SUMMARY
Pseudouridine is the most abundant RNA modification, yet except for a few well-studied cases, little is known about the modified positions and their function(s). Here, we develop Ψ-seq for transcriptome-wide quantitative mapping of pseudouridine. We validate Ψ-seq with spike-ins and de novo identification of previously reported positions and discover hundreds of novel sites in human and yeast mRNAs and snoRNAs. Perturbing pseudouridine synthases (PUSs) uncovers which PUSs modify each site and their target sequence features. mRNA pseudouridinylation depends on both site-specific and snoRNA-guided PUSs. Upon heat shock in yeast, Pus7-mediated pseudouridylation is induced at >200 sites and Pus7 deletion decreases the levels of otherwise pseudouridylated mRNA, suggesting a role in enhancing transcript stability. rRNA pseudouridine stoichiometries are conserved, but reduced in cells from dyskeratosis congenita patients, where the PUS DKC1 is mutated. Our work identifies an enhanced, transcritome-wide scope for pseudouridine, and methods to dissect its underlying mechanisms and function.
Introduction
The most abundant RNA modification is pseudouridine (Ψ). In yeast, Ψ is present in many positions in tRNAs, in 46 positions across the four rRNAs (25S, 18S, 5.8S, 5S), and in 6 positions in U1, U2 and U5 snRNA (Charette and Gray, 2000; Ofengand, 2002; Yu et al., 2011). Although ψ’s base pairing is similar to uridine, isomerization allows the potential formation of an extra hydrogen bond, and may contribute to structural stability (Durant and Davis, 1997; Kierzek et al., 2014). Pseudouridine is catalyzed by pseudouridine synthases (PUSs). In yeast, 8 non-essential PUSs directly catalyze ψ formation at specific sites in tRNA, U2 snRNA and mitochondrial rRNA (Ansmant et al., 2000; Behm-Ansmant et al., 2003; Massenet et al., 1999). In contrast, all positions in rRNA and one position in U2 snRNA are modified by the essential PUS Cbf5, which is guided to its target positions through H/ACA-box snoRNAs, based on a 10–12 nt stretch of complementarity to the sequence spanning the target (Lafontaine et al., 1998; Watkins et al., 1998). In human, 23 proteins harbor a PUS domain (Hunter et al., 2011), but most have not been studied for function or specificity.
Although the roles of individual Ψ modifications have largely remained elusive, pseudouridylation defects have profound effects. Alteration of bacterial rRNA pseudouridylation affects antibiotic sensitivity (Toh and Mankin, 2008), ablation of rRNA pseudouridylation by CBF5 deletion in S. cerevisae is lethal (Jiang et al., 1993; Zebarjadian et al., 1999), and defects in DKC1/Dyskerin, the mammalian CBF5 ortholog, cause dyskeratosis congenita (Heiss et al., 1998), a disorder characterized by failure of ribosome biogenesis and an increased risk of cancer (Hoareau-Aveilla et al., 2008). Furthermore, deletion of S. cerevisiae PUS1 results in growth defects, and mutation of human PUS1 leads to mitochondrial myopathy and sideroblastic anemia (Fujiwara and Harigae, 2013).
Recent findings that a few Ψ sites in yeast U2 snRNA are induced by nutrient deprivation or heat shock (Wu et al., 2011) and rRNA sites by rapamycin treatment (Courtes et al., 2014) raise the possibility that the pseudouridine landscape may be more complex and dynamic than previously assumed, and that Ψ might also be present in other RNA species, including mRNA. Testing this hypothesis requires a sensitive method for transcriptome-wide mapping of Ψ sites. Such an assay would also permit identification of the enzyme(s) that catalyze the modification at each site and their specificity, as well as an assessment of whether Ψ is a dynamic feature.
Here, we develop Ψ-seq, for transcriptome-wide quantitative mapping of Ψ, and use it to identify the vast majority of previously reported sites in rRNA, tRNA and snRNA and dozens of novel sites within snoRNAs and mRNAs. Combining Ψ-seq with genetic perturbations, we associate each site with its cognate PUS and/or snoRNA, finding that mRNA Ψ sites are mediated by at least 4 different PUSs, conserved from yeast to mammals. Hundreds of sites are induced by heat shock in yeast, in a Pus7-dependent manner, possibly affecting transcript stability. Finally, we find subtle, but significant, decrease in rRNA and TERC pseudouridylation in cells from patients with dyskeratosis congenita. Our results establish show that Ψ is ubiquitous in diverse RNAs, and dynamic in mRNA, and provide a transcriptome-wide assay for functional studies.
Results
Transcriptome-wide mapping of pseudouridine with Ψ-seq
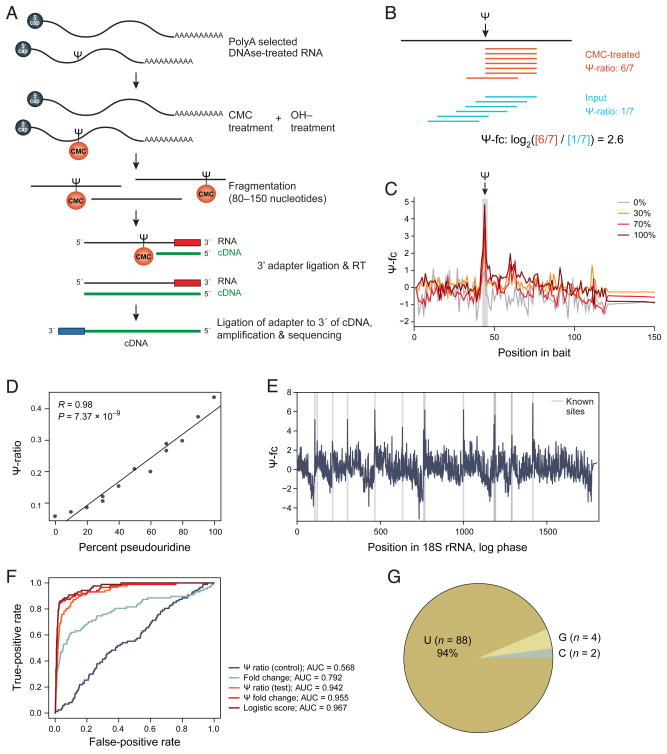
To map Ψ across the transcriptome we developed Ψ-seq, relying on the unique stability of N3-[N-cyclohexyl-N′-β-(4-methylmorpholinium)ethylcarbodiimide-Ψ (N3-CMC-Ψ) to alkaline hydrolysis, and the ability of N3-CMC-Ψ to terminate reverse transcription. This was previously used to test individual sites in primer-extension assays (Bakin and Ofengand, 1993, 1995; Bakin and Ofengand, 1998). Here, we coupled it with strand-specific RNA-seq. Specifically (Figure 1A), we (1) treat polyA-selected RNA with CMC, which covalently binds to U, G and Ψ residues (Ho and Gilham, 1971; Metz and Brown, 1969a, b); (2) incubate CMC-treated RNA at alkaline pH, leading to hydrolysis of the less stable U-CMC and G-CMC adducts; (3) fragment RNA (to 80–150 nt), ligate 3′ adapter and reverse transcribe, expecting premature termination at Ψ-CMC sites; and (4) ligate an adapter to the 3′ end of the cDNA, followed by library preparation. For each experiment, we sequenced both a treated sample and input RNA that was not treated with CMC, but otherwise handled identically (Experimental Procedures).
Figure 1. Ψ-seq quantitatively measured transcriptome-wide pseuodouridylation profiles.
(A) Ψ-seq procedure. Poly(A) selected RNA is treated with CMC, which covalently binds to U and residues, and, to limited extent, G (Ho and Gilham, 1971; Metz and Brown, 1969a, b), followed by incubation at alkaline pH, leading to hydrolysis of U-CMC adducts, which are less stable than Ψ-CMC counterparts. RNA is next fragmented to a size range of 80–150 nt, followed by adapter ligation to the 3′ end. Reverse transcription is primed off of the adapter, expected to lead to premature termination one base pair immediately downstream of pseudouridylated sites. A second adapter is ligated to the 3′ end of the cDNA and libraries are amplified and sequenced in paired end mode. (B) Scoring modified sites. Ψ-ratio (red, top) between reads terminating at a site and reads overlapping it, and Ψ-fc (blue, bottom), the fold change in Ψ-ratio in the CMC-treated sample over non-treated control. (C) Validation with a synthetic spike in. Ψ-fc (Y axis) at each position (X axis) across a synthetic spike-in harboring one pseudouridylated site at varying stoichiometries (color legend) at the indicated position (arrow). (D) Ψ-seq quantifies relative stoichiometry. Scatter plot comparing Ψ-ratios (Y axis) at the pseudouridylated site in the spike-in with pseudouridine stoichiometries (X axis). The Pearson correlation coefficient (r) and associated P value are denoted. (E) Ψ-seq detects known Ψ sites in rRNA. Ψ-fc values (Y axis, dark blue) at each position (X axis) in 18S rRNA, overlaid with all known Ψ sites in this subunit (grey vertical lines). (F) ROC curves for different metrics (color legend) for calling putative Ψ sites. Each classifier was trained based on Ψ-seq data from human rRNA, and tested on their performance in yeast rRNA. (G) Nucleotide distribution across all sites passing our detection thresholds in a sample in mid-log yeast. See also Figure S1.
We defined two metrics to analyze Ψ-seq data (Figure 1B). The Ψ-ratio is the proportion of reads supporting reverse transcription termination at a position out of all the reads overlapping it. The Ψ-fold-change (Ψ-fc) is the log2 transformed Ψ-ratio in the treated samples divided by the Ψ-ratio in the non-treated (input) samples. This controls for non-uniform coverage in RNA-seq due to mappability gaps or complex RNA processing.
We confirmed Ψ-seq’s ability to identify Ψ sites and quantify their relative stoichiometry, by spiking into a complex sample a 150-nt synthetic RNA oligoribonucleotide with a single Ψ site at position 43 (Experimental Procedures). Examining the Ψ-ratio revealed a single strong peak precisely one base downstream of the modified site, as expected (Figure 1C). To assess whether Ψ-seq could quantify Ψ’s stoichiometry, we mixed the modified oligoribonucleotide with a non-modified counterpart, at ratios ranging from 0% to 100%. There was excellent quantitative agreement between the known stoichiometry and its Ψ-seq quantification (Pearson r=0.98, Figure 1C–D). Thus, Ψ-seq can identify Ψ sites and quantify their relative stoichiometry by the Ψ-ratio. However, the Ψ-ratio does not capture the absolute stoichiometry: even with a fully pseudouridylated probe only 43% of the reads terminated at the Ψ site, suggesting partial efficiency of either CMC modification or Ψ-CMC mediated reverse transcription termination. Finally, down-sampling of the spike-in data showed that Ψ-ratios depended on sequencing depth: variability of Ψ-ratio estimates decreased with increased depth and with increased Ψ stoichiometry (Figure S1).
Ψ-seq of log-phase yeast reliably detects known Ψ sites
We applied Ψ-seq to RNA isolated from yeast at log-phase, and found excellent overlap (Figure 1E) between the experimentally determined Ψ sites and the 45 known sites within the three rRNA subunits (18S, 25S and 5.8S) (Ofengand, 2002). We had a very high discriminatory power between a gold standard of positive (known) and negative (all remaining) sites in rRNA, thresholding on either the Ψ-ratio (area under the receiver-operator curve (AUC) =0.942), Ψ-fc (AUC=0.955), or their logistic combination (AUC=0.967) (Figure 1F). Based on this analysis of known sites, we called a site as pseudouridylated in a transcriptome-wide analysis (Experimental Procedures) using the ‘pseudouridine detection thresholds’ of Ψ-fc >3 (8 fold-enrichment) and Ψ-ratio >0.1. These thresholds favor precision over sensitivity: 31/32 rRNA positions called by these criteria are known sites (precision=97%), but 14/45 known sites are not called (sensitivity=69%). Thus, all subsequent analyses may underestimate the number of true Ψ sites. Further supporting call accuracy, 88/94 called sites (94%) in log phase yeast were immediately preceded by a ‘U’ (Figure 1G), and 5/6 remaining sites were one nucleotide away from the 88 called sites, and thus read accumulation at these five sites is likely due to ‘stuttered’ termination of reverse transcription (Bakin and Ofengand, 1998). Down-sampling of a deeply sequenced sample shows dependency of site detection on sequencing depth and transcript expression (Figure S2).
Ψ-seq of deletion mutants associates Ψ sites and their cognate PUSs
Applying Ψ-seq to RNA from yeast at mid-log, we called both known target sites and ones not previously reported. To validate these putative sites, we reasoned that Ψ-seq in deletion mutants of either PUSs or relevant snoRNAs would allow us to match sites to their cognate enzymes, and increase our confidence in their accuracy. Indeed, Ψ-seq of deletion strains for snR34 or snR189 found specific reduction at the two known positions targeted by each 18S and 25S rRNA, whereas other sites remained correlated, readily identifying the cognate target sites (Figure 2A).
Figure 2. Identification of PUS-dependent Ψ sites in yeast.
(A) Ψ-seq correctly distinguishes targets of snoRNAs. A comparison of Ψ-fc scores across all known Ψ sites in three rRNAs (18S, 28S, 5.8S) measured in yeast strains in which either H/ACA box snR34 (X axis) or snR189 (Y axis) were deleted. snR34 is known to target positions 2880 and 2886 in 25S rRNA (red), snR189 targets positions 2735 in 25S and 466 in 18S rRNA (purple), all detected as outliers in these experiments. (B) Ψ-seq associated modified bases with their cognate PUS and snoRNAs. Heatmap depicting Ψ-ratios (Z scores per row, color bar) across all sites (rows, gene and position labeled on left) in the PUS-dependent collection that are dependent on a snoRNA or a non-essential site-specific PUS (columns). For tRNAs, the labeled position is with respect to a multiple alignment of tRNAs with manual correction at two sites, to allow comparison between sites. A check mark indicates that a site was both previously known to be pseudouridylated and that its catalysis was known to be mediated through the enzyme or snoRNA with the minimal Z score in the figure. (C) Ψ-seq identifies Cbf5-dependent Ψ sites. As in (B) for all sites (rows) dependent on Cbf5. (D) Distribution of Ψ sites in different RNA classes, in the Pan-Ψ collection (left) and PUS-dependent collection (right). CDS: mRNA coding sequence. (E) Ψ-ratios (Y axis) across the snoRNA snR47 (left), the mRNA GLK1 (middle), and the mRNA SPI1 (right), which depend on Cbf5, Pus1 and Pus7, respectively. Ψ-ratios are presented for CMC-treated (black) and non-treated (red) samples. Called Ψ sites are marked in vertical thick grey lines, and their ‘absence’ position (in the relevant deletion strain) is marked by a dashed grey box. See also Figure S2 and Table S1.
Next, we applied Ψ-seq to deletion strains of each of four additional snoRNAs and eight non-essential PUSs (Pus1, Pus2, Pus4, Pus5, Pus6, Pus7, Pus9, Deg1) (Figure 2B). Across the 14 experiments, we identified 291 sites with median Ψ-fc >3 and median Ψ-ratio >0.1, with 55/291 dependant on a single PUS by stringent criteria (Experimental Procedures, Figure 2B). Ψ-seq of a Cbf5 conditional knockdown detected 67 Cbf5-dependent sites (Experimental Procedures, Figure 2C).
Overall, we called 328 unique sites across these experiments (‘Pan Ψ collection’, Table S1), 108 of which were associated with one or more PUS and/or snoRNA (‘PUS-dependent Ψ collection’). These sites spanned various classes of coding and non-coding RNA (Figure 2D, 2E). While subsequent analysis focuses on the higher-confidence, PUS-dependent collection, many of the additional sites in the Pan Ψ collection are likely truly pseudouridylated, but did not pass our stringent criteria; Alternatively, Ψ at some sites may be catalyzed by more than one PUS, and thus fail to pass our stringent thresholds.
Unbiased analysis of Ψ in rRNA, tRNA and snRNA
The PUS-dependent collection associated the overwhelming majority of known sites with their correct PUS and snRNA dependencies, demonstrating the power of our approach. For rRNA, 24/24 Cbf5-dependent sites are known targets of pseudouridylation, 8/9 associations between snoRNAs and rRNA sites are known, as is the identified Pus5-mediated Ψ site in mitochondrial 21S rRNA. One unknown, snR3-dependent site at position 2140 on 25S rRNA will require further verification.
For tRNA, many known sites and their associated modifiers were identified, including Pus1-mediated Ψ sites at positions 26–28 in several tRNAs, Pus7-dependent sites at position 13 of glutamate tRNA, Pus9-dependent site in mitochondrial aspartate tRNA at position 32, and Deg1-dependent sites at positions 37–38 in several tRNAs.
For snRNAs, we correctly identified all three previously identified sites in positions 35, 42 and 44 of U2 snRNA, and their dependence on Pus7, Cbf5 and Pus1, respectively. We also detected the known site in U5 snRNA, and found it depends on Cbf5 (not previously known).
Cbf5-dependent pseudouridylation in the 5′ guiding sequence of C/D box and H/ACA box snoRNAs
To our knowledge, no Ψ site within snoRNA has been mapped to date, although Ψ content within snoRNAs has been reported (Ni et al., 1997). The PUS-dependent Ψ collection had six Cbf5-dependent snoRNA sites, 4 within C/D box snoRNAs and 2 in H/ACA box snoRNAs. All 4 Cbf5-dependent sites in C/D box snoRNAs occurred in the 5′ terminus of the snoRNA guiding sequence (Figure 3A, arrows). Pseudouridylation within this stretch is conserved, as 6/9 Ψ sites in C/D box snoRNAs identified in human HEK293 cells (below) are present within the short complementarity stretch of the snoRNA (Figure 3B). The Pan-Ψ collection had 21 additional sites in H/ACA box snoRNAs: 10/21 in H/ACA box snoRNAs with resolved hairpin (targeting) structures (Piekna-Przybylska et al., 2007), and 4/10 in the 5′ arm of the guiding sequence (none in the 3′ arm) (Figure 3C, arrows). We cannot conclusively determine that these sites are Cbf5 dependent, since our detection power is limited by the dramatic decrease in the levels of H/ACA box snoRNAs upon Cbf5 knockdown (Figure 3D), consistent with their stabilization via Cbf5 binding (Lafontaine et al., 1998).
Figure 3. Pseudouridylation in yeast snoRNAs.
(A) Complementarity regions between three C/D box snoRNAs in yeast (black) and target sites in rRNA (red). Red arrows: detected Ψ sites. (B) As in A, but in human. (C) As in A, but for H/ACA box snoRNAs. (D) SnoRNAs expression in WT strains (X axis) compared to Cbf5 knockdown strains (Y axis), showing decreased expression of H/ACA box snoRNA (red), but not C/D box snoRNAs (orange) or snRNAs (dark red), upon knockdown. (E) Cumulative distribution frequencies of minimal Z-score transformed free energies, predicted by cofolding each putative CBF5-dependent (or shuffled) site against all known H/ACA box targeting arms. Distributions are plotted separately for sites in the rRNA (green), all remaining sites in mRNA and snRNAs (blue), and their corresponding shuffled versions (yellow and red, respectively). (F) Predicted associations between three CBF5-dependent sites (red) and top-scoring H/ACA box snoRNAs (black).
PUS-dependent Ψ sites in mRNAs at consensus sequences of cognate PUSs
Whereas no Ψ sites have been previously reported in mRNAs, Ψ-seq uncovered 41 PUS-dependent sites in 41 mRNAs from yeast grown in mid-log phase. These include 34 Cbf5-dependent sites, occurring at a variety of consensus sites, suggesting they may be mediated by snoRNAs (below). Conversely, the 7 sites dependent on one of three non-essential PUSs (Pus1, Pus4, Pus7), were typically associated with distinct consensus motifs: each of the 3 PUS4 dependent sites (in TEF2, MPM1, and ICL2) resided in a ‘RRUUCNA’ motif (underlined U indicates the putative site), perfectly conforming to the known Pus4 ‘GGUUCRA’ target sequence at position 55 of tRNA (Becker et al., 1997a); one PUS7-dependent site in SPI1 was at a ‘UGUAA’ sequence, matching a ‘UNUAR’ consensus at Pus7 targets in tRNAs and in snRNAs (Behm-Ansmant et al., 2003; Decatur and Schnare, 2008) (and which we expand to ‘UGUAR’, below). With one exception, all Pus1 dependent targets (in coding and non-coding genes) were preceded by an A, suggesting partial, but lesser, sequence specificity. We did not observe any positional bias along coding sequences (we only analyzed coding regions due to very short UTRs, limited annotation, and very few Ψ sites in non-coding regions).
Cbf5-dependent Ψ sites in mRNAs and snoRNAs are likely snoRNA-guided
As the substrate specificity of Cbf5 is typically mediated through H/ACA box snoRNAs, we hypothesized that the Cbf5-dependent sites in mRNAs were also snoRNA-guided. To test this hypothesis, we compiled all known H/ACA box targeting arms and used a thermodynamics-based method to assess the folding energy of each snoRNA against each Cbf5-dependent Ψ site (Experimental Procedures). For each site, we identified the snoRNA with the minimal predicted free energy (strongest co-folding) and calculated a Z score for this association with respect to the predicted energies of association with all remaining snoRNAs. Validating our approach, we found that known rRNA Ψ sites had significantly stronger associations with H/ACA box snoRNAs than their shuffled counterparts (Figure 3E).
Overall, Cbf5-dependent sites in mRNAs and snoRNAs had stronger associations with targeting regions in H/ACA box snoRNAs than shuffled controls. This was partly evident when comparing the two distributions (one sided Wilcoxon test, P=0.07, Figure 3E), and most prominent for outliers (e.g., Figure 3F). For example, 23.2% of Cbf5-dependent sites in mRNA and snoRNAs were associated with an H/ACA box with a Z score < −3.5, compared to 11% for shuffled controls (χ2 test, P=5.8×10−8).
Dynamic Pus7-mediated pseudouridylation of mRNAs in heat shock may affect RNA levels
The PUS-dependent sites in mRNAs raised the possibility that they may be differentially regulated under different conditions. To test this hypothesis, we compared Ψ-seq profiles in log phase to those from yeast in growth saturation, cold shock, and heat shock. While only a few positions selectively acquired in Ψ stationary phase or cold shock (Figure 4A), 265 putative Ψ sites were induced in heat shock (45°C) vs. log growth (30°C) in three independent replicates (Figure 4A, Table S2). A few are in key heat shock induced genes (e.g., HSP60, HSP104, MDJ1, SSA4; Figure 4B), but most are in transcripts expressed at similar levels in heat shock and vegetative growth (Figure 4B), suggesting that they are differentially pseudouridylated, rather than differentially expressed.
Figure 4. Induction of Pus7-dependent sites in heat shock.
(A) Bar plots of the number of differentially modified sites (Y axis), in each of three conditions (X axis) relative to mid-log yeast. Sites are color-coded based on RNA class (color legend). (B) Differential modification in heat shock is not merely due to changes in expression levels. Expression levels in heat shock (X axis) or non-heat shock (Y axis) conditions for all genes harboring heat-shock induced Ψ sites. (C) Sequence motif learned across all Ψ sites induced in heat shock, is strongly enriched for the TGTAR Pus7 motif. (D) Ψ-ratios (Y axis) in CMC-treated (black) and non-treated (red) samples in five genes (columns) acquiring Ψ sites in heat shock. Ψ-ratios are shown in WT in 30° (top) and heat-shock (45°, middle), and in heat-shock in Δpus7 (bottom). Called sites are depicted in vertical grey bars in the samples in which they are called, and dashed grey boxes in the remaining samples. (E) Box plots of Ψ-ratios in WT or Δpus7 strains, grown in heat shock or 30°, for 159 heat-shock induced sites harboring a Pus7 consensus (left) and 106 sites without the consensus (right). (F) Cumulative distribution plots of fold changes of expression levels between WT and Δpus7 strains (X axis), comparing genes harboring a Pus7-induced Ψ sites (blue) to genes lacking it (red). At 45° (left), genes harboring Ψ sites are expressed at higher levels in the WT strain than in Δpus7, whereas non-pseudouridylated genes are present at similar levels. At 30° (right), these differences are marginal. P-value of the Kolmogorov-Smirnov test is depicted. (G) Pus7 transcript (left; mean TMM-normalized FPKM values from three replicates) and protein (right, data in four replicates from (Nagaraj et al., 2012)) levels (Y axis) in heat shock and non-heat shock conditions. Error bars are standard error of the mean.(H, I) Pus7 localization. Fluorescent microscopy images (H) and quantification (I) during a heat shock time course show mostly nuclear localization at 30°C (time 0 in H, and red in I), which is significantly reduced after heat shock (time 60 in H, blue in I). P-value of a Mann-Whitney test is noted. See also Figure S3 and Table S2.
The majority of heat shock acquired sites (159/265, 60%) occur in a perfect ‘UGUAR’ Pus7 consensus sequence (R=G>A) (Figure 4C), suggesting that the pseudouridylation program in heat shock is induced through Pus7. To test this hypothesis, we applied Ψ-seq to a Δpus7 strain in normal and heat shock conditions. Ψ sites harboring the Pus7 consensus were reduced to background levels in the Δpus7 strain in both conditions (Figure 4D–E). Thus, there is a widespread, Pus7-mediated, pseudouridylation program in heat shock, that may encompass the previously identified position in U2 snRNA induced in heat shock (Wu et al., 2011). Importantly, the distribution of Ψ-ratios in UGUAR sites in mRNA in heat shock is comparable to that of rRNA sites, suggesting that mRNA sites are pseudouridylated to high stoichiometries (Figure S3A).
To explore the role of pseudouridylation, we compared the expression level (from RNA-seq) between WT and Δpus7 yeast for 142 genes that harbor Pus7-dependent Ψ sites in heat shock vs. the remaining, non-pseudouridylated genes (Figure 4F), in either heat shock or normal conditions. In heat shock, pseudouridylated genes were expressed at ~25% higher levels in WT strains than in Δpus7 strains, whereas non-pseudouridylated genes were expressed at roughly equal levels in the two strains. In contrast, the two sets were comparably expressed in WT and Δpus7 at 30°C. These differences in expression only in pseudouridylated genes and only in heat shock suggest that pseudouridylation may contribute to RNA stability. We did not, however, find a significant correlation between the Ψ-ratio and the change in expression. Future studies of RNA half-life with methods compatible with dynamic responses (Miller et al., 2011) will help study this phenomenon.
We explored the basis for Pus7′s increased activity in heat shock. Surprisingly, both Pus7 RNA and protein levels are reduced in heat shock (Figure 4G), but there are significant differences in Pus7 localization: it is predominantly nuclear in 30°C, and primarily cytoplasmic in heat shock (Figure 4H, 4I). Thus, Pus7 localization may underlie its enhanced repertoire of substrates in heat shock. Supporting a functional role for Pus7 in heat shock, Δpus7 yeast show growth defects after heat shock in comparison to WT (data not shown), consistent with a previous report on their increased heat sensitivity (Sinha et al., 2008).
Finally, we explored factors that may govern modification specificity. First, in the most highly expressed genes, ~30% of UGUAG containing loci are modified, suggesting that the consensus plays a decisive role (Figure S3B). Furthermore, even among the remaining 70%, Ψ-ratios were increased in WT compared to Δpus7 in heat shock (but not at 30°C), suggesting that many are, in fact, modified (Figure S3C). Nonetheless, TGTAR loci called as modified were more likely to have an A/T at position +3, and a pyrimidine at position −4 (Figure S3D), suggesting that features beyond UGUAG also contribute to specificity.
Pseudouridylation profiles and mechanisms are conserved in human
To assess the extent of similarity between yeast and human pseudouridylation, we performed Ψ-seq in HEK293 cells and fibroblasts (below). Applying the same thresholds, we identified 396 putative Ψ sites (Table S3): 353 sites in mRNA and 43 sites in non-coding RNAs (excluding rRNA). We found no functional enrichments in pseudouridylated genes compared to an expression-matched control set and the sites were relatively uniformly distributed across coding and untranslated regions (data not shown).
To associate PUSs to Ψ sites, we examined the sequences surrounding the sites. The most enriched motif (Figure 5A, 70/396 sites), had a GUUC core and was generally characterized by ‘RGUUCNANYCY’, perfectly corresponding to the known Pus4-modified site in tRNAs, and extending the Pus4 consensus in yeast. Another enriched motif (Figure 5B), ‘UGUAG’, present in 8 sites, is consistent with a mammalian Pus7 homolog. Thus, homologs of the same two site-specific PUSs are likely active on yeast and mammalian mRNA.
Figure 5. Ψ-seq of human RNA highlights conserved features and disease-relevance.
(A–B) Sequence motifs from 70 sites harboring a Pus4 associated GUUC core (A) and 13 sites harboring a Pus7 associated UGUA core (B), of 396 modified sites in HEK293 RNA. (C) Difference in Ψ-ratios in DKC1 knockdown relative to control (Y axis) in HEK293 cells. Box plots from left to right: DKC1-dependent sites, sites harboring Pus4 (GUUC) or Pus7 (UGUAR) associated motifs, and all remaining sites. (D–F) Correlation of pseduoridylation stoichiometries in rRNA between homologous positions in S. cerevisiae and C. albicans rRNA (D), human and mouse (E), and yeast and human (F). Positions are color-coded based on rRNA subunit. (G) Distributions of Ψ-ratios across rRNA positions in fibroblasts from dsykeratosis patients (grey) and age-matched controls (pink). Left panel: 7 year old patient; Right: 11 year old patient. (H) Ψ-ratios in rRNA positions in DKC1 knockdown (grey) or control (pink) in HEK293 cells. (I) TERC modification is affected by DKC1 knockdown. Number of reads (top; indicative of RT termination) and Ψ-ratios (bottom) at each site in the TERC transcript (X axis). Reads are shown in both CMC-treated (black) and non-treated (red) samples; Ψ-ratios for the treated samples. The putative Ψ position is highlighted in a grey vertical bar. (J) Ψ-ratios for position 307 in 7-yr old patient and age matched control; Error bars: standard error. (K) RNA secondary structure of the CR4/CR5 region in TERC redrawn based on (Zhang et al., 2011). Red arrow: putative Ψ site. See also Figure S4 and Table S3.
The similarity between Ψ sites in yeast and mammals suggested that the other 318 sites, lacking a Pus4 or Pus7 consensus, may be modified by DKC1/Dyskerin, the mammalian Cbf5 ortholog. To test this hypothesis, we used siRNAs to knock down DKC1 to ~8% of WT RNA levels (data not shown), and found significant reduction (Figure 5C, Experimental Procedures) in Ψ levels at 58/396 sites. The 58 DKC1-dependent sites had significant complementarity to human snoRNAs (P<0.03, one sided K-S test vs. randomly shuffled sequences using the same approach as in yeast). In contrast, Ψ levels were only negligibly reduced upon knockdown in sites containing either a Pus4 or a Pus7 motif (Figure 5C). Many of the 262 sites that we had neither associated with DKC1 (by knockdown) nor with Pus4 nor with Pus7 (by motif) had borderline effects from DKC1 knockdown, and may thus be substrates missed by our stringent criteria.
The number of Ψ sites we report in human is likely a very conservative lower bound, due to our stringent thresholds and requirement for very deep coverage, in particular at moderately or lowly expressed transcripts (Figure S2A). Indeed, when slightly relaxing the criteria of calling a Ψ site (minimal Ψ-ratio: 0.05, minimal Ψ-fc: 2.5), we identified 176 additional Dyskerin-dependent sites. Only two sites are substantially increased upon Dyskerin/Cbf5 knockdown, suggesting a <1% false detection rate. Thus, as in yeast, mammalian mRNAs have a rich landscape of Ψ sites, mediated by at least three orthologous PUSs.
rRNA Ψ levels are conserved from yeast to human
We next examined whether the extent (stoichiometry) of pseudouridylation was conserved across species, focusing on rRNA positions that are largely conserved throughout eukaryotes and substantially covered in our data. To enhance our comparisons, we also collected Ψ-seq data from log-phase Candida albicans and from mouse bone marrow derived dendritic cells. We found that Ψ levels were quantitatively conserved between S. cerevisiae and C. albicans (R=0.78, Figure 5D), between human and mouse (R=0.78, Figure 5E), and even between yeast and human (R=0.67, Figure 5F), suggesting that both the presence and relative abundance of rRNA Ψ sites are under strong purifying selection.
Reduced rRNA Ψ levels in cells from dyskeratosis congenita patients
It has been hypothesized that aberrant rRNA pseudouridylation could underlie X-linked dyskeratosis congenita, caused by mutations in DKC1/dyskerin, but the extent to which it is altered remains controversial (Wong and Collins, 2006), (Bellodi et al., 2013), (Gu et al., 2009; Mochizuki et al., 2004).
Applying Ψ-seq to fibroblasts derived from 7 and 11 year old patients harboring two distinct mutations (del37L and A386T, respectively) and to cells from age-matched controls, we found subtle – yet significant – decreases in rRNA pseudouridylation among patients (Figure 5G), and Ψ-seq profiles further group in unsupervised analysis by disease and not by age (Figure S4A). The subtle effect, on average a reduction of 10% in Ψ-ratio per site, likely explains why these were difficult to observe with less sensitive techniques. As a reference, rRNA pseudouridylation per site is reduced on average a dramatic 55% upon DKC1 knockdown (Figure 5H).
Decreased pseudouridylation in telomerase RNA component (TERC) in dyskeratosis congenita cells
Dyskeratosis congenita can also be caused by mutations in the ncRNA telomerase component, TERC (Hoareau-Aveilla et al., 2008; Trahan and Dragon, 2009). TERC is bound and stabilized by DKC1, but is destabilized in dyskeratosis congenita (Ashbridge et al., 2009). Our findings of Ψ sites within snoRNAs (Figure 3) prompted us to examine whether TERC, which harbors an H/ACA box domain, might be pseudouridylated as well, in a DKC1-dependent manner. While TERC sites were not called in Hek293 cells, the read distribution across TERC revealed a putative Ψ site at position 307 (Figure S4B). This site was not called due to low read counts in the lowly-expressed TERC. TERC expression was even lower in the fibroblasts (Patients FPKM: 0.15 and 0.28; controls: 2.8 and 0.8), precluding any analysis.
To detect and quantify Ψ sites in TERC we first enriched it in RNA from fibroblasts of patients and age-matched controls using hybrid capture with RNA Antisense Purification (RAP) (Engreitz et al., 2014) (Experimental Procedures). This resulted in ~750 fold enrichment in the fraction of reads in the CMC-treated samples aligning to TERC compared to poly(A) selected RNA. Ψ-seq of the hybrid-captured RNA confirmed substantial pseudouridylation of position 307 (Figure 5I), a highly conserved uridine (Figure 5K) in a region essential for telomerase activity and TERT binding (Chen et al., 2002), and showed that it is modified at significantly higher levels in the control sample (Ψ-ratio=0.47) than in the patient sample (Ψ-ratio=0.33) (Figure 5J, χ2 test, P=0.006). A similar reduction was observed when comparing samples from the 11 year old patient to the age matched control (χ2 test, P=0.01; Figure S4C). Another putative Ψ site was found at position 179 albeit at substantially lower levels (Ψ-ratio=0.11). This confirms two of the six previously proposed Ψ positions in TERCs (we do not find evidence for the others) (Kim et al., 2010), suggests that TERC pseudouridylation may be compromised in dyskeratosis congenita, and provides a general way to quantify Ψ in lowly expressed genes.
Discussion
Ψ-seq profiles Ψ in an unbiased, quantitative, transcriptome wide manner at single-nucleotide resolution. It discovered pseudouridines in S. cerevisiae and human snoRNAs and mRNA, catalyzed by both snoRNA-dependent PUSs and site-specific PUSs guided by consensus sequences. We identified a dynamic program induced in yeast heat shock, where Ψ sites accumulate at hundreds of mRNAs, possibly affecting RNA levels. Ψ-seq allows us to compare Ψ stoichiometries across rRNA positions, finding that these are highly conserved, but reduced in cells from dyskeratosis congenita patients. Although we cannot exclude the possibility of some false positives, our stringent controls and excellent correspondence to prior knowledge suggest that their rate is low. The roles of PUSs and Ψ are likely more complex than previously assumed, and our genome-wide method can help study this.
Ψ-seq can be further enhanced in several ways. First, to accurately estimate Ψ levels in lowly expressed transcripts it can be combined with hybrid capture, as demonstrated for TERC, or with new methods to pre-enrich for pseudouridylated RNA. Second, Ψ-seq cannot monitor Ψ levels at 3′ transcript ends, as it relies on reverse transcription termination. To help probe Ψ in short RNA, we could couple Ψ-seq with alternative ligation protocols. Finally, Ψ-seq is theoretically limited in identifying two closely proximal pseudouridines, as reverse transcription is expected to always terminate at the downstream – and never at the upstream - site. In practice, however, this does not appear to be a major limitation: for example, we detect adjacent positions in rRNA (e.g., 25S:1052 and 1056) or in U2 snRNA (positions 42 and 44). This is likely due to a incomplete stoichiometry, incomplete CMC-labeling of Ψ bases and/or imperfect efficiency ofRT-stopping at CMC-treated sites, though Ψ quantification at such adjacent sites may nonetheless be impacted.
PUS substrate specificity
Site-specific, non-essential PUSs in S. cerevisiae were previously thought to modify only one (or a few) residues, by recognizing secondary or tertiary structures in a specific RNA substrate (Becker et al., 1997b). Our results demonstrate that such PUSs have a dramatically larger substrate repertoire than previously assumed, and suggest that many PUS-mediated modifications are guided in part by primary sequence features in their substrates (Figure 6A).
Figure 6.
(A) mRNA pseudouridylation in eukaryotes is mediated by at least four conserved PUSs, some site specific (Pus1, Pus4, Pus7) and others snoRNA mediated (yeast Cbf5/human DKC1). (B) Pus7 may orchestrate the yeast mRNA pseudouridylation program in heat shock. At 30°, Pus7 is primarily nuclear, and its localization into the cytoplasm upon heat shock may induce mRNAs pseudouridylation.
This is conserved from yeast to mammals, where the predominant consensus is identical to that in yeast Pus4, suggesting that Ψ is mediated by TruB1/TruB2, the mammalian orthologs (Zucchini et al., 2003). There are at least 23 putative PUSs in human (Hunter et al., 2011), and one cannot fully rule out the possibility of additional PUSs in yeast; perturbation experiments will help elucidate their targets, as well as resolve possible functional redundancies (Grosshans et al., 2001).
We also find evidence for mRNAs pseudouridylation via snoRNA-dependent PUSs. A more elaborate repertoire of snoRNA substrates, potentially mediated through imperfect complementarities, has previously been hypothesized (Wu et al., 2011), based on the single finding that heat-shock/nutrient-deprivation induced Ψ at position 93 in U2 snRNA is guided through imperfect pairing with snR81 (Wu et al., 2011).
Pus7-pseudouridylation program in heat shock
The Pus7-dependent induction of pseudouridylation during heat shock is an attractive system for further studies of Ψ function. The specificity of the modified site to heat shock (despite lower levels of Pus7 than in other conditions) and the similarity of distribution of Ψ-ratios to rRNA could be consistent with a regulated system, but further functional studies are warranted. This program may be mediated through Pus7 localization in the cytoplasm in heat shock (Figure 6B). Since RNA levels of pseudouridylated targets are higher in WT compared to Δpus7, it is tempting to speculate that Ψ may serve to stabilize transcript structure in heat shock. Alternatively, Ψ could target specific mRNAs to heat shock induced stress granules in a similar manner to m6A destabilization of RNA by directing it to P bodies (Wang et al., 2014). Such responses could help cells to better withstand heat shock, consistent with the increased heat sensitivity of the Pus7 deletion strain in S. cerevisiae (Sinha et al., 2008). Pseudouridylation of a stop codon was previously shown to suppress translation termination (Karijolich and Yu, 2011), suggesting that Ψ might modify the genetic code. We found only a single putative site in a stop codon in HEK293 cells and none in yeast; greater depth may revise these predictions.
Interestingly, Ψ levels at position 50 in 5S rRNA, also catalyzed by Pus7, also increase in heat shock, suggesting that it is not fully modified during exponential growth, and that its pseudouridylation might confer an advantage during heat shock. These data challenge the notion that rRNA modifications are constant across growth conditions, and suggest that the stoichiometry of rRNA modification may also change in response to external stimuli.
Pseudouridylation in dyskeratosis congenita
Pseudouridine is particularly important in dyskeratosis congenita, caused by mutations in the Cbf5 ortholog DKC1/dyskerin. Our results contribute two findings. First, we observed a subtle, yet significant, quantitative difference in Ψ levels in patient derived samples, compared to age-matched controls, which were not previously observed in some studies (Wong and Collins, 2006), but suggested in others (Bellodi et al., 2013), possibly due to less sensitive methods. Second, we detect two putative Ψ sites in TERC, one of which at very high levels and at highly conserved position 307 within the essential P6.1 hairpin (Chen et al., 2002). A Ψ site was previously proposed at that position, and shown to significantly increase stability of the loop structure in vitro (Kim et al., 2010). Our findings may unify the two roles attributed to dyskerin – of stabilizing TERC and of pseudouridylating RNA – and suggest that dyskerin may both bind to and pseudouridylate TERC, and that both processes, perhaps in a mechanistically coupled manner, may be disrupted in the disease.
Pseudouridylation in ncRNAs at sites of inter or intra-molecular interactions
It has previously been noted that Ψ sites in rRNA and snRNAs tend to occur in ‘important’ regions that directly interact with other molecules. In U1, U2, U4 and U6 snRNAs, for example, Ψ sites occur at or close to a site that basepairs with intronic RNA to facilitate RNA splicing (Charette and Gray, 2000). In rRNA there is a high density of conserved Ψ sites at the peptidyl-transferase center and at the decoding center of the 25S rRNA subunit, which interacts with mRNA and with the tRNA stem-loop (Bakin et al., 1994; Bakin and Ofengand, 1993; Lane et al., 1992). Our study extends these observations to snoRNAs, where we find Ψ sites within both C/D and H/ACA box snoRNAs, at positions involved in base-pairing with target sites. The highly conserved Ψ site in TERC is also in a region crucial for telomerase activity, involved in binding to TERT (Chen et al., 2002).
Ψ may also play a role in intramolecular interactions. For example, a Pus1 dependent Ψ site at position 273 in NME1, the RNA component of yeast RNAse MRP, is in a stem structure in the P2 region (Esakova and Krasilnikov, 2010; Esakova et al., 2013; Reilly and Schmitt, 1995), and a site in the helix 2 loop of U3, is involved in pre-ribosomal rRNA cleavage. Mapping Ψ sites in ncRNA molecules may help delineate sites involved in inter and intramolecular interactions.
Our quantitative and high-resolution methodology coupled with genetic perturbations yielded unbiased, transcriptome-wide maps of Ψ across different RNA species, identified consensus sequences recognized by the PUS enzymes, and established pseudouridine as a ubiquitous and dynamic modification of eukaryotic mRNA. Further studies will leverage this method and finding to elucidate Ψ’s function in the RNA life cycle, especially of mRNA.
Experimental Procedures
Yeast growth conditions
Unless otherwise noted, yeast were grown in YPD supplemented with .27mM uridine, heat shock was performed at 45°C for the designated time points, and collected as described in the Extended Experimental Procedures. Growth curves were measured in 96 well plates using a Tekan plate reader. Deletion strains are listed in the Extended Experimental Procedures.
Ψ-seq
Ψ-seq was performed on PolyA+ RNA treated with. CMC essentially as previously described (Bakin and Ofengand, 1993; Bakin and Ofengand, 1998), and detailed in Extended Experimental Procedures. Input controls were handled identically in all steps but without CMC treatment. Following the procedure, RNA-Seq libraries were prepared by an RNA ligation method as detailed in the Extended Experimental Procedures and sequenced on Illumina HiSeq 2500 with 30bp paired end reads.
Detection of Ψ sites
Reads were mapped to the yeast genome (sacCer3) using Bowtie, or to the human (hg19) genome using Tophat,, with parameters and scripts as described in the Extended Experimental Procedures. For rRNA and snRNA sites, we aligned reads against specific database of these two classes. Mouse reads were aligned to mouse rRNA from Refseq. C. albicans reads were aligned to the C. albicans genome (SC5314 version A21). Ψ sites were identified by first calculating a Ψ-ratio for each treated or input sample, followed by Ψ-fold change (log2 fold changes of Ψ ratios in the treated versus non-treated samples). Sites were filtered by parameters listed in the Extended Experimental Procedures. For all unique sites passing the filters in at least one sample, we calculated a Ψ-ratio and Ψ-fc in each sample and considered all sites whose median Ψ-ratio and median Ψ-fc exceeded 0.1 and 3, respectively (unless otherwise noted). We demanded that each 21 nt sequence surrounding the putative Ψ site be unique, and also filtered sites in positions other than immediately following ‘U’. To compare orthologous rRNA sites across species, we used ‘needle’ (Rice et al., 2000) for pairwise alignments of rRNAs with lift over of known Ψ sites (Machnicka et al., 2013) from human to mouse and from S. cerevisiae to C. albicans. For further details see Extended Experimental Procedures.
Synthetic spike-ins
We in vitro transcribed an RNA bait containing only a single uridine (Extended Experimental Procedures), and mixed pseudouridylated and non-pseudouridylated RNA at indicated ratios.
PUS dependent sites
We identified differentially pseudouridylated sites across deletion strains of non-essential PUSs and snoRNAs we tested both the number of reads starting and overlapping at each putatively identified position between a given sample and the background of all samples, with a procedure based on a χ2 test detailed in the Extended Experimental Procedures, and similar procedures for heat-shock, Pus7-dependent sites, Cbf5-dependent sites, and CDK1-dependent sites.
Motif analysis
We used position specific scoring matrices (PSSMs) defined by 21 bp sequences spanning the identified Ψ sites, visualized using SeqLogo in R, and displaying only regions with non-uniform sequence composition.
Prediction of association between Cbf5-dependent target sites and snoRNAs
We assessed the hybridization potential between Cbf5-dependent target sites and H/ACA box snoRNAs by calculating free energies of concatenated upstream and downstream complementary arms (Piekna-Przybylska et al., 2007), separated by a constant hairpin sequence, folded against a 20-nt region surrounding each CBF5 dependent site using RNAcofold (Gruber et al., 2008), with parameters and constraints described in the Extended Experimental Procedures. We shuffled each 20-nt region 15 times (maintaining a center U) as controls.
Pus7 localization
Pus7 localization was measured by microscopy of S. cerevisiae encoding GFP-tagged Pus7 grown at 45°C using a Nikon Eclipse Ti with oil immersion at 100X magnification. ImageJ was used to quantitate GFP intensity with DAPI staining to define nuclear boundaries, as detailed in the Extended Experimental Procedures.
Bone marrow derived dendritic cells
Cells were prepared as previously described (Amit et al., 2009). All experiments were performed in compliance with the institutional guidelines and were reviewed and approved by the Institutional Animal Use and Care Committee (IAUCC) of MIT (Protocol 0612-058-15).
DKC1 knockdown
Human HEK293 cells were plated in 6-well plates at 20% confluence. siRNAs targeting DKC1 (catalog numbers: s4111 and s4112) were transfected using Lipofectamine RNAiMAX (Life Technologies) following the manufacturer’s protocols, with boosts at 48 and 96 hours; As negative controls, we used Ambion® In Vivo Negative Control #1 siRNA (catalog number: 4457287). Cells were harvested at 144 hours.
Samples from dyskeratosis congenita patients
Fibroblasts were obtained from Coriell [(GM01774 (del37L, 7 years), AG04646 (A386T, 11 years), GM00409 - (healthy 7 year old), GM01864 - (healthy 11 year old)].
RAP-RNA enrichment of TERC
We performed RAP on intact isolated total RNA as previously described for RAP-RNA[AMT] (Engreitz et al., 2014).
Supplementary Material
Highlights.
Ψ-seq for high resolution, transcriptome-wide profiling of pseudouridine
Many novel sites in mRNA; dynamically regulated in heat shock
Sites depend on conserved cognate pseudouridine synthases in yeast and human
Reduced rRNA and TERC pseudouridine in dyskeratosis congenita patients
Acknowledgments
Work was supported by NHGRI P50HG006193, Pioneer Award, and HHMI (AR), NHGRI U54 HG003067 (ESL) Broad Institute Funds, NIH GM035010 (GRF), HFSP fellowship (SS), NIH 1F32HD075541-01 (RS), and fellowships of the Swiss National Science Foundation and the Marie Curie IOF (MJ).
Footnotes
Accession numbers
Sequencing data are in the Gene Expression Omnibus (GSE60047).
Author contributions
SS, DB, MM, ESL, GF and AR conceived and designed the study. SS led the development of experimental and computational methods. DB developed and executed all yeast experiments. MM developed and conducted all genomic assays and mammalian experiments. MJ conducted proteomics experiments. RHB, RS and BXLR analyzed data. JME and MG developed the RNA hybrid selection protocol. ESL, GF and AR guided work. SS and AR wrote the manuscript, with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science (New York, NY. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansmant I, Massenet S, Grosjean H, Motorin Y, Branlant C. Identification of the Saccharomyces cerevisiae RNA:pseudouridine synthase responsible for formation of psi(2819) in 21S mitochondrial ribosomal RNA. Nucleic acids research. 2000;28:1941–1946. doi: 10.1093/nar/28.9.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A, Lane BG, Ofengand J. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry. 1994;33:13475–13483. doi: 10.1021/bi00249a036. [DOI] [PubMed] [Google Scholar]
- Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- Bakin A, Ofengand J. Mapping of the 13 pseudouridine residues in Saccharomyces cerevisiae small subunit ribosomal RNA to nucleotide resolution. Nucleic acids research. 1995;23:3290–3294. doi: 10.1093/nar/23.16.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin AV, Ofengand J. Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods in molecular biology. 1998;77:297–309. doi: 10.1385/0-89603-397-X:297. [DOI] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Planta RJ, Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic acids research. 1997a;25:4493–4499. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Sissler M, Florentz C, Grosjean H. Major identity determinants for enzymatic formation of ribothymidine and pseudouridine in the T psi-loop of yeast tRNAs. Journal of molecular biology. 1997b;274:505–518. doi: 10.1006/jmbi.1997.1417. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Urban A, Ma X, Yu YT, Motorin Y, Branlant C. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA (New York, NY. 2003;9:1371–1382. doi: 10.1261/rna.5520403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, McMahon M, Contreras A, Juliano D, Kopmar N, Nakamura T, Maltby D, Burlingame A, Savage SA, Shimamura A, et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell reports. 2013;3:1493–1502. doi: 10.1016/j.celrep.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic acids research. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtes FC, Gu C, Wong NS, Dedon PC, Yap MG, Lee DY. 28S rRNA is inducibly pseudouridylated by the mTOR pathway translational control in CHO cell cultures. Journal of biotechnology. 2014;174:16–21. doi: 10.1016/j.jbiotec.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Schnare MN. Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs. Molecular and cellular biology. 2008;28:3089–3100. doi: 10.1128/MCB.01574-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant PC, Davis DR. The effect of pseudouridine and pH on the structure and dynamics of the anticodon stem-loop of tRNA(Lys,3) Nucleic acids symposium series. 1997:56–57. [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, Guttman M, Lander ES. RNA-RNA Interactions Enable Specific Targeting of Noncoding RNAs to Nascent Pre-mRNAs and Chromatin Sites. Cell. 2014 doi: 10.1016/j.cell.2014.08.018. in print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Krasilnikov AS. Of proteins and RNA: The RNase P/MRP family. Rna-a Publication of the Rna Society. 2010;16:1725–1747. doi: 10.1261/rna.2214510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Perederina A, Berezin I, Krasilnikov AS. Conserved regions of ribonucleoprotein ribonuclease MRP are involved in interactions with its substrate. Nucleic acids research. 2013;41:7084–7091. doi: 10.1093/nar/gkt432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Harigae H. Pathophysiology and genetic mutations in congenital sideroblastic anemia. Pediatr Int. 2013;55:675–679. doi: 10.1111/ped.12217. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Lecointe F, Grosjean H, Hurt E, Simos G. Pus1p-dependent tRNA pseudouridinylation becomes essential when tRNA biogenesis is compromised in yeast. J Biol Chem. 2001;276:46333–46339. doi: 10.1074/jbc.M107141200. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic acids research. 2008;36:W70–74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu BW, Zhao C, Fan JM, Dai Q, Bessler M, Mason PJ. Anomalous electrophoretic migration of newly synthesized ribosomal RNAs and their precursors from cells with DKC1 mutations. FEBS letters. 2009;583:3086–3090. doi: 10.1016/j.febslet.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature genetics. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- Ho NW, Gilham PT. Reaction of pseudouridine and inosine with N-cyclohexyl-N′-beta-(4-methylmorpholinium)ethylcarbodiimide. Biochemistry. 1971;10:3651–3657. [PubMed] [Google Scholar]
- Hoareau-Aveilla C, Henry Y, Leblanc T. Dyskeratosis congenita, a disease caused by defective telomere maintenance. Med Sci (Paris) 2008;24:390–398. doi: 10.1051/medsci/2008244390. [DOI] [PubMed] [Google Scholar]
- Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic acids research. 2011;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Middleton K, Yoon HJ, Fouquet C, Carbon J. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Molecular and cellular biology. 1993;13:4884–4893. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic acids research. 2014;42:3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NK, Theimer CA, Mitchell JR, Collins K, Feigon J. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic acids research. 2010;38:6746–6756. doi: 10.1093/nar/gkq525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes & development. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane BG, Ofengand J, Gray MW. Pseudouridine in the large-subunit (23 S-like) ribosomal RNA. The site of peptidyl transfer in the ribosome? FEBS letters. 1992;302:1–4. doi: 10.1016/0014-5793(92)80269-m. [DOI] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic acids research. 2013;41:D262–267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Molecular and cellular biology. 1999;19:2142–2154. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz DH, Brown GL. The investigation of nucleic acid secondary structure by means of chemical modification with a carbodiimide reagent. I. The reaction between N-cyclohexyl-N′-beta-(4-methylmorpholinium)ethylcarbodiimide and model nucleotides. Biochemistry. 1969a;8:2312–2328. doi: 10.1021/bi00834a012. [DOI] [PubMed] [Google Scholar]
- Metz DH, Brown GL. The investigation of nucleic acid secondary structure by means of chemical modification with a carbodiimide reagent. II. The reaction between N-cyclohexyl-N′-beta-(4-methylmorpholinium)ethylcarbodiimide and transfer ribonucleic acid. Biochemistry. 1969b;8:2329–2342. doi: 10.1021/bi00834a013. [DOI] [PubMed] [Google Scholar]
- Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Molecular systems biology. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki Y, He J, Kulkarni S, Bessler M, Mason PJ. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc Natl Acad Sci U S A. 2004;101:10756–10761. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, Kulak NA, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Molecular & cellular proteomics : MCP. 2012;11 doi: 10.1074/mcp.M111.013722. M111 013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS letters. 2002;514:17–25. doi: 10.1016/s0014-5793(02)02305-0. [DOI] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Decatur WA, Fournier MJ. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA (New York, NY. 2007;13:305–312. doi: 10.1261/rna.373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly TH, Schmitt ME. The yeast, Saccharomyces cerevisiae, RNase P/MRP ribonucleoprotein endoribonuclease family. Molecular biology reports. 1995;22:87–93. doi: 10.1007/BF00988711. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends in genetics : TIG. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Sinha H, David L, Pascon RC, Clauder-Munster S, Krishnakumar S, Nguyen M, Shi G, Dean J, Davis RW, Oefner PJ, et al. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics. 2008;180:1661–1670. doi: 10.1534/genetics.108.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh SM, Mankin AS. An indigenous posttranscriptional modification in the ribosomal peptidyl transferase center confers resistance to an array of protein synthesis inhibitors. Journal of molecular biology. 2008;380:593–597. doi: 10.1016/j.jmb.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA (New York, NY. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes & development. 2006;20:2848–2858. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. The EMBO journal. 2011;30:79–89. doi: 10.1038/emboj.2010.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AT, Ge J, Yu YT. Pseudouridines in spliceosomal snRNAs. Protein Cell. 2011;2:712–725. doi: 10.1007/s13238-011-1087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Molecular and cellular biology. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kim NK, Feigon J. Architecture of human telomerase RNA. Proc Natl Acad Sci U S A. 2011;108:20325–20332. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchini C, Strippoli P, Biolchi A, Solmi R, Lenzi L, D’Addabbo P, Carinci P, Valvassori L. The human TruB family of pseudouridine synthase genes, including the Dyskeratosis Congenita 1 gene and the novel member TRUB1. International journal of molecular medicine. 2003;11:697–704. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.