Abstract
The quality of structure models submitted to CASP9 is analyzed in the context of previous CASPs. Comparison methods are similar to those used in previous papers in this series, with the addition of new methods for looking at model quality in regions not covered by a single best structural template, alignment accuracy, and progress for template free models. Progress in this CASP was again modest, and statistically hard to validate. Nevertheless, there are several positive trends. There is an indication of improvement in overall model quality for the mid-range of template based modeling difficulty, methods for identifying the best model from a set generated have improved, and there are strong indications of progress in the quality of template free models of short proteins. In addition, the new examination of model quality in regions of model not covered by the best available template reveals better performance than had previously been apparent.
Keywords: Protein Structure Prediction, Community Wide Experiment, CASP
Introduction
The ninth round of CASP experiments, conducted in 2010, tested protein structure modeling methods from over 170 groups, including over 61,000 tertiary structure models. Results were analyzed by the Protein Structure Prediction Center using standard metrics established in previous experiments and by independent assessors using a combination of the standard methods, new methods of their choosing, and detailed inspection of models. The results of these analyses are discussed in papers by the assessors elsewhere in this special issue1,2. In this paper we compare the quality of the best models received for each target over the course of CASP experiments, starting in 1994, with particular emphasis to advances in the field over the most recent three CASP experiments, numbers 7, 8 and 9, held in 2006, 2008 and 2010 respectively. The analysis is based on the methods introduced in the earlier articles3–7. These include methods for assigning relative difficulty to targets, evaluating overall backbone accuracy, and measures of alignment performance. In addition, improvement in model quality over that which can be obtained from a single best template is subdivided into the contribution from correct modeling of non-template regions and the offsetting loss of model quality from alignment errors. A new standard for examining progress in the template free modeling category is introduced, following work by the CASP8 assessors8.
Target Difficulty Analysis
Different proteins may present different levels of modeling difficulty9,10. As a result, most comparison of performance on different target structures is meaningless without some form of relationship to difficulty. There is no ideal difficulty metric, and a number of factors play a role. In this and the previous comparisons of performance we use a primary difficulty scale based on the similarity of the structure and sequence of the target to those of the closest template (See Methods for details). Additional relevant factors, such as the depth and diversity of the sequence alignment of the target with other proteins and the number of related domains with known structure are not included.
For analysis, targets are divided into domains according to procedures described in Methods. Note that the domain parsing used here is slightly different to that employed by the assessors. Also, a major determinant of target difficulty is whether or not at least one related experimental structure can be identified, providing a template on which to base a model. We use the assessors’ categorization of targets into two difficulty categories, made with knowledge of the experimental structures. Template Based Modeling targets (TBM) are those where at least one useful related structure could be identified; Free Modeling cases (FM) are those, where no templates could be found or any templates were so distant that template free methods were judged necessary to generate a model11.
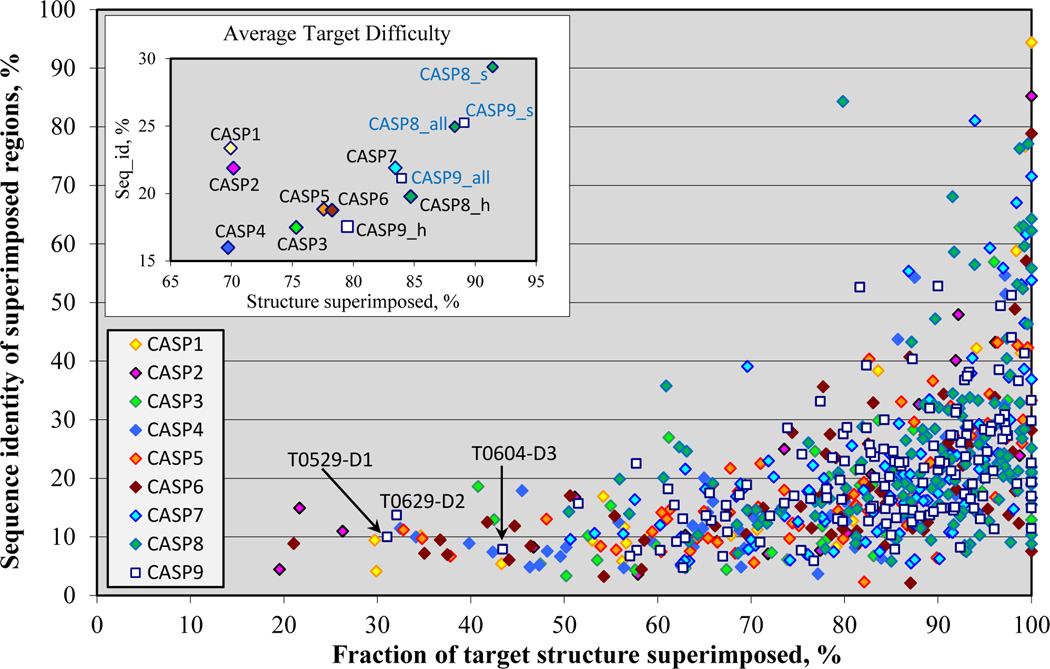
Figure 1 shows the distribution of target difficulty for all CASPs, as a function of structure and sequence similarity between the experimental structure of each target and the corresponding best available template. Targets span a wide range of structure and sequence similarity to templates, with CASP9 targets fitting the general pattern seen in all CASPs. The majority of targets in CASP9 show structure superposability with the best template above 60% and sequence identity below 40%. A notable difference to recent CASPs is that there are more targets with relatively low structural coverage, and in this respect the CASP9 target set has a greater diversity, more similar to that seen in the early experiments. Three targets (T0529_1, T0629_2 and T0604_3) have coverage below 50%. (As described in Methods, a relatively liberal definition of structural coverage is used here).
Figure 1.
Distribution of target difficulty. The difficulty of producing an accurate model is shown as a function of the fraction of each target that can be superimposed on a known structure (horizontal axis) and the sequence identity between target and template for the superimposed region (vertical axis). Average coverage and sequence identity for targets in each of the CASPs are plotted in the inset graph. The data for CASP9 and CASP8 are shown for three different target subsets: server only targets (marked with the “_s” suffix), human/server targets (“_h”), and complete set of targets (“_all”). CASP9 human/server targets are overall more difficult than those of the most recent previous CASPs, and similar in this respect to some earlier experiments in the series.
The inset in Figure 1 shows the average target difficulty for each CASP. Note that in the most recent two CASPs, targets are divided into two groups: the hardest subset, which was released for modeling by human experts and by automatic servers, and the rest released for modeling by servers only. In CASP9, after eliminating 13 canceled CASP9 targets11, and splitting the rest into appropriate domains, there are 75 human/server domain targets and 64 server only domain targets. It is clear that the CASP9 human/server target set (CASP9_h) has a lower average coverage and sequence identity than the CASP7 and CASP8 sets and therefore is likely overall more difficult to model. Closer examination of the data shows this is a general property of the targets, and not due to distortion by a few outliers. We also analyzed the difficulty of targets in all CASPs based on cumulative z-scores calculated from the distributions of average coverage and sequence identity scores in all CASPs. This analysis (Figure S1 in Supplementary Material) confirmed that CASP9 human/server targets are harder than those of CASP7 and CASP8, and approximately the same in difficulty as CASP5 and CASP6 targets. Note that in the following analysis we concentrate on the human/server targets from CASP9 and CASP8, as this selection makes the overall level of difficulty more similar to that of earlier CASPs (see the graph), and also facilitates comparison of server and human performance.
As in previous CASPs, we derive a one dimensional scale of target difficulty, using a combination of structure superposability and sequence identity (see Methods). The target categories defined by the assessors map approximately to this difficulty scale, with the template-based targets falling mostly in the easy and middle range of the scale, and template free targets concentrated at the most difficult end. More CASP9 targets than CASP8 ones lie at the difficult end of the scale, a consequence of the higher number of FM targets in CASP9 (as defined by the assessors): there were 26 human/server FM targets in CASP9, 10 in CASP8, and 19 in CASP7.
Overall Model Quality
Overall backbone accuracy of CASP models is evaluated using the GDT_TS measure12. GDT_TS represents the average percentage of residues that are in close proximity in two structures optimally superimposed using four different distance cutoffs (see Methods for a detailed definition). The use of multiple thresholds makes the measure suitable for a wide range of model accuracy. Even so, GDT_TS alone is not a reliable measure for the most difficult modeling cases, as those models are often very approximate, and below roughly 30 GDT_TS units the measure has poor discrimination of model quality. In these cases careful visual inspection of the models is needed to detect useful features.
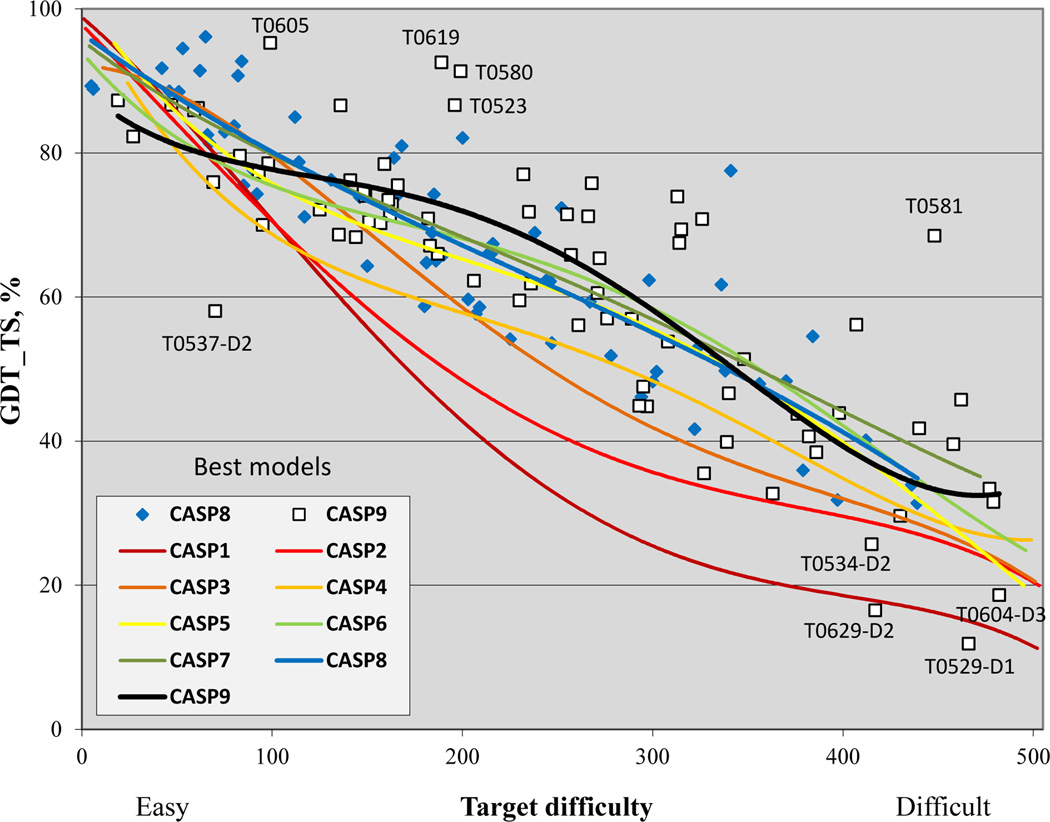
Figure 2 shows the GDT_TS score for the best model on each target as a function of target difficulty. The trend lines for successive CASPs show impressive progress over the course of the experiments, but with noticeable slowing over more recent rounds. For the CASP9 data, the trend line runs a little lower than in previous CASPs in the easiest and most difficult parts of the difficulty scale, but exhibits an easily observable bump in the mid-range of difficulty, representing apparent progress. A very similar pattern can be seen in the graphs showing average GDT_TS scores from the six best groups on each target (Figure S2 in Supplementary Material). Alternative trend lines, using moving averages (splines) and third degree polynomials, also show apparent mid-range progress (data not shown), as does a plot with target difficulty scale based on the alternative (Z-score based) difficulty ranking (Supplementary Figure S3).
Figure 2.
GDT_TS scores of submitted best models for targets in all CASPs, as a function of target difficulty. Each point represents one target. Quartic trend lines show a likely increased accuracy of modeling in the middle range of difficulty in CASP9. Other types of polynomial fit and moving average splines show a similar trend (lines not shown).
Several outliers in the CASP9 data are identified on the graph. There are four data points that are noticeably lower than expected, given their estimated difficulty. T0537_D2 is the shortest CASP9 domain target, consisting of only 31 residues, and classified by the assessors as a template free modeling target. There are templates that cover 100% of T0537_D2 length but those are very difficult to identify based on the sequence alone, as the sequence identity to the best template is 19%, equivalent to only 6 identical residues. The difficulty scale is not able to properly position such an unusual target. Domains T0529_D1 (333 residues), T0534_D2 (176 residues), T0604_D3 (205 residues) and T0629_D2 (159 residues) are four free modeling targets without good templates where all predictors submitted very poor models. As discussed later, poor performance on free modeling targets longer than about 120 residues is consistent in all CASPs, but it so happened that there are more of these in CASP9, artificially pulling down average performance for difficult targets. There are also several targets where the best model is substantially more accurate than expected for the difficulty range. An outstanding example is the free modeling target T0581, where an extraordinarily accurate model was produced. Good performance on three midrange difficulty targets, T0523, T0580 and T0619, help pull up the CASP9 trend line in the central region. All have over 90% coverage by the best template, making them unusually easy for the difficulty range. The limited number of points in any given difficulty range makes drawing fine distinctions between performances in different CASPs problematic. Nevertheless, the apparent improvement in the mid-difficulty range in CASP9 is supported by a substantial number of consistent data points.
Alignment accuracy
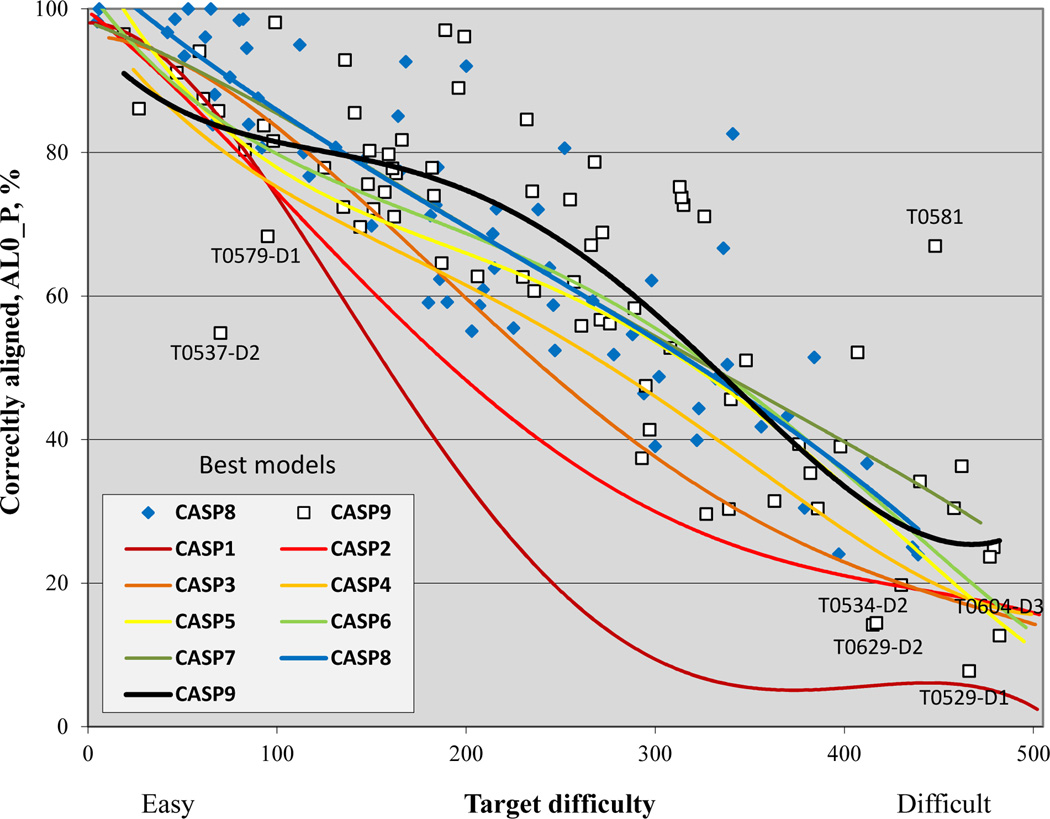
Figure 3 shows the fraction of residues correctly aligned to the best template for each target in CASPs 8 and 9 and trend lines for all CASPs as a function of target difficulty. Trends are similar to those seen in Figure 2 for GDT_TS, and are dominated by the fact that for each target, maximum alignability is determined by the fraction of residues covered by the best template. CASP9 outliers here are similar to the set seen in Figure 2, with the addition of T0579_D1, a target with a relatively low difficulty value. The easy target ranking is a consequence of a 27% sequence identity to the best template and 91% coverage of the template. Nevertheless, alignment was challenging, likely because this is a short (60 residue) target composed of two discontinuous segments. In general, discontinuous domains are more difficult to model. As in the GDT_TS trend lines, the CASP9 alignment trend for difficult targets is lower than that of recent CASPs because of the effect of the four relatively long free modeling targets already apparent in Figure 2.
Figure 3.
% of residues correctly aligned for the best model of each target in all CASPs. Trend lines are similar to those in the equivalent GDT_TS plot (Figure 2), indicating that for many targets, alignment accuracy, together with the fraction of residues that can be aligned to a single template, dominate model quality. In CASP9, the same apparent improvement for mid-range difficulty targets is present.
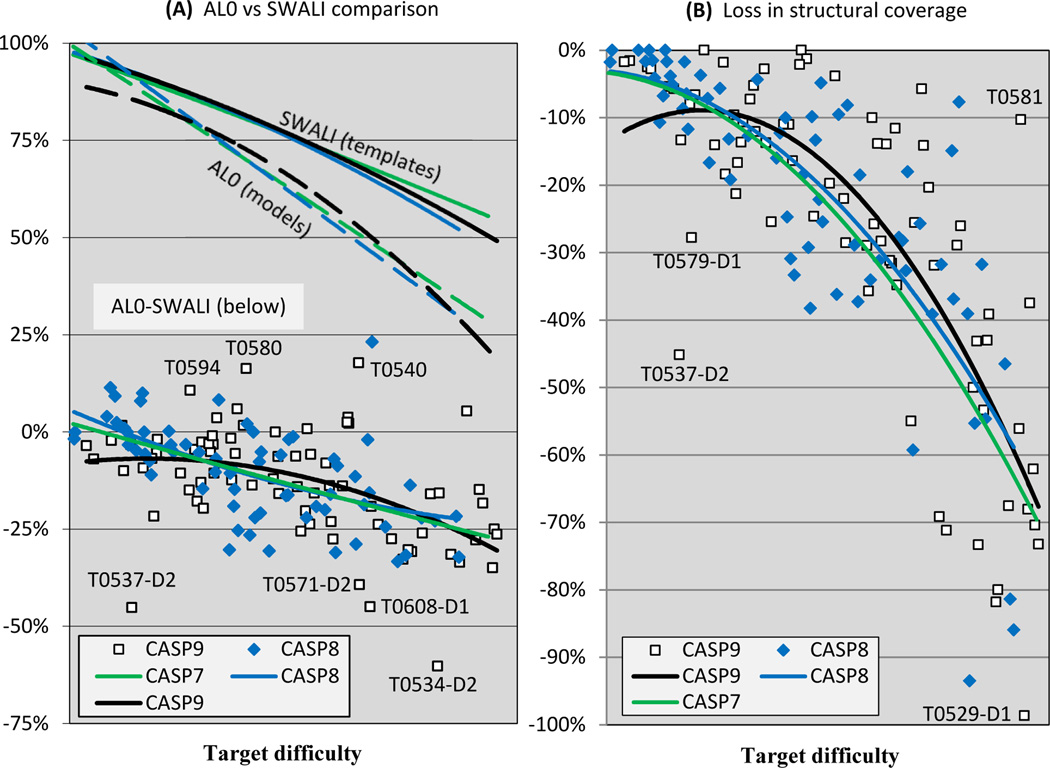
A better sense of alignment performance is provided by examining the relationship of the fraction aligned in the best model to the fraction of residues that could be aligned given knowledge of the best template structure (see Methods). Figure 4A provides two views of these data. The top part of the figure shows the trend lines as a function of target difficulty for the maximum fraction of residues that are aligned using the structure of the best template (‘SWALI’) and the fraction aligned for submitted best models (‘AL0’), for CASPs 7, and 8 and 9. The SWALI alignability curves are similar for the three CASPs, indicating that overall alignment was of approximately the same difficulty in the three experiments. The AL0 performance lines are also similar, suggesting no improvement in overall performance for this interval (although, as Figure 2 shows, there has been enormous improvement over all CASPs). The difference between the SWALI and AL0 curves at any given target difficulty provides a measure of the average fraction of residues that in principle could have been aligned in a model using the best template, but were not. The lower part of the figure shows AL0-SWALI difference curves for CASPs 7, 8 and 9, together with the individual target points for the latest two CASPs. The average fraction of residues not aligned ranges from a few % for easy targets to ~25% at the difficult end of the scale. Points with values greater than 0 indicate targets where any deficiency in alignment was more than offset by modeling residues not covered by the best template. There are three examples where the AL0 score exceeded SWALI by 10% or more, including T0580, a target where the accuracy of the best model is exceptionally high for its difficulty (see Figures 2 and 3). There are also some points with outstandingly poor alignments in CASP9 - as in Figures 2 and 3, T0537_D2, the shortest target, and T0534_D2, a long FM target. There are also two outliers not apparent in the other figures - T0608_D1 and T0571_D2, both free modeling targets.
Figure 4.
Alignment accuracy relative to the maximum that could be obtained using the single best template.
4A: Top: trend lines as a function of target difficulty for the maximum fraction of alignable residues (‘SWALI’) and for the fraction aligned for submitted best models (‘AL0’), for CASPs 7, and 8 and 9. Alignment difficulty is similar in these three CASPs, and alignment accuracy is also similar. Bottom: % difference between aligned residues (AL0) and maximum alignable residues (SWALI). The average fraction of residues not aligned ranges from a few % for easy targets to ~25% at the difficult end of the scale.
4B: % of residues that in principle could be modeled by aligning with the best template, but were not. The higher the line, the smaller the loss in the structural coverage. Trend lines are qualitatively similar for the different CASPs, and alignment accuracy falls off rapidly at the upper end of the difficulty scale.
As noted above, the AL0 alignment measure combines loss of model quality as a result of alignment errors with increase of accuracy as a result of correct positioning of non-template residues. To isolate just the effect of alignment errors, we evaluate the % of residues that in principle could be modeled using the best template, but were not. The potentially alignable residues in a target are defined as those that are within 3.8 Å of any best template Cα atom in the 5Å LGA sequence independent superposition and are in sequential order. Figure 4B shows these data for each domain target in CASPs 8 and 9 together with trend lines for CASPs 7, 8, and 9. Trend lines are qualitatively similar for CASPs 8 and 9, and alignment quality falls off sharply with increasing modeling difficulty. The CASP9 curve is below that of CASP8 in the easiest modeling range partly as a consequence of poor performance on the two outlier targets T0537_D2 and T0579_D1 discussed above. Easy targets on average have about 10% of residues misaligned, increasing to 20% for midrange targets, and up to 65% for the most difficult targets (the majority of which are template free modeling cases, where template-based methods of analysis are less reliable). There is one impressive best model for a CASP9 difficult target T0581 with ~90% of residues correctly aligned. The picture of alignment accuracy here is a little discouraging. The AL0 requirement that no other model residue be closer to a matched target residue tends to reject some mappings unreasonably. On average, this proximity condition reduces calculated alignment accuracy by about 3%, but there are cases (for example T0540 and T0558) where the difference is more than 10%.
Best model quality compared to that of a naïve model
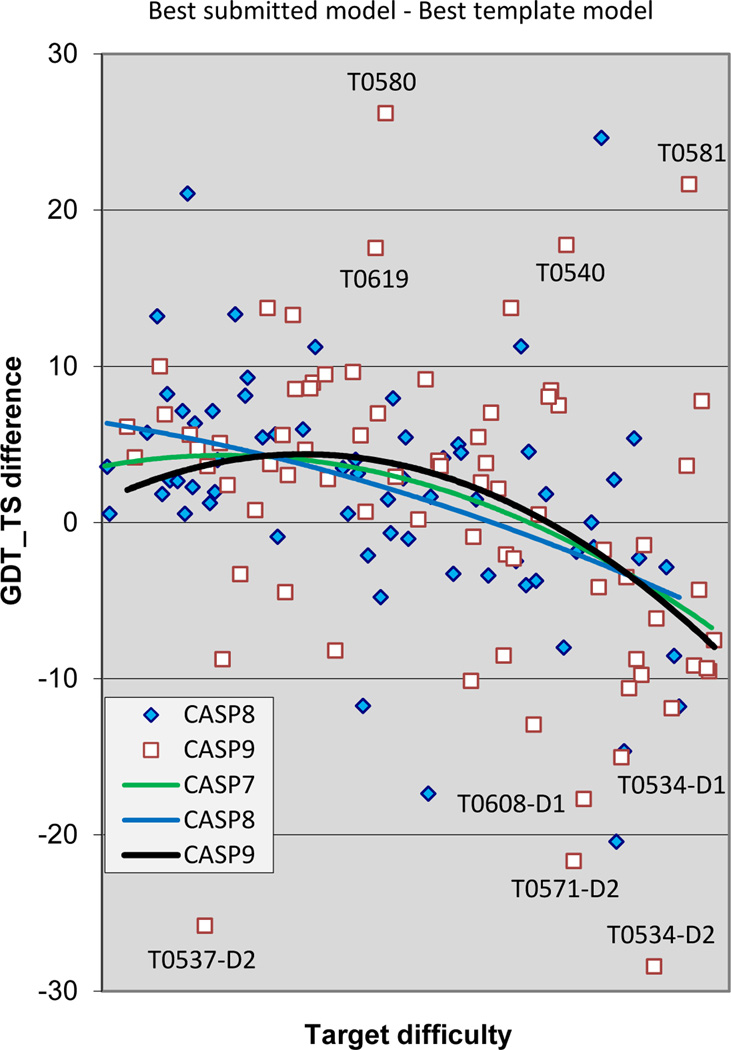
As Figures 3 and 4 demonstrate, although the alignment problem is not completely solved, very substantial progress has been made over the CASP experiments and a reasonably high model quality can often be obtained by successfully copying and aligning residues present in the best available template. In recent CASPs, emphasis has been placed on methods that can improve over such an alignment based model, both by refining an initial template based model13, and by providing accurate structure for regions not covered by the best template. To obtain a base line against which to measure such improvements, we construct a set of ‘naïve’ models, one for each target, maximally utilizing knowledge of the structure of only the best template (see Methods). These models are then evaluated exactly like the submitted ones. Figure 5 shows the difference in GDT_TS scores between these naive models and the best submitted models, with data points for the most recent two CASPs and trend lines for CASPs 7, 8 and 9. By this measure there are several targets in both CASPs 8 and 9 where the best models show outstanding improvements over the corresponding naïve models. For example, T0580 has several small loop regions not included in the best template that are accurately modeled. In both CASPs, there are also a number of best models that fall far short of what a naïve model could achieve. As noted earlier, the ‘easy’ target 537_D2 is an unusually short target. Best models substantially below trend line at the difficult end of the scale are large free modeling targets, where remote templates could not be identified. Overall, trend lines here are similar to those for best model GDT_TS scores (Figure 2), suggesting that the apparent CASP9 improvement in the midrange of target difficulty is due to additional detail in models over that which could be derived directly from the best template. As always, it should be born in mind that we are considering only best submitted models here and no single group consistently provides the best models.
Figure 5.
Difference in GDT_TS score between the best submitted model for each target, and a naïve model based on knowledge of the best single template. Values greater than zero indicate added value in the best model. By this measure, there are 7 cases in CASP9 where model improved more than 10% in GDT_TS over the naïve model, and two cases of improvements of more than 20%.
Gain of model quality due to correct structure for regions not covered by the best template
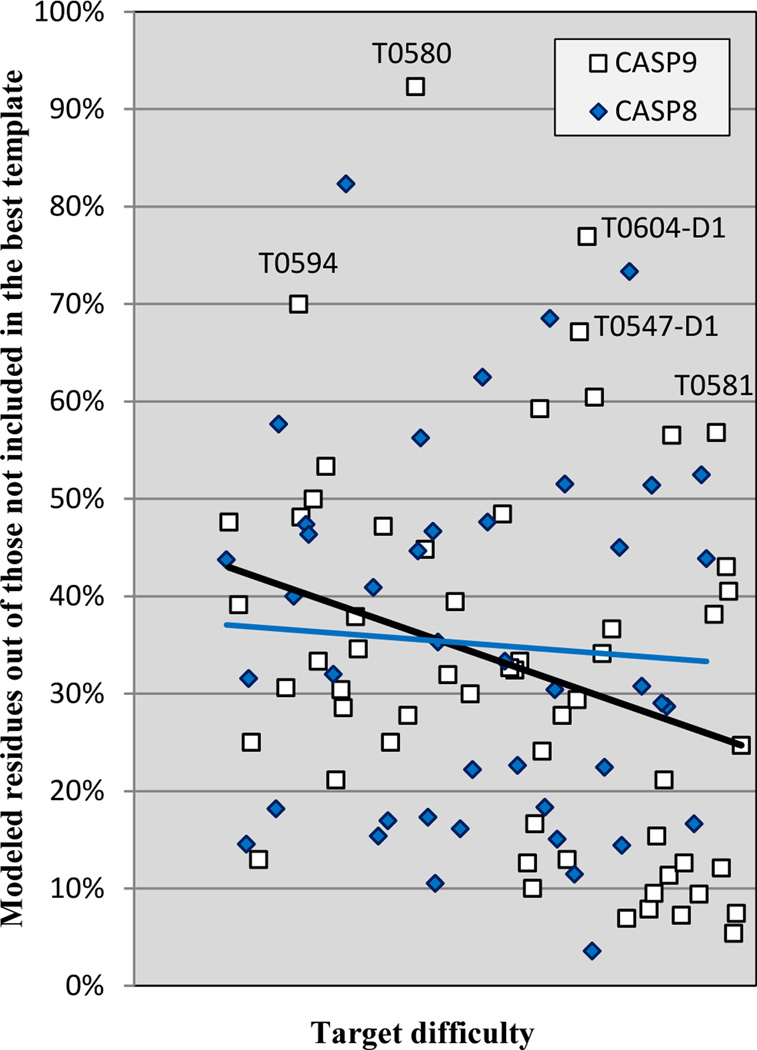
Figure 5 shows the net gain or loss of model quality over that which could be obtained using knowledge of the single best template, combining two primary effects: loss of model quality due to alignment errors, and gain of model quality due to correct positioning of residues not represented in the best template. Figure 6 provides a view of just the gain in model quality from modeling non-template regions. Data are for all CASP 8 and 9 targets in which at least 15 residues could not be aligned to the best template. For each target, the figure shows the % of residues not covered by the primary template that were correctly modeled (the Cα atom of the model is within 3.8Å of the corresponding Cα of the experimental target structure in a 4Å LGA sequence dependent superposition). Trend lines are similar for the two CASPs, and almost flat as a function of target difficulty, reflecting the fact that the fraction of residues added varies widely, and is not correlated with target difficulty. On average, about 35% of residues not covered by the best template are correctly modeled by this criterion, with a number of outstanding successes, such as T0580, for which 24 out of 26 residues missing in the best template were properly modeled (and only one residue out of the 78 present in the template was misplaced). Some of these improvements are due to the successful use of alternative templates (for example T0547_D1), some to successful template free modeling (e.g. T0581), and some perhaps to successful refinement (e.g. T0594).
Figure 6.
Gain in quality of the best model compared to that achievable with the single best template, shown as % of residues not included in the best template. An average about 35% of non-template residues are correctly modeled.
Server Performance
In recent CASP experiments, the gap between the quality of models returned by automatic servers and that of models produced with input from human experts has closed substantially. Moreover, sometimes the remaining difference in human-server performance is likely largely due to the fact that human experts have more time for the modeling, and that very frequently experts base their models on initial structures obtained from servers*. Since servers are the only option for high throughput modeling, their performance is in any case important.
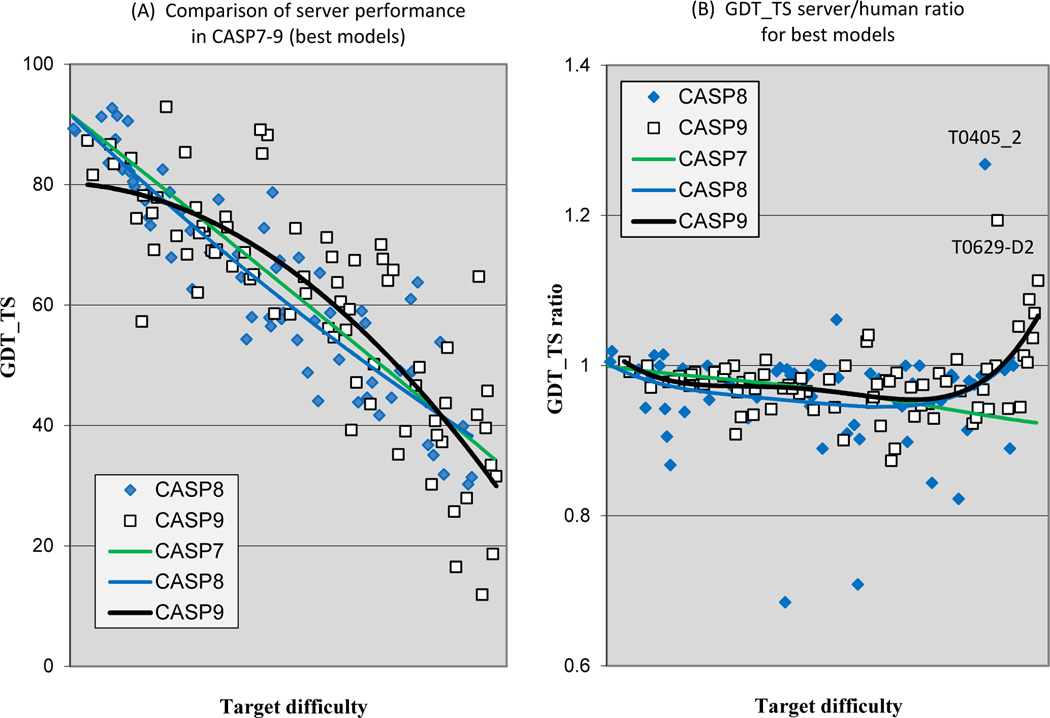
Figure 7A shows the trend lines for GDT_TS server performance as a function of target difficulty for the three most recent CASPs, together with the individual data points for CASPs8 and 9. The curves are very similar to those of Figure 2, which included both human-expert and server methods, and in particular show the same apparent improvement in CASP9 in the midrange of target difficulty. Figure 7B shows the ratio of GDT_TS for the best server model to that of the best human model for CASPs 7, 8 and 9. In CASP9, no server best model was more the 15% less accurate than the corresponding best human model by this criterion. The overall trend lines are similar for the CASPs, except for a striking upturn in the relative quality of server models for the hardest targets in CASP9. Remarkably, the eight hardest targets all have better server models than expert ones. All are relatively long template free targets, so that the absolute quality of the models tends to be low. Nevertheless, three have best server models with GDT_TS scores between 40 and 50, quite respectable for such hard targets. Evidently some server methods are now contributing meaningful features not identified by human experts.
Figure 7.
Comparison of server and human expert performance.
7A: Server performance over CASPs 7–9. The apparent improvement for mid-range difficulty targets in CASP9 is similar to that seen in Figure 2, which includes all methods.
7B: Ratio of the quality of best server models to best human models as a function of target difficulty for CASPs 7–9. The trend lines for the three CASPs are similar, except for the marked upturn in relative server performance for the most difficult CASP9 targets. In all recent CASPs, servers typically produce models only a little less accurate than those from human experts.
Selection of the best model from amongst those generated
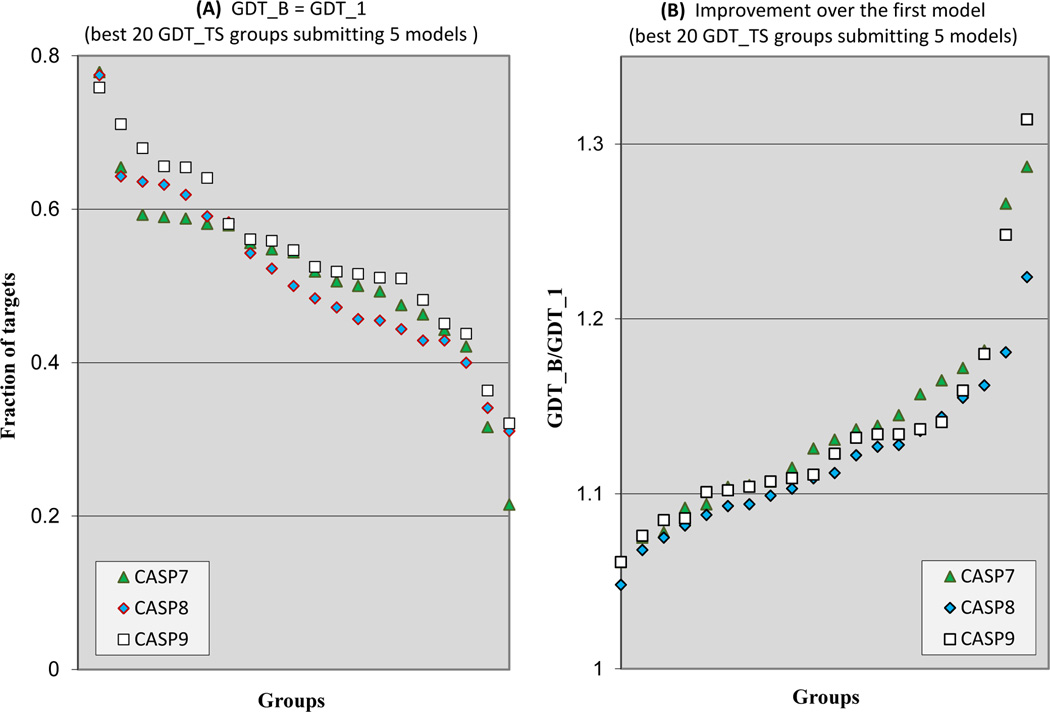
Analysis in this paper focuses mostly on the best model received for each target. However, methods are not always effective at selecting the best model from among a set that have been generated. In CASP, all groups are allowed to submit up to five models on a target, and are requested to label their models in order of their expected accuracy, i.e. the first model should be the most accurate of the five. These data provide a means of testing how well best models can be identified. Here we compare how successfully participants selected their best models in the most recent three CASPs and also evaluate the loss in model quality due to imperfect ability to identify the best model. We include all predictions where a group submitted 5 models on a target, at least four of which are non-identical, and with at least 5 GTD_TS units difference between the best and worst models. For each group that submitted at least 30 domains satisfying these criteria, we calculated the fraction of domain targets where they were able to correctly select a model within 5% of the best one, out of the five submitted. Figure 8A shows the ranking of the best 20 modeling groups (by average GDT_TS-based z-score score) in CASPs 7–9. There is a slight improvement over these three CASPs, and the most successful group is able to pick a model within 5% GDT_TS of their best model for about 75% of targets. On the other hand, this is an exceptional performance, and the least successful of the 20 only identifies such a best model for about 30% of the targets. Nevertheless, the fact that some groups are now able to identify close to best models in a reasonably high fraction of cases is encouraging – it is likely that other groups will learn from that success, and that overall performance will improve next round. Note that these results are significant. As supplementary Figure S4 shows, for all best-performing groups the average spread between best and worst models on a target is much larger then 5% GDT-TS.
Figure 8.
Selection of the most accurate models.
8A: Fraction of targets, where each of the best 20 performing groups (according to the average GDT_TS-based z-score) selected as their best model one within at most 5% in GDT_TS score of the designated first model. Groups are ordered according to their ability to select best models. There is slight improvement from CASP7 to CASP8 and again from CASP8 to CASP 9. The most proficient groups are able to select their best model for more than 75% of targets.
8B: Average GDT_TS ratio between the best submitted model and the model labeled as #1 for all targets in CASPs 7–9, for the best performing 20 groups. Although no group is able to always pick the most accurate model, these data show that for the most proficient groups, the resulting loss of model quality is usually small.
Figure 8B shows the average GDT_TS ratio between the best submitted model and the model labeled as #1 on all targets in the most recent three CASPs, again for the 20 best modeling groups who meet the criteria for inclusion. The closer the ratio to unity, the smaller the loss in modeling quality due to the selection of non-optimal models. By this measure performance is approximately the same in the three CASPs included. For the best performing groups, the loss in model quality in choosing the number one ranked model rather than the best one is only about 5%.
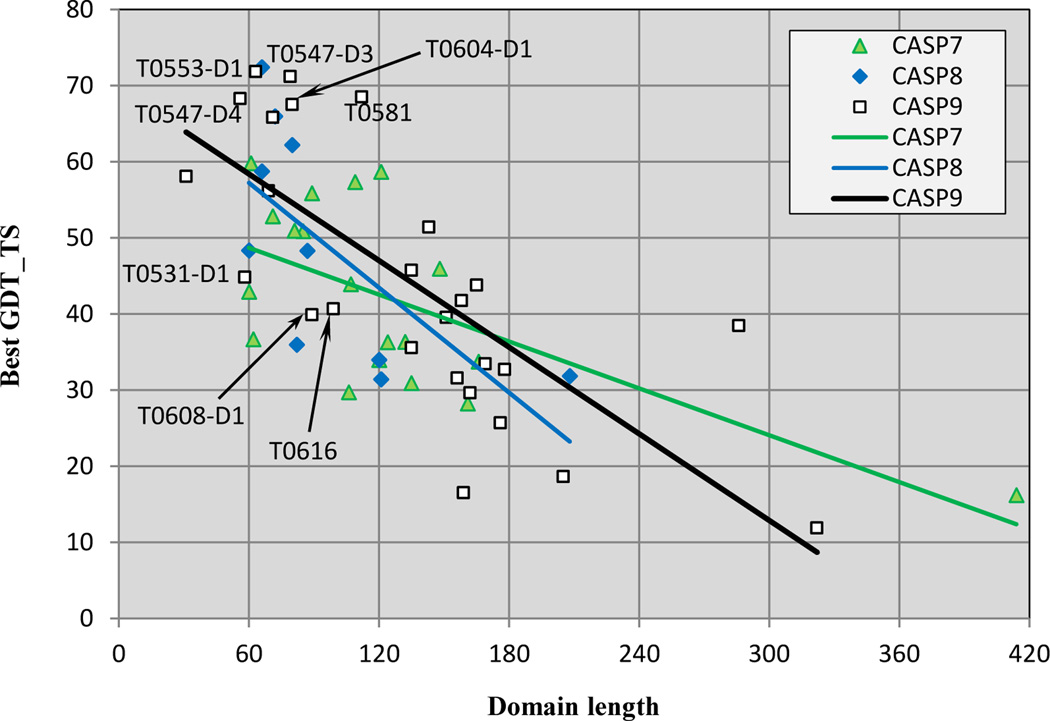
Performance on Free Modeling targets
The small number of targets in the template free modeling category always makes statistically significant comparison of performance across CASP experiments difficult. An additional complication is that the difficulty scale used for template based models (Figure 1) is not appropriate. Further although model quality in this category has improved dramatically over the CASP experiments, very high accuracy models are still the exception. Two recent insights from independent assessors have helped deal with these difficulties. The independent assessment group in CASP88, noting that method performance is very dependent on target length, adopted that as a difficulty scale. We follow that procedure here. Figure 9 shows GDT_TS best model scores for all free modeling targets in the last three CASPs as a function of length, together with linear trend lines. As expected, performance falls off dramatically with length, with almost no good quality models of greater than 120 residues. On the basis of Gaussian kernel analysis of model quality (http://prodata.swmed.edu/CASP9/evaluation/Categories.htm), another CASP assessor has observed that models with a GDT_TS score greater than 50 contain many correct features, and are generally intuitively good models. Considering only models less than 120 residues and taking a GDT_TS score of 50 as a threshold, in CASP9 there are six targets out of a total 11 with scores greater than 60, and eight out of 11 with scores greater than 50. No other CASP comes close to this – in CASP8, there are three targets out of nine with scores of greater than 60, and one other with a score greater than 50. In CASP7, there were no scores higher than 60, and seven of 12 higher than 50. (Note that differences between Figure 9 and the earlier CASP8 assessor’s plot8 are due to their inclusion of all targets rather than just the human/server targets used here, and their use of assessor trimmed domains, as opposed to the full length domains used here). Although these are small numbers, the results do suggest progress in CASP9. However, the successful methods still have a major limitation in terms of target length. It is not always apparent why some short target domains are harder. In CASP9, one of these, T608_D1, is intimately associated with the other domain of the target, and it was difficult for predictors to properly identify the boundary, affecting model quality. Further comments on target difficulty can be found in the assessment paper11.
Figure 9.
Accuracy of the best models for template free targets, as a function of target length, in CASP 7–9. For targets of less than 120 residues, the majority of best models are of reasonable quality, with the best results for CASP9. Methods are not currently effective for bigger targets.
Conclusions
After the dramatic CASP to CASP advances in the early experiments, the more modest pace of recent progress is at first glance rather discouraging. Further, because of limited statistical power in the data, and the imperfect nature of difficulty scales, only quite large increments can be reliably observed. Nevertheless, there are several positive signs in the present analysis.
First, GDT_TS (Figure 2) trend lines suggest some improvement in overall model quality in the mid-range of modeling difficulty. Second, more detailed analysis of improvement of models over that expected from using a best template shows that methods for modeling regions not covered by that template are in fact already quite effective. Third, there is clear progress in identifying the best model out of five submitted, and for the most able groups in this area, the loss of model quality as a consequence of imperfect model selection is now so small as to be inconsequential. Fourth, examining performance in the template free modeling category on the basis of target length does indicate likely progress in the quality of models for short targets. Although the current template free methods are still very ineffective on longer targets, there is an intriguing upturn in server performance compared to that of human experts for some of these.
At the CASP9 conference, there were a number of reports of new methods being aggressively pursued, particularly in the area of more effectively combining information from multiple templates, where the increasing number of experimental structures should provide improvements, and it is expected that these and other innovations will bear fruit in the near future.
METHODS
Target Difficulty
The difficulty of a target is calculated by comparing it with every structure in the appropriate release of the protein databank, using the LGA structure superposition program. For CASP9 templates were taken from the PDB releases accessible before each target deadline. Templates for the previous CASP targets are the same as those used in the earlier analyses. For each target, the most similar structure, as determined by LGA, in the appropriate version of the PDB is chosen as the representative template.
Similarity between a target structure and a potential template is measured as the number of target-template Cα atom pairs that are within 5Å in the LGA sequence independent superposition, irrespective of continuity in the sequence, or sequence relatedness. This value is a little larger than we now consider most appropriate (3.8Å), and there is some times significant superposition score between unrelated structures, particularly for small proteins. The threshold is retained in the interests of consistency. Sequence identity is defined as the fraction of structurally aligned residues that are identical, maintaining sequence order. Note that basing sequence identity on structurally equivalent regions will usually yield a higher value than obtained by sequence comparison alone. In cases where several templates display comparable structural similarity to the target (coverage differing by less than 3%), but one has clearly higher sequence similarity (7% or more) the template with the highest sequence identity was selected. There were a total of 21 such instances in each of the latest two CASPs.
Domains
Some targets may consist of several structural domains. These domains may present modeling problems of different difficulty, and independent assessment treats each identifiable domain as a separate target. As domain definitions are nearly always subjective, we subdivide targets into domains only if these divisions are likely identifiable by a predictor, i.e. in cases where domains in the same target belong to different difficulty categories and therefore require different modeling approaches (T0529, 547, 604, 608, 629), or where the domains are sequentially related to different templates (T0543, 579, 628 – human/server; T0521, 548, 589, 600 – server only). For evaluation of free modeling multi-domain targets (T0534, 537, 550, 553, 571) we treat all domains identified by the assessors as separate targets.
In CASPs 6–9 some single-domain targets were trimmed by the assessors to avoid evaluation of uncertain residues or residues strongly influenced by crystal packing. We base our analysis on the untrimmed targets following the notion that predictors had no means to establish a-priori which residues in the target will be removed by the assessors. This choice affects the results presented here as more than half of single-domain CASP9 targets were trimmed in the assessors’ analysis. We do use official (trimmed) domain definitions for some of the single-domain NMR targets, where the spread of experimental structures in the ensemble is very large (T0531, 564, 590 - human/server; T0539, 552, 555, 557, 560, 572 - server only).
Difficulty Scale
We project the two dimensional target difficulty data in Figure 1 into one dimension, using the following relationship:
Target Relative Difficulty = (RANK_STR_ALN + RANK_SEQ_ID)/2,
where RANK_STR_ALN is the rank of the target along the horizontal axis of Figure 1 (i.e. ranking by % of the template structure aligned to the target), and RANK_SEQ_ID is the rank along the vertical axis (ranking by % sequence identity in the structurally aligned regions). Only human/server targets from CASP8 and CASP9 are used in computation of the Target Relative Difficulty scale as only these targets are subsequently used in our analysis. Numbers in the inset are obtained by a simple averaging of corresponding scores within each CASP dataset.
For defining relative difficulty of the whole set of targets in each CASP (used in Figure S1), we use cumulative z-scores. First, we calculate two separate z-scores from the distributions of (1) coverage and (2) sequence identity of the best template to the corresponding target in all CASPs, then average these two scores and, lastly, multiply the result by (−1) so that the higher resulting score will identify the higher difficulty of targets in a particular CASP:
CASP Relative Difficulty = −(z_STR_ALN + z_SEQ_ID)/2.
GDT_TS
The GDT_TS value of a model is determined as follows. A large sample of possible structure superpositions of the model on the corresponding experimental structure is generated by superposing all sets of three, five and seven consecutive Cα atoms along the backbone (each peptide segment provides one super-position). Each of these initial super-positions is iteratively extended, including all residue pairs under a specified threshold in the next iteration, and continuing until there is no change in included residues. The procedure is carried out using thresholds of 1, 2, 4 and 8Å, and the final super-position that includes the maximum number of residues is selected for each threshold. Super-imposed residues are not required to be continuous in the sequence, nor is there necessarily any relationship between the sets of residues super-imposed at different thresholds. GDT_TS is then obtained by averaging over the four super-position scores for the different thresholds:
GDT_TS = ¼ [N1 + N2 +N4 +N8],
where Nn is the number of residues superimposed under a distance threshold of ‘n’Å. GDT_TS may be thought of as an approximation of the area under the curve of accuracy versus the fraction of the structure included. Different thresholds play different roles in different modeling regimes. For relatively accurate comparative models (in the ‘High Accuracy’ regime), almost all residues will likely fall under the 8Å cutoff, and many will be under 4Å, so that the 1 and 2Å thresholds capture most of the variations in model quality. In the most difficult template free modeling regime, on the other hand, few residues fall under the 1 and 2Å thresholds, and the larger thresholds capture most of the variation between models. For the bulk of the template based models, all four thresholds will often play a significant role.
Alignment quality (AL0)
AL0 score measures alignment accuracy of a model by counting the number of correctly aligned residues in the LGA 5Å superposition of the modeled and experimental structures of a target. A model residue is considered to be correctly aligned if the Cα atom falls within 3.8Å of the corresponding atom in the experimental structure, and there is no other experimental structure Cα atom nearer.
Maximum alignability (SWALI)
Maximum alignability with respect to the best single template is defined as follows: We first find all target Cα atoms that are within 3.8 Å of any template Cα atom in the 5Å LGA sequence independent superposition. Then, we use a dynamic programming procedure that determines the longest alignment between the two structures using these preselected atoms, in such a way that no atom is taken twice and all the atoms in the alignment are in the order of the sequence. The maximum alignability (the Smith-Waterman alignment score, SWALI) is then the fraction of aligned Cα atoms in the target.
Construction of naïve models
We first find an optimal template-target alignment according to the procedure used for calculation of the maximum alignability and then assign the coordinates of the template’s backbone residues to the aligned target residues. It is not always the case that the template with the best structural coverage of the target gets the highest GDT_TS score when superimposed onto the target in this way. To ensure that templates yielding the highest score possible were used, for each target, we built such “template models” based on each of the 25 templates with the best structural coverage, and then picked the template model with the highest GDT_TS score for the subsequent analysis.
Supplementary Material
Acknowledgements
This work was partly supported by NIH grant LM07085 (to KF).
Footnotes
It should be noted that the term ‘expert’ is probably misleading, since the extra time allotted for ‘human-expert’ groups is typically used to run automatic methods for some longer time or construct consensus models from the server models, rather than for a human expert to adjust an initial model.
References
- 1.Schwede T. CASP9 TBM assessment. Proteins. 2011 (Current). [Google Scholar]
- 2.Grishin N. CASP9 FM assessment. Proteins. 2011 doi: 10.1002/prot.23181. (Current). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kryshtafovych A, Fidelis K, Moult J. CASP8 results in context of previous experiments. Proteins. 2009;77(Suppl 9):217–228. doi: 10.1002/prot.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kryshtafovych A, Fidelis K, Moult J. Progress from CASP6 to CASP7. Proteins. 2007;69(Suppl 8):194–207. doi: 10.1002/prot.21769. [DOI] [PubMed] [Google Scholar]
- 5.Kryshtafovych A, Venclovas C, Fidelis K, Moult J. Progress over the first decade of CASP experiments. Proteins. 2005;61(Suppl 7):225–236. doi: 10.1002/prot.20740. [DOI] [PubMed] [Google Scholar]
- 6.Venclovas C, Zemla A, Fidelis K, Moult J. Assessment of progress over the CASP experiments. Proteins. 2003;53(Suppl 6):585–595. doi: 10.1002/prot.10530. [DOI] [PubMed] [Google Scholar]
- 7.Venclovas C, Zemla A, Fidelis K, Moult J. Comparison of performance in successive CASP experiments. Proteins. 2001;(Suppl 5):163–170. doi: 10.1002/prot.10053. [DOI] [PubMed] [Google Scholar]
- 8.Ben-David M, Noivirt O, Paz A, Prilusky J, Sussman J, Levy Y. Assessment of CASP8 structure predictions for template free targets. Proteins. 2009 doi: 10.1002/prot.22591. This issue. [DOI] [PubMed] [Google Scholar]
- 9.Tress ML, Ezkurdia I, Richardson JS. Target domain definition and classification in CASP8. Proteins. 2009;77(Suppl 9):10–17. doi: 10.1002/prot.22497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kryshtafovych A, Fidelis K, Moult J. In: CASP: A driving force in protein structure modeling. Rangwala H, Karypis G, editors. Introduction to protein structure prediction: John Wiley & Sons; 2010. pp. 15–32. [Google Scholar]
- 11.Kinch L, Shi S, Cheng H, Cong Q, Pei J, Schwede T, Grishin N. CASP9 target classification. Proteins. 2011 doi: 10.1002/prot.23190. (Current). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31(13):3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacCallum J. CASP9 refinement paper. Proteins. 2011 doi: 10.1002/prot.23131. (Current). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.