SUMMARY
Increased fatty acid synthesis is required to meet the demand for membrane expansion of rapidly growing cells. ATP-citrate lyase (ACLY) is upregulated or activated in several types of cancer, and inhibition of ACLY arrests proliferation of cancer cells. Here we show that ACLY is acetylated at lysine residues 540, 546, and 554 (3K). Acetylation at these three lysine residues is stimulated by P300/calcium-binding protein (CBP)-associated factor (PCAF) acetyltransferase under high glucose and increases ACLY stability by blocking its ubiquitylation and degradation. Conversely, the protein deacetylase sirtuin 2 (SIRT2) deacetylates and destabilizes ACLY. Substitution of 3K abolishes ACLY ubiquitylation and promotes de novo lipid synthesis, cell proliferation, and tumor growth. Importantly, 3K acetylation of ACLY is increased in human lung cancers. Our study reveals a crosstalk between acetylation and ubiquitylation by competing for the same lysine residues in the regulation of fatty acid synthesis and cell growth in response to glucose.
INTRODUCTION
Fatty acid synthesis occurs at low rates in most nondividing cells of normal tissues that primarily uptake lipids from circulation. In contrast, increased lipogenesis, especially de novo lipid synthesis, is a key characteristic of cancer cells. Many studies have demonstrated that in cancer cells, fatty acids are preferred to be derived from de novo synthesis instead of extracellular lipid supply (Medes et al., 1953; Menendez and Lupu, 2007; Ookhtens et al., 1984; Sabine et al., 1967). Fatty acids are key building blocks for membrane biogenesis, and glucose serves as a major carbon source for de novo fatty acid synthesis (Kuhajda, 2000; McAndrew, 1986; Swinnen et al., 2006). In rapidly proliferating cells, citrate generated by the tricarboxylic acid (TCA) cycle, either from glucose by glycolysis or glutamine by anaplerosis, is preferentially exported from mitochondria to cytosol and then cleaved by ATP citrate lyase (ACLY) (Icard et al., 2012) to produce cytosolic acetyl coenzyme A (acetyl-CoA), which is the building block for de novo lipid synthesis. As such, ACLY couples energy metabolism with fatty acids synthesis and plays a critical role in supporting cell growth. The function of ACLY in cell growth is supported by the observation that inhibition of ACLY by chemical inhibitors or RNAi dramatically suppresses tumor cell proliferation and induces differentiation in vitro and in vivo (Bauer et al., 2005; Hatzivassiliou et al., 2005). In addition, ACLY activity may link metabolic status to histone acetylation by providing acetyl-CoA and, therefore, gene expression (Wellen et al., 2009).
While ACLY is transcriptionally regulated by sterol regulatory element-binding protein 1 (SREBP-1) (Kim et al., 2010), ACLY activity is regulated by the phosphatidylinositol 3-kinase (PI3K)/Akt pathway (Berwick et al., 2002; Migita et al., 2008; Pierce et al., 1982). Akt can directly phosphorylate and activate ACLY (Bauer et al., 2005; Berwick et al., 2002; Migita et al., 2008; Potapova et al., 2000). Covalent lysine acetylation has recently been found to play a broad and critical role in the regulation of multiple metabolic enzymes (Choudhary et al., 2009; Zhao et al., 2010). In this study, we demonstrate that ACLY protein is acetylated on multiple lysine residues in response to high glucose. Acetylation of ACLY blocks its ubiquitinylation and degradation, thus leading to ACLY accumulation and increased fatty acid synthesis. Our observations reveal a crosstalk between protein acetylation and ubiquitylation in the regulation of fatty acid synthesis and cell growth.
RESULTS
Acetylation of ACLY at Lysines 540, 546, and 554
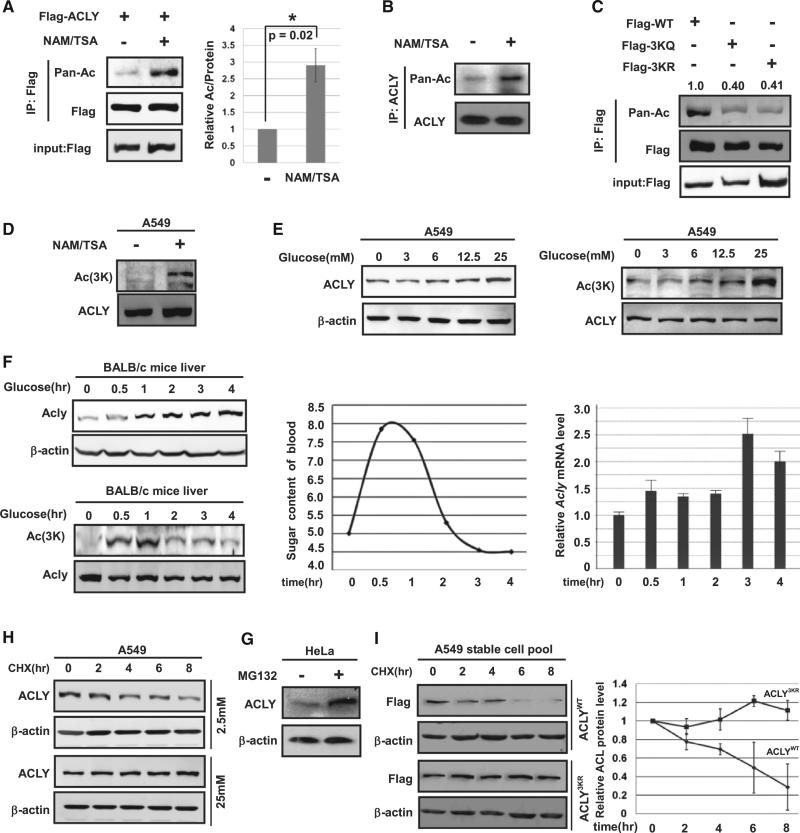
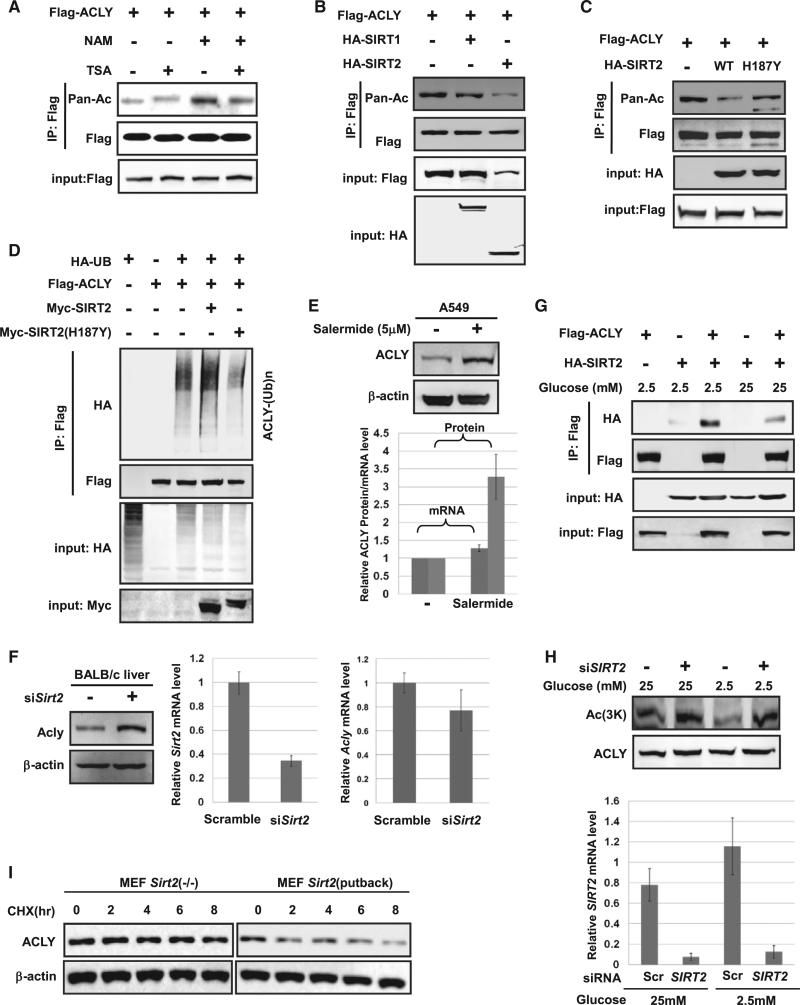
Recent mass spectrometry-based proteomic analyses have potentially identified a large number of acetylated proteins, including ACLY (Figure S1A available online; Choudhary et al., 2009, Zhao et al., 2010). To confirm the acetylation modification of ACLY, we detected the acetylation level of ectopically expressed ACLY followed by western blot using pan-specific anti-acetylated lysine antibody. This experiment showed that ACLY was indeed acetylated, and its acetylation was increased by nearly 3-fold after treatment with nicotinamide (NAM), an inhibitor of the SIRT family deacetylases, and trichostatin A (TSA), an inhibitor of histone deacetylase (HDAC) class I and class II (Figure 1A). Similar experiments with endogenous ACLY also showed that TSA and NAM treatment enhanced ACLY acetylation (Figure 1B).
Figure 1. ACLY Is Acetylated at Lysines 540, 546, and 554.
(A) Exogenous ACLY is acetylated. Ectopically expressed ACLY protein and acetylation levels were determined by western blot. Relative acetylation/protein ratios were quantified. Error bars represents ±SD for experiments performed in triplicate.
(B) Endogenous ACLY is acetylated. Endogenous ACLY protein was purified from HEK 293T cells after NAM and TSA treatment as indicated. Acetylation levels were analyzed by western blot.
(C) Mutations of 3K decrease ACLY acetylation. Acetylation of ectopically expressed ACLYWT, ACLY3KQ, and ACLY3KR was analyzed, and relative acetylation was quantified.
(D) NAM and TSA increases ACLY acetylation at 3K. A549 cells were treated with NAM and TSA. Acetylation was detected by α-Ac(3K) antibody.
(E) Glucose increases both ACLY protein and 3K acetylation levels in A549 cells. A549 cells were maintained under various glucose concentrations, and endogenous ACLY protein and acetylation levels were determined against b-actin and ACLY protein. In order to measure ACLY protein levels, the loading was normalized to total protein as indicated by the actin western blot (upper left panels). In order to measure relative ACLY acetylation, the loading was normalized to ACLY protein levels as indicated by the ACLY western blot (lower left panels).
(F) Glucose injection increases endogenous ACLY acetylation and protein levels in mice liver. BALB/c male mice were injected with PBS or 200 mg/g glucose into enterocoelia. Western blots for ACLY protein and acetylation were performed similar to that in (E). Endogenous ACLY protein levels and ACLY acetylation in mice livers at different time points were determined by western blot. ACLY mRNA levels were quantified by qPCR. Blood glucose levels were determined by ACCUCHEK Active (Roche).
(G) Endogenous ACLY is accumulated by treatment of proteasome inhibitor MG132. HeLa cells were treated with or without 10 mm MG132 for the indicated times under low glucose. Endogenous ACLY level was determined by western blot.
(H) High glucose stabilizes endogenous ACLY. A549 cells were treated with CHX maintained in 2.5 mM (upper panels) or 25 mM (lower panels) glucose for various time points. Endogenous ACLY protein level was analyzed by western blot.
(I) ACLY3KR is more stable than ACLYWT under low glucose. A549 cells with stable ACLY knockdown and re-expression of the Flag-tagged shRNA-resistant wild-type and 3KR mutant were treated with CHX for the indicated time points. Stabilities of ACLYWT and ACLY3KR protein were determined by western blot and quantified. Error bars represents ±SD for experiments performed in triplicate.
Ten putative acetylation sites were identified by mass spec-trometry analyses (Table S1). We singly mutated each lysine to either a glutamine (Q) or an arginine (R) and found that no single mutation resulted in a significant reduction of ACLY acetylation (data not shown), indicating that ACLY may be acetylated at multiple lysine residues. Three lysine residues, K540, K546, and K554, received high scores in the acetylation proteomic screen and are evolutionarily conserved from C. elegans to mammals (Figure S1A). We generated triple Q and R mutants of K540, K546, and K554 (3KQ and 3KR) and found that both 3KQ and 3KR mutations resulted in a significant (~60%) decrease in ACLY acetylation (Figure 1C), indicating that 3K are the major acetylation sites of ACLY.
To further confirm acetylation of the three residues, we then generated antibodies specific to acetylated K540, K546, and K554. The specificity of the specific anti-acetylated lysine at 540, 546, and 554 antibody (α-Ac[3K]) antibody was first confirmed by its ability to recognize the acetylated, but not unacetylated, peptide (Figures S1B–S1E). Furthermore, the α-Ac(3K) antibody recognized ectopically expressed wild-type and ACLY mutants with one of the three lysines remaining at various degrees (Figures S1C and S1F). The acetylation signal was blocked by preincubation of the antibody with antigen peptides (Figure S1E), demonstrating the specificity of the antibody. Using this antibody, we found that the acetylation of endogenous ACLY is clearly increased after treatment of cells with NAM and TSA (Figure 1D). These results demonstrate that ACLY is acetylated at K540, K546, and K554.
Glucose Promotes ACLY Acetylation to Stabilize ACLY
In mammalian cells, glucose is the main carbon source for de novo lipid synthesis. We found that ACLY levels increased with increasing glucose concentration, which also correlated with increased ACLY 3K acetylation (Figure 1E). Furthermore, to confirm whether the glucose level affects ACLY protein stability in vivo, we intraperitoneally injected glucose in BALB/c mice and found that high glucose resulted in a significant increase of ACLY protein levels (Figure 1F). Notably, ACLY acetylation increased rapidly prior to accumulation of its protein. In contrast, ACLY messenger RNA (mRNA) did not increase until 3 hr after glucose injection, whereas significant ACLY protein accumulation was observed at the 1 hr time point. These data indicate a possible correlation between ACLY acetylation and protein levels.
To determine whether ACLY acetylation affects its protein levels, we treated HeLa and Chang liver cells with NAM and TSA and found an increase in ACLY protein levels (Figure S1G, upper panel). ACLY mRNA levels were not significantly changed by the treatment of NAM and TSA (Figure S1G, lower panel), indicating that this upregulation of ACLY is mostly achieved at the posttranscriptional level. Indeed, ACLY protein was also accumulated in cells treated with the proteasome inhibitor MG132, indicating that ACLY stability could be regulated by the ubiquitin-proteasome pathway (Figure 1G). Cycloheximide (CHX) chase experiment indicated that ACLY is an unstable protein in cells grown in low-glucose medium, with a half-life around 6 hr. However, its half-life is substantially extended under high-glucose conditions (Figure 1H). We next determined its half-life in cells treated with NAM and TSA and found that blocking deacetylase activity stabilized ACLY (Figure S1H). The stabilization of ACLY induced by high glucose was associated with an increase of ACLY acetylation at K540, K546, and K554. Together, these data support a notion that high glucose induces both ACLY acetylation and protein stabilization and prompted us to ask whether acetylation directly regulates ACLY stability. We then generated ACLYWT, ACLY3KQ, and ACLY3KR stable cells after knocking down the endogenous ACLY. We found that the ACLY3KR or ACLY3KQ mutant was more stable than the ACLYWT (Figures 1I and S1I). Collectively, our results suggest that glucose induces acetylation at K540, 546, and 554 to stabilize ACLY.
Acetylation Stabilizes ACLY by Inhibiting Ubiquitylation
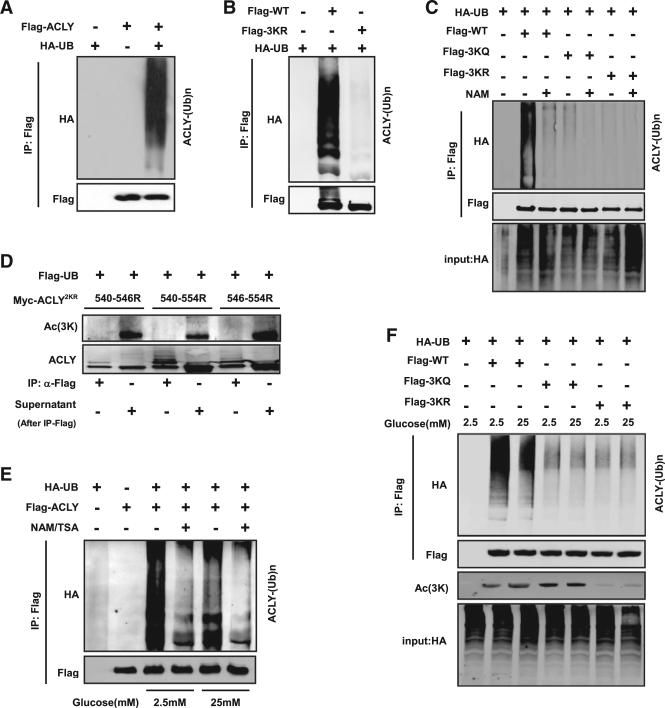
To determine the mechanism underlying the acetylation and ACLY protein stability, we first examined ACLY ubiquitylation and found that it was actively ubiquitylated (Figure 2A). Previous proteomic analyses have identified K546 in ACLY as a ubiquity-lation site (Wagner et al., 2011). In order to identify the ubiquity-lation sites, we tested the ubiquitylation levels of double mutants 540R–546R and 546–554R (Figure S2A). We found that the ubiquitylation of the 540R-546R and 546R-554R mutants is partially decreased, while mutation of K540, K546, and K554 (3KR), which changes all three putative acetylation lysine residues of ACLY to arginine residues, dramatically reduced the ACLY ubiquitylation level (Figures 2B and S2A), indicating that 3K lysines might also be the ubiquitylation target residues. Moreover, inhibition of deacetylases by NAM and TSA decreased ubiquity-lation of WT but not 3KQ or 3KR mutant ACLY (Figure 2C). These results implicate an antagonizing role of the acetylation towards the ubiquitylation of ACLy at these three lysine residues.
Figure 2. Acetylation Protects ACLY from Proteasome Degradation by Inhibiting Ubiquitylation.
(A) ACLY is ubiquitylated in vivo. Flag-tagged ACLY was cotransfected with HA-tagged ubiquitin into HEK 293T cells. Ubiquitylation of immunoprecipitated ACLY was determined.
(B) Mutations of the acetylation lysine residues decrease ACLY ubiquitylation. Flag-tagged wild-type and 3KR mutant ACLY were cotransfected with HA-tagged ubiquitin into HEK 293T cells, and ubiquitylation of purified proteins was determined.
(C) Deacetylase inhibitors NAM and TSA decrease wild-type ACLY ubiquitylation, but not 3KQ or 3KR mutants. Flag-tagged WT, 3KQ, or 3KR of ACLY was cotransfected with HA-tagged ubiquitin into HEK 293T cells with or without the treatment of NAM and TSA for indicated times. Ubiquitylation of purified proteins was analyzed by western blot.
(D) 3K residues are the potential competition sites of ubiquitylation and acetylation. K540/546R, K546/554R, and K546/554R left with one lysine were cotransfected with Flag-tagged ubiquitin. The anti-Flag-UB immunoprecipitates (input, supposed to be all ubiquitylated substrate mixture) and the remaining supernatants (output, supposed to be nonubiquitylated substrate mixture) were blotted to compare the ACLY 3K acetylation level. Acetylation at 3K was detected in the output (nonubiquitylated ACLY), but not in the input (ubiquitylated ACLY).
(E) NAM and TSA reduce ACLY ubiquitylation. Flag-tagged ACLY was cotransfected with HA-tagged ubiquitin into HEK 293T cells maintained under 2.5 mM or 25 mM glucose with or without NAM and TSA as indicated. Ubiquitylation of purified proteins was analyzed.
(F) High glucose decreases acetylation of wild-type ACLY, but not 3KQ or 3KR mutants. Flag-tagged WT, 3KQ, or 3KR of ACLY was cotransfected with HA-tagged ubiquitin into HEK 293T cells maintained under 2.5 mM or 25 mM glucose concentrations. Ubiquitylation of purified proteins and acetylation were determined by western blot.
There are two possible mechanisms by which acetylation may influence ACLY ubiquitylation. Acetylation at 3K indirectly antagonizes ACLY ubiquitylation by recruiting a factor promoting ACLY deubiquitylation. Alternatively, the three lysine residues could be the sites of ubiquitylation, with their acetylation directly blocking ACLY ubiquitylation, as both modifications are covalently linked to the same epsilon-amino group of lysine, resulting in the binding of a putative ACLY E3 ligase being affected. To distinguish between these two mechanisms, we compared the acetylation levels of the ubiquitylated and nonubiquitylated ACLY. We ectopically expressed ACLY (2KR) mutants in which two of three lysine residues (K540 and K546, K540 and K554, or K546 and K554) were mutated to arginine. The remaining K554, K546, or K540 is sufficient for acetylation, but not sufficient to block ACLY ubiquitylation (Figure 2D). We found that ACLY acetylation was only detected in the nonubiquitylated, but not the ubiquitylated (high-molecular-weight), ACLY species. This result indicates that ACLY acetylation and ubiquitylation are mutually exclusive and is consistent with the model that K540, K546, and K554 are the sites of both ubiquitylation and acetylation. Therefore, acetylation of these lysines would block ubiquitylation.
We next investigated if ubiquitylation of ACLY was regulated by glucose, which stimulates ACLY acetylation and stabilizes ACLY protein. We found that glucose upregulates ACLY acetylation at 3K and decreases its ubiquitylation (Figure S2B). High glucose (25 mM) effectively decreased ACLY ubiquitylation, while inhibition of deacetylases clearly diminished its ubiquitylation (Figure 2E). Consistently, increasing glucose concentration reduced the ubiquitylation of wild-type ACLY, but had no effect on the 3KQ or 3KR mutant ACLY (Figure 2F). From these results, we conclude that acetylation and ubiquitylation occur mutually exclusively at K540, K546, and K554 and that high-glucose-induced acetylation at these three sites blocks ACLY ubiquitylation and degradation.
UBR4 Targets ACLY for Degradation
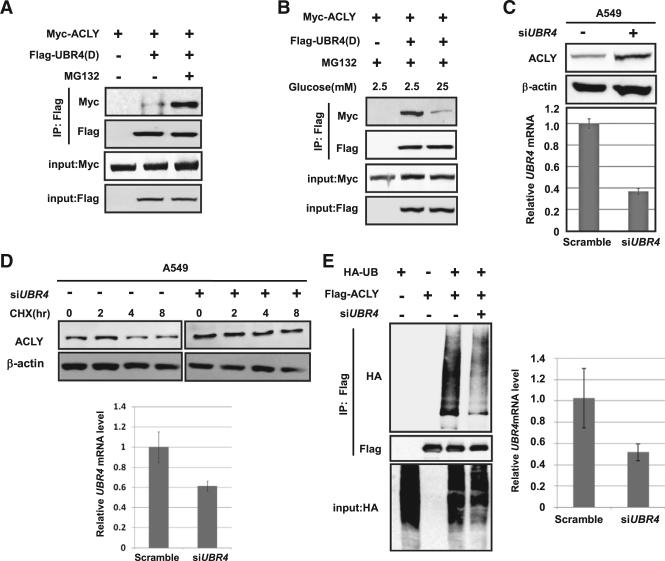
UBR4 was identified as a putative ACLY-interacting protein by affinity purification coupled with mass spectrometry analysis (data not shown). To address if UBR4 is a potential ACLY E3 ligase, we determined the interaction between ACLY and UBR4 and found that ACLY interacted with the E3 ligase domain of UBR4; this interaction was enhanced by MG132 treatment (Figure 3A). Consistently, 25 mM glucose dramatically decreased the interaction between ACLY and the E3 ligase domain of UBR4 (Figure 3B). Notably, high glucose decreases the interaction between UBR4 E3 ligase domain and wild-type ACLY, but not 3KR (Figure S3). Furthermore, UBR4 knockdown in A549 cells resulted in an increase of endogenous ACLY protein level (Figure 3C). Moreover, UBR4 knockdown significantly stabilized ACLY (Figure 3D) and decreased ACLY ubiquitylation (Figure 3E). Taken together, these results indicate that UBR4 is an ACLY E3 ligase that responds to glucose regulation.
Figure 3. UBR4 Is the E3 Ligase of ACLY.
(A) The interaction between E3 ligase domain of UBR4, noted as UBR4(D), and ACLY is enhanced by the treatment of MG132. Flag-tagged ACLY and Myc-tagged UBR4(D) was coexpressed into HEK 293T cells maintained in the presence or absence of MG132. The interaction between ACLY and UBR4(D) was determined by immunoprecipitation and western blotting (IP-western).
(B) The interaction between UBR4(D) and ACLY is decreased under high glucose. Flag-tagged ACLY and Myc-tagged UBR4(D) were coexpressed into HEK 293T cells maintained in 2.5 mM or 25 mM glucose for indicated times. The interaction between ACLY and UBR4(D) was determined by IP-western.
(C) UBR4 knockdown increases steady-state ACLY level. UBR4 in A549 cells was knocked down by siRNA, and endogenous ACLY level was determined and quantified.
(D) Knocking down UBR4 stabilizes ACLY. A549 cells with or without siUBR4 were treated with CHX for indicated time points. Stability of endogenous ACLY protein level was determined by western blot. The knockdown efficiency of UBR4 was measured by qPCR with ±SD.
(E) Knockdown of UBR4 decreases ACLY ubiquitylation. HEK 293T cells with or without siUBR4 were transfected with Flag-tagged ACLY and HA-tagged ubiquitn. The ubiquitylation of ACLY was determined by western blot. UBR4 knockdown efficiency was measured by qPCR with ±SD.
PCAF Acetylates ACLY
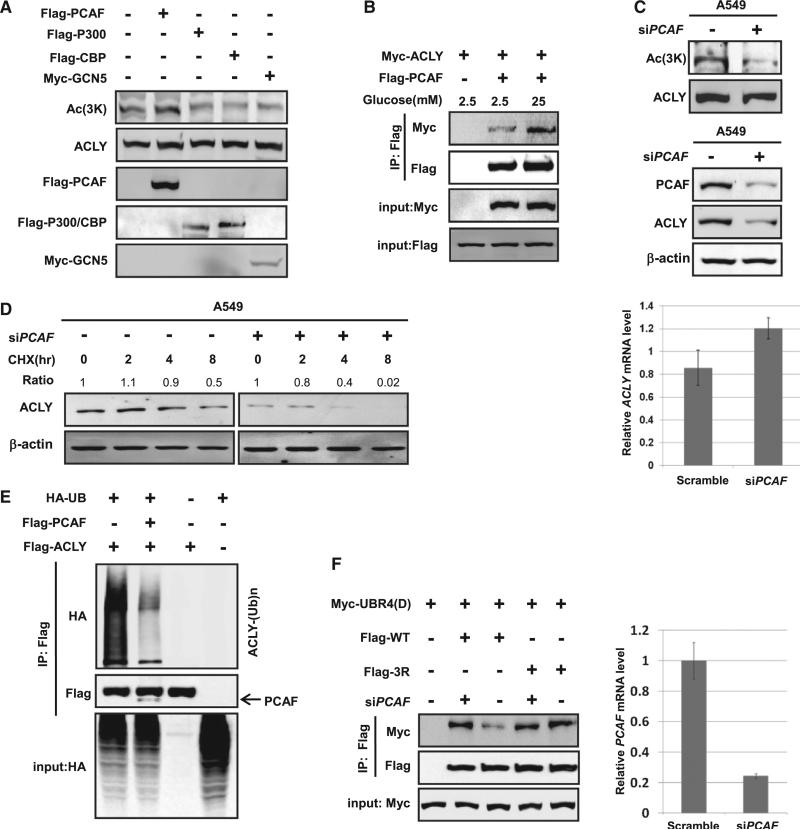
To identify the acetyltransferase responsible for ACLY acetylation at 3K, we transfected four acetyltransferases, p300 (E1A-binding protein, 300 kDa), cAMP response element-binding protein (CREB)-binding protein (CBP), PCAF (p300/CBP-associated factor, also known as lysine acetyltransferase 2B, KAT2B), and GCN5 (KAT2A), individually into human embryonic kidney (HEK) 293T cells and found that ectopic expression of PCAF increased ACLY acetylation at 3K, whereas the other acetyltransferases had little effect (Figures 4A and S4). Notably, glucose increased the interaction between ACLY and PCAF (Figure 4B). PCAF knockdown significantly reduced acetylation of 3K, indicating that PCAF is a potential 3K acetyltransferase in vivo (Figure 4C, upper panel). Furthermore, PCAF knockdown decreased the steady-state level of endogenous ACLY, but not ACLY mRNA (Figure 4C, middle and lower panels). Moreover, we found that PCAF knockdown destabilized ACLY (Figure 4D). In addition, overexpression of PCAF decreases ACLY ubiquitylation (Figure 4E), while PCAF inhibition increases the interaction between UBR4 E3 ligase domain and wild-type ACLY, but not 3KR (Figure 4F). Together, our results indicate that PCAF increases ACLY protein level, possibly via acetylating ACLY at 3K.
Figure 4. PCAF Is the Acetylase of ACLY.
(A) Overexpression of PCAF, but not other acetyltransferases (HATs), increases endogenous ACLY acetylation at 3K. Flag-tagged PCAF, P300, or CBP or Myc-tagged GCN5 was tranfected individually into HEK 293T cells. Endogenous acetylation levels of ACLY were determined by anti-3K (Ac) antibody.
(B) ACLY-PCAF interaction is enhanced under high glucose. Myc-tagged ACLY and Flag-tagged PCAF were coexpressed into HEK 293T cells maintained in 2.5 mM or 25 mM glucose for indicated times. The interaction between ACLY and PCAF was determined by IP-western.
(C) PCAF knockdown decreases endogenous ACLY acetylation at 3K and its protein level. A549 cells were transfected with siPCAF or control as previously described, and endogenous ACLY acetylation at 3K was measured by western blot against ACLY protein level (upper panel). ACLY transcription level was measured by qPCR (lower panel). Endogenous ACLY levels with or without PCAF knockdown were determined by anti-ACLY antibody against b-actin.
(D) PCAF knockdown decreases ACLY protein stability. A549 cells were transfected with siPCAF or control as previously described and were treated with CHX for the indicated times. The endogenous ACLY protein was determined and quantified by western blot against b-actin.
(E) Overexpression of PCAF decreases ACLY ubiquitylation. Flag-tagged PCAF was cotransfected with Flag-tagged ACLY and HA-tagged ubiquitin. The ubiquitylation of ACLY was determined by western blot.
(F) PCAF inhibition increases the interaction between UBR4(D) and wild-type ACLY, but not 3KR. Myc-tagged UBR4(D) and Flag-tagged wild-type and 3KR of ACLY were cotransfected into HEK 293T cells treated with high (25 mM) glucose. The interaction between ACLY and UBR4(D) was determined by western blot. PCAF knockdown efficiency is shown on the right.
SIRT2 Deacetylates ACLY
To identify the ACLY deacetylase, we first took advantage of TSA, a class I, II, and IV deacetylase inhibitor, and NAM, a NAD(+)-dependent class III deacetylase inhibitor, to determine whether SIRTs or HDACs are involved in ACLY deacetylation. Treatment of cells with SIRT inhibitor NAM, but not HDAC inhibitor TSA, increased ACLY acetylation (Figures 5A and S5A), indicating that a SIRT family member is preferentially involved in ACLY deacetylation. We coexpressed ACLY with SIRT1, SIRT2, or other SIRT deacetylases and found that SIRT2, but not SIRT1 (Figure 5B) or other SIRTs (data not shown), decreased ACLY acetylation. We then performed coimmunoprecipitation experiments and found that ACLY readily coprecipitated SIRT2, but not SIRT1 (Figure S5B). Ectopic expression of SIRT2 decreased acetylation of wild-type, but not the 3KR mutant, of ACLY (Figure S5C). Furthermore, expression of wild-type SIRT2, but not the catalytically inactive H187Y mutant, led to a decrease of ACLY acetylation (Figure 5C). Importantly, coexpression of wild-type, but not H187Y mutant, SIRT2 increased ACLY ubiquitylation (Figures 5D and S5D). In addition, we found that treatment of cells with salermide, a class III histone deacetylases inhibitor (Lara et al., 2009), increased endogenous ACLY protein level (Figure 5E). Similarly, knockdown of SIRT2 by small hairpin RNA (shRNA) also stabilized the ACLY level (Figure S5E). We next tested the regulation of ACLY level by SIRT2 in vivo by mouse tail vein injection of shRNA against Sirt2. This experiment showed that Sirt2 knockdown significantly enhanced ACLY protein level in mice livers (Figure 5F). We then examined the effect of glucose on the SIRT2-ACLY interac tion. We found that high glucose decreased the interaction between ACLY and SIRT2 (Figure 5G) and suppressed the deacetylation of ACLY by SIRT2 (Figure 5H). Finally, we found that restoration of SIRT2 destabilizes ACLY in Sirt2 −/− mouse embryonic fibroblast (MEF) cells (Figure 5I). Together, these results demonstrate that SIRT2 is the primary deacetylase for ACLY, and the interaction between SIRT2 and ACLY is maintained in cells grown in low glucose but significantly reduced when cells are grown in high glucose.
Figure 5. SIRT2 Decreases ACLY Acetylation and Increases Its Protein Levels In Vivo.
(A) NAM, but not TSA, increases ACLY acetylation. Flag-tagged ACLY was transfected into HEK 293T cells with or without sirtuin deacetylase inhibitor NAM and HDAC inhibitor TSA. Acetylation of ACLY was measured by western blot.
(B) SIRT2 overexpression decreases ACLY acetylation. Flag-tagged ACLY was cotransfected with either HA-tagged SIRT1 or SIRT2 into HEK 293T cells. Acetylation of purified proteins was determined by western blot.
(C) Catalytic activity of SIRT2 is required for the deacetylation of ACLY. Flag-tagged ACLY was cotransfected with HA-tagged SIRT2 WT or catalytically inactive mutant H187Y into HEK 293T cells. Acetylation was determined by western blot.
(D) SIRT2, but not its catalytic mutant, promotes ACLY ubiquitylation. Flag-tagged ACLY and HA-tagged ubiquitin were cotransfected with Myc-tagged SIRT2 or SIRT2H187Y into HEK 293T cells. Ubiquitylation of purified proteins was detected.
(E) Inhibition of SIRT2 with inhibitor increases endogenous ACLY protein level. HEK 293T cells were treated with or without SIRT2 inhibitor salermide. Endogenous ACLY protein levels were determined by western blot, and relative ACLY mRNA levels were quantified by qPCR.
(F) Sirt2 knockdown increases endogenous ACLY protein level in mice liver. Livers from BALB/c mice with tail vein injection of control and Sirt2 shRNA were prepared. The Sirt2 knockdown efficiency was determined by qPCR. Endogenous ACLY protein levels were determined by western blot.
(G) High glucose decreases the interaction between ACLY and SIRT2. Flag-tagged ACLY were cotransfected with HA-tagged SIRT2 into HEK 293T cells. Protein interactions were determined.
(H) SIRT2 is required for glucose-regulated ACLY acetylation. HEK 293T cells transfected with or without siSIRT2 were cultured in medium containing 2.5 mM or 25 mM glucose. Endogenous ACLY acetylation was determined by western blot. SIRT2 knockdown efficiency was determined by qPCR.
(I) Restoration of SIRT2 destabilizes ACLY. MEF cells with Sirt2 −/− or Sirt2 −/− MEF cells stably re-expressing Sirt2 were treated with CHX at the indicated time points. The stability of endogenous ACLY protein level was determined by western blot.
Acetylation of ACLY Promotes Cell Proliferation and De Novo Lipid Synthesis
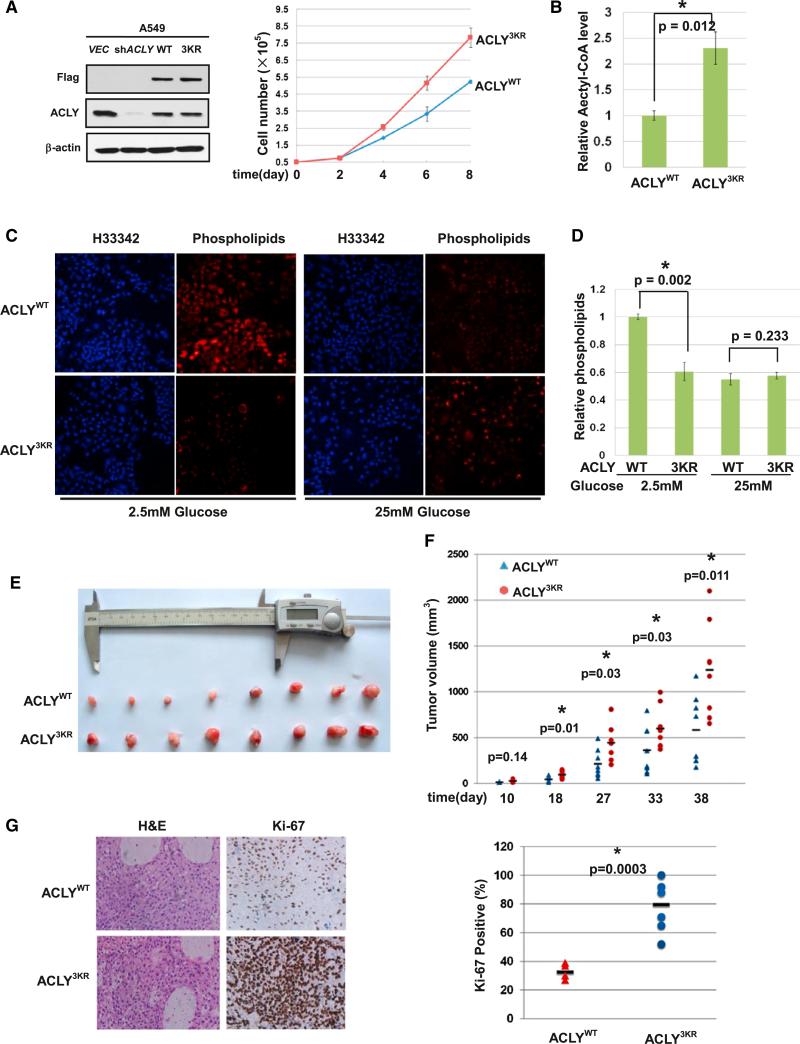
Given the high expression of ACLY in cancer cells (Kuhajda, 2000; Milgraum et al., 1997; Swinnen et al., 2004; Yahagi et al., 2005), we next examined the effect of ACLY acetylation on cell proliferation and tumor growth. To this end, we generated stable A549 cell lines in which the endogenous ACLY was knocked down by shRNA and the shRNA-resistant ACLY wild-type, 3KQ, 3KR, or H760A (which is a catalytically inactive mutant) was stably expressed. Western blotting analysis demonstrated that the endogenous ACLY was effectively knocked down and that wild-type, 3KQ, 3KR, or H760A mutant ACLY were expressed at levels similar to that of endogenous ACLY (Figures 6A and S6A). We found that cells expressing ACLY3KQ and ACLY3KR mutant proliferated faster, while H760A mutant proliferated slower than the cells expressing wild-type ACLY under a low-glucose concentration. Moreover, the ACLY expression levels in both WT and mutant stable cells were determined after maintaining cells under 6 mM glucose culture medium for 8 days. The protein levels of ACLY 3KQ and 3KR were accumulated to a level higher than the wild-type cells upon extended culture in low-glucose medium (Figure S6A, right panel), indicating a growth advantage conferred by ACLY stabilization resulting from the disruption of both acetylation and ubiquitylation at K540, K546, and K554. Cellular acetyl-CoA assay showed that cells expressing 3KQ or 3KR mutant ACLY produce more acetyl-CoA than cells expressing the wild-type ACLY under low glucose (Figures 6B and S6B), further supporting the conclusion that 3KQ or 3KR mutation stabilizes ACLY.
Figure 6. Acetylation of ACLY at 3K Promotes Lipogenesis and Tumor Cell Proliferation.
(A) ACLY3KR mutant promotes cell proliferation. ACLY was stably knocked down with shRNA in A549 cells. shRNA-resistant wild-type and 3KR mutant of ACLY were stably re-expressed in the knockdown cells to a level that is compatible with endogenous ACLY. ACLY knockdown efficiency and re-expression level were determined by western blot. ACLYWT or ACLY3KR cells were seeded at the same number in each well. Cell numbers were counted every 48 hr. Error bars represent cell numbers ±SD for experiments performed in triplicate.
(B) ACLY3KR increases cellular acetyl-CoA level under low glucose. Flag-tagged ACLYWT and ACLY3KR were transfected into Chang shACLY stable cell line maintained in 2.5 mM glucose for the indicated times. Total cellular acetyl-CoA was measured using PicoProbe Acetyl-CoA Assay Kit (Abcam). Error bars represent ±SD for experiments performed in triplicate.
(C) The ACLY3KR mutant-expressing cells show reduced uptake of extra phospholipids. Intracellular accumulation of phospholipids was measured using phospholipids conjugated to fluorescent dyes.
(D) Intracellular accumulation of phospholipids is quantified. The fluorescence of intracellular phospholipids was measured at optical density 594 (OD594) on a plate reader. Error bars represent ±SD.
(E) ACLY3KR mutant promotes xenograft tumor growth. A549 cell lines characterized in (A) were injected subcutaneously into the flanks of nude mice. At 7 weeks after injection, tumors from 8 mice were extracted and photographed.
(F) Tumor growth curves in nude mice. Tumor diameters were measured at the indicated time points, and tumor volumes were calculated. Significant difference curves were evaluated by t test.
(G) ACLY3KR expression promotes tumor cell proliferation in vivo. Tumor sections from xenografts were prepared for IHC. All eight pairs of hematoxylin and eosin (H&E)- and Ki-67-stained ACLYWT and ACLY3KR were analyzed.
ACLY is a key enzyme in de novo lipid synthesis. Silencing ACLY inhibited the proliferation of multiple cancer cell lines, and this inhibition can be partially rescued by adding extra fatty acids or cholesterol into the culture media (Zaidi et al., 2012). This prompted us to measure extracellular lipid incorporation in A549 cells after knockdown and ectopic expression of ACLY. Cells were seeded in medium containing phospholipids coupled with fluorescent dyes for monitoring the uptake of phospholipids. Consistent with a previous report (Migita et al., 2008), we found that ACLY knockdown dramatically increased the uptake of extracellular phospholipids, as determined by LipidTOX staining (Figure S6C), supporting the notion that deficiency in the endogenous lipid metabolism would stimulate cells to increase the uptake of extracellular lipid. Next, we compared phospholipid uptake by wild-type, 3KQ, or 3KR mutant ACLY-expressing cells cultured in different glucose concentrations. We found that when cultured in low glucose (2.5 mM), cells expressing wild-type ACLY uptake significantly more phospholipids compared to cells expressing 3KQ or 3KR mutant ACLY (Figures 6C, 6D, and S6D). When cultured in the presence of high glucose (25 mM), however, cells expressing either the wild-type, 3KQ, or 3KR mutant ACLY all have reduced, but similar, uptake of extracellular phospholipids (Figures 6C, 6D, and S6D). The above results are consistent with a model that acetylation of ACLY induced by high glucose increases its stability and stimulates de novo lipid synthesis.
To determine whether the 3KR mutant of ACLY also rendered growth advantage to tumor cells in vivo, we performed xenograft studies. A549 cells with stable knockdown of endogenous ACLY and ectopic expression of wild-type or 3KR mutant ACLY were injected into nude mice, and tumor cell growth was monitored over a period of 7 weeks. Sustained expression of wild-type and 3KR mutant ACLY in xenograft tumors was verified by western blot (Figure S6E, left panel). We also found that in four pairs of xenografts, ACLY was accumulated in the 3KR xenograft, but not in the WT xenograft (Figure S6E, right panel). The tumor volumes in mice injected with cells expressing 3KR mutant ACLY were significantly (p < 0.05) larger, while H760A mutants were dramatically smaller than those from mice injected with cells expressing wild-type ACLY (Figures 6E, 6F, and S6F). Ki-67 staining demonstrated that the tumors derived from 3KR-expressing cells were more proliferative than those derived from cells expressing wild-type ACLY (Figures 6G and S6G). Taken together, these results demonstrate that stabilization of ACLY by the disruption of three ubiquitylation sites increased cell proliferation and tumor growth.
3K Acetylation of ACLY Is Increased in Lung Cancer
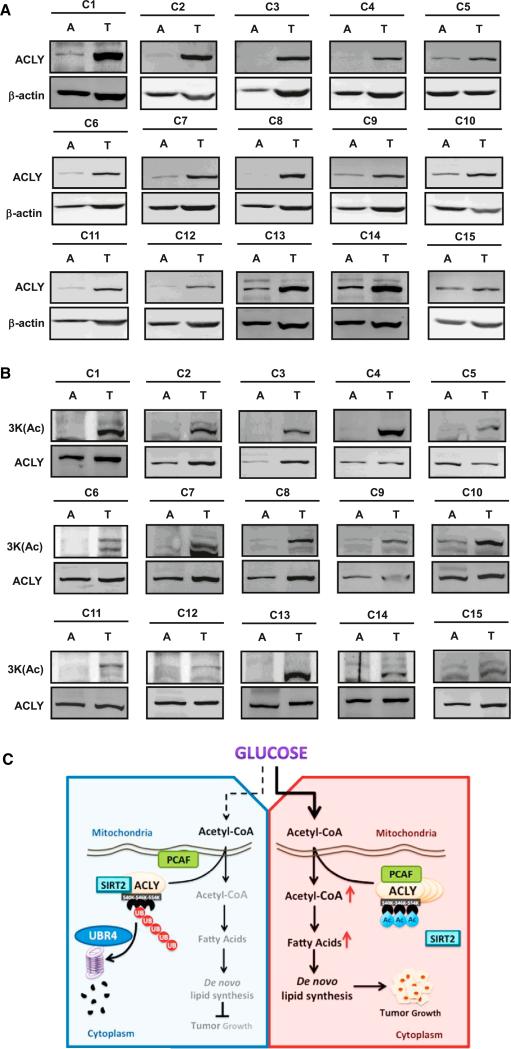
ACLY is reported to be upregulated in human lung cancer (Migita et al., 2008). Many small chemicals targeting ACLY have been designed for cancer treatment (Zu et al., 2012). The finding that 3KQ or 3KR mutant increased the ability of ACLY to support A549 lung cancer cell proliferation prompted us to examine 3K acetylation in human lung cancers. We collected a total of 54 pairs of primary human lung cancer samples with adjacent normal lung tissues and performed immunoblotting for ACLY protein levels. This analysis revealed that, when compared to the matched normal lung tissues, 29 pairs showed a significant increase of total ACLY protein using b-actin as a loading control (Figures 7A and S7A). To determine whether 3K aectylation correlates with the increase of ACLY protein in these 29 pairs of lung cancer tissues, we further determined acetylated ACLY at 3K by normalizing to ACLY protein levels. Among the 29 pairs, 15 pairs of samples showed relatively higher levels of 3K acetylation in the tumor tissues than the matched normal tissues (Figures 7B and S7B and Table S2). The tumor sample analyses demonstrate that ACLY protein levels are elevated in lung cancers, and 3K acetylation positively correlates with the elevated ACLY protein. These data also indicate that ACLY with 3K acetylation may be potential biomarker for lung cancer diagnosis.
Figure 7. Acetylation of ACLY at 3K Is Upregulated in Human Lung Carcinoma.
(A) Lung cancer clinical cases with an increase in ACLY protein. Human lung carcinoma samples paired with carcinoma tissue (shown as T) and adjacent normal tissue (shown as A) were lysed. The ACLY protein levels were compared against b-actin under western blot.
(B) Clinical cases with increased ACLY acetylation at 3K in ACLY-upregulated lung cancer. ACLY-upregulated human lung carcinoma samples paired with carcinoma tissue (shown as T) and adjacent normal tissue (shown as A) were lysed. The acetylation level of ACLY at 3K was compared against ACLY protein by western blot.
(C) Shown is a working model depicting how acetylation at 3K protects ACLY from ubiquitin-mediated proteasome degradation in response to high glucose. This upregulation of ACLY protein level contributes to de novo lipogenesis and promotes tumor cell proliferation.
DISCUSSION
Dysregulation of cellular metabolism is a hallmark of cancer (Hanahan and Weinberg, 2011; Vander Heiden et al., 2009). Besides elevated glycolysis, increased lipogenesis, especially de novo lipid synthesis, also plays an important role in tumor growth. Because most carbon sources for fatty acid synthesis are from glucose in mammalian cells (Wellen et al., 2009), the channeling of carbon into de novo lipid synthesis as building blocks for tumor cell growth is primarily linked to acetyl-CoA production by ACLY. Moreover, the ACLY-catalyzed reaction consumes ATP. Therefore, as the key cellular energy and carbon source, one may expect a role for glucose in ACLY regulation. In the present study, we have uncovered a mechanism of ACLY regulation by glucose that increases ACLY protein level to meet the enhanced demand of lipogenesis in growing cells, such as tumor cells (Figure 7C). Glucose increases ACLY protein levels by stimulating its acetylation.
Upregulation of ACLY is common in many cancers (Kuhajda, 2000; Milgraum et al., 1997; Swinnen et al., 2004; Yahagi et al., 2005). This is in part due to the transcriptional activation by SREBP-1 resulting from the activation of the PI3K/AKT pathway in cancers (Kim et al., 2010; Nadler et al., 2001; Wang and Dey, 2006). In this study, we report a mechanism of ACLY regulation at the posttranscriptional level. We propose that acetylation modulated by glucose status plays a crucial role in coordinating the intracellular level of ACLY, hence fatty acid synthesis, and glucose availability. When glucose is sufficient, lipogenesis is enhanced. This can be achieved, at least in part, by the glucose-induced stabilization of ACLY. High glucose increases ACLY acetylation, which inhibits its ubiquitylation and degradation, leading to the accumulation of ACLY and enhanced lipogenesis. In contrast, when glucose is limited, ACLY is not acetylated and thus can be ubiquitylated, leading to ACLY degradation and reduced lipogenesis. Moreover, our data indicate that acetylation and ubiquitylation in ACLY may compete with each other by targeting the same lysine residues at K540, K546, and K554. Consistently, previous proteomic analyses have identified K546 in ACLY as a ubiquitylation site (Wagner et al., 2011). Similar models of different modifications on the same lysine residues have been reported in the regulation of other proteins (Grönroos et al., 2002; Li et al., 2002, 2012). We propose that acetylation and ubiquitylation have opposing effects in the regulation of ACLY by competitively modifying the same lysine residues. The acetylation-mimetic 3KQ and the acetylation-deficient 3KR mutants behaved indistinguishably in most biochemical and functional assays, mainly due to the fact that these mutations disrupt lysine ubiquitylation that primarily occurs on these three residues.
In rapidly proliferating cancer cells, a high level of de novo lipogenesis provides precursors for membrane biogenesis. However, this is a process that demands a high level of energy (Rysman et al., 2010). Many studies have shown that de novo lipid synthesis is dramatically increased in cancer cells (Medes et al., 1953; Menendez and Lupu, 2007; Ookhtens et al., 1984; Sabine et al., 1967). It has been reported that ACLY inhibition with chemical inhibitors or RNAi can suppress tumor cell proliferation (Bauer et al., 2005; Hatzivassiliou et al., 2005). Our studies show that stabilization of ACLY by the mutation at K540, K546, and K554 (3KR) significantly enhances lipogenesis, increases cell proliferation, and promotes in vivo tumor growth. These results support a critical role for ACLY acetylation in the coordination of glucose availability and de novo lipid synthesis and the regulation of cell growth and tumorigenesis. They also suggest that the possibility of drugs inhibiting the ACLY acetylation may merit exploration as a therapeutic agent for cancer. Notably, ACLY is increased in lung cancer tissues compared to adjacent tissues. Consistently, ACLY acetylation at 3K is also significantly increased in lung cancer tissues. These observations not only confirm ACLY acetylation in vivo, but also suggest that ACLY 3K acetylation may play a role in lung cancer development. Our study reveals a mechanism of ACLY regulation in response to glucose signals.
EXPERIMENTAL PROCEDURES
Immunoblotting
Cells were lysed in a NP40 buffer, and western blot analysis was carried out according to standard methods. Antibodies specific to Flag (Sigma), HA (Santa Cruz), Myc (Santa Cruz), b-actin (Sigma), Ki-67 (Epitomics), PCAF (Epitomics), and ACLY (Abcam) were commercially obtained. The anti-pan-acetylated lysine antibodies were generated in our lab (Zhao et al., 2010). Antibodies specifically recognizing acetylation at lysines 540, 546, and 554 (Ac[3K]) were prepared commercially by immunizing rabbits at Shanghai Genomics, Inc.
In Vivo Ubiquitylation Assay
In vivo ubiquitylation assay was performed following the protocol as previously described (Jiang et al., 2011). Briefly, at ~24–48 hr after transfection, cells were collected and lysed in 0.1% SDS (Tris [pH 7.5], 0.5 mM EDTA, 1 mM dithiothreitol [DTT]) in Tris-HCl buffer with inhibitors. Analyses of ubiquitylation were performed by western blot.
siRNA Transfection and RNAi
For RNAi experiments, small interfering RNA (siRNA) oligos of SIRT2 and UBR4 were carried out using commercial synthetic siRNA oligonucleotides (Shanghai GenePharma), and each target gene employed two effective sequences. siSIRT2-1: 5′-GAGGCCAUCUUUGAGAUCAdTdT-3′; siSIRT2-2: 5′-AUGACAACCUAGAGAAGUAdTdT-3′; siUBR4-1: 5′-GCAAAGAGUUGU UGGAAUAdTdT-3′; siUBR4-2: 5′-AAUGAUGAGCAGUCAUCUAdTdT-3′.
The siRNA oligos of PCAF were carried out using a commercial siRNA assay kit (Invitrogen). All siRNA transfections were performed with Lipofectamine 2000 (Invitrogen), and the knockdown efficiency was verified by quantitative PCR (qPCR) or western blot.
For murine RNAi, synthetic siRNA oligonucleotides of Sirt2 (Shanghai GenePharma) was prepared for tail vein injection of each mouse using the hydrodynamic transfection method (Hamar et al., 2004). All animal-related procedures were performed under Division of Laboratory Animal Medicine regulations of Fudan University. Briefly, 50 uM synthetic modified siSirt2 (diluted in 1 ml of PBS) or 1 ml of PBS solution was rapidly injected (within 10 s) into tail veins of 8- to 10-week-old male BALB/c mice. Mice livers were removed 24 hr after injection, and the homogenates were prepared using NP40 buffer for western blot. Liver mRNA was isolated for qPCR to test the knockdown efficiency.
Knocking Down and Putting Back Stable Cell Lines
shRNA constructs, including shACLY, employed two effective sequences as follows: 5′-ATCAAACGTCGTGGAAAA-3′; 5′-GAGGAAGCTGATGAATAT-3′.
For retroviral production, A549 cells were infected with pMKO-shACLY and pMKO-shVEC retrovirus. ACLY knockdown of the stable cell line was collected after drug selection (knocking down). Then, pQC-XIH-H760A, pQC-XIH-WT, pQC-XIH-3KQ, and pQC-XIH-3KR retroviruses were added into the A549 shACLY stable cell line as previously described. Positive cells stably expressing Flag-tagged wild-type, H760A, 3KQ, and 3KR of ACLY (putting back) were collected and verified by western blotting after double drug selection.
Cell Proliferation Analysis
A total of 5 × 104 A549 stable cells were seeded in triplicate in each well of a 6-well plate, and the cell numbers were counted every 2 days over an 8-day period.
Lipid Staining
The assays were carried out with the HCS LipidTOX Phospholipidosis and Steatosis Detection Kit according to the manufacturer's instructions (Invitrogen).
Xenograft Analysis
Nude mice (nu/nu, 6- to 8-week-old males) were injected subcutaneously with 1.5 × 105 A549 stable cells. The diameters of tumors were measured every 10 days. Around 7 weeks after injection, the tumors were dissected and analyzed.
Immunohistochemistry
Paraffin-cut sections of xenograft tumors were prepared, and immunohisto-chemistry (IHC) was performed as previously described (Lei et al., 2006).
Lung Cancer Samples
Lung cancer samples were acquired from the Tongji University Affiliated East Hospital. A physician obtained informed consent from the patients. The procedures related to human subjects were approved by the Ethics Committee of the Institutes of Biomedical Sciences (IBS), Fudan University. Direct immuno-blotting was performed as above.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Fudan Molecular and Cell Biology laboratory for discussions throughout this study and Beezly Groh for critical reading of the manuscript. This work was supported by 973 grants (2011CB910600 and 2009CB918401), NSFC (31071192 and 81225016), NCET-09-0315, Shanghai Key basic Research Program (12JC1401100), 100 Talents Program of Shanghai Health (XBR2011041), the Dawn Program of Shanghai Education Commission, and the Program for the Shanghai Outstanding Academic Leader (13XD1400600) to Q.Y.L. This work was supported by NSFC (81071744) to T.R. This work was also supported by the 985 Program, the Shanghai Leading Academic Discipline Project (B110), 973 grant (2012CB910101), and NIH grants (to Y.X. and K.-L.G.) This work is dedicated to the memory of Zhen Yu, who prepared the 3K acetylation antibody.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2013.07.002.
REFERENCES
- Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J. Biol. Chem. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Grönroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell. 2002;10:483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- Hamar P, Song E, Kökény G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc. Natl. Acad. Sci. USA. 2004;101:14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Icard P, Poulain L, Lincet H. Understanding the central role of citrate in the metabolism of cancer cells. Biochim. Biophys. Acta. 2012;1825:111–116. doi: 10.1016/j.bbcan.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Shin HT, Seo YH, Byun HO, Yoon SH, Lee IK, Hyun DH, Chung HY, Yoon G. Sterol regulatory element-binding protein (SREBP)-1-mediated lipogenesis is involved in cell senescence. J. Biol. Chem. 2010;285:29069–29077. doi: 10.1074/jbc.M110.120386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000;16:202–208. doi: 10.1016/s0899-9007(99)00266-x. [DOI] [PubMed] [Google Scholar]
- Lara E, Mai A, Calvanese V, Altucci L, Lopez-Nieva P, Martinez-Chantar ML, Varela-Rey M, Rotili D, Nebbioso A, Ropero S, et al. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene. 2009;28:781–791. doi: 10.1038/onc.2008.436. [DOI] [PubMed] [Google Scholar]
- Lei Q, Jiao J, Xin L, Chang CJ, Wang S, Gao J, Gleave ME, Witte ON, Liu X, Wu H. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9:367–378. doi: 10.1016/j.ccr.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- Li H, Wittwer T, Weber A, Schneider H, Moreno R, Maine GN, Kracht M, Schmitz ML, Burstein E. Regulation of NF-kB activity by competition between RelA acetylation and ubiquitination. Oncogene. 2012;31:611–623. doi: 10.1038/onc.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAndrew PF. Fat metabolism and cancer. Surg. Clin. North Am. 1986;66:1003–1012. doi: 10.1016/s0039-6109(16)44037-5. [DOI] [PubMed] [Google Scholar]
- Medes G, Thomas A, Weinhouse S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 1953;13:27–29. [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Migita T, Narita T, Nomura K, Miyagi E, Inazuka F, Matsuura M, Ushijima M, Mashima T, Seimiya H, Satoh Y, et al. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–8554. doi: 10.1158/0008-5472.CAN-08-1235. [DOI] [PubMed] [Google Scholar]
- Milgraum LZ, Witters LA, Pasternack GR, Kuhajda FP. Enzymes of the fatty acid synthesis pathway are highly expressed in in situ breast carcinoma. Clin. Cancer Res. 1997;3:2115–2120. [PubMed] [Google Scholar]
- Nadler ST, Stoehr JP, Rabaglia ME, Schueler KL, Birnbaum MJ, Attie AD. Normal Akt/PKB with reduced PI3K activation in insulin-resistant mice. Am. J. Physiol. Endocrinol. Metab. 2001;281:E1249–E1254. doi: 10.1152/ajpendo.2001.281.6.E1249. [DOI] [PubMed] [Google Scholar]
- Ookhtens M, Kannan R, Lyon I, Baker N. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am. J. Physiol. 1984;247:R146–R153. doi: 10.1152/ajpregu.1984.247.1.R146. [DOI] [PubMed] [Google Scholar]
- Pierce MW, Palmer JL, Keutmann HT, Hall TA, Avruch J. The insulin-directed phosphorylation site on ATP-citrate lyase is identical with the site phosphorylated by the cAMP-dependent protein kinase in vitro. J. Biol. Chem. 1982;257:10681–10686. [PubMed] [Google Scholar]
- Potapova IA, El-Maghrabi MR, Doronin SV, Benjamin WB. Phosphorylation of recombinant human ATP:citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of ATP:citrate lyase by phosphorylated sugars. Biochemistry. 2000;39:1169–1179. doi: 10.1021/bi992159y. [DOI] [PubMed] [Google Scholar]
- Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D, Daniëls VW, Machiels J, et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70:8117–8126. doi: 10.1158/0008-5472.CAN-09-3871. [DOI] [PubMed] [Google Scholar]
- Sabine JR, Abraham S, Chaikoff IL. Control of lipid metabolism in hepatomas: insensitivity of rate of fatty acid and cholesterol synthesis by mouse hepatoma BW7756 to fasting and to feedback control. Cancer Res. 1967;27:793–799. [PubMed] [Google Scholar]
- Swinnen JV, Heemers H, van de Sande T, de Schrijver E, Brusselmans K, Heyns W, Verhoeven G. Androgens, lipogenesis and prostate cancer. J. Steroid Biochem. Mol. Biol. 2004;92:273–279. doi: 10.1016/j.jsbmb.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Swinnen JV, Brusselmans K, Verhoeven G. Increased lipogenesis in cancer cells: new players, novel targets. Curr. Opin. Clin. Nutr. Metab. Care. 2006;9:358–365. doi: 10.1097/01.mco.0000232894.28674.30. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics. 2011;10:M111, 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dey SK. Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet. 2006;7:185–199. doi: 10.1038/nrg1808. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahagi N, Shimano H, Hasegawa K, Ohashi K, Matsuzaka T, Najima Y, Sekiya M, Tomita S, Okazaki H, Tamura Y, et al. Co-ordinate activation of lipogenic enzymes in hepatocellular carcinoma. Eur. J. Cancer. 2005;41:1316–1322. doi: 10.1016/j.ejca.2004.12.037. [DOI] [PubMed] [Google Scholar]
- Zaidi N, Royaux I, Swinnen JV, Smans K. ATP citrate lyase knockdown induces growth arrest and apoptosis through different cell- and environment-dependent mechanisms. Mol. Cancer Ther. 2012;11:1925–1935. doi: 10.1158/1535-7163.MCT-12-0095. [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu XY, Zhang QH, Liu JH, Cao RX, Zhong J, Yi GH, Quan ZH, Pizzorno G. ATP citrate lyase inhibitors as novel cancer therapeutic agents. Recent Patents Anticancer. Drug Discov. 2012;7:154–167. doi: 10.2174/157489212799972954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.