Abstract
Receptor/gene-mediated effects of corticosteroids on hepatic tyrosine aminotransferase (TAT) were evaluated in normal rats. A group of normal male Wistar rats were injected with 50 mg/kg methylprednisolone (MPL) intramuscularly at the nadir of their plasma corticosterone (CST) rhythm (early light cycle) and sacrificed at various time points up to 96 h post-treatment. Blood and livers were collected to measure plasma MPL, CST, hepatic glucocorticoid receptor (GR) mRNA, cytosolic GR density, TAT mRNA, and TAT activity. The pharmacokinetics of MPL showed bi-exponential disposition with two first-order absorption components from the injection site and bioavailability was 21%. Plasma CST was reduced after MPL dosing, but resumed its daily circadian pattern within 36 h. Cytosolic receptor density was significantly suppressed (90%) and returned to baseline by 72 h resuming its biphasic pattern. Hepatic GR mRNA follows a circadian pattern which was disrupted by MPL and did not return during the study. MPL caused significant down-regulation (50%) in GR mRNA which was followed by a delayed rebound phase (60–70 h). Hepatic TAT mRNA and activity showed up-regulation as a consequence of MPL, and returned to their circadian baseline within 72 and 24 h of treatment. A mechanistic receptor/gene-mediated pharmacokinetic/pharmacodynamic model was able to satisfactorily describe the complex interplay of exogenous and endogenous corticosteroid effects on hepatic GR mRNA, cytosolic free GR, TAT mRNA, and TAT activity in normal rats.
Keywords: Methylprednisolone, Corticosteroids, Pharmacokinetics, Pharmacodynamics, Tyrosine aminotransferase, Glucocorticoid receptors
Introduction
Corticosteroids (CS) are effective anti-inflammatory and immuno-modulatory agents, which are widely and frequently used for the treatment of various diseases. Acute, high doses of CS are used in the treatment of septic shock, acute kidney transplant rejection, and diffuse alveolar hemorrhage. Chronic, low doses of these drugs are used in the treatment of chronic diseases such as lupus erythematosus and rheumatoid arthritis [1]. Despite their beneficial effects, long-term therapy causes severe metabolic disorders similar to those observed with abnormalities in endogenous hormones [2–5] leading to Cushing’s syndrome, hypertension, hyperglycemia, muscle atrophy, and dyslipidemia.
Receptor/gene-mediated effects of acute and chronic doses of CS on TAT dynamics in (ADX) rats have been studied extensively in our lab [6–11]. Due to absence of endogenous CST, studies in adeenalectomized ADX rats facilitates assessment of various pharmacodynamic/toxicodynamic endpoints governed by synthetic CS without confounding factors such as non-stationary baselines of biomarkers. Although the practical advantages in using this model are substantial, adrenalectomy may lead to undesirable physiological changes such as increased sensitivity of hepatic cAMP in response to catecholamines [12], decrease in body weight [13], delay in onset of puberty in female rats, changes in expression of various neuropeptide genes [14], and—most importantly—multi-factorial changes in the gluconeogenic pathway [15]. Despite the complexities that may be involved in using normal rats, they are more physiologically relevant especially when evaluating the inter-relationships between carbohydrate, protein, and lipid metabolism.
In this report, we attempted to quantitatively describe the receptor/gene-mediated dynamic interplay of endogenous and exogenous CS in normal rats to regulate one of the most highly studied biomarkers of CS, hepatic TAT (mRNA and activity).
Materials and methods
Animals
Normal male Wistar rats weighing between 125 and 175 g were purchased from Harlan-Sprague-Dawley Inc. (Indianapolis, IN). The animals were housed in our University Laboratory Animal Facility and acclimatized under constant temperature (22°C) and humidity (72%) with controlled 12 h/12 h light/dark cycle for at least 2 weeks with lights on at 6:15 AM and lights off at 6:15 PM. All rats had access to rat chow and drinking water. Our protocol adhered to the Principles of Laboratory Animal Care (National Institute of Health publication 85–23, revised 1985) and was approved by the University at Buffalo Institutional Animal Care and Use Committee.
Experimental
After 2–3 weeks of acclimatization, rats weighing 250–325 g received 50mg/kg methylprednisolone succinate (Solu-Medrol®, Pharmacia & Upjohn Company, Kalamazoo, MI) by intramuscular (IM) injection in the left hind haunch (gluteus muscle) at the nadir of the circadian pattern of their endogenous CST (between 7:45 and 9:45 AM). Rats were weighed, anesthetized with ketamine/xylazine, and sacrificed by aortic exsanguinations at 0.25, 0.5, 0.75, 1, 2, 4, 5, 6, 7, 8, 12, 24, 36, 48, 60, 72, 84 and 96 h (n = 3 per time point). Six rats injected with IM saline and sacrificed at 12 and 24 h (n = 3 per time point) served as controls. Blood was collected from the abdominal aorta; plasma was harvested by centrifuging blood at 2,000g at 4°C for 15 minutes and stored at −80°C until analysis.
For TAT enzyme assays approximately 1 g of liver was rapidly excised for preparation of crude supernatants in ice-cold potassium chloride buffer. Remaining liver tissue was flash-frozen in liquid nitrogen and stored at −80°C until further analyses.
Plasma steroid assays
Plasma was thawed and kept on ice until steroid extraction. The CST and MPL concentrations were determined by a normal phase high-performance liquid chromatography (HPLC) method [16]. The lower limits of quantification for both of these steroids are 10 ng/ml. Inter-day and intra-day coefficients of variation (CV) for this assay were less than 10%.
Glucocorticoid receptors
A previously developed [10,17] radio-ligand binding assay was used to quantify the hepatic free cytosolic GR density with some modifications. Liver tissues stored at −80°C were ground in liquid nitrogen chilled mortars and pestles. The ground liver tissue, 1.5g, was thawed in 9ml of ice-cold assay buffer (50nM Trizma base, 0.2 mM sodium EDTA, and 10mM sodium molyb-date at pH = 7.5) for 20min. Livers were homogenized and centrifuged at 10,000g (4°C) for 30min. The supernatant was passed through two layers of cheesecloth to obtain “crude” cytosol. Then 0.5 ml of dextran-coated activated charcoal in icecold assay buffer (5%) was added to this supernatant (0.5 ml charcoal/5 ml of supernatant) and spun at 10,000g at 4°C for 15 min to remove any free MPL or CST. The resulting supernatant was spun at 90,000g (4°C) for 90min in a Beckman L7–55 ultracentrifuge (Beckman Instruments, Inc., Fullerton, CA) to obtain the cytosol used in this assay. This step removes materials which increase the non-specific binding of the 3H-labeled dexamethasone (DEX) in the crude cytosol. Aliquots of 300µl cytosol were incubated at 4°C for 18 h with 75µl of 3H-labeled DEX (39 Ci/mmol; Amersham, Arlington Heights, IL) in 1.5 ml polypropylene microfuge tubes (VWR Scientific, Rochester, NY). Assay concentrations of DEX ranged from 0.63 to 50 nM. Parallel incubations were set up in the presence and absence of 75 µl excess unlabeled DEX (Sigma Chemical Co., St. Louis, MO; assay concentration, 16.7 µM). Final assay volumes were 450µl. Two 50µl aliquots of cytosol were counted by liquid scintillation (Packard Instrument Co., Meriden, CT) to determine the total concentrations of 3H-DEX added into the assay. After 18h of incubation, 150µl of dextran-coated charcoal in ice-cold assay buffer was added to the cytosol to remove any free 3H-DEX. After centrifugation, 400µl aliquots of the supernatants were counted to determine total binding (DT) and nonspecific binding (DNS). Cytosolic free GR density (Bmax) was estimated using the ADAPT II program (Biomedical Simulations Resource, Los Angeles, CA) by solving simultaneously:
| (1) |
| (2) |
where KD is the equilibrium dissociation constant for DEX for glucocorticoid receptor binding and KNS is the non-specific binding constant. The receptor density values were normalized by the protein content in the cytosol preparation [18]. The intra- and inter-assay CV for this method was less than 10%.
Hepatic GR and TAT mRNA Measurements
RNA preparation
Total RNA in the liver (ground liver stored at −80°C) was extracted in TRIzol (In-vitrogen Corp, Carlsbad, CA) reagent according to manufacturer’s instructions. To account for variable extraction yields, an external cRNA standard (GRG 1-cRNA) was added to each liver sample before extraction. The integrity of extracted RNA was confirmed by agarose formaldehyde gel electrophoresis. The quantity of extracted RNA was determined optical densities at 260. Total RNA samples were diluted to desired concentrations in nuclease free water (Ambion, Austin, TX) and stored at −80°C until further analysis. The yield of RNA extraction was determined for each liver sample by comparing the quantity of external cRNA standard added before the extraction into liver tissue with that recovered after extraction [19].
Absolute quantification of GR and TAT mRNA by real-time quantitative RT-PCR
To quantify the TAT, GR and GRG-1 mRNAs we used QRT RT-PCR assays which use gene specific TAQMAN probes and primers (Stratagene, La Jolla, CA) and in vitro-synthesized cRNA as standards. The cRNA was prepared using conventional cloning methods [19,20] and then in vitro transcribed to cRNA using T7 MEGAs-cript In Vitro Transcription Kits (Ambion, Austin, TX). For QRT RT-PCR assays, specific TaqMan probes and primers were designed using PrimerExpress software (Applied Biosystems, Foster City, CA); sequences sharing homology with other rat genes were excluded. Primers/probes were custom synthesized by Biosearch Technologies, Inc. (Novato, CA). The probes were synthesized with the fluorescent reporter FAM (for TAT and GR) or HEX (GRG) attached to the 5′ end and the appropriate BHQ attached to the 3′ end. Forward and reverse primer design allowed positioning of the two oligonucleotides as close to one another without overlapping the probe.
The assay was performed using the Brilliant 1-Step Quantitative RT-PCR Core Reagent Kit (Strategene) according to manufacturer’s instructions. A cRNA standard curve was generated for each assay; standards were run in duplicate whereas the samples were run in triplicate. A reverse transcriptase minus control (non-amplification control) for each sample to test for the possibility of genomic DNA contamination in extracted RNA and non-template controls (NTC) were run with each assay. In all cases these controls gave no significant amplification signal. Construction of cRNA standard curves allowed quantification of mRNA in moles of message. The optimum concentrations for the various forward and reverse primers, probes, dNTP and magnesium chloride along with the oligonucleotide sequences are listed in Table 1. As a time saving measure, a multiplex assay for GR and GRG-1 message was run in a single tube without reduction in sensitivity. The intra- and inter-assay CV for all transcripts of interest were under 15%.
Table 1.
Reaction conditions and primer/probe sequences used for quantitative RT-PCR assays
| Genes | Oligonucleotide | Cone. nM | Sequence 5′-3′ | MgCl2 mM |
|---|---|---|---|---|
| Tyrosine aminotransferase | Forward primer | 300 | GACATGGTGTTTTCAGATTGCAA | 3 |
| Reverse primer | 300 | CAGGACAGGATGGGAACATTG | ||
| FAM-labeled probe | 100 | TACGAACCACTGGCCAACCTCAGCA | ||
| Glucocorticoid receptor | Forward primer | 300 | AACATGTTAGGTGGGCGTCAA | 5 |
| Reverse primer | 300 | GGTGTAAGTTTCTCAAGCCTAGTATCG | ||
| FAM-labeled probe | 100 | TGATTGCAGCAGTGAAATGGGCAAAG | ||
| GRG-1 (external standard) | Forward primer | 300 | CGGTTCTGGTGTAATGCTAAAGCT | 5 |
| Reverse primer | 300 | AGTTCGCCAAGGGCTTCTC | ||
| HEX-labeled probe | 100 | CCCTTCGAAATTCCAAGCCAAGTATGTCAT |
TAT activity
Hepatic TAT activity was determined by the colorimetric assay of Diamondstone [21] and reported as the change in absorbance (ABS) at 331 nm over time (dABS/dt). These values were normalized for protein content in the crude liver supernatant using the Lowry assay [18].
Additional data source
Information about the circadian rhythm of various genes and protein markers had been obtained from a circadian rhythm study in our lab [22,23] where two groups of rats were acclimatized to a strict 12 h/12 h light/dark regimen and sacrificed at various time points to characterize the circadian variation in various glucocorticoid regulated genetic and protein biomarkers such as glucocorticoid receptor (GR) mRNA, free cytosolic GR levels, and TAT mRNA and activity.
The MPL concentrations after IV dosing were obtained from a pharmacokinetic study where four male Wistar rats were given MPL via jugular vein cannula [24]. Blood samples were collected at 0.25, 0.5, 1, 2, 3, 4 and 6 h.
Pharmacokinetic/pharmacodynamic modeling
Methylprednisolone pharmacokinetics
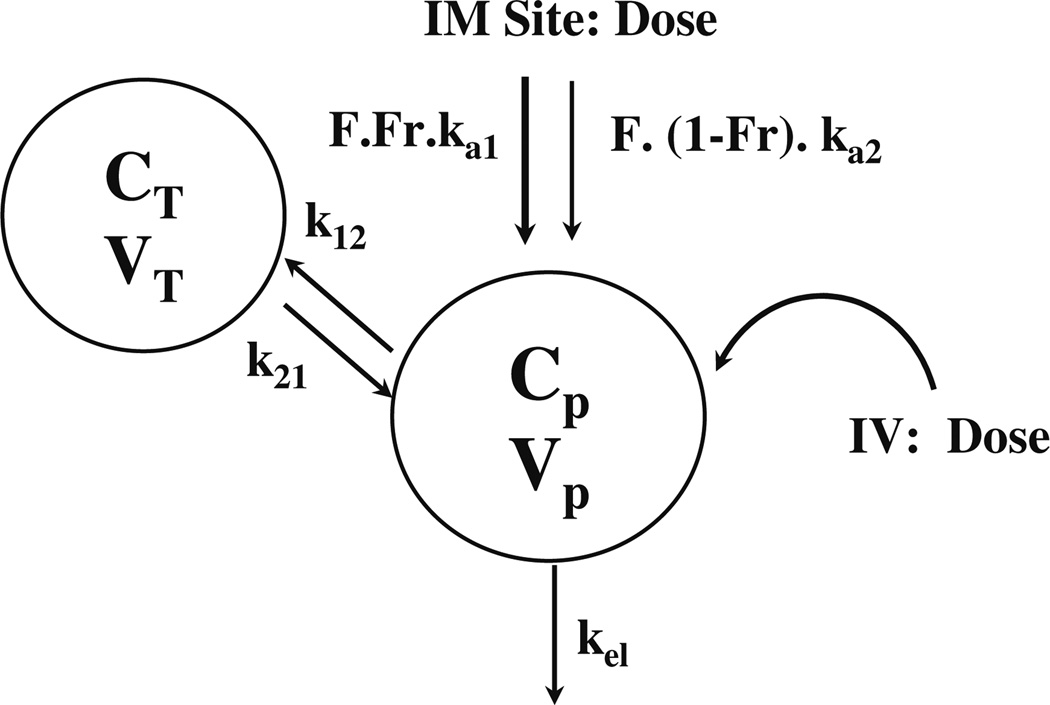
A two-compartment mammillary model with two absorption components from the injection site (Fig. 1) was used to describe the plasma PK after 50 mg/kg MPL. The equations describing the model are:
| (3) |
| (4) |
| (5) |
| (6) |
where A, C and D are the amount, concentration and dose in the corresponding compartments designated by the subscripts P and T representing plasma and tissue (distribution) compartments; kel is the first-order elimination rate constant from the central compartment, Vc is the central volume of distribution, k12 and k21 are the first-order inter-compartmental distribution rate constants, Fr and (1 – Fr) are the fractions of dose absorbed through two absorption pathways described by two first-order rate constants, ka1 and ka2. The overall IM bioavailability (F) was determined by simultaneously fitting plasma MPL concentrations after 50mg/kg IV and IM dosing.
Fig. 1.
Pharmacokinetic model for methylprednisolone after IV and IM administration (50 mg/kg) in normal rats
Corticosterone dynamics
The circadian rhythm of endogenous plasma corticoste-rone (CST) in normal untreated male Wistar rats was described by an indirect response model where CST is synthesized by a time-dependent synthesis rate, described by two harmonic functions [22,23] and degraded by a first-order loss rate constant, kd,CST,
| (7) |
The time-dependent synthesis rate of CST, ks,CST, can be described as:
| (8) |
where a0, a1, a2, b1 and b2 are the Fourier coefficients which were obtained by fitting endogenous plasma CST profiles from untreated rats [22,23]. Subsequently, the inhibition of CST by MPL was described as:
| (9) |
where, IC50,MPL is the concentration of plasma MPL responsible for 50% inhibition of CST. Zero-order secretion of CST (CSTstress/τ), independent of natural production of plasma CST due to stress, is incorporated to describe early stress-induced secretion of CST where CSTstress, the maximum concentration of plasma CST due to stress, and τ, time when the stress-induced secretion of CST stops, were estimated as model parameters. Thus, the term (CSTstress/τ) operates when τ ≥ t ≥ tinj (time of injection). The initial conditions for both Eqs. 7 and 9 were fixed to 33.6ng/ml, which was obtained from the circadian control animals [23].
Pharmacodynamics
Mechanistic basis of pharmacodynamics
Corticosteroids are highly protein bound in plasma [1]. Due to their moderate lipo-philicity, the unbound fraction is known to passively diffuse into the cell [10,25] and bind to their high affinity, specific nuclear receptor (a ligand activated transcription factor) known as glucocorticoid receptor (GR). Upon binding to the ligand, the receptor dissociates from heat-shock proteins, hsp 90, 70 and 56 [26,27], undergoes conformational change, becomes phosphorylated and activated [28]. The activated drug–receptor complex rapidly translocates into the nucleus where it binds to specific palindromic sequences in the target gene, known as glucocorticoid response elements (GRE) causing either stimulation or inhibition of transcription of various genes, such as TAT. Changes in transcription rates result in alteration in specific mRNA levels, which subsequently can result in changes in proteins leading to various physiological effects exerted by CS [3,10,29]. It had been postulated that part of these receptors may undergo degradation and the rest may recycle back to the cytosol to bind to new ligand [10,30,31]. Although much less is known about CS inhibitory (negative GREs) effects compared to their stimulatory (positive GREs) effects [32], CS are known to inhibit their own receptor gene by a process called homologous down-regulation [30,31]. In addition to rapid down-regulation of thecytosolic free receptors caused by the translocation process, inhibition of GR mRNA transcription by activated nuclear drug–receptor complex further reduces the free cytosolic GR. rats [10], the general structure of the fifth generation model [9] of receptor dynamics was followed. However, more complex regulation features of GR mRNA in homeostasis and post-treatment conditions in normal rats were incorporated.
Receptor dynamics
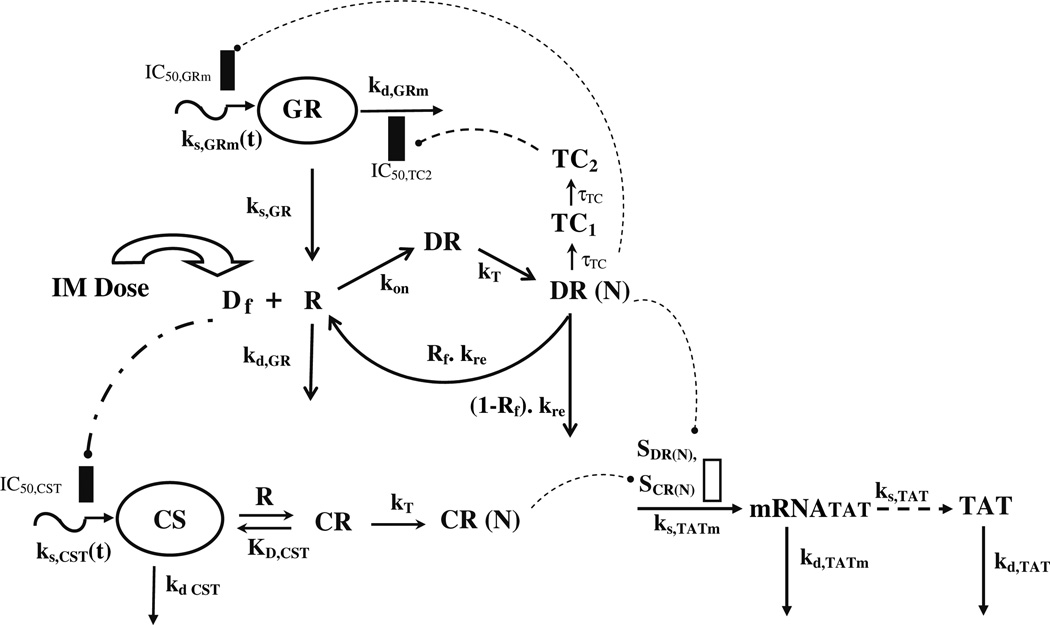
An integrated PK/PD model of CS receptor/gene-mediated effects in normal rats is shown in Fig. 2. Since the pattern of cytosolic receptor density was identical to ADX rats [10], the general structure of the fifth generation model [9] of receptor dynamics was followed. However, more complex regulation features of GR mRNA in homeostasis and post-treatment conditions in normal rats were incorporated.
Fig. 2.
Pharmacodynamic/pharmacogenomic model of receptor/gene-mediated corticosteroid effects on hepatic tyrosine aminotransferase in normal rats
Since the free cytosolic receptors in the control rats and from the previously performed circadian rhythm study [22,23] did not show any circadian variation, it was postulated that the down-regulation of receptors is mediated by MPL only and not by endogenous CST. In the absence of the drug, the receptors are produced from their GR mRNA by a first-order rate constant, ks,GR, and degraded by a first-order rate constant kd,GR. Free cytosolic receptors interact with free MPL to form drug–receptor complex in the cytosol (DR), which rapidly translocates into the nucleus with a first-order rate constant kT, forming DR(N). Part of DR(N) may either recycle back (Rf) to the cytosol or may degrade with a rate constant of (1 – Rf) ·kre. The equations describing this chain of events are:
| (10) |
| (11) |
| (12) |
The receptor density was determined in two groups of control animals sacrificed at 12 and 24 h, and the initial condition for Eq. 10 was fixed to the mean values from those two groups (476 fmol/g). The initial conditions for Eqs. 11 and 12 were set to zero. The free fraction (f) of MPL was fixed to 0.23 [33].
The regulation of GR mRNA was more complex in normal rats than ADX rats. An obvious circadian rhythm was observed in hepatic GR mRNA from our circadian rhythm study [22,23]. However, this variation was not in agreement with general theory of CS-mediated down-regulation of GR mRNA [8–11,23,31] and was described by an indirect response model with time-dependent production rate (independent of endogenous CST circadian rhythm), kin,GRm, and a first-order loss rate constant, kd,GRm:
| (13) |
The time-dependent expression rate, kin,GRm(t), of GR mRNA in control rats was described by two harmonic functions as:
| (14) |
where a0, GR, a1, GR, a2,GR, b1, GR, and b2,GR are Fourier coefficients obtained by fitting the GR mRNA circadian rhythm data from control animals using FOURPHARM [34].
The down-regulation followed by rebound in GR mRNA caused by MPL was described by DR(N)-mediated inhibition of ks,GRm(t) and subsequent inhibition of kd,GRm by a transduction signal generated from DR(N),
| (15) |
| (16) |
| (17) |
where TC1 and TC2 are two transit compartments, τDR(N) is the mean transit time for signal transduction from DR(N), IC50, DR(N) is the concentration of DR(N) at which the synthesis rate of GR mRNA is reduced to 50% of its baseline, and IC50,TC2 is the concentration of TC2 responsible for 50% inhibition of the loss rate for GR mRNA. Because stationarity was not assumed for normal rats, the ks,GR parameter was estimated with other parameters. The initial conditions for Eqs. 15 and 16 were set to zero, whereas the initial condition for Eq. 17 was fixed to the measured average values of GR mRNA from the control rats sacrificed at 24 h. The GR mRNA and free hepatic cytosolic receptor density from the treated animals were fitted simultaneously using the equations described above and the fitted parameters were fixed for further analysis of TAT dynamics.
Tyrosine aminotransferase dynamics
Corticosteroids can affect TAT dynamics by increasing transcription of TAT mRNA. TAT showed circadian variation in normal untreated rats [22,35] in both mRNA and activities which was postulated to be mediated by endogenous CST Regulation of TAT mRNA (Eq. 20) and activity (Eq. 21) after MPL in normal rats was assumed to be mediated by both endogenous CST as well as MPL. In the model depicted in Fig. 2, free CST (CSTf) was allowed to interact with the free cytosolic receptors, R, (given in Eq. 10) the dynamics of which are governed by MPL, to form drug–receptor complex in the cytosol (CR), which rapidly translocates to the nucleus forming DR(N). Both nuclear drug–receptor complexes act non-competitively on TAT mRNA production rate as given by,
| (18) |
| (19) |
| (20) |
where Kd,CST is the dissociation constant for CST binding to free receptors (R) and was calculated from a quantitative structure–property relationship (QSPR) model [36]; kT is the translocation constant to account for rapid entry of CR to form CR(N). The TAT mRNA is produced at a zero-order rate (ks,TATm) and dissipates with a first-order rate constant, kd,TATm. Nuclear drug–receptor complexes from both CST and MPL were assumed to act on the same process (ks,TATm) in a non-competitive manner [37] with their respective linear stimulation constants, STAT,MPL and STAT,CST. The free fraction of CST was fixed to 0.017 [23,38].
The TAT activity was assumed to be proportional to the TAT protein and was translated from its mRNA. The production and loss of the protein were described by two first-order rate constants dependent on the TAT mRNA (ks,TAT) and TAT activity (kd,TAT). The amplification factor γ indicated that one copy of the mRNA is able to translate multiple copies of the protein. The equation describing the enhanced expression of TAT activity is:
| (21) |
Data analysis
The animals in the treatment group were dosed with MPL between 1.5 and 3.5 h after “lights on”. For simplicity, we assumed the treatment time to be at 2.5 h in the circadian time in order to compare the data from our circadian rhythm study [22,23] which served as an additional set of controls for this experiment. All the figures with temporal profiles of dynamic measurements (CST, GR mRNA, GR, TAT mRNA, and TAT activity) have been plotted with respect to circadian time where the dosing was initiated at 2.5h. However, all times specified in this report will refer to post-treatment time unless mentioned otherwise.
Data from multiple animals were pooled and ADAPT II with the maximum likelihood method was applied for all fittings. The variance models for MPL and CST PK (Eq. 22) and for the PD (Eq. 23) are given by,
| (22) |
| (23) |
where Y represents the predicted value; σ 1 and σ2 are the variance parameters which were fitted, and θ represents the structural parameters. The goodness of the fit was assessed by model convergence, visual inspection, Akaike Information Criterion (AIC), Schwarz Criterion (SC), estimator criterion value, and examination of residuals.
Simulations of the driving forces for PD effects
In order to facilitate the understanding of the relationship between various driving forces and the consequent pharmacodynamic effects, simulations were performed using Eqs. 3–21 using ADAPT II. The parameter values used for these simulations are listed in Tables 2–6 and graphs are shown in Fig. 7.
Table 2.
Pharmacokinetic parameters for methylprednisolone
| Parameters | Definitions | Value (CV%) |
|---|---|---|
| kel (h−1) | Elimination rate constant | 5.57 (28.6) |
| Vc (ml/kg) | Central volume | 718.7 (39.5) |
| CL (l/h/kg) | Clearance | 4.0 (15.9) |
| k12 (h−1) | Distribution rate constant | 3.61 (50.8) |
| k21 (h−1) | Distribution rate constant | 2.84 (21.3) |
| ka1 (h−1) | Absorption rate constant | 1.255 (23.2) |
| ka2 (h−1) | Absorption rate constant | 0.219 (53.6) |
| F | Bioavailability | 0.214 (16.4) |
| Fr | Fraction absorbed by ka1 | 0.725 (11.3) |
Table 6.
Dynamic parameters for hepatic tyrosine aminotransferase
| Parameters | Definitions | Value (CV%) |
|---|---|---|
| ks,TATm (fmol/g/h) | Synthesis rate for TAT mRNA | 0.115 (51.4) |
| kd,TATm (h−1) | Loss rate for TAT mRNA | 0.232 (26.1) |
| SDR(N) (nM/mg protein)−1 | DR(N) stimulation constant | 0.0125 (54.4) |
| SCR(N) (nM/mg protein)−1 | CR(N) stimulation constant | 0.0025 (94.4) |
| kd,TAT (h−1) | Synthesis rate for TAT | 0.087 (44.2) |
| kd,TAT (h−1) | Loss rate for TAT | 0.31 (41.4) |
| γ | Amplification factor | 2.46 (25.8) |
| KD,CST (nM) | Dissociation constant for CST | 5.13 (fixed) |
| mRNATAT (0) | Initial value for TAT mRNA | 0.259 (fixed) |
| TAT (0) | Initial value for TAT activity | 0.1256 (fixed) |
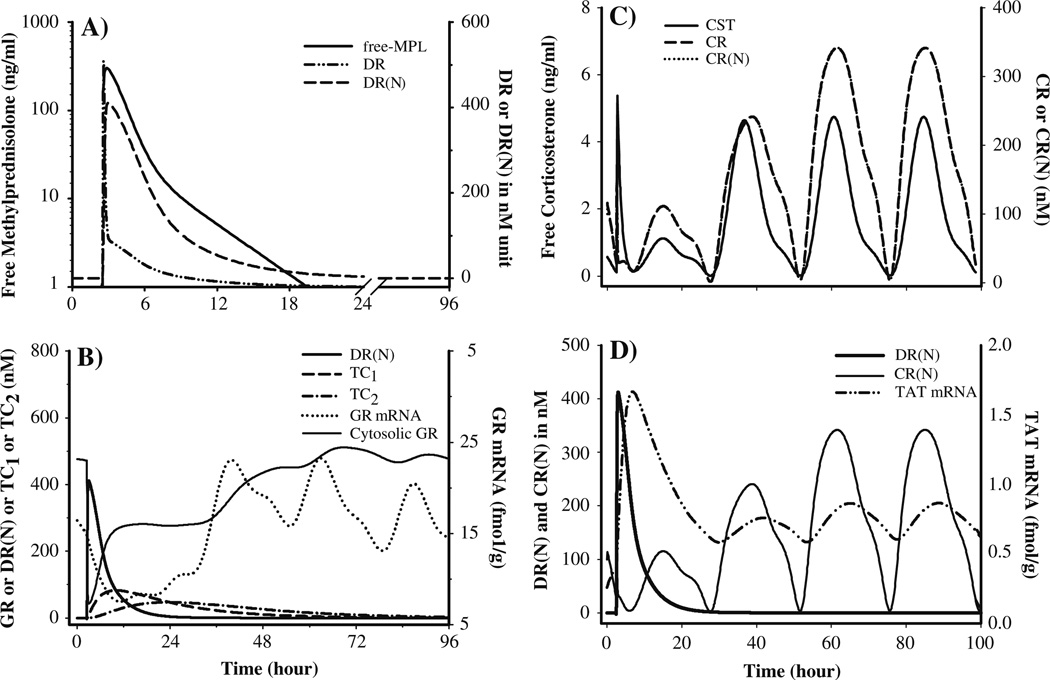
Fig. 7.
Simulated profiles of the driving forces controlling GR and TAT regulation after 50mg/kg IM MPL based on the model given in Fig. 2 and parameters given in Tables 2–6. (A) MPL, DR and DR(N) dynamics; (B) DR(N), hepatic GR mRNA and free cytosolic GR dynamics; (C) CST, CR and CR(N) dynamics and (D) CR(N), hepatic TAT mRNA and activity dynamics
Results
Pharmacokinetics
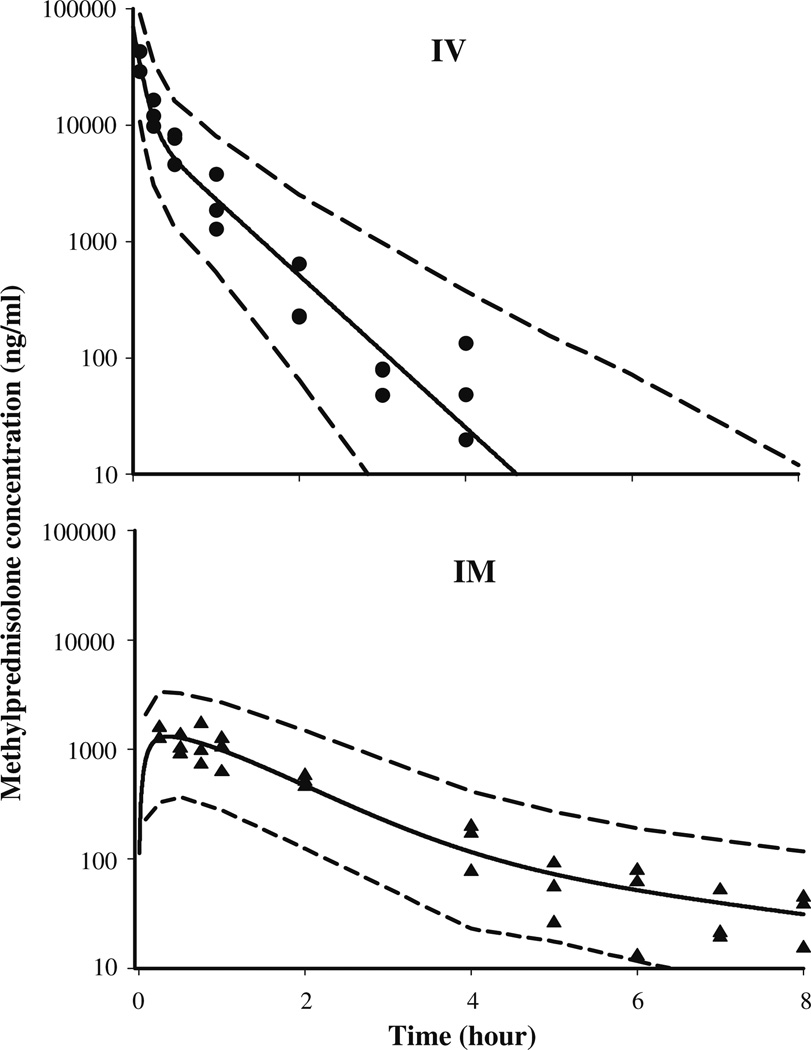
The time course of plasma MPL concentrations after 50 mg/kg IM administration is shown in Fig. 3. Comparison with the IV MPL kinetics from the ADX [10] and normal rats [22] indicated a longer half-life after IM dosing MPL was quantifiable in all rats up to 8 h in the IM group compared to only 4 h in the IV group indicating absorption rate limited elimination (flip-flop kinetics). Simultaneous fittings of both IV and IM data at the 50 mg/kg dose of MPL allowed us to resolve all kinetic parameters with reasonable variability (Table 2). Two first-order absorption processes, one with a faster rate (1.255 h−1, 23.2% CV) than the other (0.219 h−1, 53.6% CV) were optimal to describe the release of MPL from the injection site. The value of Fr indicates that about 72.5% of the drug in the muscle site can be released in the circulation via the faster absorption pathway (1.255 h−1) whereas the remaining fraction is released more slowly (0.219 h−1). The overall bioavailability of IM MPL, 0.214 (16.4% CV) in rats is lower compared to humans [39,40]. It is also lower than another CS, dexamethasone, following IM dosing in rats [41]. The clearance (4.0l/h/kg, calculated from kel · Vc) and Vc (0.719l/kg, 39.5% CV) values were similar to previously reported values (CL: 3.48l/h/kg and Vc: 0.73l/kg) in ADX rats after single doses of MPL [10].
Fig. 3.
Plasma pharmacokinetic profile of methylprednisolone after 50 mg/kg IV (top) and IM (bottom) dosing. The symbols represent individual data from rats and the solid line represents the model (Fig. 1, Table 2) fitted lines. The dashed lines depict 95% confidence intervals of model predictions
Corticosterone pharmacodynamics
The temporal patterns of plasma CST in control and treated animals are shown in Fig. 4. A simulated CST profile from the circadian rhythm study [22,23] is also shown for comparison. The parameters describing the circadian rhythm of CST are given in Tables 3 and 4.
Fig. 4.
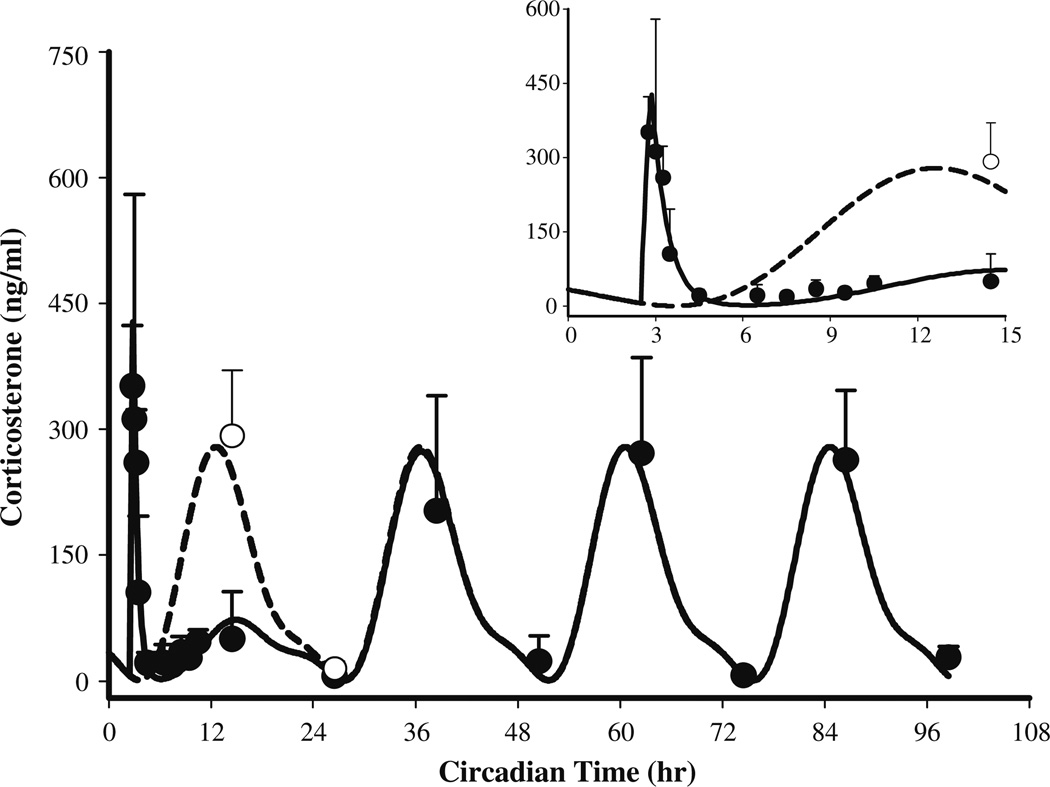
Plasma corticosterone profile in normal male Wistar rats (●: mean observed data ± SD after 50 mg/kg IM MPL, ○: controls sacrificed at 14.5 and 26.5 h circadian time, solid line shows model fittings to Eq. 9 (parameters are given in Tables 3 and 4) and the broken line depicts simulated profile of CST from the circadian rhythm study [23]. The inset represents a similar plot truncated at 15h
Table 3.
Dynamic parameters for corticosterone after MPL
| Parameters | Definitions | Value (CV%) |
|---|---|---|
| kd,CST (h−1) | Loss rate constant for CST | 1.83 (17.2) |
| IC50,CST (ng/ml) | Half-maximal inhibition constant |
6.0 (27.5) |
| τ (h) | Time to CSTmax | 0.35 (13.2) |
| CSTmax (ng/ml) | Maximum CST due to stress |
597.4 (14.1) |
Table 4.
Circadian rhythm parameters for CST and GR mRNA
| Parameters | Definitions | CST | GR mRNA |
|---|---|---|---|
| T (h) | Biorhythmic Period | 24 | 24 |
| a0 | 115.7 | 14.273 | |
| a1 | −120.9 | −1.528 | |
| a2 | Fourier Coefficients | 38.8 | 0.554 |
| b1 | −41.1 | −3.036 | |
| b2 | 1.58 | 1.188 |
Although MPL was given at the nadir of CST (early light cycle), significantly higher CST concentrations were observed (15 min post-dosing) compared to the controls from the “circadian rhythm study”, perhaps due to stress during dosing. An empirical zero-order secretion rate of CST independent of daily CST production rate was able to capture these early high concentrations of CST. The latter averaged 597.4ng/ml (14.1% CV) over a period of 21 min (13.2% CV). MPL however, caused marked adrenal suppression which persisted up to 6–7 h post-dosing and CST slowly returned to its daily rhythm by 24 h. Even after 12h, CST concentrations were visibly lower compared to the controls. Our model describing the inhibitory effect of MPL on the endogenous secretion rate of CST (Eq. 9) was able to capture the CST profile well with reasonable variability of the estimated parameters (Table 4). The IC50,CST value of ~6.0ng/ml (27.5% CV) indicated strong inhibitory effects of MPL on CST. The turnover rate constant of 1.83 h−1 (17.2% CV) was higher than reported literature values for cortisol or corticosterone [42,43].
GR mRNA dynamics
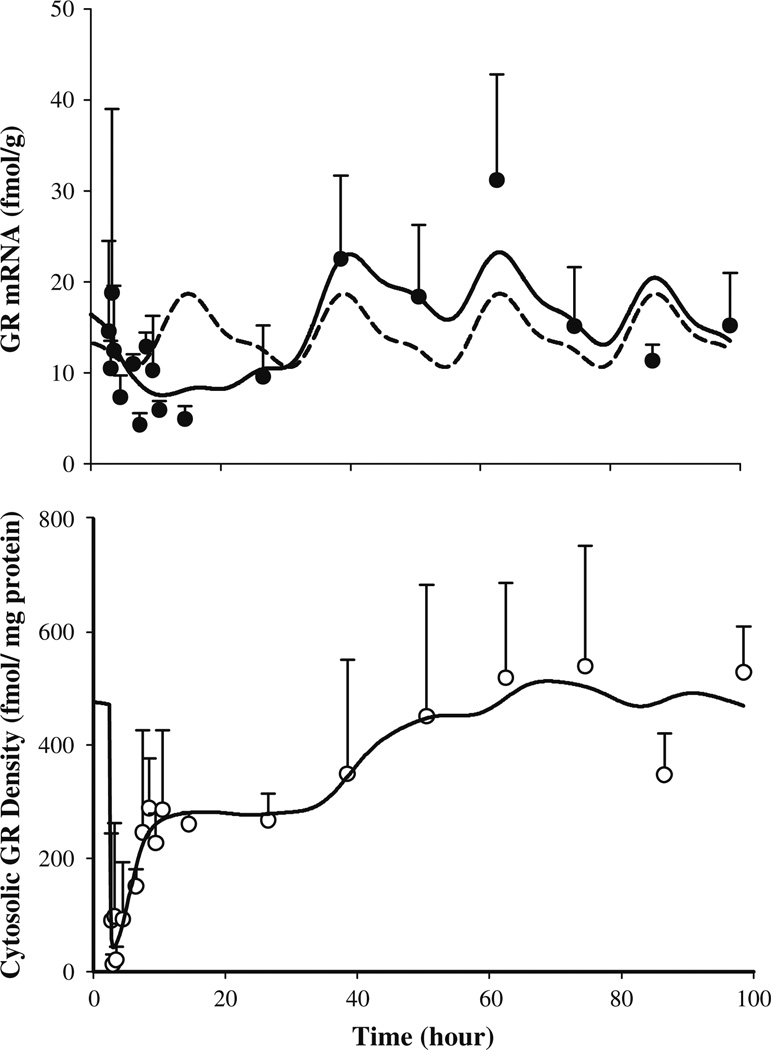
The dynamics of hepatic GR mRNA after MPL dosing along with the model fittings and simulations based on the parameters obtained from fitting GR mRNA profiles from the circadian rhythm study are shown in Fig. 5 (top panel). The parameters describing the circadian rhythm of GR mRNA are given in Table 4. MPL causes significant perturbation in GR mRNA, which unlike CST, does not return to its regular daily rhythm within the time frame of our study. The GR mRNA declined from the baseline (~16–18 fmol/g) to the trough (25–30%) in 8–12 h after MPL followed by a rebound phase with mRNA rising significantly higher than its baseline (approximately 30–40% above the baseline at 48 and 60 h). Although suppression of GR mRNA up to 45–50% of baseline after 50 mg/kg IV dosing of MPL was reported in ADX rats [8,10,11] such rebound was not observed. The estimated parameters for Eqs. 15–17 are shown in Table 5. The estimated kd,GRm, (0.12 h−1,20.4%CV) and IC50,GRm value (15.2 nM, 129% CV) closely resembled previously reported values (kd,GRm: 0.11 h−1, IC50, GRm: 26.2–50.7nM) in ADX rats [10,11]. The generation of a late rebound peak in the GR mRNA required two transduction compartments with estimated mean transit time of 15.6 h (68.4% CV). Although our model was unable to completely match the rebound phase, the down-regulation of GR mRNA was captured quite well.
Fig. 5.
Hepatic GR mRNA (top panel) and free cytosolic GR density (bottom panel) in normal rats after 50mg/kg IM MPL. The symbols represent the mean±SD and the solid lines depict model (Fig. 2) fitted lines. The dashed line in the top panel shows the simulated circadian rhythm of GR mRNA based on results obtained from our previous circadian rhythm study (parameters are given in Tables 4 and 5)
Table 5.
Dynamic parameters for glucocorticoid receptors
| Parameters | Definitions | Value (CV%) |
|---|---|---|
| kd,GRm (h−1) | Loss rate for GR mRNA | 0.12 (20.4) |
| IC50,GRm (nM) | Half-maximal inhibition ks | 15.2 (129.7) |
| IC50,TC2 (nM) | Half-maximal inhibition of kd | 60.5 (75.6) |
| τTC (h) | Transduction delay | 15.6 (68.4) |
| ks,GR (nM/h) (fmol/g)−1 | Synthesis rate for GR | 1.4 (74.6) |
| kd,GR (h−1) | Loss rate for GR | 0.05 (80.8) |
| kon (nM−1 h−1) | Association constant | 0.016 (32.9) |
| kre (h−1) | Loss rate for DR(N) | 1.31 (45.7) |
| Rf | Recycling factor | 0.93 (3.0) |
| GRm(0) (fmol/g) | Initial value for GR mRNA | 16.34 (fixed) |
| R(0) (fmol/mg protein) | Initial value for GR density | 476.0 (fixed) |
GR density
The time course of free hepatic cytosolic receptor density in normal rats along with the model (shown in Fig. 2) fittings are given in Fig. 5 (bottom panel). The estimated parameters for Eqs. 10–14 are listed in Tables 5 and 6. Free GR readily disappeared from the cytosol (within 15 min) after MPL, followed by a biphasic return as reported in ADX rats after various doses of IV MPL [10,11]. Almost 50% of GR (constituting the first return phase) was recovered within 5 h post-treatment, which was much faster than predicted by simulations (data not shown) using parameters from the fourth-generation model of steroid receptor dynamics [10]. The estimated recycling factor (Rf) of 0.925 (3% CV) indicating almost 93% recovery of receptors from the nuclear to the cytosolic compartment and the higher value of kre (1.31 h−1, 45.7% CV) compared to the ADX rats (Rf: 0.49 and kre: 0.57 h−1) accounted for the faster initial return phase in normal IM MPL treated rats. The second return phase closely followed the return and rebound phases of GR mRNA. It took almost 60 h for complete recovery of GR density. Similar to GR mRNA, GR turnover rate constants, ks,GR (1.4 fmol/g/h, 74.6% CV) and kd,GR (0.05 h−1, 80.8% CV) also closely resembled the reported values (ks,GR: 1.2nMg/h/fmol; kd,GR: 0.0572 h−1) for ADX rats [10]. Although obvious circadian variation was observed in GR mRNA, subtle if any daily variation was seen in cytosolic receptors and our model was able to capture these features well. However, it should be noted that most of the receptor dynamic parameters were associated with relatively higher CV%. Lack of stationary baselines (unlike in ADX rats) for both the GR mRNA and GR may have resulted in over-parameterization of the system, thus yielding poor precision of the estimated parameters.
TAT dynamics
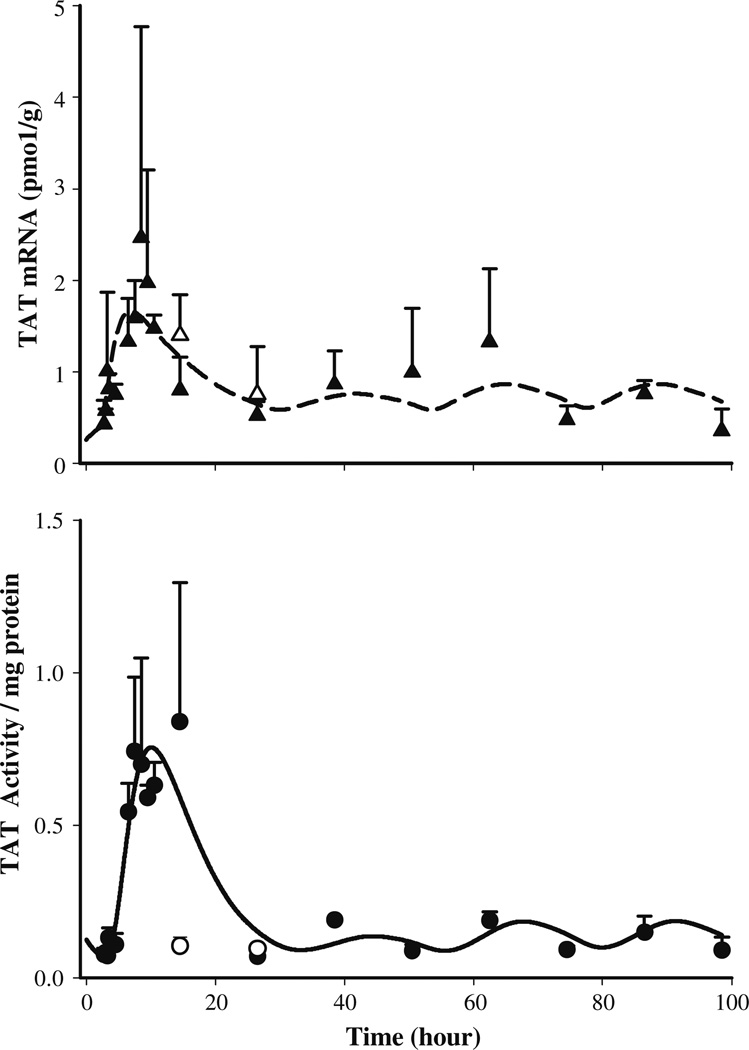
The TAT mRNA in untreated normal rats followed a circadian pattern varying between 0.13 and 0.5 pmol/g [22]. After MPL the TAT message started to increase at 1 h post-treatment reaching 2.0–2.5 pmol/g at 6–7 h (ca. 8.5–9.5 h) and returned to the baseline by 24 h. The circadian rhythm of TAT mRNA did not seem to resume until 72 h. Compared to the baseline message in normal rats from the circadian rhythm study [22], 0.3–0.4 pmol/g between circa time of 2.5 (early light cycle) and 14.5 (early dark cycle) hours the baseline values in the present study were higher ranging between 0.4 and 0.7 pmol/g at equivalent “circa time” between 74.5 and 86.5 h. The higher baseline values in the MPL treated rats was described by estimated higher ks,TATm (0.115 pmol/g/h, 51.4%) compared to the TAT mRNA synthesis rate (0.0256 pmol/g/h) found for untreated rats from the circadian rhythm study [22]. A relatively higher linear stimulation constant for DR(N) (0.0125 nM−1 mg protein, 54.4% CV) compared to CR(N) (0.0025nM−1 mg protein, 94.4% CV) was necessary to describe the effects of the exogenous and endogenous corticosteroids. The temporal profile of TAT mRNA along with the model predictions are shown in Fig. 6 (top panel). The values of the estimated TAT dynamic parameters are given in Table 6.
Fig. 6.
Hepatic TAT mRNA (top panel) and TAT activity (bottom panel) dynamics after 50 mg/kg IM MPL. The symbols (▲, treated; ∆, control) represent the mean ± SD and solid or dashed lines depict model (Fig. 2) predictions based on the parameters shown in Tables 5 and 6
The TAT activity profile, as shown in Fig. 6 (bottom), followed a similar increase as TAT mRNA with a lag time of 1.5–2 h found in ADX rats [10,11]. The TAT activity reached a peak of 0.7–0.84/mg protein at 6–8 h (post-treatment) which was almost 7–8 fold higher than the controls (0.09–0.1/mg protein), and resumed its circadian pattern by 24 h. The return of a circadian pattern in TAT activity was more apparent compared to TAT mRNA. Perhaps due to similar reasons as GR dynamics, i.e., non-stationarity in TAT mRNA and activity baselines and consequent over-parameterization of our model, precision for the estimated parameters describing TAT dynamics were high (>40%). Although the baseline for TAT mRNA in the IM MPL group of rats was higher compared to controls from the circadian rhythm study [22], baselines for TAT activities were comparable. Due to this discrepancy, simultaneous analyses of these two datasets was not done.
Simulations of driving forces for PD effects
To understand the basis of corticosteroid regulation of GR and TAT mRNA dynamics, the driving force for these effects were simulated using Eqs. 11, 12, 15, 16, 18 and 19 and parameters obtained from Tables 2 to 6. Figure 7A shows simulated profiles for free MPL concentrations, cytosolic DR, and nuclear DR(N) profiles. The formation and dissipation (translocation half-life: 5 min) of the DR is quite rapid as has previously been reported in various in vitro studies [44,45] based on which the cytosolic-nuclear translocation constant of DR was fixed to 58.5 h−1 [46]. The DR(N) was used as the driving force to regulate the down-regulation of GR mRNA. Figure 7B shows the temporal profiles for the transduction compartments used to describe the rebound phase of GR mRNA as well the model-predicted profiles for GR mRNA and density. The initial down-regulation and first return phase of GR density (within 10 h) was closely followed by up-regulation of DR(N) indicating influence of the translocation process in determining this initial phase of GR dynamics. On the other hand, the second return phase of the receptors to the baseline was a slower process and followed the return phase of GR mRNA.
Figure 7C shows the simulated CST, CR and CR(N) profiles. Since the profiles for CR and CR(N) are practically super-imposable, perhaps the concentration of CR could be used to drive further PD effects. Because steroids are known to regulate transcription of genes after their nuclear translocation and efforts in estimating a different translocation constant for CR resulted in poor precision of the parameters, we fixed the kT value for CR also as 58.5 h −1. Figure 7D shows the simulated concentrations of CR(N), DR(N) as well as their regulation of TAT mRNA.
Discussion
Investigation of various receptor/gene-mediated metabolic effects of CS has been of great interest to us. Several generations of increasingly mechanistic models have been developed to describe the dynamics of one of the most highly studied biomarkers of CS, hepatic TAT (for both mRNA and protein) with acute and chronic doses of corticosteroids in ADX rats. In this report we sought to extend our understanding of dynamic effects of CS to intact rats, a more complex, however, more physiologically relevant animal model. Several factors, related to both PK and PD, needed careful attention due to the presence of endogenous CST such as stress related surges in CST and non-stationarity of the dynamic markers. Despite all the complications that may be involved in using this rat model, our objective was to quantitatively understand the interplay of endogenous and exogenous CS by means of PK/PD modeling.
In our previous studies with ADX rats, animals underwent jugular vein cannulation for IV dosing which causes stress related persistent increases in CST in normal rats (unpublished results). Therefore, switching to normal rats required use of a less stressful parenteral route of administration producing similar exposure as the IV route. Simultaneous analysis of plasma as well as muscle site PK of MPL in rats revealed incomplete bioavailability and a complex release pattern [24] from the injection site (flip-flop kinetics with two absorption rates) compared to humans [39,40] and even compared to other CS in rats [41]. Despite these complexities in PK via the IM route, examination of TAT dynamics revealed similar IM and IV response profiles [24]. Therefore, the more convenient IM dosing was chosen.
The dose of MPL was given IM at the nadir of the CST circadian rhythm [22,23], i.e., between 1.5 and 3.5 h after the light cycle started with the intention of keeping adrenal suppression minimal. However, the stress-related surge in CST (comparable to the highest CST at early dark period) was quite apparent due to handling of the rats during dosing. This surge was not observed in the circadian rhythm study where the animals were also handled in order to anesthetize with IM ketamine/xylazine for sacrifice. Therefore, it was assumed that the stress induced CST surge is not instantaneous and the time (21 min) to achieve the maximum CST (~600 ng/ml) was close to the first sampling time point of 15 min. Exogenous corticosteroids are known to inhibit endogenous CST secretion rapidly by a negative feedback loop on the HPA axis in humans as well as in rodents [43,47,48]. The IM MPL caused marked adrenal suppression indicated by CST at 12 h in the MPL group (50.3 ± 50.0 ng/ml) compared to the controls (292.1 ±78.2 ng/ml). However, this suppression was not prolonged, and the daily rhythm of CST was resumed within 24 h. Although the choice of the highest level of stress-induced plasma CST (based on the observed data) and the time to achieve this concentration was empirical, overall our model of CST dynamics was able to fit the observed data well.
The GR mRNA in normal rat liver exhibits circadian variation, reaching its peak at similar times as CST, i.e., at early dark cycle and nadir at early light cycle [38]. This pattern is contrary to the inhibitory effects of exogenous CS on GR mRNA, suggesting differential regulation of GR message in homeostatic conditions. Therefore, CST was not used as a driving force to regulate the circadian rhythm of GR mRNA and an empirical two-harmonic function was used to describe its rhythm.
Despite low bioavailability (21%) of MPL via the IM route, it caused substantial down-regulation in GR mRNA (~50%) as reported in ADX rats after IV MPL [10,11]. Autologous down-regulation of GR mRNA in various tissues is a well-known phenomenon, the exact mechanism of which remains unclear. Steroid-mediated decreased transcription due to the presence of specific DNA sequences (perhaps negative GRE) [30], interaction of steroid-bound receptors with other positive regulators of GR mRNA [49], and destabilization of GR mRNA by CS [50] have been postulated to be possible mechanisms. Interestingly, following down-regulation of GR mRNA, we observed a subtle rebound phase. Although such biphasic regulation of GR mRNA was reported in neuronal culture following dexamethasone treatment, apparently due to the presence of two different types of GR (type I: CST preferring and type II) in the brain [49], such phenomenon has not been reported for any other tissues. Perhaps prolonged and profound deviation from its tightly controlled baseline may cause negative feedback regulation. However, we were unable to use the changes in GR mRNA baseline to drive the rebound phase due to poor precision of the estimated parameters (results not shown) compared to the present model where a signal transduction compartment derived from DR(A) was assumed to inhibit the loss rate of GR mRNA.
The circadian rhythm of GR mRNA did not produce variation in receptor concentrations. Rather a constant level of receptors was observed in the control rats (average of 476 fmol/mg protein from 6 control rats) suggesting a slower turn-over process (ks,GR: 1.4nM/fmol/g GR mRNA/h; kd,GR: 0.05 h−1) for hepatic receptors. The estimated receptor half-life from our model (~13.9 h) closely resembled previously reported values (control: 19.9 h; treated: 9.7 h) from rat pituitary cell lines after exposure to triamcinolone acetonide [51]. The IM MPL produced rapid and significant down-regulation (within 15 min of dosing) in free cytosolic receptor density in normal rats followed by a biphasic return to its baseline, a pattern which closely resembled the reported GR profile effects in ADX rats after IV MPL. An integrated model, similar to our fifth-generation model in ADX rats [9,10], was used to describe the dynamics of GR mRNA and cytosolic receptor simultaneously where only binding of MPL (not CST) to the GR was assumed to inhibit the synthesis of GR mRNA. Ideally we should have been able to use the well-known Gaddum equation [52] to describe the competitive binding of MPL and CST to their receptors (with their respective dissociation constants) and use the resultant steroid-bound receptor to drive the down-stream dynamic effects. However, neither GR mRNA nor free cytosolic GR showed any apparent CST regulation. Consequently, simulated GR patterns (results not shown) using such competitive interaction resulted in underestimation of free receptors in rat liver.
The TAT gene contains a positive GRE sequence in the promoter region. Therefore, CS are major regulators of this transcription. In addition, the TAT gene is also regulated by cAMP and insulin [53,54]. The TAT mRNA and activity followed a marked circadian variation which was postulated to be mediated by the circadian rhythm of CST [22]. Similar to previous studies in ADX rats, we observed a significant increase in TAT mRNA and activity following MPL and TAT activity resumed its circadian pattern faster than did TAT mRNA. The presence of a daily rhythm in GR mRNA contrary to the regulation of exogenous CS on this gene and absence of any rhythm in hepatic GR suggested a different CS regulation pathway for GR compared to TAT. For modeling purposes we simply assumed that GR, both at mRNA and protein levels, was insensitive to changes in plasma CST concentrations. Therefore, Eqs. 10–17 describing GR dynamics did not have any CST component. On the other hand, since we observed resumption of a daily rhythm in TAT enzyme, it was assumed that free CST binds to the available free cytosolic receptors (modulated by MPL) to form nuclear drug–receptor complex, CR(N), which in turn maintains the daily rhythm of TAT after MPL effects dissipate. The linear stimulation factors for DR(N) and CR(N) to stimulate the transcription of TAT mRNA differed significantly (SDR(N): 0.0125nM−1 mg protein, SCR(N): 0.0025 nM−1 mg protein) from each other indicating different efficacy for MPL and CST. This difference can be explained by comparing the simulated concentrations of nuclear drug–receptor complex for MPL and CST (shown in Fig. 7D). The simulated maximum circadian concentration of CR(N) (~325 nM), based on a fixed in vitro kD of 5.13 nM and 1.7% fraction unbound CST, were quite similar to the highest DR(N) concentrations (~400nM) derived from the our model shown in Fig. 2. Clearly, the ability of CR(N) to stimulate TAT mRNA was much less than DR(N), as reflected by their estimated stimulation constant values. Although this difference could also be described by a higher in vivo kD value for CST, use of the former approach yielded better fitting results.
Ideally, simultaneous fitting of TAT dynamics from the circadian rhythm study and IM MPL treated normal rats should be possible. However, marked differences in TAT mRNA baselines in these two groups, yielding similar TAT activities imply that further complexities are involved. Our model was able to capture the general trend of GR and TAT dynamics quite well. However, possible over-parameterization of the model yielded less than superior precision of the estimated parameters.
In summary, the regulation by MPL of GR as well as TAT dynamics was similar in normal and ADX rats. However, interaction of exogenous and endogenous steroids in regulating various biomarkers was far more complicated and dynamic than our expectations based on previous models. Our current model of receptor/gene-mediated effects of corticosteroids in normal rats was based on our highly mechanistic receptor/gene-mediated [9,10] and QSPR-pharmacodynamic models [7] and was able to describe the complex interplay of exogenous and endogenous corticosteroid effects on the dynamics of hepatic GR mRNA, cytosolic free GR, TAT mRNA, and TAT activity in normal rats.
Acknowledgements
Supported by Grant GM24211 from the National Institute of Health.
Contributor Information
Anasuya Hazra, Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, State University of New York at Buffalo, 565 Hochstetter Hall, Buffalo, NY 14260, USA; Clinical Pharmacology, Pfizer Inc., Groton, CT 06340, USA.
Nancy Pyszczynski, Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, State University of New York at Buffalo, 565 Hochstetter Hall, Buffalo, NY 14260, USA.
Debra C. DuBois, Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, State University of New York at Buffalo, 565 Hochstetter Hall, Buffalo, NY 14260, USA Department of Biological Sciences, State University of New York at Buffalo, Buffalo, NY 14260, USA.
Richard R. Almon, Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, State University of New York at Buffalo, 565 Hochstetter Hall, Buffalo, NY 14260, USA Department of Biological Sciences, State University of New York at Buffalo, Buffalo, NY 14260, USA.
William J. Jusko, Email: wjjusko@buffalo.edu, Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, State University of New York at Buffalo, 565 Hochstetter Hall, Buffalo, NY 14260, USA.
References
- 1.Czock D, Keller F, Rasche FM, Haussler U. Pharmacokinetics and pharmacodynamics of sys-temically administered glucocorticoids. Clin Pharmacokinet. 2005;44:61–98. doi: 10.2165/00003088-200544010-00003. [DOI] [PubMed] [Google Scholar]
- 2.Bagdade JD, Porte D, Jr, Bierman EL. Steroid-induced lipemia. A complication of high-dosage corticosteroid therapy. Arch Int Med. 1970;125:129–134. doi: 10.1001/archinte.125.1.129. [DOI] [PubMed] [Google Scholar]
- 3.Schacke H, Docke WD, Asadullah K. Mechanisms involved in the side effectsof glucocorticoids. Pharmacol Ther. 2002;96:23–43. doi: 10.1016/s0163-7258(02)00297-8. [DOI] [PubMed] [Google Scholar]
- 4.Staels B. When the clock stops ticking, metabolic syndrome explodes. Nat Med. 2006;12:54–55. doi: 10.1038/nm0106-54. [DOI] [PubMed] [Google Scholar]
- 5.Wilcke JR, Davis LE. Review of glucocorticoid pharmacology. Vet Clin North America - Small Anim Pract. 1982;12:3–17. doi: 10.1016/s0195-5616(82)50001-0. [DOI] [PubMed] [Google Scholar]
- 6.Haughey DB, Jusko WJ. Receptor-mediated methylprednisolone pharmacodynamics in rats: steroid-induced receptor down-regulation. J Pharmacokinet Biopharm. 1992;20:333–355. doi: 10.1007/BF01062462. [DOI] [PubMed] [Google Scholar]
- 7.Mager DE, Pyszczynski NA, Jusko WJ. Integrated QSPR-pharmacodynamic model of genomic effects of several corticosteroids. J Pharm Sci. 2003;92:881–889. doi: 10.1002/jps.10343. [DOI] [PubMed] [Google Scholar]
- 8.Ramakrishnan R, DuBois DC, Almon RR, Pyszczynski NA, Jusko WJ. Pharmacodynamics and pharmacogenomics of methylprednisolone during 7-day infusions in rats. J Pharmacol Exp Ther. 2002;300:245–256. doi: 10.1124/jpet.300.1.245. [DOI] [PubMed] [Google Scholar]
- 9.Ramakrishnan R, DuBois DC, Almon RR, Pyszczynski NA, Jusko WJ. Fifth-generation model for corticosteroid pharmacodynamics: applicationtosteady-state receptor down-regulation and enzyme induction patterns during seven-day continuous infusion of methylprednisolone in rats. J Pharmacokinet Pharmacodyn. 2002;29:1–24. doi: 10.1023/a:1015765201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun YN, DuBois DC, Almon RR, Jusko WJ. Fourth-generation model for corticosteroid phar-macodynamics: a model for methylprednisolone effects on receptor/gene-mediated glucocorticoid receptor down-regulation and tyrosine aminotransferase inductionin rat liver. J Pharmacokinet Biopharm. 1998;26:289–317. doi: 10.1023/a:1023233409550. [DOI] [PubMed] [Google Scholar]
- 11.Sun YN, DuBois DC, Almon RR, Pyszczynski NA, Jusko WJ. Dose-dependence and repeated-dose studies for receptor/gene-mediated pharmacodynamics of methylprednisolone on glucocorticoid receptor down-regulation and tyrosine aminotransferase induction in rat liver. J Pharmacokinet Bio-pharm. 1998;26:619–648. doi: 10.1023/a:1020746822634. [DOI] [PubMed] [Google Scholar]
- 12.Wolfe BB, Harden TK, Molinoff PB. Beta-adrenergic receptors in rat liver: effects of adrena-lectomy. Proc Natl Acad Sci USA. 1976;73:1343–1347. doi: 10.1073/pnas.73.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meijs-Roelofs HM, Kramer P. Effects of adrenalectomy on the release of follicle-stimulating hormone and the onset of puberty in female rats. J Endocrinol. 1977;75:419–426. doi: 10.1677/joe.0.0750419. [DOI] [PubMed] [Google Scholar]
- 14.Savontaus E, Conwell IM, Wardlaw SL. Effects of adrenalectomy on AGRP, POMC, NPY and CART gene expression in the basal hypothalamus of fed and fasted rats. Brain Res. 2002;958:130–138. doi: 10.1016/s0006-8993(02)03674-0. [DOI] [PubMed] [Google Scholar]
- 15.Exton JH. Regulation of gluconeogenesis by glucocorticoids. Monogr Endocrinol. 1979;12:535–546. doi: 10.1007/978-3-642-81265-1_28. [DOI] [PubMed] [Google Scholar]
- 16.Haughey DB, Jusko WJ. Analysis of methylprednisolone, methylprednisone and corticosterone for assessment of methylprednisolone disposition in the rat. J Chromatogr. 1988;430:241–248. doi: 10.1016/s0378-4347(00)83159-x. [DOI] [PubMed] [Google Scholar]
- 17.Boudinot FD, D’Ambrosio R, Jusko WJ. Receptor-mediated pharmacodynamics of predniso-lone in the rat. J Pharmacokinet Biopharm. 1986;14:469–493. doi: 10.1007/BF01059656. [DOI] [PubMed] [Google Scholar]
- 18.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.DuBois DC, Almon RR, Jusko WJ. Molar quantification of specific messenger ribonucleic acid expression in northern hybridization using cRNA standards. Anal Biochem. 1993;210:140–144. doi: 10.1006/abio.1993.1164. [DOI] [PubMed] [Google Scholar]
- 20.DuBois DC, Xu ZX, McKay L, Almon RR, Pyszcznski N, Jusko WJ. Differential dynamics of receptor down-regulation and tyrosine aminotransferase induction following glucocorticoid treatment. J Steroid Biochem Mol Biol. 1995;54:237–243. doi: 10.1016/0960-0760(95)00139-q. [DOI] [PubMed] [Google Scholar]
- 21.Diamondstone TI. Assay of tyrosine aminotransferase by conversion of p-hydroxypheny-lpyruvate to p-hydroxybenzaldehyde. Anal Biochem. 1966;16:395–401. [Google Scholar]
- 22.Hazra A, Jusko WJ, Almon RR, DuBois DC. Pharmacodynamics of circadian rhythm of corti-costerone effects on tyrosine aminotransferase in normal rats. AAPS J. 2005;6 Abstract T3355. [Google Scholar]
- 23.Yao Z, Dubois DC, Almon RR, Jusko WJ. Modeling circadian chythms of glucocorticoid receptor and glutamine synthetase expression in rat skeletal muscle. Pharm Res. 2006;23:670–679. doi: 10.1007/s11095-005-9608-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazra A, Pyszczynski NA, Dubois DC, Almon RR, Jusko WJ. Pharmacokinetics of methylpred-nisolone after intravenous and intramuscular administration in rats. Biopharm Drug Disp. 2007;28:263–277. doi: 10.1002/bdd.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vamvakopoulos NO. Tissue-specific expression of heat shock proteins 70 and 90: potential implication for differential sensitivity of tissues to glucocorticoids. Mol Cell Endocrinol. 1993;98:49–54. doi: 10.1016/0303-7207(93)90235-c. [DOI] [PubMed] [Google Scholar]
- 26.Czar MJ, Owens-Grillo JK, Dittmar KD, Hutchison KA, Zacharek AM, Leach KL, Deibel MR, Jr, Pratt WB. Characterization of the protein-protein interactions determining the heat shock protein (hsp90.hsp70.hsp56) heterocomplex. J Biol Chem. 1994;269:11155–11161. [PubMed] [Google Scholar]
- 27.Hutchison KA, Dittmar KD, Czar MJ, Pratt WB. Proof that hsp70 is required for assembly of the glucocorticoid receptor into a heterocomplex with hsp90. J Biol Chem. 1994;269:5043–5049. [PubMed] [Google Scholar]
- 28.Orti E, Hu LM, Munck A. Kinetics of glucocorticoid receptor phosphorylation in intact cells. Evidence for hormone-induced hyperphosphorylation after activation and recycling of hyperphospho-rylated receptors. J Biol Chem. 1993;268:7779–7784. [PubMed] [Google Scholar]
- 29.Schaaf MJ, Cidlowski JA. Molecular mechanisms of glucocorticoid action and resistance. J Steroid Biochem Mol Biol. 2002;83:37–48. doi: 10.1016/s0960-0760(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 30.Burnstein KL, Jewell CM, Cidlowski JA. Human glucocorticoid receptor cDNA contains sequences sufficient for receptor down-regulation. J Biol Chem. 1990;265:7284–7291. [PubMed] [Google Scholar]
- 31.Oakley RH, Cidlowski JA. Homologous down regulation of the glucocorticoid receptor: the molecular machinery. Crit Rev Eukar Gene Express. 1993;3:63–88. [PubMed] [Google Scholar]
- 32.Beato M, Chalepakis G, Schauer M, Slater EP. DNA regulatory elements for steroid hormones. J Steroid Biochem. 1989;32:737–747. doi: 10.1016/0022-4731(89)90521-9. [DOI] [PubMed] [Google Scholar]
- 33.Kong AN, Jungbluth GL, Pasko MT, Beam TR, Jusko WJ. Pharmacokinetics of methylpred-nisolone sodium succinate and methylprednisolone in patients undergoing cardiopulmonary bypass. Pharmacotherapy. 1990;10:29–34. [PubMed] [Google Scholar]
- 34.Krzyzanski W. FOURPHARM user’s guide: a computer program applying Fourier analysis to biorhythmic data. Buffalo, NY: 2000. [Google Scholar]
- 35.Cahill AL, Ehret CF. Circadian variations in the activity of tyrosine hydroxylase, tyrosine ami-notransferase, and tryptophan hydroxylase: relationship to catecholamine metabolism. J Neurochem. 1981;37:1109–1115. doi: 10.1111/j.1471-4159.1981.tb04660.x. [DOI] [PubMed] [Google Scholar]
- 36.Wolff ME, Baxter JD, Kollman PA, Lee DL, Kuntz ID, Bloom E, Matulich DT, Morris J. Nature of steroid-glucocorticoid receptor interactions: thermodynamic analysis of the binding reaction. Biochemistry. 1978;17:3201–3208. doi: 10.1021/bi00609a005. [DOI] [PubMed] [Google Scholar]
- 37.Earp JC, Krzyzanski W, Chakraborty A, Zamacona MK, Jusko WJ. Assessment of drug interactions relevant to pharmacodynamic indirect response models. J Pharmacokinet Pharmacodyn. 2004;31:345–380. doi: 10.1007/s10928-004-8319-4. [DOI] [PubMed] [Google Scholar]
- 38.Hazra A. Pharmacokinetic/pharmacodynamic modeling of selected receptor/gene mediated effects of corticosteroids. Ph.D. Thesis. Buffalo, NY: State University of New York at Buffalo; 2007. [Google Scholar]
- 39.Antal EJ, Wright CE3rd, Gillespie WR, Albert KS. Influence of route of administration on the pharmacokinetics of methylprednisolone. J Pharmacokinet Biopharm. 1983;11:561–576. doi: 10.1007/BF01059057. [DOI] [PubMed] [Google Scholar]
- 40.Daley-Yates PT, Gregory AJ, Brooks CD. Pharmacokinetic and pharmacodynamic assessment of bioavailability for two prodrugs of methylprednisolone. Br J Clin Pharmacol. 1997;43:593–601. doi: 10.1046/j.1365-2125.1997.00598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samtani MN, Jusko WJ. Comparison of dexamethasone pharmacokinetics in female rats after intravenous and intramuscular administration. Biopharm Drug Dispos. 2005;26:85–91. doi: 10.1002/bdd.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty A, Krzyzanski W, Jusko WJ. Mathematical modeling of circadian cortisol concentrations using indirect response models: comparison of several methods. J Pharmacokinet Biopharm. 1999;27:23–43. doi: 10.1023/a:1020678628317. [DOI] [PubMed] [Google Scholar]
- 43.Samtani MN, Pyszczynski NA, Dubois DC, Almon RR, Jusko WJ. Modeling glucocorticoid-mediated fetal lung maturation: I. Temporal patterns of corticosteroids in rat pregnancy. J Pharmacol Exp Ther. 2006;317:117–126. doi: 10.1124/jpet.105.095851. [DOI] [PubMed] [Google Scholar]
- 44.Hache RJ, Tse R, Reich T, Savory JG, Lefebvre YA. Nucleocytoplasmic trafficking of steroid-free glucocorticoid receptor. J Biol Chem. 1999;274:1432–1439. doi: 10.1074/jbc.274.3.1432. [DOI] [PubMed] [Google Scholar]
- 45.Htun H, Barsony J, Renyi I, Gould DL, Hager GL. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci USA. 1996;93:4845–4850. doi: 10.1073/pnas.93.10.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hazra A, Dubois DC, Almon RR, Jusko WJ. Assessing the dynamics of nuclear glucocorticoid-receptor complex: addingflexibility togene expression modeling. J Pharmacokinet Pharmacodyn Epub. 2007 doi: 10.1007/s10928-007-9049-1. (ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rix M, Birkebaek NH, Rosthoj S, Clausen N. Clinical impact of corticosteroid-induced adrenal suppression during treatment for acute lymphoblastic leukemia in children: a prospective observational study using the low-dose adrenocorticotropin test. J Pediatrics. 2005;147:645–650. doi: 10.1016/j.jpeds.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Yamada K, Satoh T. In vivo effects of glucocorticoids on serum corticosterone levels in rats. Res Commun Mol Pathol Pharmacol. 1985;47:441–444. [PubMed] [Google Scholar]
- 49.Pepin MC, Beaulieu S, Barden N. Differential regulation by dexamethasone of glucocorticoid receptor messenger RNA concentrations in neuronal cultures derived from fetal rat hypothalamus and cerebral cortex. Cell Mol Neurobiol. 1990;10:227–235. doi: 10.1007/BF00734576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vedeckis WV, Ali M, Allen HR. Regulation of glucocorticoid receptor protein and mRNA levels. Cancer Res. 1989;49:2295s–2302s. [PubMed] [Google Scholar]
- 51.McIntyre WR, Samuels HH. Triamcinolone acetonide regulates glucocorticoid-receptor levels by decreasing the half-life of the activated nuclear-receptor form. J Biol Chem. 1985;260:418–427. [PubMed] [Google Scholar]
- 52.Gaddum JH. The quantitative effect of antagonistic drugs. J Physiol (Lond) 1937;89:7–9. [Google Scholar]
- 53.Granner DK, Hargrove JL. Regulation of the synthesis of tyrosine aminotransferase: the relationship to mRNATAT. Mol Cell Biochem. 1983;53–54:113–128. doi: 10.1007/BF00225249. [DOI] [PubMed] [Google Scholar]
- 54.Schmid E, Schmid W, Jantzen M, Mayer D, Jastorff B, Schutz G. Transcription activation of the tyrosine aminotransferase gene by glucocorticoids and cAMP in primary hepatocytes. Eur J Biochem. 1987;165:499–506. doi: 10.1111/j.1432-1033.1987.tb11467.x. [DOI] [PubMed] [Google Scholar]