Abstract
Macroautophagy is a physiological cellular response to nutrient stress, which leads to the engulfment of cytosolic contents by a double-walled membrane structure, the phagophore. Phagophores seal to become autophagosomes, which then fuse with lysosomes to deliver their contents for degradation. Macroautophagy is regulated by numerous cellular factors, including the Class III PI3K (phosphoinositide 3-kinase) Vps34 (vacuolar protein sorting 34). The autophagic functions of Vps34 require its recruitment to a complex that includes Vps15, Beclin-1 and Atg14L (autophagy-related 14-like protein) and is known as Vps34 Complex I. We have now identified NRBF2 (nuclear receptor-binding factor 2) as a new member of Vps34 Complex I. NRBF2 binds to complexes that include Vps34, Vps15, Beclin-1 and ATG- 14L, but not the Vps34 Complex II component UVRAG (UV radiation resistance-associated gene). NRBF2 directly interacts with Vps15 via the Vps15 WD40 domain as well as other regions of Vps15. The formation of GFP–LC3 (light chain 3) punctae and PE (phosphatidylethanolamine)-conjugated LC3 (LC3-II) in serum-starved cells was inhibited by NRBF2 knockdown in the absence and presence of lysosomal inhibitors, and p62 levels were increased. Thus NRBF2 plays a critical role in the induction of starvation-induced autophagy as a specific member of Vps34 Complex I.
Keywords: autophagy, human vacuolar protein sorting 34 (hVps34), macroautophagy, mass spectrometry, nuclear receptor-binding factor 2 (NRBF2), vacuolar protein sorting 34 (Vps34), vacuolar protein sorting 15 (Vps15)
INTRODUCTION
Macroautophagy (subsequently referred to as autophagy) is a regulated process in which cells degrade cytoplasmic components during times of nutrient stress [1–3]. During autophagy, bulk cytosol, as well as damaged organelles and some selective targets, are enclosed in a double-membrane structure called the phagophore. The phagophore seals to become an autophagosome and then fuses with the lysosome, delivering its contents for degradation. Autophagy is also activated as part of the innate immune response to pathogens, and is important in the maintenance of neuronal integrity [4–6]. Pathological problems such as autoimmune diseases, developmental disorders, metabolic diseases and cancer have been found to be associated with defective regulation of autophagy [7–12].
The Class III phosphoinositide kinase Vps34 (vacuolar protein sorting 34) is important in both the induction of autophagosomal particles and their eventual fusion with lysosomes [13–15]. In both yeast and higher organisms, Vps34 forms distinct protein complexes that perform distinct functions [1–3]. Vps34 Complex I is an important component regulating autophagy, and includes the putative protein kinase Vps15, the coiled-coil-and BH3-containing protein Beclin-1 as well as the autophagy-specific adapter Atg14L (autophagy-related 14-like protein). Vps34 Complex II includes Vps15, Beclin-1 and UVRAG (UV radiation resistance-associated gene), and is implicated in regulation of endosomal maturation and trafficking. Additional regulatory proteins that may associate with both complexes include BIF1 (Bax-interacting factor 1), Rubicon (RUN domain and cysteine-rich domain containing Beclin-1-interacting protein) and AMBRA1 (autophagy/Beclin-1 regulator 1) [1–3]. In addition, theVps34/Vps15 complexes in early and late endosomes interact with Rab5, Rab7 and the PI3P (phosphatidylinositol 3-phosphate) phosphatases MTM1 (myotubularin 1) and MTMR2 (myotubularin-related protein 2) [16–19]. However it is not clear whether these interactions involve just Vps34/Vps15 or the larger complexes. Vps34/Vps15 are known to have Beclin/Atg6-independent functions during pheromone signalling in yeast [20].
In the present study we have used a proteomic approach to identify novel regulators of Vps34 signalling. We have identified NRBF2 (nuclear receptor-binding factor 2) as a Vps34/Vps15-binding protein. NRBF2 was previously identified as a Vps34-interacting protein in a large proteomics screen [21], but this interaction has not been further investigated. NRBF2 was originally described as a regulator of nuclear receptors such as PPARα (peroxisome-proliferator-activated receptor α), RAR (retinoic acid receptor) and RXRα (retinoid X receptor α) [22, 23]. NRBF2 binds to the AF-2 (activation function-2) domains of the nuclear receptors and decreases, without completely repressing, the activity of PPARα and RXRα. NRBF2 was also found to be a transcriptional activator when tethered to a heterologous DNA-binding domain in both mammalian cells and yeast [22]. Although its previously described functions are nuclear, NRBF2 is also localized to the cytoplasm [22]. In the present study, we show that NRBF2 is a new member of Vps34 Complex I, and regulates the induction of autophagy in response to serum starvation.
MATERIALS AND METHODS
Cell lines and constructs
T-REx™-293 cells (Life Technologies) were transfected with a tetracycline-regulated FLAG–Vps34 construct in the pcDNA4/TO vector (Life Technologies). HEK (human embryonic kidney)-293A cells stably expressing GFP–LC3 (light chain 3) (provided by Dr Sharon Tooze, Cancer Research UK, London, U.K.) were infected with control or NRBF2-shRNA lentivirus and selected with puromycin. Expression constructs for in vitro translation of Vps15, Vps15-ΔWD40, and Vps15-WD40 have been described previously [18]. The expression construct for NRBF2 was from Dr Brian J. Aneskievich (University of Connecticut, Storrs, CT, U.S.A.).
Inhibitors and antibodies
The lysosome inhibitors NH4Cl and leupeptin (Fisher Scientific) were used together at 20 mM and 200 µM respectively for 4 h. The lysosomal inhibitor concanamycin A (Sigma–Aldrich) was used at 1 µM for 30 min. Primary antibodies used for immunoprecipitation and Western blotting were as follows: anti-NRBF2 (Cell Signaling Technology), anti-Vps34 for immunoprecipitation [24], anti-Vps34 for Western blotting [25], anti-Vps15 [18], anti-Beclin-1 (BD Biosciences), anti- Atg14L (MBL International), anti-UVRAG (Cell Signaling Technology), anti-LC3 (Cell Signaling Technology), anti-p62 (MBL International), anti-V5 for immunoprecipitation (Thermo Scientific); anti-V5 for Western blotting (Life Technologies), anti-FLAG (Sigma–Aldrich); anti-actin (Sigma–Aldrich), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase; MBL International) and anti-(rabbit IgG) (Jackson ImmunoResearch). Anti-HA (haemagglutinin) and anti-Myc antibodies were produced in-house.
Sample preparation for silver stain and MS
T-REx™-293-Flag-Vps34 cells were induced with 0.01 µg/ml tetracycline for 12 h. Untransfected T-REx™-293 cells were used as controls. Cells were washed with PBS and lysed in 137 mM NaCl, 20 mM Tris/HCl (pH 7.5), 1 mM MgCl2, 1 mM CaCl2, 10% (v/v) glycerol and 1% (v/v) NP40 (Nonidet P40) with protease and phosphatase inhibitors. Cell lysates were incubated with Sepharose beads at 4 °C for 30 min followed by anti-FLAG M2 affinity gel (Flag-beads) (Sigma–Aldrich) for 2 h. After incubation, Flag-beads were washed with Wash Buffer 400N [50 mM Tris/HCl (pH 8.0), 400 mM NaCl, 0.1 mM EDTA, 10% (v/v) glycerol and 0.5% NP40] with 1 mM DTT and protease and phosphatase inhibitors, and then Wash Buffer 150N [50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 0.1 mM EDTA, 10% (v/v) glycerol and 0.1% NP40] with 1 mM DTT and protease and phosphatase inhibitors. Proteins bound to the Flag-beads were eluted with 0.1 mg/ml FLAG peptide (Sigma–Aldrich). A portion of the eluate was used for silver stain gel examination, and the remainder was used for analysis of protein composition by MS (Bioproximity).
GST or GST–NRBF2-coupled glutathione beads preparation
GST and GST–NRBF2 (human)were expressed in BL21 bacterial cells. Proteins were purified with glutathione beads (Thermo Scientific), analysed by SDS/PAGE and Coomassie Blue staining, and used for pulldown experiments.
Pulldown assays with in vitro-translated Vps15
Vps15, Vps15-ΔWD40 and Vps15-WD40 were synthesized using the TNT Quick Coupled Transcription/Translation Systems (Promega) and Expre35s35s, [35S]-Protein Labeling Mix (PerkinElmer). GST and GST–NRBF2 beads were incubated with the labelled proteins at 4 °C overnight. After four washes in 50 mM Tris/HCl (pH 8.0), 400 mM NaCl and 0.5% NP40 and one wash in 50 mM Tris/HCl (pH 8.0), 150 mM NaCl and 0.1% NP40, proteins bound to the beads were analysed by SDS/PAGE and autoradiography.
Imaging
Acid-washed coverslips were coated with 0.5 mg/ml poly-l-lysine (Sigma–Aldrich) at room temperature for 1 h. Control and NRBF2-knockdown HEK-293A-GFP–LC3 cells were seeded on to coverslips 48 h before imaging. Cells were incubated without or with serum for 16 h, followed by a 30-min incubation in the absence or presence of 1 µM concanamycin A. The cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) and images were obtained using a Nikon Eclipse E400 microscope with ×60 1.4 NA (numerical aperture) objective and a Roper CoolSNAP HQ CCD (charge-coupled device) camera. Punctae were counted manually and the data are pooled from three separate experiments with ~100 cells counted per condition in each experiment. Results are means ± S.D.
Western blot analysis
Western blots were performed according to standard protocols. LC3-II and p62 blots were quantified by scanning densitometry. The data were pooled from three separate experiments, each of which was normalized to the level of LC3-II or p62 seen in control cells after starvation. Results are means ± S.E.M.
Statistics
All experiments were repeated two to four times. For the quantitative Figures, statistical significance was defined using the two-tailed Student’s t test using Vassar Stats (http://vassarstats.net/).
RESULTS
We used an MS-based approach to identify novel members of Vps34 complexes. Tetracycline-regulated expression of FLAG– Vps34, followed by purification using anti-FLAG beads and LC– MS/MS analysis of FLAG peptide eluates, led to the identification of all of the core Vps34 Complex I and Complex II proteins, including Vps15, Beclin-1, Atg14L and UVRAG (Table 1). Surprisingly, we detected more peptides derived from NRBF2 than from any associated protein except for Vps15. NRBF2 has previously been identified as a Vps34-interacting protein in a large-scale proteomic analysis of the autophagic system [21]. Other than its possessing an N-terminal MIT (microtubule interacting and trafficking) domain, which has been identified in trafficking proteins such as Vps4 [26], its known functions are nuclear [22, 23] and unrelated to trafficking or autophagy.
Table 1. Peptides identified by LC–MS/MS analysis of eluate from an anti-FLAG column.
Hits refer to the aggregate number of sequences obtained from the peptides for each sample and reflect protein abundance. Control (non-transfected) against FLAG–Vps34-transfected cells were compared.
| Gene name | Description | Control peptides |
Sample peptides |
Control hits |
Sample hits |
|---|---|---|---|---|---|
| PIK3C3 | Vps34 | 0 | 78 | 0 | 1919 |
| PIK3R4 | Vps15 | 0 | 60 | 0 | 372 |
| NRBF2 | Nuclear receptor-binding factor 2 | 0 | 13 | 0 | 63 |
| BECN1 | Beclin-1 | 0 | 19 | 0 | 57 |
| ATG14L | Atg14L | 0 | 11 | 0 | 41 |
| UVRAG | UV radiation resistance-associated | 0 | 14 | 0 | 38 |
| HSPA9 | Heat-shock 70 kDa protein 9 (mortalin) | 0 | 6 | 0 | 6 |
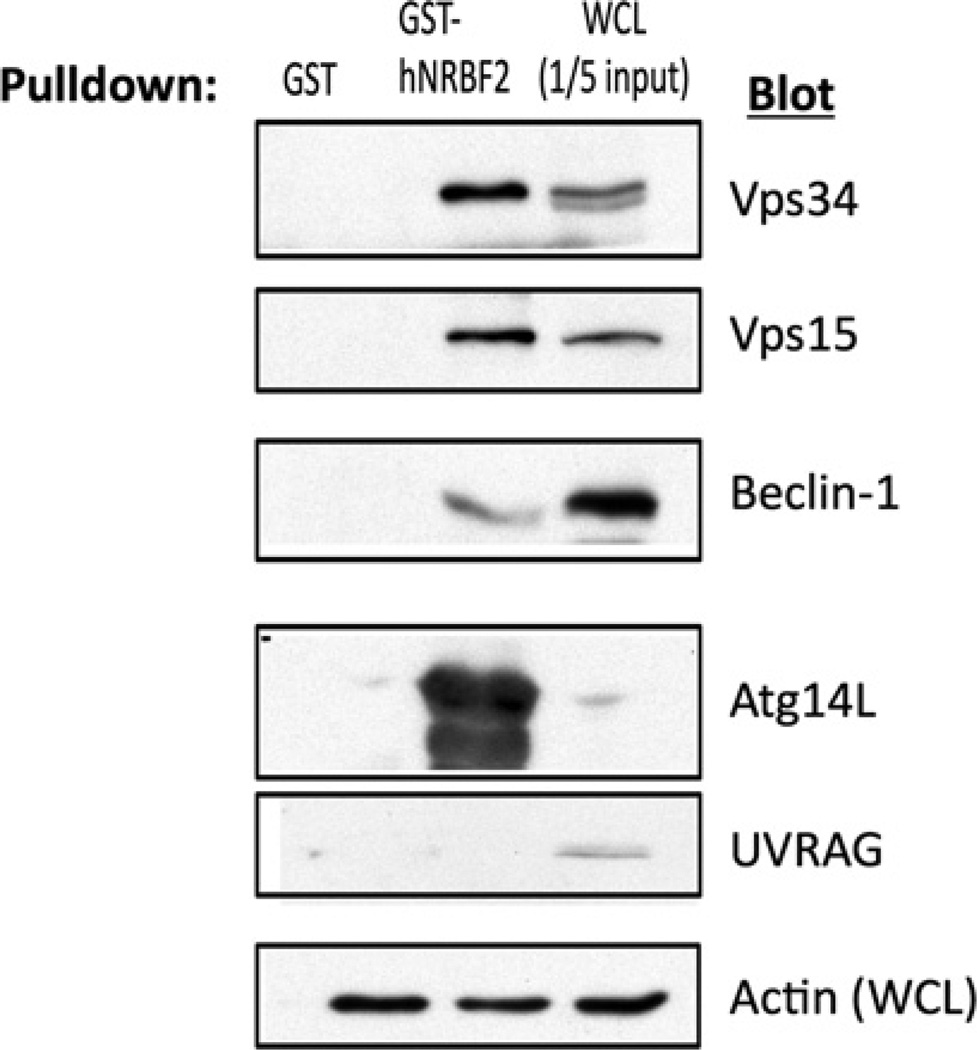
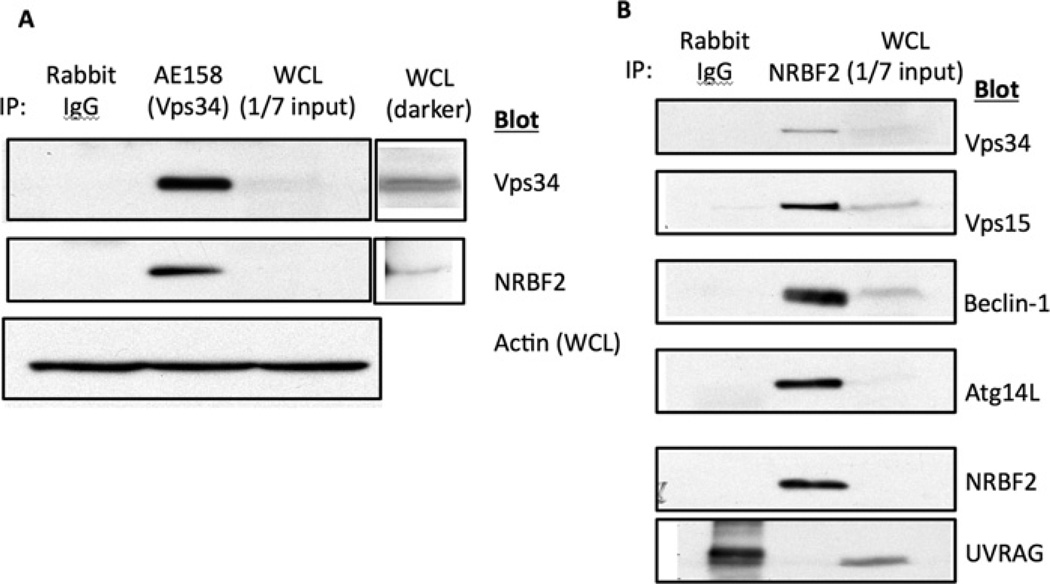
To characterize NRBF2–Vps34 interactions in more detail, we performed a pulldown assay with recombinant GST–NRBF2 fusion proteins. We were able to detect Vps34, Vps15, Beclin-1 and Atg14L, but not the Complex II protein UVRAG in GST–NRBF2 pulldowns (Figure 1). To determine whether the interaction occurred with endogenous proteins, we blotted anti-Vps34 immunoprecipitates for NRBF2 and vice versa. NRBF2 was easily detectable in the Vps34 immunoprecipitates (Figure 2A), and all the members of Vps34 Complex I, but not the Complex II member UVRAG, were detected in anti-NRBF2 immunoprecipitates (Figure 2B).
Figure 1. Recombinant NRBF2 binds Vps34 Complex I.
GST or GST–NRBF2 (10 µg), bound to glutathione–Sepharose beads, was incubated with lysates from HEK-293T cells for 6 h. The beads were washed and analysed by SDS/PAGE and Western blotting for Vps34, Vps15, Beclin-1, Atg14L and UVRAG. WCL, whole-cell lysate.
Figure 2. Co-immunoprecipitation of endogenous NRBF2 and Vps34.
(A) HEK-293T cells were lysed and control IgG or anti-Vps34 immunoprecipitates were blotted with anti-NRBF2 or anti-Vps34 antibody. (B) Anti-NRBF2 immunoprecipitates were blotted for Vps34, Vps15, Beclin-1, Atg14L, NRBF2 and UVRAG. IP, immunoprecipitation; WCL, whole-cell lysate.
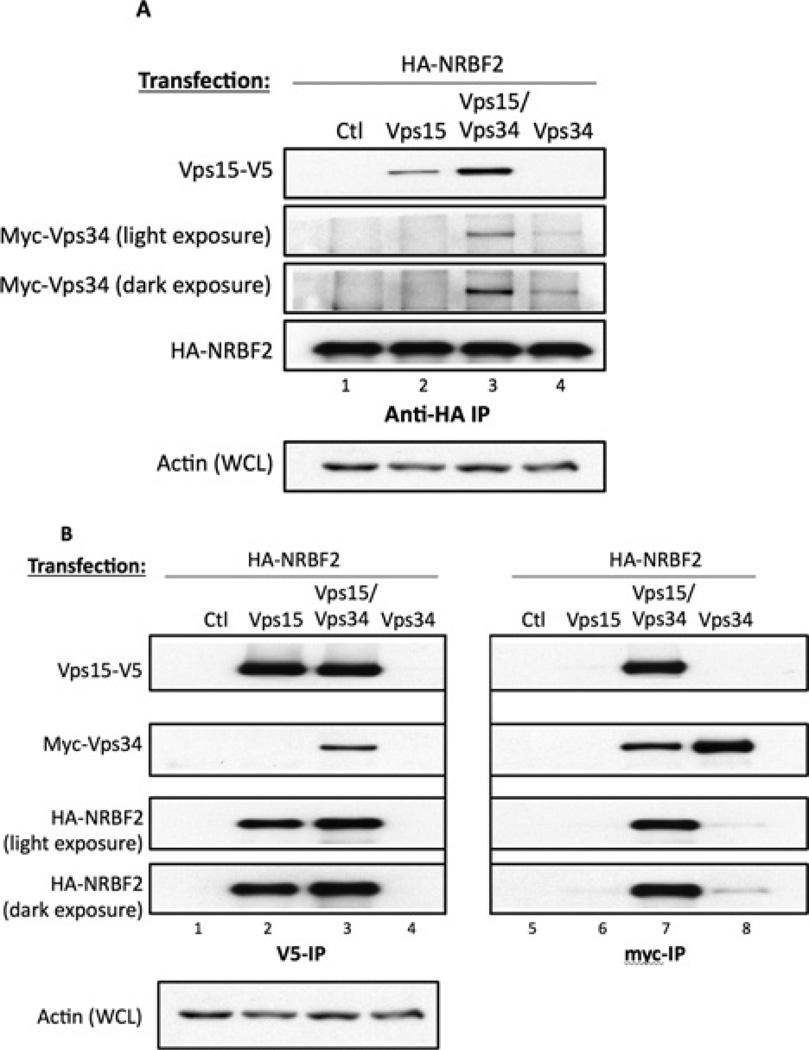
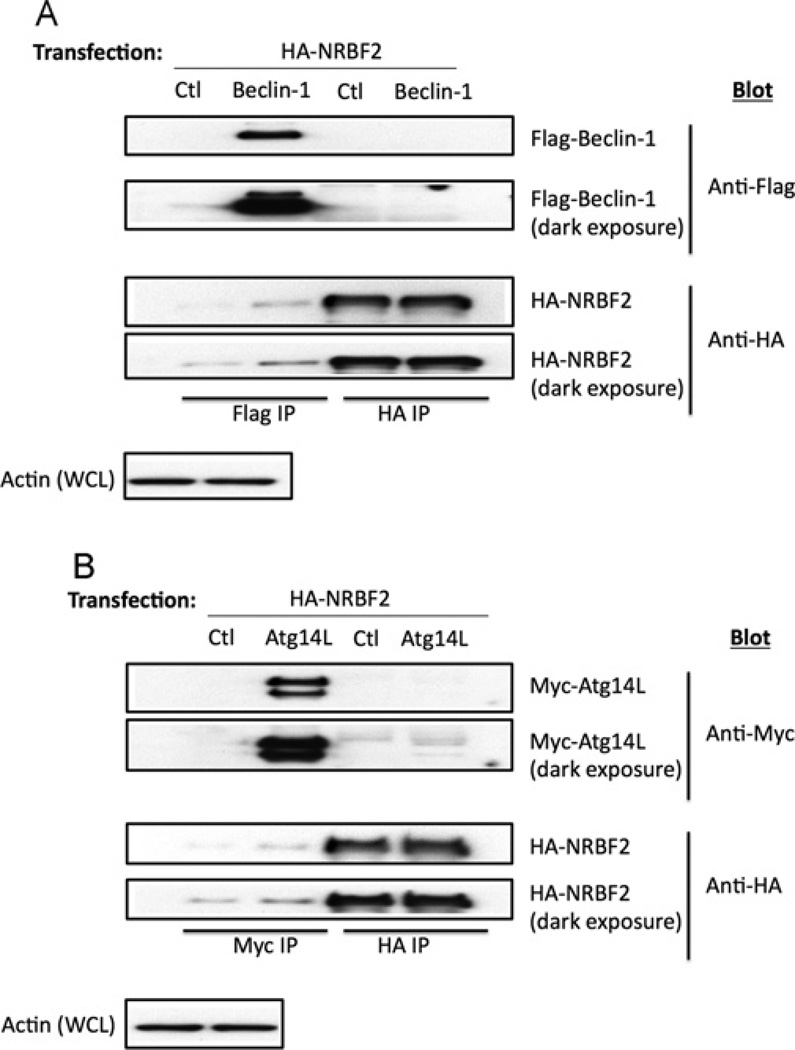
To test whether Vps34 and NRBF2 interacted directly, we expressed Myc-tagged Vps34 and HA-tagged NRBF2 in HEK-293T cells. Surprisingly, in the anti-HA immunoprecipitate we could only weakly detect Myc–Vps34 (Figure 3A, lane 4). However, if we included V5-tagged Vps15 in the transfection, Vps15 and Vps34 were then robustly detected in the anti-HA– NRBF2 immunoprecipitate (Figure 3A, lane 3). Vps15 also coimmunoprecipitated with HA–NRBF2 in cells transfected with these proteins alone (Figure 3A, lane 2). These data suggest that the primary interaction may involve Vps15 and NRBF2, and not Vps34. To test this hypothesis, we expressed HA–NRBF2 and Vps15–V5, alone or with Myc–Vps34, using a bicistronic expression system. NRBF2 could be detected in the anti-Vps15–V5 immunoprecipitates in the absence or presence of Vps34 (Figure 3B, lanes 2 and 3), whereas NRBF2 was only detected in the anti-Myc–Vps34 immunoprecipitates when Vps15 was present (Figure 3B, lane 7 compared with lane 8). We also tested whether NRBF2 bound directly to other members of Vps34 Complex I. Co-expression experiments with HA–NRBF2 and FLAG–Beclin-1 or Myc–Atg14L failed to detect interactions between the pairs of proteins (Figure 4). Taken together, these data suggest that NRBF2 interacts primarily with Vps15.
Figure 3. NRBF2 binds directly to Vps15 but not Vps34.
(A) HEK-293T cells were transfected with HA–NRBF2 and Vps15–V5, Myc–Vps34, or both Vps15–V5 and Myc–Vps34. Anti-HA immunoprecipitates were blotted with anti-Myc, anti-V5 or anti-HA antibodies. (B) HEK-293T cells were transfected as above and immunoprecipitated (IP) with anti-V5 or anti-Myc/Ctl antibodies. The immunoprecipitates were blotted as above. Ctl, control; WCL, whole-cell lysate.
Figure 4. NRBF2 does not directly bind to Beclin-1 or Atg14L.
(A) HEK-293T cells were transfected with HA–NRBF2 and FLAG–Beclin-1. Anti-FLAG and anti-HA immunoprecipitates were blotted with anti-FLAG and anti-HA antibodies. (B) HEK-293T cells were transfected with HA–NRBF2 and Myc–Atg14L. Anti-Myc and anti-HA immunoprecipitates were blotted with anti-Myc and anti-HA antibodies. Ctl, control; IP, immunoprecipitation; WCL, whole-cell lysate.
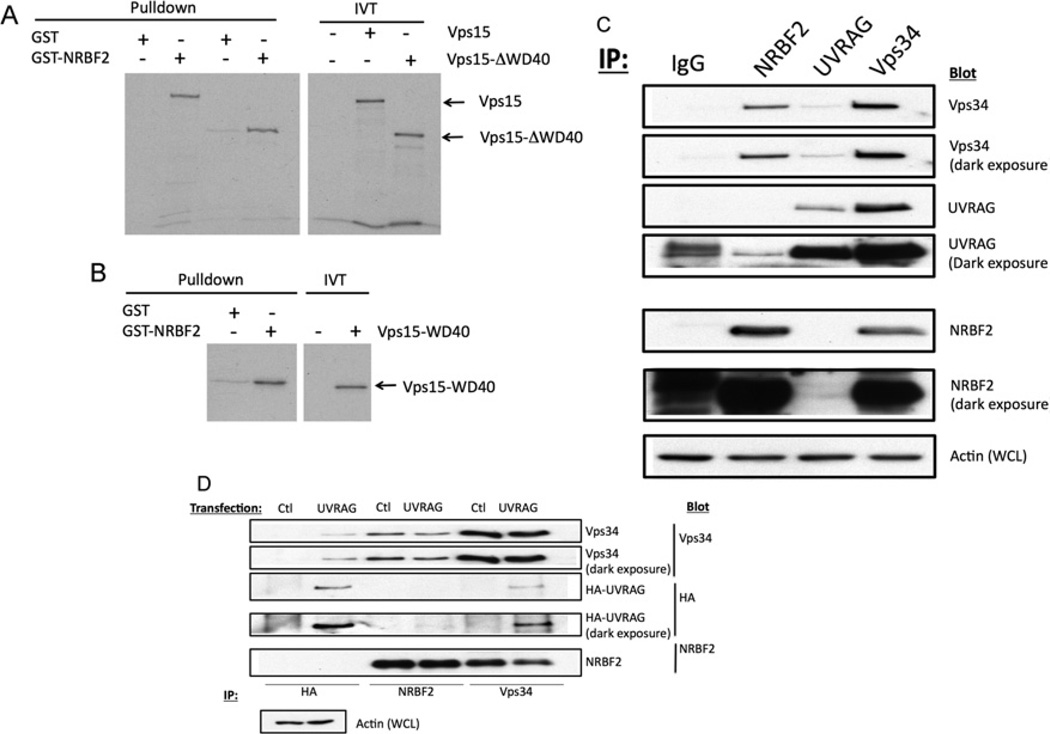
To confirm that NRBF2 interacts directly with Vps15, we incubated in vitro-translated Vps15 with GST or GST–NRBF2. Specific binding of 35S-labelled Vps15 to GST–NRBF2 was readily detected (Figure 5A). Previous studies have shown that the N-terminus of Vps15 binds to Vps34, whereas the C-terminal WD40 repeats bind to upstream regulators such as Rab5 [18, 27]. In fact, the WD40 domain of Vps15 was sufficient to bind to NRBF2 (Figure 5B). However, a truncated Vps15 lacking the WD40 domain still bound NRBF2 (Figure 5A), suggesting that other regions of Vps15 are also involved.
Figure 5. NRBF2 binds directly to Vps15, but does not interact with Vps34 Complex II.
(A)GST or GST–NRBF2 were incubated with 35S-labelled in vitro-translated Vps15 or Vps15-ΔWD40. The beads were washed and analysed by SDS/PAGE and autoradiography. (B) GST or GST–NRBF2 was incubated with in vitro-translated Vps15 WD40 domain, and analysed as above. (C) HEK-293T cells were immunoprecipitated with anti-NRBF2, anti-UVRAG and anti-Vps34 antibodies. The immunoprecipitates were blotted for endogenous Vps34, UVRAG and NRBF2. (D) HEK-293T cells were transfected with HA–UVRAG. Anti-NRBF2, anti-HA and anti-Vps34 immunoprecipitates were blotted with anti-HA, anti-Vps34 and anti-NRBF2 antibodies. Ctl, control; IP, immunoprecipitation; WCL, whole-cell lysate.
The absence of UVRAG in the anti-NRBF2 immunoprecipitates suggested that this protein specifically interacts with Vps15 in Complex I. To examine this in more detail, we blotted anti-NRBF2 immunoprecipitates with anti-UVRAG and vice versa. Although Vps34 was detected in all the immunoprecipitates, we could not detect co-immunoprecipitation of endogenous NRBF2 and UVRAG (Figure 5C); a small amount of UVRAG was seen in the non-specific IgG immunoprecipitate. To increase the sensitivity of the assay, we overexpressed HA–UVRAG, and blotted anti-HA immunoprecipitates for endogenous NRBF2 and anti-NRBF2 immunoprecipitates for HA–UVRAG. Once again, we were unable to detect any interactions (Figure 5D). These data suggest that NRBF2 specifically interacts with Vps15 in the context of Vps34 Complex I, but not Complex II.
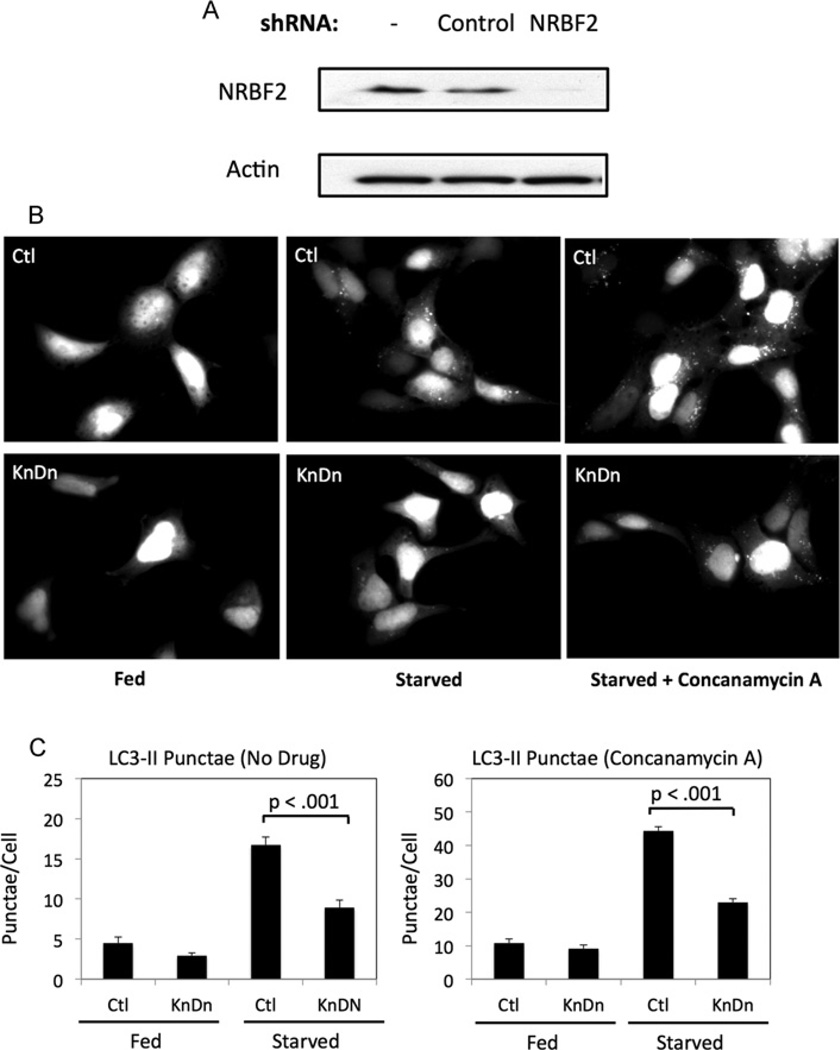
To test whether NRBF2 regulates autophagy, we knocked down NRBF2 in HEK-293A cells stably expressing GFP–LC3 (Figure 6A). We then stimulated autophagy by removing serum for 16 h. Although the accumulation of GFP punctae was clearly visible in the control cells (Figure 6B, upper panels), it was suppressed in NRBF2-knockdown cells (Figure 6B, lower panels). To more quantitatively assess the effects of NRBF2 knockdown on autophagy, we measured the number of GFP–LC3 punctae in both fed and starved cells in the absence or presence of the lysosomal inhibitor concanamycin A. This experiment distinguishes a reduction in punctae due to a reduced rate of formation as opposed to an increased rate of clearance. We found that the number of punctae in the serum-starved NRBF2-knockdown cells was significantly lower than in the control cells (Figure 6C). Treatment with concanamycin A increased the number of punctae in both wild-type and knockdown cells, but the number of punctae in the knockdown cells was still lower than in the controls (Figure 6C).
Figure 6. NRBF2 is required for autophagy in response to serum starvation.
(A) Anti-NRBF2 blots from stable GFP–LC3 HEK-293A cells infected with control or NRBF2-shRNA lentivirus. (B) Control and NRBF2-knockdown GFP–LC3 HEK-293A cells were incubated for 16 h in the presence or absence of 10% FBS, and then incubated for an additional 30 min in the absence or presence of 1 µM concanamycin A. Cells were fixed and imaged. (C) The number of GFP–LC3 punctae per cell was counted. Results are means ± S.D. from three experiments with approximately 100 cells/condition in each experiment. Ctl, control; KnDn, knockdown.
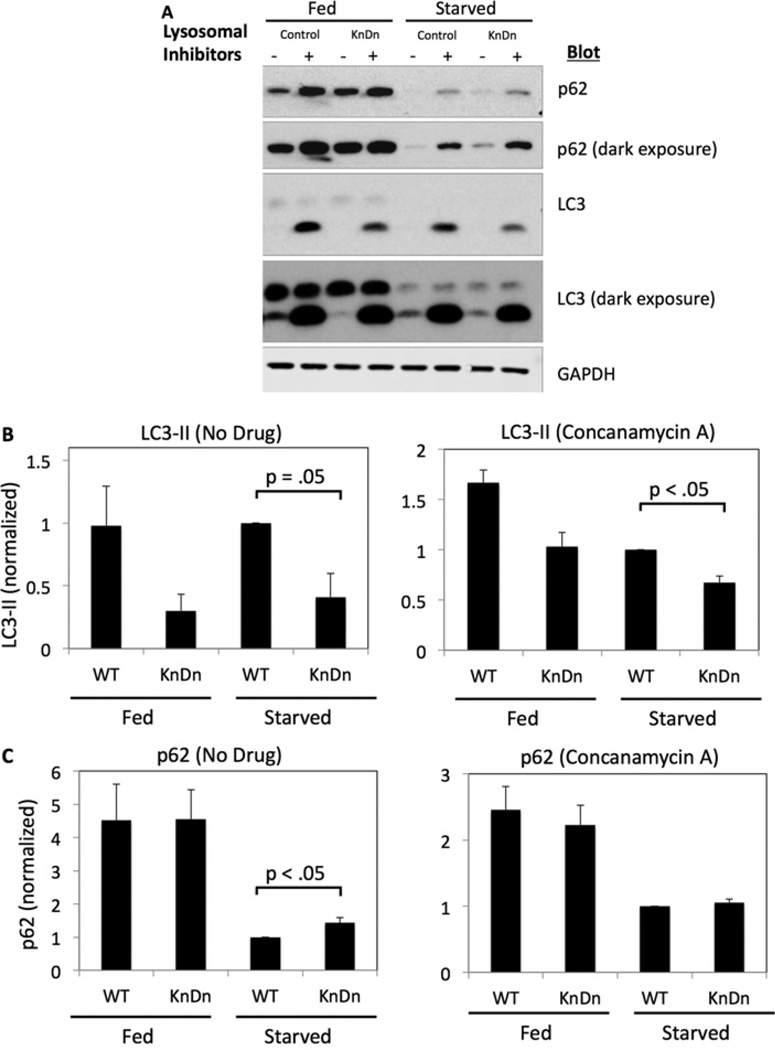
We also measured the starvation-induced production of LC3-II by Western blotting. The levels of LC3-II were reduced in the NRBF2-knockdown cells as compared with the control cells after overnight starvation (Figures 7A and 7B). Although treatment with lysosomal inhibitors caused an increase in LC3-II in both control and knockdown cells, the levels in knockdown cells were still reduced as compared with the control (Figures 7A and 7B). Finally, we measured the levels of p62, which is degraded through macroautophagy. p62 levels were significantly increased in the NRBF2-knockdown cells as compared with the controls (Figures 7A and 7C). Both the control and NRBF2-knockdown cells displayed an increase in p62 to similar levels following treatment with lysosomotropic agents. This is the expected result, since blockade of lysosomal degradation should eliminate differences in p62 levels due to lysosomal delivery. Taken together, our data show that NRBF2 is required for autophagosome formation.
Figure 7. NRBF2 knockdown inhibits autophagy induction.
(A) Control or NRBF2-knockdown GFP–LC3 cells were incubated for 16 h in the absence or presence of 10%FBS. For the last 4 h, cells were incubated without or with the lysosomal inhibitors NH4Cl (20 mM) and leupeptin (200 µM). The cells were lysed and blotted for p62 and LC3. (B) LC3-II levels in each condition were quantified. The data are normalized for the levels of LC3-II in control serum-starved cells, and are means ± S.E.M. from three separate experiments. (C) p62 levels in each condition were quantified as above. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KnDn, knockdown; WT, wild-type.
DISCUSSION
We have identified NRBF2 as a component of Vps34 Complex I and a regulator of starvation-induced autophagy. Our co-immunoprecipitation experiments suggest that NRBF2 interacts directly with Vps15; interactions with Vps34, Beclin-1 and Atg14L were weak and NRBF2–Vps15 binding was reconstituted with in vitro-translated material. Given its binding to Vps15, which is present in both Vps34 Complex I and Complex II, it is not clear why NRBF2 is excluded from complexes containing UVRAG. One potential explanation is based on the observation that UVRAG plays important roles in the Rab5 and Rab7 endocytic compartments [28, 29]. Vps15 is targeted to endosomes by the binding of its WD40 domain to Rab5 and Rab7 [18, 30]. Given that NRBF2 also binds to the Vps15 WD40 domain, it might decrease the fraction of Vps34/Vps15 available for interactions with UVRAG in Rab5/Rab7 endosomes, therefore increasing the fraction available for binding to autophagosomal structures as components of Complex I. The fact that Vps15 binding to NRBF2 is multivalent, involving both the WD40 domain and other regions of Vps15,would presumably increase its binding affinity to NRBF2 as compared with Rab5/Rab7. Future studies defining the other sites of interaction between Vps15 and NRBF2 and measuring the relative affinity of Vps15–Rab5 compared with Vps15–NRBF2 binding will test this model.
Our in vivo data suggest that NRBF2 is a positive regulator of autophagy, as autophagosome formation is depressed in NRBF2-knockdown cells. The mechanistic role of NRBF2 is not yet clear. Preliminary data do not support a direct effect on Vps34 lipid kinase activity. We have previously reported that the activity of Vps34, both total and Beclin-1-associated, is decreased by nutrient starvation [31]. Recent studies have suggested that Atg14L-associated Vps34 is regulated differently from the rest of the Vps34 pool, in that its activity is increased during nutrient starvation [32–34]. This Atg14L-specific regulation occurs via stimulatory inputs from the AMPK (AMP-activated protein kinase) and ULK1 (unc-51-like autophagy-activating kinase 1) kinases, and a loss of inhibitory inputs from the mTORC1 (mammalian target of rapamycin complex 1) kinase. We have been unable to observe increases in Atg14L-associated Vps34 under the starvation conditions used in the present study, and NRBF2 knockdown does not appear to affect Atg14L-associated Vps34 lipid kinase activity (results not shown). Furthermore, we see minimal changes in the levels of Vps34, Vps15 and Beclin-1 in anti-Atg14L immunoprecipitates from NRBF2-knockdown cells, suggesting that the formation of Complex I is not affected (results not shown). Although we cannot rule out the presence of low levels of residual NRBF2 in the knockdown cells, our data suggest that NRBF2 does not act like the MIT-domain-containing yeast protein, which enhances Complex I formation [35]. We do not yet understand the mechanism of NRBF2 action, but we suspect that it occurs at the levels of Vps34 targeting. This would be consistent with NRBF2 binding to the Vps15 WD40 domain, which is known to target Vps34 to endosomes [18]. This will be an important area for future studies on this new member of Vps34 Complex I.
ACKNOWLEDGEMENTS
We thank Dr Sharon Tooze for the GFP–LC3 HEK-293A cells, Dr Brian Aneskievich for the HA–NRBF2 construct and Dr Peng Guo for expert assistance in confocal imaging and data analysis and the Analytical Imaging Facility (AIF) at Albert Einstein College of Medicine supported by an NCI (National Cancer Institute) cancer center support grant [grant number P30CA013330].
FUNDING
The work was supported by the NIH (National Institutes of Health) [grant numbers AG039632 (to J.M.B) and AR064420 (to J.M.B. and J.E.P)].
Abbreviations
- Atg14L
autophagy-related 14-like protein
- HA
haemagglutinin
- HEK
human embryonic kidney
- LC3
light chain 3
- NP40
Nonidet P40
- NRBF2
nuclear receptor-binding factor 2
- PPARα
peroxisome-proliferator-activated receptor α
- RXRα
retinoid X receptor α
- UVRAG
UV radiation resistance-associated gene
- Vps
vacuolar protein sorting
Footnotes
AUTHOR CONTRIBUTION
Yanyan Cao planned and performed all experiments, with the exception of the LC3-II analysis, and contributed to data analysis and writing the paper; Yichen Wang performed the LC3-II and p62 measurements; Widian Abi Saab contributed to Western blot analyses; Fajun Yang contributed to planning the proteomic experiments; Jeffrey Pessin contributed to planning the autophagy experiments; and Jonathan Backer contributed to experimental planning, data analysis and writing the paper.
REFERENCES
- 1.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid. Redox Signal. 2013;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Codogno P. The mechanism and physiological function of macroautophagy. J. Innate Immun. 2013;5:427–433. doi: 10.1159/000351979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventruti A, Cuervo AM. Autophagy and neurodegeneration. Curr. Neurol. Neurosci. Rep. 2007;7:443–451. doi: 10.1007/s11910-007-0068-5. [DOI] [PubMed] [Google Scholar]
- 7.Gianchecchi E, Delfino DV, Fierabracci A. Recent insights on the putative role of autophagy in autoimmune diseases. Autoimmun. Rev. 2013;13:231–241. doi: 10.1016/j.autrev.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Lee KM, Hwang SK, Lee JA. Neuronal autophagy and neurodevelopmental disorders. Exp. Neurobiol. 2013;22:133–142. doi: 10.5607/en.2013.22.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satriano J, Sharma K. Autophagy and metabolic changes in obesity-related chronic kidney disease. Nephrol. Dial. Transplant. 2013;28:iv29–iv36. doi: 10.1093/ndt/gft229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierdominici M, Barbati C, Vomero M, Locatelli SL, Carlo-Stella C, Ortona E, Malorni W. Autophagy as a pathogenic mechanism and drug target in lymphoproliferative disorders. FASEB J. 2013;28:524–535. doi: 10.1096/fj.13-235655. [DOI] [PubMed] [Google Scholar]
- 11.Gukovsky I, Li N, Todoric J, Gukovskaya A, Karin M. Inflammation, autophagy, and obesity: common features in the pathogenesis of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1199–1209. doi: 10.1053/j.gastro.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leone RD, Amaravadi RK. Autophagy: a targetable linchpin of cancer cell metabolism. Trends Endocrinol. Metab. 2013;24:209–217. doi: 10.1016/j.tem.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wirth M, Joachim J, Tooze SA. Autophagosome formation: the role of ULK1 and Beclin1–PI3KC3 complexes in setting the stage. Semin. Cancer Biol. 2013;23:301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 15.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao C, Backer JM, Laporte J, Bedrick EJ, Wandinger-Ness A. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol. Biol. Cell. 2008;19:3334–3346. doi: 10.1091/mbc.E08-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao C, Laporte J, Backer JM, Wandinger-Ness A, Stein MP. Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic. 2007;8:1052–1067. doi: 10.1111/j.1600-0854.2007.00586.x. [DOI] [PubMed] [Google Scholar]
- 18.Murray JT, Panaretou C, Stenmark H, Miaczynska M, Backer JM. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic. 2002;3:416–427. doi: 10.1034/j.1600-0854.2002.30605.x. [DOI] [PubMed] [Google Scholar]
- 19.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip S-C, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 20.Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein α subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 21.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasumo H, Masuda N, Furusawa T, Tsukamoto T, Sadano H, Osumi T. Nuclear receptor binding factor-2 (NRBF-2), a possible gene activator protein interacting with nuclear hormone receptors. Biochim. Biophys. Acta. 2000;1490:189–197. doi: 10.1016/s0167-4781(99)00244-4. [DOI] [PubMed] [Google Scholar]
- 23.Flores AM, Li L, Aneskievich BJ. Isolation and functional analysis of a keratinocyte-derived, ligand-regulated nuclear receptor comodulator. JInvest. Dermatol. 2004;123:1092–1101. doi: 10.1111/j.0022-202X.2004.23424.x. [DOI] [PubMed] [Google Scholar]
- 24.Siddhanta U, McIlroy J, Shah A, Zhang YT, Backer JM. Distinct roles for the p110a and hVPS34 phosphatidylinositol 3_-kinases in vesicular trafficking, regulation of the actin cytoskeleton, and mitogenesis. J. Cell Biol. 1998;143:1647–1659. doi: 10.1083/jcb.143.6.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan Y, Flinn RJ, Wu H, Schnur RS, Backer JM. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem. J. 2009;417:747–755. doi: 10.1042/BJ20081865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwaya N, Takasu H, Goda N, Shirakawa M, Tanaka T, Hamada D, Hiroaki H. MIT domain of Vps4 is a Ca2+-dependent phosphoinositide-binding domain. J. Biochem. 2013;153:473–481. doi: 10.1093/jb/mvt012. [DOI] [PubMed] [Google Scholar]
- 27.Budovskaya YV, Hama H, DeWald D, Herman PK. The C-terminus of the Vps34p PI 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J. Biol. Chem. 2002;277:287–294. doi: 10.1074/jbc.M109263200. [DOI] [PubMed] [Google Scholar]
- 28.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang C, Sir D, Lee S, Ou JH, Jung JU. Beyond autophagy: the role of UVRAG in membrane trafficking. Autophagy. 2008;4:817–820. doi: 10.4161/auto.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein MP, Feng Y, Cooper KL, Welford AM, Wandinger-Ness A. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 2003;4:754–771. doi: 10.1034/j.1600-0854.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- 31.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 32.Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9 doi: 10.4161/auto.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J. Cell Biol. 2013;203:299–313. doi: 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]