.4) |
11 | 4 (1–24) |
24 | 2 (0–10) |
14 | 2.5 (1–6) |
9 | 2 (0–10) |
23 |
| T1/2 (hrs) |
9.6 (7–70.4) |
8 | 12.3 (3.2–40.8) |
6 | 9.7 (3.2–70.4) |
14 | 22.2 (6–71.1) |
10 | 17.7 (3.4–28.8) |
6 | 22.2 (3.4–71.1) |
16 |
Data are presented as median (range).
AUC, area under the observed concentration vs. time curve;
Cmax, maximum observed concentration;
CL/F, apparent clearance/oral bioavailability (F);
Tmax, time of observed maximum concentration;
T1/2, terminal phase half-life (note half-lives were too long to be determined for some patients);
n, the number of concentration vs. time profiles used for each median value;
AUC Ratio, the ratio of AUCs calculated as (AUC OSI-420)/(AUC OSI-774).
OSI-420 was undetectable in one patient treated on Arm B
Pharmacogenetics
Erlotinib is metabolized primarily by CYP3A4/5 and CYP1A1/216,17 and actively excreted via ABCB1 and ABCG2.23,24 Genetic polymorphisms in these genes were evaluated in the first 26 patients on study. Additional genotyping was completed for CYP3A4, CYP3A5, and ABCB1 in 16 additional patients. These data are summarized in Table 5.
Table 5.
Pharmacogenetics of Select Genes Relevant for Erlotinib Disposition
| Genotype* | Allele | Freq** | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP ID | M/M | n | M/m | n | m/m | n | M | m | m |
| CYP1A1 | rs1048943 | A/A | 25 | A/G | 1 | G/G | 0 | 51 | 1 | 0.019 |
| rs2606345 | C/C | 20 | C/A | 5 | A/A | 1 | 45 | 7 | 0.135 | |
| rs4646421 | C/C | 10 | C/T | 9 | T/T | 1 | 29 | 11 | 0.275 | |
| CYP1A2 | rs2069526 | T/T | 20 | T/G | 2 | G/G | 2 | 42 | 6 | 0.125 |
| rs762551 | C/C | 8 | C/A | 3 | A/A | 5 | 19 | 13 | 0.406 | |
| rs2470890 | C/C | 9 | C/T | 1 | T/T | 0 | 19 | 1 | 0.050 | |
| CYP3A4 | rs2740574 | G/G | 9 | G/A | 14 | A/A | 3 | 32 | 20 | 0.385 |
| rs35599367 | C/C | 41 | C/T | 1 | T/T | 0 | 83 | 1 | 0.012 | |
| CYP3A5 | rs776746 | A/A | 10 | A/G | 20 | G/G | 9 | 40 | 38 | 0.487 |
| rs41303343 | -/- | 22 | -/T | 4 | T/T | 0 | 48 | 4 | 0.077 | |
| T12952C | T/T | 22 | T/C | 3 | C/C | 0 | 47 | 3 | 0.060 | |
| C31611T | C/C | 5 | C/T | 14 | T/T | 6 | 24 | 26 | 0.520 | |
| ABCB1 | rs1128503 | C/C | 11 | C/T | 2 | T/T | 2 | 24 | 6 | 0.200 |
| rs1045642 | C/C | 18 | C/T | 13 | T/T | 2 | 49 | 17 | 0.258 | |
| rs1564481 | C/C | 11 | C/T | 9 | T/T | 0 | 31 | 9 | 0.225 | |
| ABCG2 | rs2622605 | T/T | 18 | T/C | 7 | C/C | 1 | 43 | 9 | 0.173 |
| rs2622624 | A/A | 11 | A/G | 7 | G/G | 1 | 29 | 9 | 0.237 | |
| rs3114018 | A/A | 8 | A/C | 10 | C/C | 2 | 26 | 14 | 0.350 | |
The number of patients (n) with genotypes for major (M) and minor (m) alleles.
Freq is the frequency of minor allele occurrence.
Thirty of 39 (77%) patients evaluated were homozygous (*1/*1) or heterozygous (*1/*3) for the CYP3A5 rs776746 A allele, suggesting they had functional expression of CYP3A5. Of the 9 patients homozygous for the G allele, only 3 had pharmacokinetic data for both cycles, thus limiting statistical comparison of this polymorphism and erlotinib or OSI-420 pharmacokinetics. Notably, one of the 3 patients with the G/G genotype had no measureable OSI-420 during either cycle 1 or 2, potentially reflecting dysfunctional CYP3A5 and limited formation of the metabolite. AUC0–24 ratios of OSI-420/OSI-774 were low to moderate for the other two (e.g. 0.022 and 0.060 for cycle 1) compared to remaining patients with PK data (range 0.005 – 0.129).
Recently, the CYP3A4 rs35599367 (*22) polymorphism was shown to associate significantly with pharmacokinetics and dose titration of statins, tacrolimus and cyclosporine.25–28 Data from these studies suggest this polymorphism, which reduces CYP3A4 activity, is less prevalent in African Americans compared to other populations studied. Consistent with previous data, all but one of 42 patients evaluated for the rs35599367 polymorphism were homozygous for the major C allele. This patient was a male former smoker with EGFR gene amplification who received 150 mg erlotinib. He was also one of 9 patients homozygous for the CYP3A5*3 G allele. Pharmacokinetic data was not available for this patient. He achieved grade 1 rash during cycle 1, and his best response was progressive disease.
Erlotinib is also a known substrate of CYP1A1/2 and multidrug resistance transporters, P-glycoprotein (P-gp, ABCB1) and breast cancer resistance protein (BCRP, ABCG2). A total of 13 polymorphic variants were evaluated for these genes. Frequencies for these variants were similar to those previously reported (see Table 5).20 No significant associations were identified with these genes.
We also examined relationships between individual polymorphisms and outcomes from therapy, including rash and response. No trends or significant associations were found.
Population pharmacokinetics and covariate analysis
The final structural population pharmacokinetic model constructed in this study differed from previously published models. It utilized one-compartment models for both erlotinib and OSI-420, and it included inter-individual variability (IIV) parameters on erlotinib clearance (CL), volume of distribution (V1), and OSI-420 clearance (CLM). A shared ETA approach was used for CL and CLM, as these were observed to be highly correlated (0.86 correlation coefficient). We also included a single inter-occasion variability (IOV) parameter, which was estimated on CL. The final residual error model included a single exponential term for each, erlotinib and OSI-420. The erlotinib first-order absorption rate constant (Ka) was estimated with poor precision (coefficient of variation, CV > 80%) and was therefore fixed to 1.04 h−1 based on estimated values from the model and comparison with other reported values.29,30 Table 6 lists estimated parameter values for the base model. Notable results include high precision for all base structural model parameters (less than 16% CV), except V2 which had a 62.8% CV. However, IIVCL was estimated with relatively poor precision with 80.8% CV and a lower confidence interval (CI) boundary of 0.005. Also, while the shared ETA scale factor had moderate precision (37.0% CV), the base model bootstrap CI included zero (−148.10, 16.11). Ultimately, when re-evaluating the shared ETA in the covariate model, it maintained acceptable precision (56.5 CV%), and its bootstrap confidence interval no longer contained zero. The shared ETA scale factor was therefore retained in the model to address the high correlation between CL and CLM.
Table 6.
Population PK parameter estimates for erlotinib and OSI-420
| Model Term | Base model (n=26) | Covariate model (LBM, n=26) | ||
|---|---|---|---|---|
| Estimate (CV %) | Bootstrap median (95% CI) |
Estimate (CV %) | Bootstrap median (95% CI) |
|
| OBJ | 968.48 | -- | 963.26 | -- |
| θKa (h−1) | 1.04 | -- | 1.04 | -- |
| θCL (L/h) | 6.22 (8.5) | 6.24 (4.72, 8.11) | 6.4 (9.0) | 6.37 (4.99, 8.09) |
| θV1 (L) | 284 (15.7) | 275.00 (193.00, 448.05) | 283 (37.1) 2 | 74.00 (195.00, 447.03) |
| θFM | 1.41 (12.8) | 1.22 (0.47, 3.33) | 0.88 (26.4) | 1.03 (0.23, 3.49) |
| θCLM (L/h) | 21.5 (8.9) | 23.60 (4.76, 47.91) | 32.6 (10.4) | 27.80 (4.04, 55.60) |
| θV2 (L) | 7.18 (62.8) | 7.67 (0.09, 21.91) | 11.6 (12.1) | 8.99 (0.04, 26.11) |
| Shared ETA scale factor, θSE |
3.46 (37.0) | 3.18 (−148.10, 16.11) | 5.96 (56.5) | 6.42 (2.29, 577.30) |
| θLBM | -- | -- | 1.71 (27.8) | 1.75 (0.69, 3.08) |
| IIVCL | 0.06 (80.8) | 0.24 (0.005, 0.44) | 0.02 (127.1) | 0.13 (0.001, 0.35) |
| IIV V1 | 0.92 (50.5) | 0.90 (0.40, 1.33) | 0.95 (50.7) | 0.92 (0.43, 1.37) |
| IOV CL | 0.38 (55.5) | 0.58 (0.29, 0.89) | 0.30 (67.9) | 0.51 (0.15, 0.85) |
| Residual error for erlotinib, ε1 |
1.2 (37.2) | 1.08 (0.66, 1.47) | 1.2 (37.1) | 1.07 (0.65, 1.48) |
| Residual error for OSI-420, ε2 |
0.78 (23.3) | 0.87 (0.37, 1.06) | 0.78 (24.2) | 0.87 (0.67, 1.06) |
OBJ, objective function value; LBM, lean body mass; IIV, inter-individual variability; IOV, inter-occasion variability; CV%, standard error of estimate expressed as coefficient of variation (%); The shared eta scale factor was estimated as described in the text. IIV, IOV and residual errors are expressed as variances.
Covariate analysis was conducted to identify sources of variability between patients. Several covariates were evaluated for effect on model parameters, including age, weight, sex, smoking status, lean body mass (LBM), pharmacogenetics, and co-medications. Only LBM was found to significantly reduce the objective function value (OBJ). Only 2 patients received a CYP3A4/5 inhibitor (voriconazole) or inducer (phenytoin) during therapy. Ten patients received CYP1A2 inducers (6 omeprazole, 5 insulin, 1 received both). Several patients received CYP1A1/2 and CYP3A4/5 substrates (the most common of these were acetaminophen and oxycodone). There were no evident relationships between co-medications and pharmacokinetics in this study. The population PK parameter estimates in bootstrap analyses were comparable with those generated using the original data set, indicating acceptable accuracy and stability of the structural and covariate models (see Table 6). One exception to this was IIVCL, for which the bootstrap median was approximately 6-fold higher compared to the model estimated value for the original dataset (0.13 vs. 0.02, respectively). Again, as for the base model, the standard error was relatively high for this estimate (127.1% CV), and the confidence interval had a lower boundary near zero (0.001).
Tumor Genetics
Tissue in sufficient quantities for evaluation of one or more molecular aberrations was available from 53 patients. Tumor genetic data is summarized in Table 7. EGFR exon 19 insertions or deletions and exon 21 mutations were successfully evaluated in 45 patients, and only one patient had an exon 19 short in-frame deletion. Notably, an additional EGFR (T790M) mutation31 was present in this patient at a biopsy obtained at progression after two years on erlotinib. A second patient had an EGFR R776L mutation. Seventeen of 47 patients (36%) scored positive for EGFR gene amplification (defined as 4–10 EGFR gene copies in >20% of the tumor cells independent of EGFR to CEP7 ratio). Interestingly, the ALK gene was translocated in 5 of 39 patients evaluated (13%), and one additional patient had ALK amplification (>10 copies). KRAS mutations were identified in 8 of 46 patients (17%); of these KRAS mutations, 6 were of the G12 type32, and two were of uncertain significance (C/T genotypes in the 5’-UTR, i.e., −9 and −6 positions).
Table 7.
Tumor Genetics
| Tested | Positive (%) | No. of DC at 3m | |
|---|---|---|---|
| EGFR mutations | 45 | 2 (4) | 1 |
| EExon 19 indel | F | 1 | 1 |
| Exon 21 mutation | 0 | 0 | |
| R776L mutation | 1 | 0 | |
| EGFR gene amplification | 47 | 17 (36) | 3 |
| ALK gene translocation | 39 | 5 (13) | 0 |
| ALK gene amplification | 39 | 1 | 1 |
| KRAS mutation | 46 | 8 (17) | 2 |
| G12C (c.32 G>T) | 4 | 0 | |
| G12D (c.33 G>A) | 1 | 0 | |
| G12V (c.33 G>T) | 1 | 1 | |
| 5’-UTR (−9C>T, −6 C>T) | 2 | 1 | |
| Total | 32 | 7 | |
There were no clear associations between DC and absence (5 patients) or presence (7 patients) of most molecular abnormalities (see Table 7). Notably, four of the five patients with ALK translocation had progressive disease as best response and the fifth was not evaluable due to death before a restaging scan could be performed. Four of these patients were current smokers.
DISCUSSION
African Americans suffer from a heavier burden of NSCLC compared to other racial and ethnic groups within the U.S., and standard treatment regimens produce less favorable outcomes in African Americans for reasons that are not clear. The approval of EGFR tyrosine kinase inhibitors has yielded increased availability of treatment choices for patients with NSCLC, although there is paucity of prospective data regarding the benefits of these choices for African Americans. To address this gap we set out to prospectively evaluate the efficacy and toxicity of erlotinib at standard doses and with an alternative dose-to-rash strategy, as well as to characterize drug disposition, pharmacogenetics, and tumor/host factors as potential contributors to resistance for these agents in African Americans with NSCLC.
Erlotinib-related rash has been heralded as a potentially predictive factor for erlotinib activity. In theory, higher dosing leading to higher erlotinib exposure may lead to higher EGFR inhibition and pharmacodynamic effects, such as rash and tumor response in patients with EGFR driven tumors. Interestingly, rash was not correlated with erlotinib exposure, although it did directly associate with OSI-420 exposure. Absence of rash was correlated with shorter TTP in our study, although presence of rash was not associated with improvements in overall survival.
With regard to the dose to rash strategy employed in this study, 18 of the 27 enrolled patients in Arm B experienced rash during the first cycle, and one additional patient achieved rash beyond cycle 1. However, only 3 of the 9 potential patients for dose escalation received higher than standard doses of erlotinib. Five patients were not treated on cycle 2, and 1 patient had started cycle 1 at the maximum allowable dose of 200 mg. Two of the 3 patients who were dose escalated also reached the maximum allowable dose of 200 mg without rash, and further escalation was not permitted per protocol design and IND restrictions at the time the study was initiated.
The available PK data shows erlotinib exposure in patients receiving 175 mg tended to be similar to that of patients receiving the 150 mg dose (see Table S1). This is likely a function of the relatively small number of patients receiving the 175 mg dose level, the relatively small difference between the doses, and the wide variability observed with TKI therapies. The data also show that toxicities were overall mild in this group of patients, similar to those reported in other studies using either 21 day cycles or continuous 150 mg erlotinib doses.in 81 Caucasian and 1242 Asian NSCLC patients12,33 However, in these previous studies, dose reductions due to adverse events were more frequent (14% and 23%, respectively) compared to no required dose reductions in our study. Although we did not monitor plasma drug levels in patients beyond cycle 2, noncompliance was low according to patient dose diaries. Collectively, this information suggests that further dose escalation may have been tolerated and that inclusion of plasma AUC for dose escalation decisions (even beyond the 200 mg level) may have been a more productive strategy.
Pharmacogenetic factors more prevalent within the African American population that are associated with altered CYP1A and CYP3A activities are well documented and likely alter PK of drugs available for treatment of NSCLC, such as erlotinib. Pharmacogenetic factors observed in our study population were in fact suggestive of high CYP3A activity, consistent with most published literature. Only 2% and 26% of subjects harbored the most functionally relevant poor metabolizing genotypes, CYP3A4*22 T allele (rs35599367) and CYP3A5*3 A/A genotype (rs776746), respectively. Similarly low prevalence of CYP1A1/2 alleles with reduced enzymatic activities were also observed in our study population. Only 17% of 24 subjects carry the CYP1A2*1K allele (rs2069536), and on the contrary 50% of 16 subjects carry the CYP1A2*1F allele (rs762551) which is highly inducible (http://www.cypalleles.ki.se/, accessed 11 December 2013).
While overall non-compartmental single dose and steady state PK data in this study was generally similar to that reported in other studies30,34, estimated erlotinib clearance from nonlinear mixed-effects modeling was higher in our study (6.22 ± 1.52 L/hr) compared to those reported in any other study (range of population estimates for CL/F, 3.29–5.82 L/hr)13,30,34–36. Notably, the highest population estimate of CL/F in the previous studies was obtained by Kraut and colleagues, where erlotinib was combined with docetaxel, a known inducer of CYP3A enzyme activity37,38.
African Americans also have a historically higher incidence of tobacco smoking,39 which is associated with higher erlotinib clearance and has therefore been argued to contribute to the observed outcomes disparities.39,40 Despite randomization in our study, an unequal distribution of smokers was observed between the control and experimental arms of the study. Current smokers comprised 48% and 29% of the experimental Arm B and Control Arm A, respectively. Pharmacokinetics analysis within the limits of the small number of patients evaluated (n = 26) did show differences (albeit not statistically significant) between current- and former/never-smokers (Figure 1). These data may indicate the intent to increase erlotinib exposure in patients treated on the experimental arm was further inhibited by an increased proportion of smokers randomly assigned to that arm.
Outcomes data indicated DCR at 3 months was similar to what would be expected with standard erlotinib doses in a population of previously treated patients with largely EGFR wild type tumors.41,42,13 As reported by others43 the prevalence of EGFR mutations was low in this population with high prevalence of tobacco use. However, a surprisingly high number of patients were found to have ALK translocation and could have benefitted from now clinically available ALK inhibitors. 44
This is the first study to prospectively evaluate tumor genetics, pharmacogenetics, erlotinib pharmacokinetics, rash, and clinical outcomes in African Americans with NSCLC receiving single agent TKI therapy. A relatively high incidence of tumor genetics (KRAS mutations, EML4-ALK) associated with erlotinib resistance was found. As with other ethnicities, assessment of tumor genetics prior to treatment is recommended. Furthermore, smoking history and pharmacogenetic factors consistent with high erlotinib metabolism are predominantly present in African Americans. The observed apparent higher erlotinib clearance and mild toxicity data suggest that standard erlotinib dosing in African American patients may be associated with lower than desired exposure, and would support the consideration of higher doses from the outset as well as the ability to escalate beyond 200 mg. Overall, the data from this study supports the hypothesis that African Americans may have, on average, increased erlotinib metabolism compared to other patient populations. A larger study directly comparing erlotinib PK in African Americans and non-African Americans receiving the same dose and confirming compliance with drug monitoring will be necessary to better quantify this potential difference.
MATERIALS AND METHODS
Patient eligibility
The Cancer Institutional Review Boards of The Ohio State University and the University of North Carolina approved this study (clinicaltrials.gov NCT00230126). All patients provided written informed consent prior to study entry. Eligible patients included African Americans 18 years or older with measurable, stage IIIB or IV NSCLC, who had received no more than two chemotherapy regimens. ECOG performance status ≤2, normal organ and marrow function, and tumor tissue samples from archived or fresh biopsies were required. Exclusions included pregnancy, prior treatment with EGFR inhibitors, major surgery within 21 days, severe pulmonary insufficiency (pO2 <90%; CO2 >50mmHg) and uncontrolled inter-current illnesses. Patients receiving HIV combination anti-retroviral therapy were also excluded. Patients were asked to keep a protocol specific diary and to record any medications taken during the study.
Randomization, treatment and response evaluation
Patients were randomized 1:1 into two treatment arms by The Ohio State University Biostatistics Core using fixed-block, non-stratified randomization with a block size of 10. In Arm A patients received standard 150 mg daily doses of oral erlotinib throughout treatment, unless dose reductions were required due to toxicity. At the initiation of this trial, erlotinib had not been fully approved by the U.S. Food and Drug Administration, and the maximal dose tested in clinical studies and allowed by the manufacturer at that point was 200 mg. Patients in Arm B received initial daily oral erlotinib doses of 150 mg, 175 mg, or the maximum allowable 200 mg based on body weights of <80 kg, 80–90 kg, and >90 kg, respectively. These doses were maintained throughout cycle 1 (28 days) unless reduction was required due to toxicity. For cycle 2, doses remained unchanged for all patients who developed skin rash (grade 1 or greater) during cycle 1, or who started cycle 1 at the maximum study dose of 200 mg daily. For the remaining patients, doses were increased 25 mg weekly until the dose reached 200 mg or until rash was observed. A CONSORT diagram and randomization and dosing scheme are available as supplementary data. Patients completed a daily drug diary to assess compliance, and bottles were returned at the end of each cycle for pill counts. Treatment was available for continuation until disease progression or unacceptable toxicity.
Disease response and progression were evaluated using Response Evaluation Criteria in Solid Tumors (RECIST) every 3 cycles (12 weeks).45 Toxicities were graded according to National Cancer Institute (NCI) Common Terminology Criteria for Adverse Events version 3 (CTCAE v3.0). Erlotinib was withheld for patients developing grade 4 non-hematological toxicity, grade 4 myelosuppression, grade 3 or greater nausea or vomiting unresponsive to maximal antiemetic therapy, grade 3 or greater asthenia/fatigue or diarrhea, and pulmonary insufficiency, and discontinued if symptoms did not resolve after 2 weeks.
Pharmacogenetics
Genomic DNA extracted from peripheral blood mononuclear cells was used in multiplexed analysis of 189 known polymorphisms in drug transporters and metabolizing enzymes as previously described.46 Additional genotyping was completed for CYP3A4, CYP3A5 and ABCB1 using standard methods as published previously.28,47
Pharmacokinetics
Pharmacokinetics of erlotinib and active metabolite OSI-420 were determined for a subset of patients. Plasma samples were collected up to 24 hours after the initial erlotinib dose on cycle 1, day 1, and again following the first dose on cycle 2, day 1. Both compounds were simultaneously quantified in each plasma sample using a validated liquid chromatography-mass spectrometry assay as previously described.30
Non-compartmental pharmacokinetic parameter estimates were generated using WinNonlin v. 5.2 (Pharsight, Mountain View, CA). Population pharmacokinetic modeling and covariate analysis were completed with NONMEM v 7.1.2 using methods similar to those described previously29,30,35. A base structural model was built with first order conditional estimation considering proportional and additive residual error models, IIV, and IOV on all estimated parameters. A one compartment model with first order absorption and elimination rate constant was used for parent drug (i.e., erlotinib), parameterized with the use of Ka, CL and V1. The erlotinib model was linked to another one compartment model with first order elimination for active metabolite (i.e., OSI-420), parameterized in terms of metabolite clearance (CLM) and volume of distribution (V2). The model included an estimate for fraction (FMET) of erlotinib converted to OSI-420, which was coded as in equation 1, where FM is an estimated parameter.
| (eq.1) |
For covariate analyses, continuous covariates, including weight, age and LBM, were normalized to median values and evaluated as described in equation 2:
| (eq. 2) |
CONji is the value of the continuous covariate j for individual i, CONj-med is the population median value of covariate j for all individuals, θi is the individual PK parameter estimate, θpop is the population parameter estimate for the typical individual, and ηi is the IIV parameter. Lean body mass was estimated for 25 patients using equation 348
| (eq. 3.1) |
| (eq. 3.2) |
where W is body weight in kilograms and H is body height in centimeters. For a male patient who was a bilateral amputee whose record of original height was not available, we used “assumed height” to estimate LBM. This was achieved using linear regression to fit a line for the relationship between height and weight for males.
Pharmacogenetic covariates were dichotomized and evaluated in the model both with heterozygous genotypes included in the homozygous major and minor allele groups (i.e. M/m genotypes were coded both as 0 and 1 with M/M always in the 0 category and m/m always in the 1 category). Similarly, co-medications were dichotomized as 0 (no relevant co-medications) or as 1 (CYP1A1/2 and CYP3A4/5 substrates, inhibitors, or inducers). Categorical covariates, including sex, previous smoking status, and current smoking status, were evaluated as in equation 4:
| (eq. 4) |
CATji is the value of the categorical covariate j for individual i, and Θj is an estimated parameter. Using a cutoff of p ≤ .05 for the likelihood-ratio test, corresponding to a decrease in OBJ of 3.84 or greater, individual covariates were evaluated with forward inclusion, backward deletion, and forward selection followed by backward elimination to finalize the covariate model. Model selection in multivariate analysis was based on reduction of OBJ by ≥ 3.84 for forward inclusion, reduction of OBJ ≥ 6.64 (P ≤ 0.01) for backward deletion, and reduction in residual error and/or IIV of the evaluated PK parameter.
Accuracy and bias of the base and covariate model estimates were evaluated using the nonparametric bootstrap procedure in the Wings for NONMEM software. The median and 95% CI were computed for all population pharmacokinetic parameters using sets of parameter estimates from 1000 bootstrap runs.
Tumor genetics
Formalin fixed, paraffin embedded tissues were obtained from each patient, and genomic DNA was extracted from sections using standard protocols. Extracted DNA was evaluated for KRAS (promoter region and G12, G13 codon positions) and EGFR (exons 19 and 21 and codon positions T790 and R776) mutations by PCR. EML4-ALK fusion and EGFR gene amplification were evaluated by fluorescence in-situ hybridization. Assays were conducted at The Ohio State University Molecular Pathology Core Facility according to previously published methods.49–51 Additional details on these methods are available as supplement.
Data Analyses
The trial utilized a Simon 2-stage minimax design with arms A and B treated as independent phase II studies. The primary endpoint was disease control rate (DCR = complete response (CR), partial response (PR) or stable disease (SD) using RECIST criteria) at 3 months. Early stopping criteria in each arm was set to DCR probability <25% (p0) with alpha and beta both equal to 0.10. This equated to a first stage accrual of 11 and total accrual of 26 for each treatment arm. Therefore, if disease control was observed in 3 or more of the first 11 patients treated, the arm would be expanded to accrue 15 additional patients. Furthermore, DCR probability ≥50% (p1) was required for recommendation of further study. Setting alpha and beta to 0.10, equated to observation of DCR in a minimum of 10 of the 26 patients treated for each arm.
Time to progression (TTP) was calculated from date of the first treatment to disease progression. Patients who had not progressed were censored at the last visit. Deaths occurring prior to documented progression were also censored. Overall survival (OS) was determined from the date of the first treatment to death from any cause. Survival curves were estimated using the method of Kaplan-Meier. Differences between survival curves were assessed by log-rank test. Correlations between clinical outcomes (response, PR+SD vs. PD) and rash (Yes vs. No) and tumor genetics (KRAS mutation, ALK translocation, and EFGR amplification) were studied using Fisher’s exact test. Secondary evaluations to identify associations between tumor genetics, pharmacokinetics, pharmacogenetics, smoking status, and outcomes used parametric and non-parametric tests. Analyses were performed with the R statistical system52 and SAS version 9.2 (SAS Institute).
Supplementary Material
STUDY HIGHLIGHTS.
What is the current knowledge on the topic?
African Americans have a higher incidence of lung cancer and less favorable outcomes from approved therapies compared to other patient populations. The reasons for this are unclear, and prospective studies conducted to better understand these disparities are lacking.
What question this study addressed?
The current study was conducted to determine if an alternative dose-to-rash strategy for the EGFR inhibitor, erlotonib, could produce more favorable outcomes in African Americans with NSCLC.
What this study adds to our knowledge?
Results in 52 evaluable African American patients indicated the dose-to-rash strategy produced minimal toxicity, modest improvement in time to progression, and no change in overall survival. Pharmacokinetic data suggests these patients had relatively high clearance of erlotinib, which is supported by associated pharmacogenetic features. Tumor EGFR genetic abnormalities were rare, and EML4-ALK translocations were more prevalent than expected.
How this might change clinical pharmacology and therapeutics?
Prospective tumor genotyping will improve therapy selection, and pharmacokinetically guided dosing will increase erlotinib exposure and potentially improve outcomes in African Americans with NSCLC.
ACKNOWLEDGEMENTS
We would like to thank Dr. Kenneth K. Chan and Dr. Wolfgang Sadee for their intellectual contributions to the design and conduct of this study. This work was supported by a V Foundation/American Association for Cancer Research Translational Research Grant, The Schnipke Family Research Fund, NIH grants U01GM092655 and 5KL2RR025754, and Genentech, Inc.
Abbreviations
- AUC
area under the concentration vs. time curve
- Cmax
maximum observed plasma concentration
- CI
Confidence Interval
- CL
erlotinib plasma clearance
- CR
complete response
- DC
disease control
- DCR
disease control rate (DCR=CR+PR+SD)
- OS
Overall Survival
- PD
progressive disease
- PR
partial response
- SD
stable disease
- TTP
Time To Progression
Footnotes
CONFLICTS OF INTEREST
No authors have conflicts of interest to disclose. The sponsors had no role in the design and conduct of the study, collection, management, analysis, and interpretation of the data, or preparation, review, or approval of the manuscript.
REFERENCES
- 1.LAG R, MP E, CL K, et al., editors. SEER Cancer Statistics Review, 1975–2000. Bethesda, MD: National Cancer Institute; 2003. [Google Scholar]
- 2.Gadgeel SM, Severson RK, Kau Y, et al. Impact of race in lung cancer: analysis of temporal trends from a surveillance, epidemiology, and end results database. Chest. 2001;120:55–63. doi: 10.1378/chest.120.1.55. [DOI] [PubMed] [Google Scholar]
- 3.Ward E, Jemal A, Cokkinides V, et al. Cancer disparities by race/ethnicity and socioeconomic status. CA Cancer J Clin. 2004;54:78–93. doi: 10.3322/canjclin.54.2.78. [DOI] [PubMed] [Google Scholar]
- 4.Hunt JD, Strimas A, Martin JE, et al. Differences in KRAS mutation spectrum in lung cancer cases between African Americans and Caucasians after occupational or environmental exposure to known carcinogens. Cancer Epidemiol Biomarkers Prev. 2002;11:1405–1412. [PubMed] [Google Scholar]
- 5.Wu X, Zhao H, Amos CI, et al. p53 Genotypes and Haplotypes Associated With Lung Cancer Susceptibility and Ethnicity. J Natl Cancer Inst. 2002;94:681–690. doi: 10.1093/jnci/94.9.681. [DOI] [PubMed] [Google Scholar]
- 6.Bach PB, Cramer LD, Warren JL, et al. Racial differences in the treatment of early-stage lung cancer. N Engl J Med. 1999;341:1198–1205. doi: 10.1056/NEJM199910143411606. [DOI] [PubMed] [Google Scholar]
- 7.Blackstock AW, Herndon JE, 2nd, Paskett ED, et al. Outcomes among African-American/non-African-American patients with advanced non-small-cell lung carcinoma: report from the Cancer and Leukemia Group B. J Natl Cancer Inst. 2002;94:284–290. doi: 10.1093/jnci/94.4.284. [DOI] [PubMed] [Google Scholar]
- 8.Felip E, Rosell R. Clinical experience with erlotinib in non-small-cell lung cancer. Drugs Today (Barc) 2006;42:147–156. doi: 10.1358/dot.2006.42.3.957358. [DOI] [PubMed] [Google Scholar]
- 9.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 10.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 11.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 12.Janne PA, Wang X, Socinski MA, et al. Randomized phase II trial of erlotinib alone or with carboplatin and paclitaxel in patients who were never or light former smokers with advanced lung adenocarcinoma: CALGB 30406 trial. J Clin Oncol. 2012;30:2063–2069. doi: 10.1200/JCO.2011.40.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudin CM, Liu W, Desai A, et al. Pharmacogenomic and pharmacokinetic determinants of erlotinib toxicity. J Clin Oncol. 2008;26:1119–27. doi: 10.1200/JCO.2007.13.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heist RS, Christiani D. EGFR-targeted therapies in lung cancer predictors of response and toxicity. Pharmacogenomics. 2009;10:59–68. doi: 10.2217/14622416.10.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Soler R. Phase II clinical trial data with the epidermal growth factor receptor tyrosine kinase inhibitor erlotinib (OSI-774) in non-small-cell lung cancer. Clin Lung Cancer. 2004;6(Suppl 1):S20–S23. doi: 10.3816/clc.2004.s.010. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Zhao M, He P, et al. Differential metabolism of gefitinib and erlotinib by human cytochrome P450 enzymes. Clin Cancer Res. 2007;13:3731–3737. doi: 10.1158/1078-0432.CCR-07-0088. [DOI] [PubMed] [Google Scholar]
- 17.Rakhit A, Pantze MP, Fettner S, et al. The effects of CYP3A4 inhibition on erlotinib pharmacokinetics: computer-based simulation (SimCYP) predicts in vivo metabolic inhibition. Eur J Clin Pharmacol. 2008;64:31–41. doi: 10.1007/s00228-007-0396-z. [DOI] [PubMed] [Google Scholar]
- 18.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 19.He P, Court MH, Greenblatt DJ, et al. Genotype-phenotype associations of cytochrome P450 3A4 and 3A5 polymorphism with midazolam clearance in vivo. Clin Pharmacol Ther. 2005;77:373–387. doi: 10.1016/j.clpt.2004.11.112. [DOI] [PubMed] [Google Scholar]
- 20.Information NCfB. dbSNP Short Genetic Variations. 2012 Jun 26;:2012. [Google Scholar]
- 21.Rotunno M, Yu K, Lubin JH, et al. Phase I metabolic genes and risk of lung cancer: multiple polymorphisms and mRNA expression. PLoS One. 2009;4:e5652. doi: 10.1371/journal.pone.0005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright CM, Larsen JE, Colosimo ML, et al. Genetic association study of CYP1A1 polymorphisms identifies risk haplotypes in nonsmall cell lung cancer. Eur Respir J. 2010;35:152–9. doi: 10.1183/09031936.00120808. [DOI] [PubMed] [Google Scholar]
- 23.Shi Z, Peng XX, Kim IW, et al. Erlotinib (Tarceva, OSI-774) antagonizes ATP-binding cassette subfamily B member 1 and ATP-binding cassette subfamily G member 2-mediated drug resistance. Cancer Res. 2007;67:11012–11020. doi: 10.1158/0008-5472.CAN-07-2686. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Cusatis G, Brahmer J, et al. Association of variant ABCG2 and the pharmacokinetics of epidermal growth factor receptor tyrosine kinase inhibitors in cancer patients. Cancer Biol Ther. 2007;6:432–438. doi: 10.4161/cbt.6.3.3763. [DOI] [PubMed] [Google Scholar]
- 25.Elens L, Becker ML, Haufroid V, et al. Novel CYP3A4 intron 6 single nucleotide polymorphism is associated with simvastatin-mediated cholesterol reduction in the Rotterdam Study. Pharmacogenet Genomics. 2011;21:861–866. doi: 10.1097/FPC.0b013e32834c6edb. [DOI] [PubMed] [Google Scholar]
- 26.Elens L, Bouamar R, Hesselink DA, et al. A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin Chem. 2011;57:1574–1583. doi: 10.1373/clinchem.2011.165613. [DOI] [PubMed] [Google Scholar]
- 27.Elens L, van Schaik RH, Panin N, et al. Effect of a new functional CYP3A4 polymorphism on calcineurin inhibitors’ dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenomics. 2011;12:1383–1396. doi: 10.2217/pgs.11.90. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Guo Y, Wrighton SA, et al. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11:274–286. doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White-Koning M, Civade E, Geoerger B, et al. Population analysis of erlotinib in adults and children reveals pharmacokinetic characteristics as the main factor explaining tolerance particularities in children. Clin Cancer Res. 2011;17:4862–4871. doi: 10.1158/1078-0432.CCR-10-3278. [DOI] [PubMed] [Google Scholar]
- 30.Kraut EH, Rhoades C, Zhang Y, et al. Phase I and pharmacokinetic study of erlotinib (OSI-774) in combination with docetaxel in squamous cell carcinoma of the head and neck (SSCHN) Cancer Chemother Pharmacol. 2011;67:579–586. doi: 10.1007/s00280-010-1332-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts PJ, Stinchcombe TE, Der CJ, et al. Personalized medicine in non-small-cell lung cancer: is KRAS a useful marker in selecting patients for epidermal growth factor receptor-targeted therapy? J Clin Oncol. 2010;28:4769–4777. doi: 10.1200/JCO.2009.27.4365. [DOI] [PubMed] [Google Scholar]
- 33.Mok T, Wu YL, Au JS, et al. Efficacy and safety of erlotinib in 1242 East/South-East Asian patients with advanced non-small cell lung cancer. J Thorac Oncol. 5:1609–1615. doi: 10.1097/JTO.0b013e3181e15d55. [DOI] [PubMed] [Google Scholar]
- 34.Scheffler M, Di Gion P, Doroshyenko O, et al. Clinical pharmacokinetics of tyrosine kinase inhibitors: focus on 4-anilinoquinazolines. Clin Pharmacokinet. 2011;50:371–403. doi: 10.2165/11587020-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 35.Lu JF, Eppler SM, Wolf J, et al. Clinical pharmacokinetics of erlotinib in patients with solid tumors and exposure-safety relationship in patients with non-small cell lung cancer. Clin Pharmacol Ther. 2006;80:136–145. doi: 10.1016/j.clpt.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Thomas F, Rochaix P, White-Koning M, et al. Population pharmacokinetics of erlotinib and its pharmacokinetic/pharmacodynamic relationships in head and neck squamous cell carcinoma. Eur J Cancer. 2009;45:2316–2323. doi: 10.1016/j.ejca.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Nallani SC, Genter MB, Desai PB. Increased activity of CYP3A enzyme in primary cultures of rat hepatocytes treated with docetaxel: comparative evaluation with paclitaxel. Cancer Chemother Pharmacol. 2001;48:115–122. doi: 10.1007/s002800100283. [DOI] [PubMed] [Google Scholar]
- 38.Fujitaka K, Oguri T, Isobe T, et al. Induction of cytochrome P450 3A4 by docetaxel in peripheral mononuclear cells and its expression in lung cancer. Cancer Chemother Pharmacol. 2001;48:42–46. doi: 10.1007/s002800100291. [DOI] [PubMed] [Google Scholar]
- 39.Waller LL, Miller AA, Petty WJ. Using erlotinib to treat patients with non-small cell lung cancer who continue to smoke. Lung Cancer. 2010;67:12–16. doi: 10.1016/j.lungcan.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Hughes AN, O’Brien ME, Petty WJ, et al. Overcoming CYP1A1/1A2 mediated induction of metabolism by escalating erlotinib dose in current smokers. J Clin Oncol. 2009;27:1220–1226. doi: 10.1200/JCO.2008.19.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 42.Yoshioka H, Hotta K, Kiura K, et al. A phase II trial of erlotinib monotherapy in pretreated patients with advanced non-small cell lung cancer who do not possess active EGFR mutations: Okayama Lung Cancer Study Group trial 0705. J Thorac Oncol. 2010;5:99–104. doi: 10.1097/JTO.0b013e3181c20063. [DOI] [PubMed] [Google Scholar]
- 43.D’Angelo SP, Pietanza MC, Johnson ML, et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J Clin Oncol. 2011;29:2066–2070. doi: 10.1200/JCO.2010.32.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwak EL, Bang Y-J, Camidge DR, et al. Anaplastic Lymphoma Kinase Inhibition in Non-Small-Cell Lung Cancer. New England Journal of Medicine. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 46.Dai Z, Papp AC, Wang D, et al. Genotyping panel for assessing response to cancer chemotherapy. BMC Med Genomics. 2008;1:24. doi: 10.1186/1755-8794-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang D, Johnson AD, Papp AC, et al. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics. 2005;15:693–704. [PubMed] [Google Scholar]
- 48.Hume R. Prediction of lean body mass from height and weight. J Clin Pathol. 1966;19:389–391. doi: 10.1136/jcp.19.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 50.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 51.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.