Abstract
The use of infectious agents as vaccine adjuvants has shown utility in both prophylactic and therapeutic vaccinations. Listeria monocytogenes has been used extensively as a vaccine vehicle due to its ability to initiate both CD4+ and CD8+ immune responses. Previous work from this laboratory has used transgenic Listeria to deliver vaccine constructs. A chimeric protein composed of tumor antigen and a non-hemolytic variant of the Listeria protein, listeriolysin O (LLO), has demonstrated effective tumor protection beyond that of antigen alone expressed in the same system. To address the question of how fusion with LLO improves vaccine efficacy, we constructed a number of DNA plasmid vaccines to isolate this effect in the absence of other endogenous Listeria effects. Here we have analyzed the ability of these vaccines to induce the regression of previously established tumors. A vaccine strategy using DNA vaccines bearing the tumor antigen either alone or in combination with LLO in addition to plasmids encoding MIP-1α and GM-CSF was examined. Further, LLO was used either as a chimera or in a bicistronic construct to address the importance of fusion between these elements. Notably, the strategies employing both chimeric and bicistronic vaccines were effective in reducing tumor burden suggesting that LLO can act as an adjuvant that does not require fusion with the tumor antigen to mediate its effect.
Keywords: DNA vaccine, Listeriolysin O, HPV-16 E7, CD4+ T cell, CD8+ T cell, Adjuvant
Introduction
Since the discovery by Dr. William Coley in the late nineteenth century that cancer patients suffering from bacterial infection demonstrated improvement in their tumor burden, it has been shown that immune activation against a pathogen may be beneficial in oncology treatment [1]. Since then the use of live attenuated and heat killed microorganisms has been explored to generate more specific anti-tumor immunotherapy.
Listeria monocytogenes is a gram-positive bacterium that has been used for the delivery of antigens to the immune system due to its ability to generate both CD4+ and CD8+ T cell responses in infected individuals. These immune responses were a direct result of this bacteria’s life cycle in which they are initially taken up by phagocytosis, but then escape these vesicles by the expression of virulence genes such as the Listeria hemolysin protein, listeriolysin O (LLO), and phospholipases. The production of LLO is regulated by the virulence gene PrfA in response to the acidic environment of the phagosome. LLO is thought to function by association with cholesterol and subsequent oligomerization and permeabilization of the membrane allowing for escape of Listeria into the cytosol. This lifecycle exposes Listeria’s proteins to both MHC class II presentation following degradation in the lysosome as well as MHC class I presentation of proteins following escape of Listeria into the cytosol (reviewed in [2]). Because of these properties Listeria has been explored as a vector for viral and tumor antigens for over a decade. However, it has also been observed to act as a Coley’s toxin in non-specifically slowing tumor growth [3, 4].
For the analysis of Listeria-based vaccines we used the HPV-16 E7 protein as a model antigen. HPV-16 is associated with >50% of cervical cancer cases [5]. E6 and E7 are early viral proteins expressed by HPV-16 that are known to be sufficient for the transformation of infected cells and are expressed constitutively in HPV-associated tumors [6]. A murine tumor model for HPV-16, TC-1, has been developed by immortalizing C57BL/6 lung epithelial cells by transfection with genes encoding HPV-16 E6 and E7 followed by transformation with c-Ha-ras [7]. This transplantable tumor allows for the analysis of immunotherapeutic vaccines in vivo [7]. Previous work from our laboratory has shown that the fusion of HPV-16 E7 with a truncated (non-hemolytic) form of LLO enhances the immunogenicity and anti-tumor efficacy of the tumor antigen when delivered by Listeria (Lm-LLO-E7) [8] or Vaccinia [9]. Moreover, the fusion protein expressed by live Listeria has been shown to induce dendritic cell (DC) maturation by upregulating MHC class II molecules, CD80, CD86 and CD40 costimulatory molecule surface expression [10]. We have studied the anti-tumor elements responsible for the enhanced efficacy of Lm-LLO-E7. Although both Lm-LLO-E7 and Listeria expressing the tumor antigen alone (Lm-E7) induce good CD8+ T cells in the spleen, only those induced by Lm-LLO-E7 appear to traffic to and penetrate the tumor [11, 12].
To further explore how LLO mediates vaccine efficacy, we have constructed eukaryotic expression plasmids for use as DNA vaccines. DNA vaccines, rather than recombinant L. monocytogenes permits the isolation of specific factors controlling the immune response without the background of endogenously expressed L. monocytogenes genes. In contrast to live vaccines, DNA vaccines offer many advantages. For example, DNA vaccines are relatively stable, and can be easily prepared and harvested in large quantities. Additionally, plasmid DNA is relatively safe and can be repeatedly administered without adverse effects [13, 14]. One of the concerns about DNA vaccines is their low immunogenicity. Several studies have shown that plasmid cytokines can augment the immunogenicity of plasmid DNA vaccines. For example, plasmid GM-CSF has been shown to enhance DNA vaccine elicited cellular immune responses specific for a variety of antigens [15–17]. McKay et al. [18] reported that co-administration of plasmid GM-CSF with the DNA vaccine resulted in the recruitment of macrophages to the site of inoculation and specifically augmented vaccine-elicited CD4+ T lymphocyte responses. In contrast, co-administration of plasmid MIP-1α with the DNA vaccine resulted in the recruitment of DCs to the injection site and enhanced vaccine-elicited CD8+ T lymphocyte responses. Interestingly, co-administration of both plasmid GM-CSF and plasmid MIP-1α with the DNA vaccine recruited both macrophages and DCs and led to a synergistic and sustained augmentation of CD4+ and CD8+ T lymphocyte responses [18].
In this study, we compared the immunogenicity and anti-tumor efficacy of different forms of E7 DNA vaccines administered with plasmids encoding GM-CSF and MIP-1-α in order to determine which elements mediated the greatest efficacy. We found that the inclusion of GM-CSF and MIP-1-α was required for optimal vaccine efficacy, consistent with the findings of others [15, 18, 19]. More importantly, we found that LLO acts as an adjuvant to enhance both CD4+ and CD8+ T cell responses. Fusion of LLO to E7 is required to induce E7-specific CD8+ T cell responses consistent with our hypothesis that the PEST region of LLO enhances antigen processing by targeting the antigen to the ubiquitin-proteosome pathway [12]. However, fusion of LLO to E7 is not necessary to augment E7-specific CD4+ T cell responses, as similar levels of CD4+ T cells are induced if the LLO adjuvant is given either as a mixture of plasmid LLO and plasmid E7 or as a plasmid expressing a bicistronic message LLO-IRES-E7. Delivering the LLO and E7 plasmids at distinct sites did not improve the CD4+ T cell response over that achieved using E7 alone. Thus, for maximal CD4+ T cell responses it appears necessary for the LLO plasmid to be expressed in the same cells as the E7 plasmid.
Materials and methods
Mice
Six to eight week-old C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA).
Cell lines
The C57BL/6 syngeneic TC-1 tumor cell has been immortalized with HPV-16 E6 and E7 and transformed with the c-Ha-ras oncogene [7]. TC-1 tumor cells express low levels of E6 and E7 and are highly tumorigenic. TC-1 cells were grown in RPMI 1640 medium, 10% FCS, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 100 mM non-essential amino acids, 1 mM sodium pyruvate, 50 mM 2-ME at 37°C with 10% CO2.
Plasmid DNA constructs and preparation
To generate the LLO-E7-expressing plasmid (pcDNA3.1-LLO-E7), LLO-E7 DNA was amplified from plasmid pGG55, previously described [8], by PCR using primers designed to generate NheI and NotI restriction sites at the 5′ and 3′ ends of the amplified fragments, respectively. The amplified LLO-E7 DNA was then cloned into the unique NheI and NotI cloning sites of the pcDNA3.1(−) expression vector (Invitrogen, Carlsbad, CA) downstream of the cytomegalovirus promoter. A single plasmid expressing both LLO and E7 proteins (pcDNA3.1-LLO-IRES-E7) was generated from the LLO-E7-expressing parent plasmid by inserting a PCR-amplified IRES element into a unique XhoI site located between the LLO and E7 elements. The IRES element was amplified from MIGR1, generously provided by Dr. W. Pear (University of Pennsylvania). Similarly, a plasmid expressing LLO alone (pcDNA3.1-LLO) was generated from the same parent plasmid by excising the E7 element and inserting a linker encoding a stop codon. pcDNA-E7 was a gift from Dr. T-C. Wu (Department of Molecular Microbiology and Immunology, The Johns Hopkins Medical Institutes, Baltimore, MD). pcDNA-GM-CSF and pcDNA-MIP-1α were gifts from Dr. David Weiner, (Department of Pathology, University of Pennsylvania, Philadelphia, PA), respectively. Briefly, E7 DNA was cloned into the unique BamHI and HindIII cloning sites of the pcDNA3.1(−). Murine GM-CSF DNA was cloned into the unique EcoRI and XbaI cloning sites of the pcDNA3.1. Human MIP-1α DNA was cloned into the unique KpnI and BamHI cloning sites of the pcDNA3.1.
Plasmid LLO-E7 and E7 were purified by Puresin Inc. (Malvern, PA). Plasmid LLO-IRES-E7, LLO, GM-CSF and MIP-1α were purified using Qiagen plasmid mega kits (Qiagen Sciences, MD). DNA concentration was determined by the absorbance measured at 260 nm. The presence of the insert was confirmed by restriction enzyme digestion and gel electrophoresis. Fifty micrograms of each plasmid DNA was injected i.m..
Verification of LLO and E7 expression
Cell lysates and RNA were isolated from transiently transfected 293FT cells 48 h following the transfection. RT-PCR of actin, LLO and E7 using purified first strand synthesis product was performed. Western blot for LLO protein expression from transiently transfected 293FT cell lysates was probed with rabbit polyclonal sera that was custom made to peptide sequence 1–29 of LLO by AnaSpec Inc., San Jose, CA. The secondary antibody was HRP-conjugated goat anti-rabbit (Amersham Pharmacia Biotech, Little Chalfont, UK). Blots were developed with Amersham ECL detection reagents and exposed to Hyperfilm (Amersham Pharmacia Biotech).
Immunization protocol
C57BL/6 mice were immunized i.m. with 50 μg of each DNA plasmid used. A booster was given after 7 days. Splenocytes were harvested 7 or 9 days after the booster injection. Total splenocytes or splenocytes depleted of CD4+ T cells or CD8+ T cells using magnetic beads coated with anti-CD4 or anti-CD8 monoclonal antibodies (Miltenyi Biotec, Auburn, CA) were used in the experiments.
Flow cytometric analysis
C57BL/6 mice were immunized i.m. with E7 + GM-CSF + MIP-1α or LLO-E7 + GM-CSF + MIP-1α and boosted 7 days later. Three-color flow cytometry for CD8 (53–6.7, FITC conjugated), CD62 ligand (CD62L; MEL-14, APC conjugated), and E7 H-2Db tetramer-PE conjugated was performed using a FACSCalibur flow cytometer with CellQuest software (Becton Dickinson, Mountain View, CA). Splenocytes harvested 9 days after the boost were stained at room temperature with H-2Db tetramers loaded with the E7 peptide (RAHYNIVTF). Tetramers were provided by the National Institute of Allergy and Infectious Diseases Tetramer Core Facility. The tetramers were used at a 1/200 dilution. Cells were analyzed as described above comparing tetramer+, CD8+, CD62Llow cells generated by DNA immunization.
ELISPOT analysis
The 96-well filtration plates (Millipore, Bedford, MA) were coated with 15 μg/ml rat anti-mouse IFN-γ antibody (clone AN18, MABTECH, Mariemont, OH) in 100 μl of PBS. After overnight incubation at 4°C, the wells were washed and blocked with culture medium containing 10% fetal bovine serum. Splenocytes from each vaccinated mice group (1 × 105 or 2 × 104per well) were added to the wells along with E7 protein (5 μg/ml) or E7-specific H-2Db CTL epitope (5 μg/ml) plus IL-2 (5 U/ml). Cells were incubated at 37°C for 24 h. Then the plate was washed and followed by incubation with 1 μg/ml biotinylated IFN-γ antibody (clone R4-6A2, MABTECH, Mariemont, OH) in 100 μl PBS at 4°C overnight. After washing, 1:100 streptavidin-horseradish peroxidase in 100 μl PBS were added and incubated for 1 h at room temperature. Spots were developed by adding 100 μl of substrate after washing and incubated at room temperature for 15 min. Then color development was stopped by washing extensively in tap water. The spots were counted on an ELISPOT reader.
Measurement of tumor growth
Tumor growth was observed every 3 days with calipers spanning the shortest and longest surface diameters. The mean of these two measurements was calculated as the mean tumor diameter in millimeters. Tumors with a diameter of <3 mm could not be measured and were classified as a tumor-free size. Mice were sacrificed when the tumor diameter reached >20 mm. Data are shown as the percentage of mice tumor free for each vaccine group at various time points after tumor challenge.
Statistics
For Figs. 3 and 4, statistical significance was calculated by unpaired Student’s t test. For Tables 1 and 2, statistical significance between groups was calculated by Student’s t test for paired two samples. For both tests P values of <0.05 were considered significant.
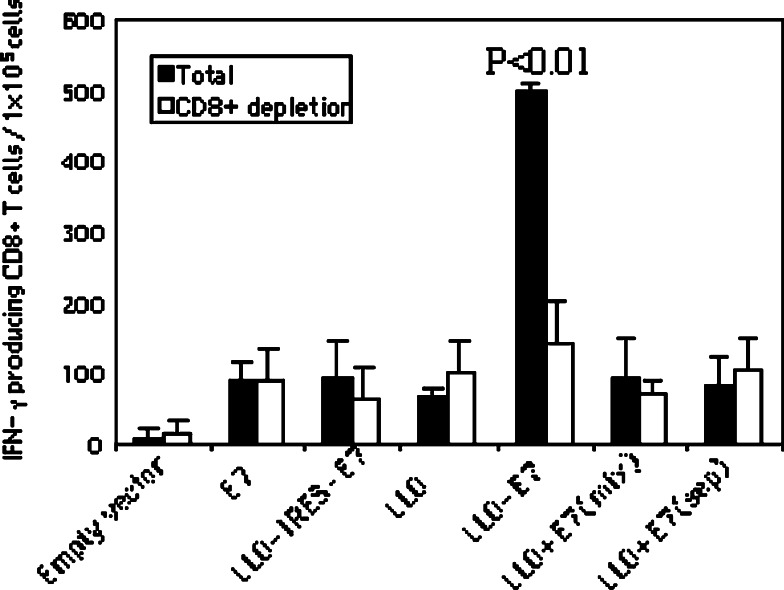
Fig. 3.
LLO-E7 induces E7-specific CD8+ T cell responses. Mice were immunized with 50 μg of E7 DNA vaccines i.m. as indicated in the figure. All vaccines were mixed with two cytokine plasmids encoding GM-CSF and MIP-1α before injection. Mice were boosted with the same vaccines 7 days after the primary injection. Splenocytes were harvested on day 7 after a booster injection. Either total splenocytes or CD8+ depleted splenocytes were cultured with the E7-specific, immunodominant, H-2Db CTL epitope and IL-2 (5 U/ml). The number of IFN-γ producing E7-specific CD8+ T cells was determined using the ELISPOT assay. The spot numbers were the mean of triplicates ± STD in each vaccinated group. The data presented in this figure are representative for two experiments with similar results
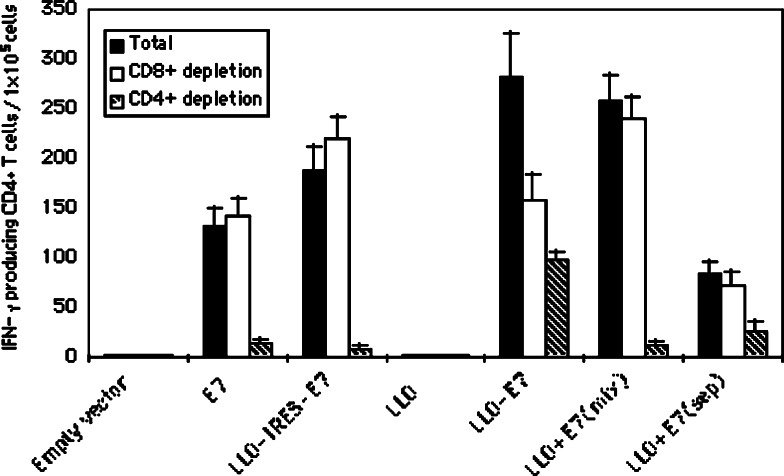
Fig. 4.
Fusion of LLO to E7 is not required to induce E7-specific CD4+ T cell responses. Mice were immunized with 50 μg of E7 DNA vaccines i.m. as indicated in the figure. All vaccines were mixed with two cytokine plasmids GM-CSF and MIP-1α before injection. Mice were boosted with the same vaccines 7 days after the primary injection. Splenocytes were harvested at day 7 after the booster injection. Either total splenocytes or CD8+ depleted splenocytes or CD4+ depleted splenocytes were cultured with E7 protein (5 μg/ml) plus IL-2 (5 U/ml). The number of IFN-γ producing E7-specific CD4+ T cells was determined using the ELISPOT assay. The spot numbers were the mean of triplicates ± STD in each vaccinated group. The data presented in this figure are representative for three experiments with similar results
Table 1.
Plasmids encoding GM-CSF and MIP-1α enhance E7-specific anti-tumor responses induced by LLO-E7
| Experiment 1 | |||||
|---|---|---|---|---|---|
| Days | Untreated | E7 | E7 + GM-CSF + MIP-1α | LLO-E7 | LLO-E7 + GM-CSF + MIP-1α |
| 14 | 0 | 0 | 13 | 13 | 75 |
| 19 | 0 | 0 | 0 | 13 | 63 |
| 26 | 0 | 0 | 0 | 13 | 63 |
| 33 | 0 | 0 | 0 | 13 | 63 |
| Experiment 2 | |||||
|---|---|---|---|---|---|
| Days | Untreated | GM-CSF + MIP-1α | LLO-E7 + GM-CSF | LLO-E7 + MIP-1α | LLO-E7 + GM-CSF + MIP-1α |
| 10 | 25 | 38 | 50 | 75 | 88 |
| 17 | 0 | 0 | 50 | 25 | 50 |
| 24 | 0 | 0 | 50 | 13 | 50 |
| 31 | 0 | 0 | 38 | 13 | 50 |
C57BL/6 mice (eight per group) received 2 × 104 TC-1 cells by s.c. injection on the left flank. Mice were treated on days 3 and 10 following tumor challenge with 50 μg of DNA as indicated in the table or were left untreated. The average tumor diameter was measured with calipers. Mice were sacrificed when tumor diameter reached approximately 2.0 cm. The data presented in this table are representative of two experiments with similar results. Values are percentage of mice TC-1 tumor free after treatment
Table 2.
LLO genetically fused to E7 induces stronger anti-tumor therapy against established TC-1 tumors than LLO and E7 separately
| Days post-tumor challenge | Untreated | Empty vector | E7 | LLO-E7 | LLO + E7 (sep) | LLO-IRES-E7 | LLO | LLO + E7 (mix) |
|---|---|---|---|---|---|---|---|---|
| Percentage of mice TC-1 tumor free after treatment | ||||||||
| 10 | 38 | 75 | 50 | 88 | 50 | 88 | 50 | 63 |
| 17 | 0 | 0 | 25 | 75 | 13 | 63 | 0 | 50 |
| 24 | 0 | 0 | 0 | 63 | 0 | 38 | 0 | 38 |
| 31 | 0 | 0 | 0 | 63 | 0 | 38 | 0 | 38 |
| Percentage of mice B16F10 tumor free after treatment | ||||||||
| 13 | 25 | 38 | ||||||
| 20 | 0 | 0 | ||||||
| 27 | 0 | 0 | ||||||
C57BL/6 mice (eight per group) received 2 × 104 TC-1 cells or B16F10 cells (as a control for specificity) by s.c. injection on the left flank. Mice were treated on days 3 and 10 following tumor challenge with 50 μg of DNA encoding for GM-CSF, MIP-1α and the test construct indicated in the table or were left untreated. The data shown in the table represent the percentage of tumor free mice for each group. Student’s t test for paired two samples was performed for the TC-1 tumor experiment on day 31 and for B16F10 tumor on day 20. The following groups were statistically different:
LLO-E7 versus LLO + E7 (mix), P < 0.05
LLO-E7 versus LLO-IRES-E7, P < 0.05
LLO-E7 versus LLO + E7 (sep), P < 0.01
LLO-E7 versus E7, P < 0.01
There was no statistical difference between TC-1 tumor size between the groups treated with LLO-IRES-E7 and LLO + E7 (mix) or between B16F10 tumor size between the untreated and the LLO-E7 treated groups
Results
Fusion of LLO to E7 and the addition of cytokine expressing plasmids enhance anti-tumor immunity
Previous Listeria-based vaccine constructs demonstrated variability of efficacy dependent upon whether the antigen was fused to LLO [8]. To compare the anti-tumor efficacy of E7 DNA and LLO-E7 DNA, tumor regression experiments were performed. Because cytokines have been shown to enhance T cell responses to DNA plasmid delivered antigens [15–18], we first examined E7 and LLO-E7 DNA vaccines with and without plasmids encoding GM-CSF plus MIP-1α. TC-1 cells were first injected into C57BL/6 mice s.c. at a dose of 2 × 104 cells/mouse in the left flank. Three days later, mice (n = 8) were injected i.m. with 50 μg of either E7 or LLO-E7 plasmid either with or without plasmids encoding GM-CSF plus MIP-1α. Mice were boosted with the same vaccines 7 days after priming. Mice immunized with the E7 or LLO-E7 plasmids alone did not show any anti-tumor response but co-administration of cytokine plasmids with the LLO-E7 DNA vaccine significantly augmented E7-specific anti-tumor responses (Table 1; experiment 1). We next examined whether both GM-CSF and MIP-1α were required to augment the efficacy of LLO-E7 DNA. Groups of mice (n = 8) were treated on days 3 and 10 after TC-1 challenge with LLO-E7 DNA plus either GM-CSF, or MIP-1α or a combination of the two cytokines or with the cytokines alone. As shown in Table 1, experiment 2, cytokines alone had no effect on tumor growth compared with the untreated group. The inclusion of GM-CSF was more efficacious than MIP-α. However, the use of both cytokines was the most effective vaccine strategy in that LLO-E7 DNA with two cytokines induced the complete regression of tumors in 63% of mice (five out of eight) in experiment 1 and 50% of mice (four out of eight) in experiment 2. Thus, for the remainder of these studies, mice were coinjected with E7 or LLO-E7 DNA vaccine mixed with the two cytokine plasmids (GM-CSF and MIP-1α).
Fusion of LLO to E7 induces E7-specific CD8+ T cells
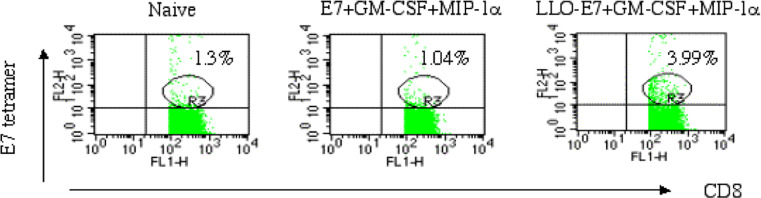
Next we compared the abilities of the two E7 DNA vaccines to induce E7-specific CD8+ T cells. Mice were immunized and boosted with the plasmid mixture E7 + cytokine expressing plasmids or LLO-E7 + cytokine expressing plasmids. Spleens were removed on day 9 after the second injection and splenocytes were stained with H-2Db tetramers loaded with the E7 peptide. As shown in Fig. 1, E7 plasmid did not induce any detectable tetramer-positive CD8+ T cells compared with naïve mice. However, the LLO-E7 plasmid induced 3.9% of tetramer-positive CD8+ T cells, which is consistent with its anti-tumor effect.
Fig. 1.
LLO-E7 enhances the induction of E7 tetramer+ CD8+ T cells. C57BL/6 mice were immunized and boosted 9 days later with E7 + GM-CSF + MIP-1α or LLO-E7 + GM-CSF + MIP-1α. Ex vivo splenocytes were stained with an H-2Db E7 tetramer, anti-CD8, and anti-CD62L. The population analyzed in the figure is CD8+CD62Llow. A Db/GP33 control tetramer was also included in the experiment. The percentage of Db/GP33 positive cells was less than 1% in all experimental groups (data not shown)
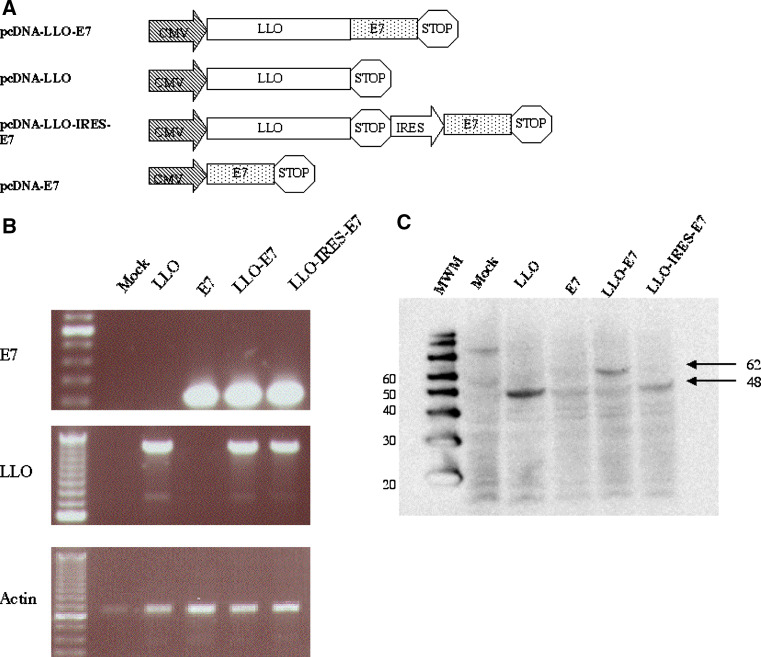
Construction of LLO-IRES-E7 DNA vaccine
We next examined whether fusion of E7 to LLO was required for maximum anti-tumor efficacy. pcDNA3.1 plasmids were constructed for the expression of either LLO alone or LLO and E7 from the same mRNA transcript by way of an internal ribosomal entry site (IRES). The composition of these vectors is outlined in Fig. 2a. Construction of the bicistronic LLO-IRES-E7 enables us to compare the efficacy of a vaccine where these two proteins are generated in a single cell to DNA vaccines where each are administered as a mixture to the same site, or separately to different sites entirely. The ability of these vectors to mediate transcription and translation of the appropriate DNA elements was tested by transient transfection into 293FT cells followed by RT-PCR and Western blotting. The data, shown in Fig. 2b, demonstrates the recovery of mRNA specific for these constructs. Figure 2c shows the expression of LLO proteins of appropriate molecular weight for each DNA vaccine. Unfortunately the commercially available anti-E7 monoclonal antibody could not detect the E7 protein in a Western blot. However, as we show below, the generation of E7 specific CD4+ T cell responses confirms E7 expression by the E7, LLO-E7 and LLO-IRES-E7 plasmids.
Fig. 2.
Construction of the bicistronic message LLO-IRES-E7 DNA vaccine. a Variations of the mammalian expression construct pcDNA3.1-LLO-E7 were used to elucidate the effect of LLO in tumor vaccines. These vectors all drive the eukaryotic expression of proteins under the control of the CMV promoter, except for pcDNA3.1-LLO-IRES-E7 that also uses the IRES element derived from the MigR1 vector to drive E7 expression. b RT-PCR demonstrates the expression of LLO and E7 from the expression plasmid. RNA was isolated from the lysate of transiently transfected 293 FT cells 48 h following the transfection. RT-PCR of actin, LLO and E7 mRNA was performed using purified first strand synthesis product. c Western blot demonstrates the expression of LLO protein from the expression plasmids. Western blot for LLO protein expression from transiently transfected 293FT cell lysates probed with a rabbit polyclonal anti-LLO antibody
Fusion of LLO to E7 induces stronger anti-tumor responses than LLO-IRES-E7 and a mixture of LLO and E7
To compare the anti-tumor effect of LLO-E7 and LLO-IRES-E7, mice (n = 8) were injected s.c. with TC-1 cells. Three days later, mice were injected i.m. with vaccines as indicated in Table 2. A booster was given 10 days after TC-1 challenge. As shown in Table 2, empty vector, E7, LLO, and LLO + E7 (two plasmids injected at separate sites) failed to induce tumor regression. In contrast, LLO-IRES-E7, similar to LLO + E7 (mixed and administered together), rendered three out of eight mice tumor-free (38%). However, LLO-E7 rendered five out of eight mice tumor-free (63%). These differences are reproducible and statistically significant (P < 0.05). To test for non-specific tumor effects we also examined the ability of the most effective vaccine in the TC-1 model (LLO-E7) to impact on the growth of a tumor that does not express the E7 antigen. We chose B16F10, since this is also a C57BL/6 derived tumor. As shown in Table 2, LLO-E7 had no ability to induce the regression of B16F10 tumors and they grew out at the same rate as unvaccinated mice (data not shown).
Fusion of LLO to E7 is required to induce E7-specific CD8+ T cell responses
CD8+ T cells play a crucial role in anti-tumor immunity. To compare CD8+ T cell responses induced by E7 DNA vaccines, mice were immunized with various forms of E7 DNA vaccines i.m. as indicated in Fig. 3. All vaccines were mixed with two cytokine plasmids, GM-CSF and MIP-1α, before injection. Mice were boosted with the same vaccines 7 days after the primary immunization. Splenocytes were harvested at day 7 after the booster injection. Either total splenocytes or CD8+ depleted splenocytes were cultured with the E7-specific H-2Db CTL epitope, RAHYNIVTF. The number of IFN-γ producing E7-specific T cells was determined using the ELISPOT assay. As shown in Fig. 3, E7, LLO-IRES-E7 or a mixture of LLO and E7 induced an insignificant number of IFN-γ-producing T cells compared to the mice immunized with just LLO. However, LLO-E7 fusion DNA dramatically enhanced IFN-γ-producing T cells (P < 0.01 compared to each of the other vaccine groups). When CD8+ cells were depleted by magnetic beads coated with anti-CD8 monoclonal antibodies, the number of CD8+ T cells secreting IFN-γ were profoundly diminished in the mice immunized with LLO-E7 but were unchanged in all of the other groups (Fig. 3). Thus the RAHYNIVTF stimulated splenocytes induced by LLO-E7, detected as E7-specific IFN-γ producing-cells in total splenocytes, were CD8+ T cells.
Fusion of LLO to E7 is not required to enhance E7-specific CD4+ T cell response
To determine E7-specific CD4+ T cell responses generated by these DNA vaccines, mice were immunized twice as indicated in Fig. 4. Splenocytes were harvested and ELISPOT analyses were performed after stimulating the cells with exogenous E7 protein. Figure 4 shows that empty vector and LLO did not induce any IFN-γ secreting cells. The mean numbers of E7-specific IFN-γ secreting-cells induced by E7 DNA and LLO + E7 injected at separate sites were 575 (±62) and 396 (±31) SFC per 105 spleen cells, respectively, which were not statistically different but were increased compared to the empty vector (P < 0.01). However, LLO-IRES-E7, LLO + E7 (injected as mixed plasmids) and LLO-E7 significantly enhanced E7-specific IFN-γ secreting-cells compared to E7 DNA alone or LLO + E7 injected at separate sites (P < 0.05). A significantly increased level of IFN-γ secreting T cells was induced by LLO-E7 plasmid compared to LLO-IRES-E7, LLO + E7 (injected as mixed plasmids) (P < 0.05). However, when CD8+ cells were depleted by magnetic beads coated with anti-CD8 monoclonal antibodies, the number of IFN-γ secreting-cells did not differ between the groups of mice immunized with LLO + E7 (injected separately), LLO + E7 (injected as a mixture) or LLO-IRES-E7. However, there was a reduced response in the group of mice immunized with LLO-E7, which suggests that CD8+ T cells in LLO-E7 immunized mice also produced IFN-γ in response to E7 protein stimulation, probably through cross-presentation by professional antigen presenting cells present in the splenocytes. When CD4+ cells were depleted by magnetic beads coated with anti-CD4+ monoclonal antibodies, the number of IFN-γ secreting-cells produced by each vaccine group, except the LLO-E7 group, decreased to the same levels as the empty vector group. The responses in the group of mice immunized with LLO-E7 decreased to a level commensurate with the residual CD8 response (Fig. 4). These data indicate that most of the E7-specific IFN-γ secreting-cells were CD4+ T cells. Taken together, our data suggest that LLO augments E7-specifc CD4+ T cell responses either in the form of bicistronic message or as a mixture of E7 and LLO or LLO-E7 fusion DNA.
Fusion of LLO to E7 does not induce an E7-specific antibody response
To investigate the ability of the vaccines to induce an HPV-16 E7-specific humoral response, mice were immunized twice with empty vector, E7 or LLO-E7 DNA together with plasmid GM-CSF and MIP-1α. Blood was collected at day 14 after the second injection. Anti-E7 ELISA was performed by coating E7 protein on 96-well plates. No anti-E7 IgG could be detected in the sera of any vaccinated mice (data not shown). The positive control (a commercial anti-E7 monoclonal antibody) was well detected by the ELISA. This suggests that CD4+ T cells induced by LLO-E7 are CD4+ Th1 cells.
Discussion
In this study, we demonstrate that LLO can dramatically enhance the potency of HPV-16 E7-expressing DNA vaccines in a vaccine strategy that included plasmids encoding GM-CSF and MIP-1α. LLO acts as an adjuvant to augment E7-specific CD4+ T cell responses. However, fusion of LLO to E7 is required to induce E7-specific CD8+ T cell responses. Mice immunized with LLO-E7 DNA showed the greatest anti-tumor response in vivo, consistent with its ability to induce both improved E7-specific CD4+ and CD8+ T cell responses.
We previously developed two L. monocytogenes based cancer vaccines, Lm-E7 and Lm-LLO-E7, that induce immunity to the HPV-16 oncoprotein E7 [8, 12]. Lm-E7 secretes recombinant protein E7, but Lm-LLO-E7 secretes a fusion protein consisting of a truncated, non-membrane-active LLO joined at the C-terminus to E7 [8]. Although both L. monocytogenes recombinants secrete the E7 tumor antigen, they induce radically different anti-tumor responses in vivo. Previous work from our laboratory has shown that Lm-LLO-E7 could induce complete regression of established TC-1 tumors in syngeneic mice, whereas Lm-E7 only slowed the growth of such tumors [8, 12]. Furthermore, Lm-LLO-E7 is more effective than Lm-E7 at inducing DC maturation [10]. Therefore, the difference in anti-tumor efficacy of the two vaccines may be due to the ability of LLO to render immature DCs effective antigen presenting cells.
To address the question of how LLO mediates vaccine efficacy, we constructed eukaryotic expression plasmids for use as DNA vaccines. DNA vaccines, rather than recombinant Listeria, permits the isolation of specific factors controlling the immune response without the background of endogenously expressed Listeria genes. DNA vaccines have inherently low immunogenicity, however several strategies have been developed to enhance the potency of DNA vaccines, including some that improve MHC class I and class II presentation of the antigen, and others aimed at prolonging DC life by targeting inhibitors of apoptosis such as Bcl-xl [14, 20–25]. One method for overcoming this obstacle is to formulate DNA vaccines together with DNA expressing cytokines shown to enhance their efficacy. Here, we demonstrate that the Listeria protein, LLO, also has adjuvant properties that increase the efficacy of associated vaccines.
It has been shown that DNA vaccines targeting E7 to subcellular compartments, using proteins such as Mycobacterium tuberculosis heat-shock protein 70 (HSP70), calreticulin (CRT), or the sorting signal of the lysosome-associated membrane protein 1 (LAMP-1), enhances DNA vaccine potency [21, 24, 25]. This study demonstrates that fusion of LLO to E7 also augments the efficacy of a DNA vaccine against HPV-16 E7. In addition we show that the inclusion of LLO in the LLO-IRES-E7 vaccine or administering it as a mixture of plasmids in the same injection site as plasmid E7, enhances E7-specific IFN-γ secreting CD4+ T cells. However, when E7 and LLO DNA were injected into opposite leg muscles, LLO did not enhance either CD4+ or CD8+ T cell responses. We, thus hypothesize that LLO improves MHC class II antigen presentation by upregulating co-stimulatory molecules and MHC.
In contrast to our findings of the CD4+ T cell response to the LLO plus E7 plasmid vaccines, only LLO genetically fused to E7 (LLO-E7) increased E7-specific IFN-γ secreting CD8+ T cells. In addition, fusion of LLO to E7 appears to result in more efficient priming of CD8+ T cells in response to exogenous E7 protein (Fig. 3). A possible reason may be the presence of a 19-amino acid sequence within LLO called a PEST sequence. PEST regions (P, proline; E, glutamic acid; S, serine; T, threonine) are hydrophilic amino acid sequences that reside near the NH2 or COOH termini of certain enzymes [26, 27]. These sequences are thought to target proteins for rapid degradation by the cellular proteasome. It has been shown that the PEST region of LLO is vital for the survival of Listeria in the host possibly because it causes the rapid degradation of LLO itself before it damages the host cell’s plasma membrane [26]. Similar data demonstrating that the PEST sequence induces proteolysis of LLO supports this conclusion [27]. On the basis of these data, we have explored the role of the PEST sequence in the immune enhancing properties of LLO. We found that the inclusion of the PEST element of LLO in a fusion with E7 protein in a live Listeria vaccine results in increased efficiency in clearing TC-1 tumors compared to vaccines consisting of E7 alone or a fusion protein that lacks the PEST domain [12]. Thus the presence of the PEST region in LLO in our DNA vaccine may enhance its efficacy by causing rapid degradation and presentation of the tumor antigen by antigen-presenting cells.
Mice immunized even with LLO-E7 DNA, the most effective vaccine, exhibited rather poor anti-tumor immunity in the absence of plasmids encoding cytokines MIP-1α and GM-CSF (Table 1). It has been previously shown that these plasmid cytokines can augment the immunogenicity of plasmid DNA vaccines [18–21]. In our study the inclusion of both GM-CSF and MIP-1α plasmids in the vaccine protocol confirmed that co-injection of these cytokine plasmids together with a plasmid encoding the antigen DNA induced stronger antigen-specific immune responses.
In summary, our results indicate that LLO acts as an adjuvant to enhance E7-specific CD4+ T cell responses but that fusion of LLO to E7 is required to generate stronger E7-specific CD8+ T cell responses and effective anti-tumor immunity. These data, together with our previous observations of DC maturation in response to vaccines including fusion with LLO suggest this may be a valuable component of an effective DNA vaccine.
Acknowledgments
This work was supported by grant number CA69632 from the National Institute of Health and American Cancer Society grant number TURSG LIB-01-168-01. Yvonne Paterson wishes to disclose that she has a financial interest in Advaxis Inc., a vaccine and therapeutic company that has licensed or has an option to license all patents from the University of Pennsylvania that concern the use of Listeria or listerial products as vaccines.
Abbreviations
- LLO
Listeriolysin O
- GM-CSF
Granulocytes macrophage-colony stimulating factor
- MIP-1α
Macrophage inflammatory protein-1α
- DC
Dendritic cells
References
- 1.Hoption Cann SA, van Netten JP, van Netten C. Dr. William Coley and tumour regression: a place in history or in the future. Postgrad Med J. 2003;79:672. [PMC free article] [PubMed] [Google Scholar]
- 2.Pamer EG. Immune responses to Listeria monocytogenes . Nat Rev Immunol. 2004;4:812. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 3.Pan ZK, Weiskirch LM, Paterson Y. Regression of established B16F10 melanoma with a recombinant Listeria monocytogenes vaccine. Cancer Res. 1999;59:5264. [PubMed] [Google Scholar]
- 4.Yoshimura K, Jain A, Allen HE, Laird LS, Chia CY, Ravi S, Brockstedt DG, Giedlin MA, Bahjat KS, Leong ML, Slansky JE, Cook DN, Dubensky TW, Pardoll DM, Schulick RD. Selective targeting of antitumor immune responses with engineered live-attenuated Listeria monocytogenes . Cancer Res. 2006;66:1096. doi: 10.1158/0008-5472.CAN-05-2307. [DOI] [PubMed] [Google Scholar]
- 5.Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) study group. J Natl Cancer Inst. 1995;87:796. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 6.Choo CK, Rorke EA, Eckert RL. Differentiation-independent constitutive expression of the human papillomavirus type 16 E6 and E7 oncogenes in the CaSki cervical tumour cell line. J Gen Virol. 1994;75:1139. doi: 10.1099/0022-1317-75-5-1139. [DOI] [PubMed] [Google Scholar]
- 7.Lin KY, Guarnieri FG, Staveley-O’Carroll KF, Levitsky HI, August JT, Pardoll DM, Wu TC. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56:21. [PubMed] [Google Scholar]
- 8.Gunn GR, Zubair A, Peters C, Pan ZK, Wu TC, Paterson Y. Two Listeria monocytogenes vaccine vectors that express different molecular forms of human papilloma virus-16 (HPV-16) E7 induce qualitatively different T cell immunity that correlates with their ability to induce regression of established tumors immortalized by HPV-16. J Immunol. 2001;167:6471. doi: 10.4049/jimmunol.167.11.6471. [DOI] [PubMed] [Google Scholar]
- 9.Lamikanra A, Pan ZK, Isaacs SN, Wu TC, Paterson Y. Regression of established human papillomavirus type 16 (HPV-16) immortalized tumors in vivo by vaccinia viruses expressing different forms of HPV-16 E7 correlates with enhanced CD8(+) T-cell responses that home to the tumor site. J Virol. 2001;75:9654. doi: 10.1128/JVI.75.20.9654-9664.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng X, Hussain SF, Paterson Y. The ability of two Listeria monocytogenes vaccines targeting human papillomavirus-16 E7 to induce an antitumor response correlates with myeloid dendritic cell function. J Immunol. 2004;172:6030. doi: 10.4049/jimmunol.172.10.6030. [DOI] [PubMed] [Google Scholar]
- 11.Hussain SF, Paterson Y. What is needed for effective anti-tumor immunotherapy? Lessons learned using Listeria monocytogenes as a live vector for HPV associated tumors. Cancer Immunol Immunother. 2005;54:577. doi: 10.1007/s00262-004-0600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sewell DA, Shahabi V, Gunn GR, Pan ZK, Dominiecki ME, Paterson Y. Recombinant Listeria vaccines containing PEST sequences are potent immune adjuvants for the tumor-associated antigen human papillomavirus-16 E7. Cancer Res. 2004;64:8821. doi: 10.1158/0008-5472.CAN-04-1958. [DOI] [PubMed] [Google Scholar]
- 13.Nagata T, Aoshi T, Uchijima M, Suzuki M, Koide Y. Cytotoxic T-lymphocyte-, and helper T-lymphocyte-oriented DNA vaccination. DNA Cell Biol. 2004;23:93. doi: 10.1089/104454904322759902. [DOI] [PubMed] [Google Scholar]
- 14.Donnelly JJ, Ulmer JB, Shiver JW, Liu MA. DNA vaccines. Annu Rev Immunol. 1997;15:617. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 15.Ou-Yang P, Hwang LH, Tao MH, Chiang BL, Chen DS. Co-delivery of GM-CSF gene enhances the immune responses of hepatitis C viral core protein-expressing DNA vaccine: role of dendritic cells. J Med Virol. 2001;66:320. doi: 10.1002/jmv.2148. [DOI] [PubMed] [Google Scholar]
- 16.Barouch DH, Santra S, Tenner-Racz K, Racz P, Kuroda MJ, Schmitz JE, Jackson SS, Lifton MA, Freed DC, Perry HC, Davies ME, Shiver JW, Letvin NL. Potent CD4+ T cell responses elicited by a bicistronic HIV-1 DNA vaccine expressing gp120 and GM-CSF. . J Immunol. 2002;168:562. doi: 10.4049/jimmunol.168.2.562. [DOI] [PubMed] [Google Scholar]
- 17.Haddad D, Ramprakash J, Sedegah M, Charoenvit Y, Baumgartner R, Kumar S, Hoffman SL, Weiss WR. Plasmid vaccine expressing granulocyte-macrophage colony stimulating factor attracts infiltrates including immature dendritic cells into injected muscles. J Immunol. 2000;165:3772. doi: 10.4049/jimmunol.165.7.3772. [DOI] [PubMed] [Google Scholar]
- 18.McKay PF, Barouch DH, Santra S, Sumida SM, Jackson SS, Gorgone DA, Lifton MA, Letvin NL. Recruitment of different subsets of antigen-presenting cells selectively modulates DNA vaccine-elicited CD4+ and CD8+ T lymphocyte responses. Eur J Immunol. 2004;34:1011. doi: 10.1002/eji.200324840. [DOI] [PubMed] [Google Scholar]
- 19.Choo AY, Choo DK, Kim JJ, Weiner DB. DNA vaccination in immunotherapy of cancer. Cancer Treat Res. 2005;123:137. doi: 10.1007/0-387-27545-2_6. [DOI] [PubMed] [Google Scholar]
- 20.Kim TW, Lee JH, He L, Boyd DA, Hung CF, Wu TC. DNA vaccines employing intracellular targeting strategies and a strategy to prolong dendritic cell life generate a higher number of CD8+ memory T cells and better long-term antitumor effects compared with a DNA prime-vaccinia boost regimen. Hum Gene Ther. 2005;16:26. doi: 10.1089/hum.2005.16.26. [DOI] [PubMed] [Google Scholar]
- 21.Kim TW, Hung CF, Juang J, He L, Kim TW, Armstrong DK, Pai SI, Chen PJ, Lin CT, Boyd DA, Wu TC. Comparison of HPV DNA vaccines employing intracellular targeting strategies. Gene Ther. 2004;11:1011. doi: 10.1038/sj.gt.3302252. [DOI] [PubMed] [Google Scholar]
- 22.Kim TW, Hung CF, Boyd DA, He L, Lin CT, Kaiserman D, Bird PI, Wu TC. Enhancement of DNA vaccine potency by co-administration of a tumor antigen gene and DNA encoding serine protease inhibitor-6. Cancer Res. 2004;64:400. doi: 10.1158/0008-5472.CAN-03-1475. [DOI] [PubMed] [Google Scholar]
- 23.Kim TW, Hung CF, Boyd D, Juang J, He L, Kim JW, Hardwick JM, Wu TC. Enhancing DNA vaccine potency by combining a strategy to prolong dendritic cell life with intracellular targeting strategies. J Immunol. 2003;171:2970. doi: 10.4049/jimmunol.171.6.2970. [DOI] [PubMed] [Google Scholar]
- 24.Cheng WF, Hung CF, Chai CY, Hsu KF, He L, Ling M, Wu TC. Tumor-specific immunity and antiangiogenesis generated by a DNA vaccine encoding calreticulin linked to a tumor antigen. J Clin Invest. 2001;108:669. doi: 10.1172/JCI200112346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CH, Wang TL, Hung CF, Yang Y, Young RA, Pardoll DM, Wu TC. Enhancement of DNA vaccine potency by linkage of antigen gene to an HSP70 gene. Cancer Res. 2000;60:1035. [PubMed] [Google Scholar]
- 26.Decatur AL, Portnoy DA. A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science. 2000;290:992. doi: 10.1126/science.290.5493.992. [DOI] [PubMed] [Google Scholar]
- 27.Lety MA, Frehel C, Dubail I, Beretti JL, Kayal S, Berche P, Charbit A. Identification of a PEST-like motif in listeriolysin O required for phagosomal escape and for virulence in Listeria monocytogenes . Mol Microbiol. 2001;39:1124. doi: 10.1111/j.1365-2958.2001.02281.x. [DOI] [PubMed] [Google Scholar]