Abstract
In neurons, entry of extracellular calcium (Ca2+) into synaptic terminals through Cav2.1 (P/Q-type) Ca2+ channels is the driving force for exocytosis of neurotransmitter-containing synaptic vesicles. This class of Ca2+ channel is, therefore, pivotal during normal neurotransmission in higher organisms. In response to channel opening and Ca2+ influx, specific Ca2+-binding proteins associate with cytoplasmic regulatory domains of the P/Q channel to modulate subsequent channel opening. Channel modulation in this way influences synaptic plasticity with consequences for higher-level processes such as learning and memory acquisition. The ubiquitous Ca2+-sensing protein calmodulin (CaM) regulates the activity of all types of mammalian voltage-gated Ca2+ channels, including the P/Q class, by direct binding to specific regulatory motifs. More recently, experimental evidence has highlighted a role for additional Ca2+-binding proteins, particularly of the CaBP and NCS families in the regulation of P/Q channels. NCS-1 is a protein found from yeast to humans and that regulates a diverse number of cellular functions. Physiological and genetic evidence indicates that NCS-1 regulates P/Q channel activity, including calcium-dependent facilitation, although a direct physical association between the proteins has yet to be demonstrated. In this study, we aimed to determine if there is a direct interaction between NCS-1 and the C-terminal cytoplasmic tail of the Cav2.1 α-subunit. Using distinct but complementary approaches, including in vitro binding of bacterially expressed recombinant proteins, fluorescence spectrophotometry, isothermal titration calorimetry, nuclear magnetic resonance, and expression of fluorescently tagged proteins in mammalian cells, we show direct binding and demonstrate that CaM can compete for it. We speculate about how NCS-1/Cav2.1 association might add to the complexity of calcium channel regulation mediated by other known calcium-sensing proteins and how this might help to fine-tune neurotransmission in the mammalian central nervous system.
Cav2.1 P/Q-type channels are responsible for the entry of Ca2+ into synaptic terminals in many brain regions.1 P/Q-type function is therefore pivotal in the regulation of neurotransmitter release and communication throughout the central nervous system (CNS). Indeed, mutations in the gene encoding the human P/Q α1 subunit are responsible for a number of clinically relevant neurological disorders.2 An interesting feature of many Cav proteins (Cav1.x and Cav2.x), including the P/Q channel, is that channel activity is itself regulated by Ca2+.3 Ca2+-dependent channel modulation manifests as two distinct phenomena termed Ca2+-dependent inactivation (CDI)4 and Ca2+-dependent facilitation (CDF).5 CDI represents a mechanism by which channels become refractory to further opening in the presence of a sustained stimuli, whereas CDF manifests as the opposite process whereby Ca2+ augments channel currents.6 Specific Ca2+-binding proteins can respond to the entry of Ca2+ through Cav channels by interacting directly with regulatory motifs present in the intracellular domains of the α-subunit.7−10 These interactions, by currently poorly understood mechanisms, alter channel behavior to elicit either CDI or CDF. CDI and CDF both have a direct impact on synaptic neurotransmitter release, the strength of synaptic signaling, and ultimately synaptic plasticity. Understanding the basic modes of Cav regulation at the molecular level therefore generates insights into more abstract, higher-level, processes, including learning, memory acquisition, and reasoning.
Of the characterized Ca2+-binding proteins that control Cav activity, the ubiquitous small EF-hand-containing protein calmodulin (CaM) has been most intensively studied. CaM has been shown to interact with a consensus isoleucine-glutamine “IQ” motif present in the carboxy terminus of the channel, although the precise details of the interaction differ for Cav1.x and Cav2.x subtypes.11,12 A second Ca2+-dependent CaM-binding motif immediately C-terminal to the IQ domain has also been characterized in Cav2.1 P/Q-type channels.13 This CaM-binding domain (CBD) has been found to influence P/Q channel activity in some studies13 but not others,7 and its true significance remains somewhat controversial.
A number of CaM-related Ca2+-binding proteins are enriched in the mammalian CNS and have been found to modulate the properties of Cav channels. In all instances thus far characterized, channel regulation is distinct from that observed with CaM, suggesting nonredundant functions that have been thought to permit complex modes of channel regulation and neuronal signaling.12 At present, two CaM-related Ca2+-binding proteins, Ca2+-binding protein-1 (CaBP1) and visinin-like protein-2 (VILIP-2), have been shown to directly interact with P/Q-type α-subunits to exert unique regulatory outcomes.10,14 Large numbers of additional CaM-related small EF-hand-containing proteins are also expressed in the mammalian CNS. Neuronal Ca2+ sensor-1 (NCS-1) is a CaM-related protein that is evolutionarily conserved from yeast to humans15 and has been implicated in specific human neuronal disorders.16−19 Evidence of a regulatory role of NCS-1 on P/Q-type channels has been reported on the basis of physiological experiments in mammalian cells, including an effect on CDF,20−22 and a genetic study in Drosophila,23 although no direct interaction between the two proteins has thus far been demonstrated.
In this paper, we aimed to use exclusive yet complementary analytical methods to rigorously determine if there is a direct interaction between NCS-1 and the α-subunit of the P/Q-type channel and to provide the first evidence of an interaction between a defined region of the C-terminal domain of the P/Q α-subunit and NCS-1. We show that a segment of the P/Q α-subunit encoding the IQ motif and CBD interacts with NCS-1 in a Ca2+-dependent manner. Using mutagenesis and NMR experiments, we further refine the NCS-1-binding site to the IQ motif and show that the interaction is competitive with CaM. We provide further evidence using an ex vivo HeLa cell model that NCS-1 and the C-terminal tail of Cav2.1 interact. These novel observations expand the repertoire of P/Q-type interacting proteins and add to the potential variety of modes by which such channels can be controlled.
Experimental Procedures
Cloning
DNA constructs and molecular biology GST-NCS-124 and NCS-1-mCherry18 plasmids were as previously described. The coding sequence for human CaM was amplified by polymerase chain reaction (PCR) from oligo-dT(15)-primed HeLa cell RNA and reverse-transcribed first-strand cDNA template with the following primer pair (based on GenBank accession number M19311) containing restriction endonuclease sites (underlined) for subcloning into N-terminal GST tag vector pGex6P-1 (Amersham Biosciences): sense (BamHI), 5′-ATATGGATCCATGGCTGACCAACTGACTG-3′; antisense (XhoI), 5′-ATATCTCGAGTCACTTTGCTGTCATCATTTG-3′. GST-P/Q, encoding residues 1898–2035 of the rat P/Q channel α1 subunit (GenBank accession number NM_012918) was amplified by PCR from a rat brain single-strand cDNA template with the following primer pair containing restriction endonuclease sites (underlined) for subcloning into pGex6P-1: sense (BamHI), 5′-ATATGGATCCAAGTCCACGGACCTGACAGTG-3′; antisense (XhoI), 5′-ATATCTCGAGCTAGGGGAGGTAGTGTTCGCTGTC-3′. GST-CBD (residues 1950–2035) of the rat P/Q channel α1 subunit was amplified by PCR from the GST-P/Q template using the P/Q reverse primer and a sense primer containing a BamHI site (underlined) for subsequent subcloning into pGex6P-1 (5′-ATATGGATCCGAGGGAGGACCCAGCCAAAAC-3′). GST-P/QIM→EE was generated by site-directed mutagenesis of the GST-P/Q plasmid using the QuikChange reagents and protocol (Agilent Technologies) and the following primer pair: sense, 5′-AAGATCTACGCAGCCATGATGGAGGAGGAGTACTACCGGCAGAGCAAG-3′; antisense, 5′-CTTGCTCTGCCGGTAGTACTCCTCCTCCAGCAGGGCTGCGTAGATCTT-3′.
SUMO-tagged residues 1909–2035 of the rat P/Q channel α1 subunit were amplified by PCR from the GST-P/Q template encoding residues 1898–2035 with the following primer pair for subcloning into the pOPINS vector: sense, 5′-GCGAACAGATCGGTGGTGCAGCCATGATGATCATGGAG-3′; antisense, 5-ATGGTCTAGAAAGCTTTAGGGGAGGTAGTGTTCGCTGT-3′. Ligation of the PCR product and KpnI/HindIII-digested vector employed the infusion reaction method (Clontech) according to the manufacturer’s protocol.
Myristoylated PQ (residues 1898–2035, myrPQ-YFP) was constructed as follows. A DNA fragment encoding the myristoylation sequence taken from ref (25) was inserted between the XhoI and HindIII sites of pEYFP-N1 (Clontech) to generate pmyrEYFP-N1. PQ1898–2035 was amplified by PCR from the existing pGex template with the following primer pair containing restriction endonuclease sites (underlined) for subsequent subcloning into pmyrEYFP-N1: sense (HindIII), 5′-ATAT AAG CTT AAG TCC ACG GAC CTG ACA GTG-3′; antisense (SacII), 5′-ATAT CCG CGG GGG GAG GTA GTG TTC GCT GTC-3′. All constructs were verified by dideoxy sequencing (The Sequencing Service, University of Dundee, Dundee, U.K.).
Cell Culture and Plasmid Transfections
HeLa cells were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum, 1% (v/v) penicillin/streptomycin, and 1% (v/v) nonessential amino acids. All cells were maintained in a humidified 95% air/5% CO2 atmosphere at 37 °C. Cells were plated onto sterile 13 mm round coverslips at a density of 0.25 × 106 cells/well. After 24 h, cells were transiently transfected with the indicated expression vectors using GeneJuice transfection reagent (Novagen) according to the manufacturer’s protocol. For single and double transfections, 1 μg of each plasmid was used.
Cell Fixation and Confocal Imaging Analysis
Twenty-four hours post-transfection, cells on coverslips were washed with phosphate-buffered saline [137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, and 2 mM NaH2PO4 (pH 7.4)] and then fixed with 4% (v/v) formaldehyde in PBS for 6 min at room temperature. Coverslips were subsequently air-dried and mounted onto microscope slides using Prolong antifade glycerol (Life Technologies). Fixed cells were imaged using a Leica TCS-SP2 confocal system (Leica Microsystems, Heidelberg, Germany) with a pinhole set to 1 Airy unit and a 63× oil immersion objective with a numerical aperture of 1.3. Images were exported as TIFF files and compiled, processed, and analyzed with ImageJ and CorelDraw X6 applications.
In Vitro Binding Assays
For the In vitro protein binding assays, 5 μM GST fusion protein (GST-P/Q, GST-P/QIM→EE, or GST-CBD) was immobilized onto 30 μL of glutathione-agarose resin (Thermo Scientific) that had been prewashed in binding buffer (BB) [150 mM KCl, 20 mM HEPES (pH 7.4), 10% (v/v) glycerol, 5 mM NTA, 5 mM EGTA, 1 mM DTT, and 0.1% (v/v) NP-40] or BB supplemented with 1 μM free Ca2+ (BB+Ca2+) by incubation for 30 min with constant agitation at 4 °C. GST-free NCS-1 and CaM were added to samples at concentrations of 5 μM (total final binding assay volumes of 100 μL) and incubations continued for 1 h with constant agitation at 4 °C. For competition binding assays, 5 μM GST-P/Q was prebound to glutathione-agarose resin in BB+Ca2+ by incubation for 30 min at 4 °C with constant agitation; 5 μM NCS-1 was then added to samples in the presence of increasing concentrations of CaM ranging from 0 to 10 μM. Binding reactions were continued for 1 h at 4 °C while the mixtures were constantly agitated. For the Ca2+ dose dependency of the NCS-1/GST-P/Q interaction, 1 μM GST-P/Q was incubated with 1 μM NCS-1 in the presence of increasing concentrations of free Ca2+. For all binding assays, glutathione-agarose pellets were collected by centrifugation (3000 rpm for 1 min at 4 °C) and washed three times with 1 mL of BB or BB+Ca2+ and bound proteins were extracted by boiling of final bead pellets for 5 min in 50 μL of SDS dissociation buffer [125 mM HEPES (pH 6.8), 10% (w/v) sucrose, 10% (v/v) glycerol, 4% (w/v) SDS, 1% (v/v) β-mercaptoethanol, and 2 mM EDTA]. Proteins were resolved on 4 to 12% Tris-glycine gradient gels (Novex, Life Technologies) and transferred to nitrocellulose membranes for Western blotting by transverse electrophoresis.
Western Blots
Nitrocellulose filters were blocked by incubation in a blocking solution [3% (w/v) skim milk powder in PBS] for 1 h at room temperature. Filters were subsequently incubated with a primary antibody [rabbit anti-NCS-1 (1:100026) or rabbit anti-calmodulin (1:500) (AbCam)] diluted in a blocking solution overnight at 4 °C with constant agitation. Filters were washed three times in PBS supplemented with 0.05% (v/v) Tween 20 (PBST) and twice with PBS before being incubated with a HRP-conjugated species specific secondary antibody (1:400, anti-rabbit HRP, Sigma) in a blocking solution for 1 h at room temperature. Filters were washed three times with PBST and twice with PBS prior to application of ECL reagents and visualization of immunoreactivity using a Chemidoc automated gel/blot documentation system (Bio-Rad). Densitometry analysis of developed Western blots was performed using Quantity-1 (Bio-Rad). For the Ca2+ dose dependency of the NCS-1/GST-P/Q interaction, densitometry data were analyzed by applying nonlinear curve fitting with OriginPro8 (OriginLab).
Synthetic Peptide
The peptide used corresponds to residues 1903–1929 of the human P/Q receptor. The synthetic P/Q peptide, TVGKIYAAMMIMEYYRQSKAKKLQAMR (hereafter termed the PQIQ peptide), was purchased from GenicBio. The peptide was delivered >95% pure.
Protein Expression and Purification
NCS-1 was expressed in Escherichia coli strain BL21(DE3) (Novagen) and purified as previously described.27 Expression was induced overnight at 18 °C; cells were harvested, resuspended into lysis buffer [50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5 mM CaCl2, and Complete EDTA Free Protease Inhibitor (Roche Applied Science)], lysed, centrifuged, filtered, and loaded onto the Hiprep 16/10 Phenyl FF High Sub (GE Healthcare) column that was pre-equilibrated with buffer A [50 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 5 mM CaCl2]. The column was extensively washed with buffer A and NCS-1 eluted using Milli-Q water. The eluted protein was buffer exchanged into 50 mM Tris (pH 7.4) and 500 mM NaCl, and the N-terminal His tag was removed by incubating the protein overnight at 4 °C using TEV protease (1:20 TEV protease:NCS-1 molar ratio). His-tagged uncleaved NCS-1 and TEV protease were separated for cleaved NCS-1 using a HisTrap FF 5 mL affinity column (GE Healthcare). The cleaved NCS-1 was further purified using a Superdex 75 Hiload 26/60 (Amersham Biosciences) size-exclusion column [50 mM Tris-HCl (pH 7.5) and 150 mM NaCl]. The purity of the sample was assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and deemed to be >95% pure. The eluted peak was concentrated and stored at −80 °C.
Calmodulin was cloned into a pET-15b (Novagen) vector (gift from A. Kitmitto, University of Manchester, Manchester, U.K.) and expressed in E. coli strain BL21(DE3) (Novagen) at 37 °C. Harvested cells were resuspended in lysis buffer [50 mM Tris (pH 7.5) and 2 mM DTT], disrupted using a French press (Sim Aminco) at 1000 psi, and centrifuged, and the supernatant was recovered, CaCl2 added to a final concentration of 2 mM, and the protein solution loaded onto a 20 mL HiPrep phenyl sepharose hydrophobic column (GE Healthcare), pre-equilibrated with CaM buffer A [50 mM Tris (pH 7.5), 200 mM NaCl, 2 mM CaCl2, and 0.5 mM DTT]. The column was washed with CaM buffer A, followed by buffer B [50 mM Tris (pH 7.5), 0.5 mM CaCl2, and 0.5 mM DTT], and pure calmodulin eluted with CaM buffer C [50 mM Tris (pH 7.5), 1 mM EGTA, and 0.5 mM DTT]. The protein was further purified using a 26/60 Superdex 75 (GE Healthcare) size-exclusion column pre-equilibrated with CaM gel filtration (GF) buffer [50 mM Tris (pH 7.5), 200 mM NaCl, 10 mM CaCl2, and 0.5 mM DTT]. The sample was loaded before isocratic elution. The purity of the sample was assessed by SDS–PAGE and deemed to be >95% pure. CaM was dialyzed against water and lyophilized before being stored at −20 °C.
15N-labeled SUMO-tagged PQIQ 1909–2035 was expressed in E. coli BL21(DE3) in 2M9 medium with overnight incubation 18 °C, and cells were harvested and resuspended in lysis buffer [50 mM Tris-HCl (pH 7.4) and 500 mM NaCl] supplemented with Compete EDTA Free Protease Inhibitor (Roche Applied Science). The cells were lysed; 250 μg of bovine pancreas deoxyribonuclease I (Sigma) was added, the sample centrifuged, and the supernatant collected, filtered through 0.22 μm acrodisc, and loaded onto a HisTrap 5 mL FF column (GE Healthcare) pre-equilibrated with buffer A [50 mM Tris-HCl (pH 7.4) and 500 mM NaCl]. Unbound proteins were removed through an extensive wash with buffer A, with further successive washes with buffer A containing 10, 25, and 50 mM imidazole. SUMO-tagged PQIQ 1909–2035 was eluted with buffer A containing 250 mM imidazole and buffer exchanged into SUMO cleavage buffer [50 mM Tris (pH 7.4) and 150 mM NaCl], and SUMO protease was added and the solution incubated overnight. Cleaved PQIQ 1909–2035 was separated from the His-SUMO tag by reloading the sample onto the HisTrap 5 mL FF column. PQIQ 1909–2035 was further purified using a Superdex 57 26/60 (GE Healthcare) size-exclusion column. The eluted protein was dialyzed against Milli-Q water and stored as lyophilized samples. Confirmation of the identity of purified PQIQ 1909–2035 was achieved via matrix-assisted laser desorption ionization time-of-flight mass spectrometry performed on unlabeled PQIQ 1909–2035 prepared using a procedure identical to that described above.
Isothermal Titration Calorimetry
ITC experiments were performed using a MicroCal ITC200 instrument. Because of limitations in PQIQ peptide solubility, NCS-1 was titrated into the peptide. Ca2+/NCS-1 at 1 mM was prepared by buffer exchange using a PD10 column equilibrated in 50 mM Tris (pH 7.5), 50 mM NaCl, and 5 mM CaCl2, and the PQIQ peptide solution at 100 μM was prepared using the same buffer. If necessary, minor adjustments were made to the pH of the peptide solution. Experiments were conducted using 200 μL of 100 μM PQIQ peptide in the cell and 60 μL of 1 mM NCS-1 in the syringe at 25 °C. One injection of 0.2 μL, followed by 20 injections of 2 μL, was made with a 180 s spacing to allow the baseline to return after each injection. All experiments were performed in triplicate. The data were analyzed with a one-site (three-parameter) curve fitting conducted using the MicroCal-supported ITC module within Origin version 7.
Spectrofluorimetry
To monitor the intrinsic tryptophan fluorescence of NCS proteins,28,29 purified recombinant NCS-1 at a concentration of 1 μM in a buffer [50 mM Tris (pH 7.5), 50 mM NaCl, and 5 mM CaCl2] was excited at room temperature with 280 nm wavelength light and the emission measured between 290 and 410 nm with a slit width of 20 nm using a CARY Eclipse spectrofluorimeter. The PQIQ peptide (stocks ranging from 50 μM to 1 mM) was then added to give an incremental increase in peptide concentration, and emission spectra were acquired after each addition. Experiments were performed in triplicate. The data for the measured tryptophan fluorescence change at each peptide concentration were fit to a logistic equation using nonlinear curve fitting in OriginPro version 9.0.
NMR Spectroscopy
NCS-1 was prepared in Tris buffer (pH 6.8) in the presence of 5 mM CaCl2. NMR spectra were recorded at 298 K on Bruker Avance II 800 and 600 MHz spectrometers equipped with cryoprobes. Data were processed using Bruker Software TopSpin and analyzed using CCPN.30 Sequence specific assignment of the PQIQ peptide was achieved using homonuclear two-dimensional TOCSY, COSY, and NOESY data. 13C- and 15N-filtered TOCSY and NOESY experiments were used to assign the peptide resonances in complex with [13C,15N]NCS-1.
Results
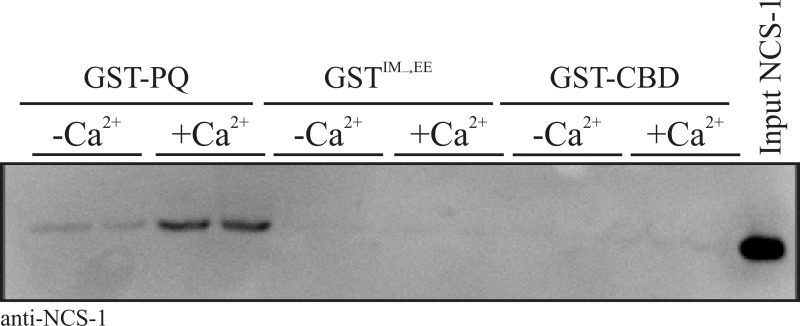
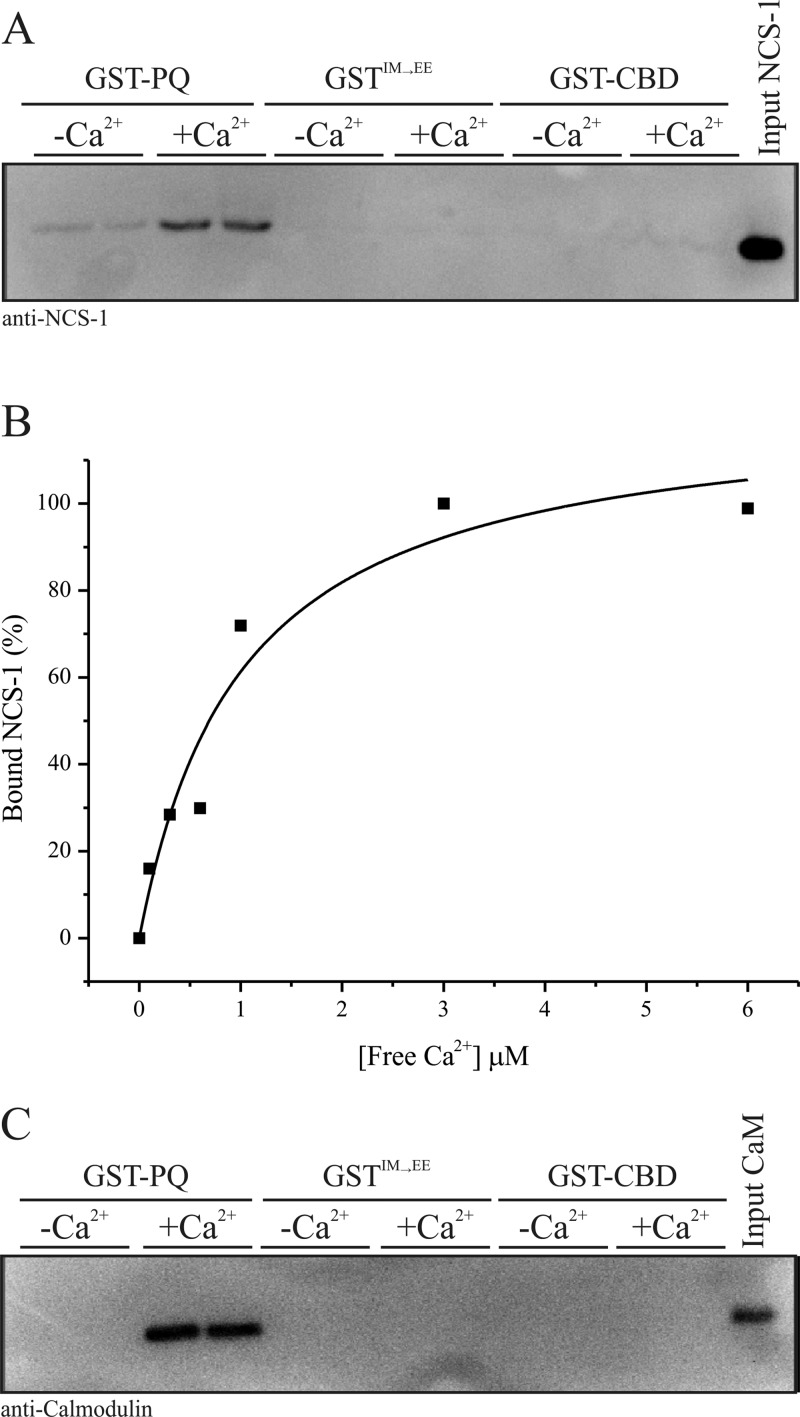
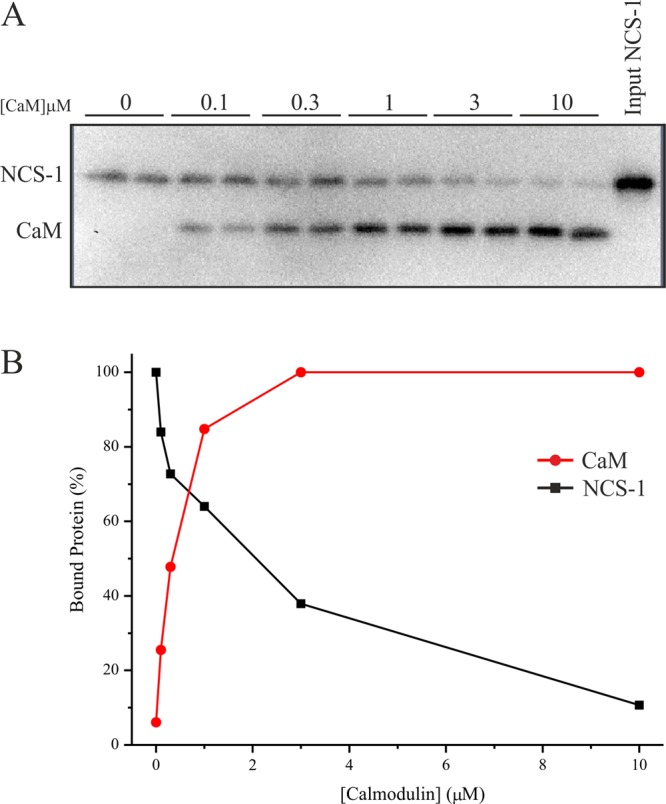
The aim of this study was to test, using a range of complementary methods, the possibility that the small calcium-sensing protein NCS-1 directly interacts with the Cav2.1 P/Q-type Ca2+ channel as has been indirectly suggested in prior functional studies.20−23 With the knowledge that other known small EF-hand Ca2+-binding proteins interact with the α-subunit of the P/Q channel predominantly through motifs located in the C-terminal tail, we reasoned that the same domains represent the most likely sites for an NCS-1 interaction. In initial experiments, we generated a recombinant, GST-tagged, form of the P/Q α1 subunit C-terminal tail domain corresponding to residues 1898–2035 (Figure 1, GST-PQ). This fragment encompasses both the IQ motif (IM in the P/Q-type channel) and the CaM-binding domain (CBD).10 Two further constructs derived from this precursor were the CBD alone [residues 1950–2035 (Figure 1, GST-CBD)] and an IQ motif mutant in which the P/Q channel IM residues at positions 1913 and 1914 were mutated to glutamic acid to generate an IM → EE double mutant (Figure 1, GSTIM→EE). This mutation has been previously shown to abolish CaM-dependent CDF and CDI of the human P/Q-type channel.7 We used these constructs to investigate whether recombinant NCS-1 could associate with P/Q-type C-terminal regulatory domains in vitro. NCS-1 exhibited Ca2+-dependent binding to GST-PQ but was not observed to bind to either GST-CBD or GSTIM→EE under any tested conditions (Figure 1A), suggesting a specific interaction with residues in the IQ region of the channel. Averaged densitometry data from three independent binding experiments are listed in Table 1.
Figure 1.
Interactions of NCS-1 and CaM with rat PQ α1 subunit C-terminal binding motifs. (A) Recombinant NCS-1 was incubated with GST-PQ (residues 1898–2035 of the rat PQ α1 subunit, encompassing both the IQ and CBD motifs), GSTIM→EE (equivalent to the IQ motif present in Cav1.x channels), and GST-CBD (residues 1950–2035 of the rat PQ α1 subunit) in the presence or absence of 1 μM free Ca2+. Bound NCS-1 was detected by Western blotting with a specific antibody. This blot is a representative result, and averaged data from three independent binding assays are listed in Table 1. (B) Densitometry quantification of an NCS-1/GST-PQ binding Ca2+ dose response. Recombinant NCS-1 (1 μM) was incubated with 1 μM GST-PQ in the presence of increasing concentrations of free Ca2+ and bound protein detected by Western blotting with an anti-NCS-1 specific antibody. Western blot signal intensities were quantified by densitometry and plotted as a function of free Ca2+ concentration present in the binding reaction mixture. (C) Recombinant CaM was incubated with GST-PQ, GSTIM→EE, and GST-CBD in the presence or absence of 1 μM free Ca2+. Bound CaM was detected by Western blotting with a specific antibody.
Table 1. Average Densitometry Data for Three Independent NCS-1/GST PQ Binding Assays Described in Figure 1Aa.
| condition | average NCS-1 bound (%) | SEM (n = 3) |
|---|---|---|
| GST-PQ with Ca2+ | 100 | – |
| GST-PQ without Ca2+ | 31.2 | 3.7 |
| GST-CBD with Ca2+ | 20.3 | 10.2 |
| GST-CBD without Ca2+ | 4.9 | 4.9 |
| GSTIM→EE with Ca2+ | 10.3 | 3.9 |
| GSTIM→EE without Ca2+ | 2.8 | 2.0 |
Maximal binding (NCS-1 bound to GST-PQ in the presence of calcium for all experiments) was set to 100% binding. Minimal binding for each assay (NCS-1 bound to either GST-CBD or GSTIM→EE in the absence of calcium depending on the experiment) was set to 0% binding. All other data were normalized in this range.
A Ca2+ titration of binding of NCS-1 to GST-PQ confirmed the Ca2+ dependency of the interaction and allowed us to derive a Kd of 0.7 μM based on densitometry quantification of Western blot data (Figure 1B). For comparison, we performed the same binding analysis with CaM (Figure 1C). Similar to our results with NCS-1, CaM displayed Ca2+-dependent binding to GST-PQ but no detectable binding under any conditions to either GST-CBD or GSTIM→EE.
Our initial data suggested that both NCS-1 and CaM could interact with the same IQ motif region of the rat P/Q-type channel, and therefore, we extended our binding studies to evaluate the potential existence of a common, overlapping, binding site for both proteins. Competitive association of both proteins with GST-PQ was assessed by binding of NCS-1 to GST-PQ in the presence of increasing concentrations of recombinant CaM (Figure 2). Western blot results from these experiments highlighted a clear displacement of NCS-1 from GST-PQ that was first apparent at a CaM concentration between 0.3 and 1 μM (Figure 2A). Densitometry analysis of the Western blot data confirmed a competitive association between NCS-1 and CaM for GST-PQ (Figure 2B). Our initial experiments indicated that the IQ-like motif of the P/Q-type channel is important for interaction with both NCS-1 and CaM, and therefore, in subsequent NMR and ITC analyses, we focused on only this region of the channel.
Figure 2.
Competitive interaction between NCS-1 and CaM for binding to GST-PQ. (A) NCS-1 (5 μM) was incubated with 5 μM GST-PQ in the presence of varying concentrations of CaM. Bound NCS-1 and CaM were visualized by Western blotting with specific antibodies. (B) Data from panel A were quantified by densitometry, and NCS-1/CaM bound to GST-PQ was plotted as a function of the CaM concentration present in the binding assay.
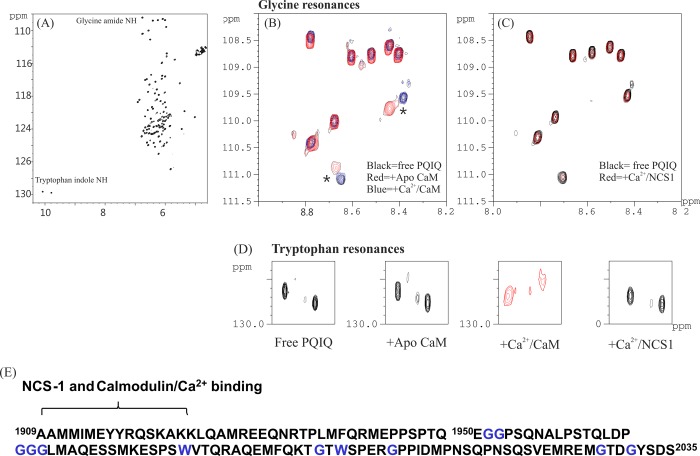
Further testing of the ability of NCS-1 to bind the P/Q-type channel and to delineate the NCS-1- and CaM-binding regions of the P/Q-type channel was performed using NMR experiments. A sample of 15N-labeled P/Q spanning residues 1909–2035 (encompassing much of the IQ motif and the CBD) was prepared. Addition of CaM shows chemical shift perturbations of glycine and tryptophan residues, whereas NCS-1 has no effect on either group of residues (Figure 3). Glycine and tryptophan residues are located in the CBD region but not in or around the IQ segment. Hence, it is clear that NCS-1 does not bind to the CBD of the P/Q channel, although there is an indication that there is weak binding of CaM to this site.
Figure 3.
Delineation of binding of NCS-1 to PQ 1909–2035. 1H–15N HSQC spectra of 15N-labeled PQ1909–2035 in the presence (A) and absence of CaM (B) and of NCS-1 (C), showing the glycine amide resonance, and (D) tryptophan NεH resonances. Panel A shows the full 15N–1H spectrum; expanded regions show resonances from only glycine amide (B and C) and tryptophan indole (D) groups. (E) The amino acid sequence of PQ 1909–2035 is shown with glycine and tryptophan residues colored blue. The likely CaM- and NCS-1-binding region is indicated.15
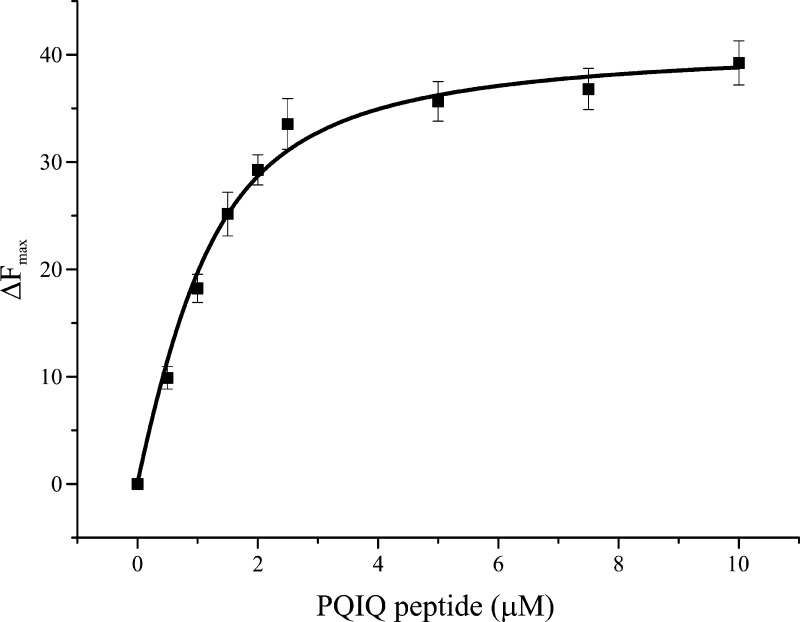
Knowing from the experiments described above that the major site of binding of NCS-1 on the P/Q C-terminal domain was likely to be the IQ motif, we used a synthetic PQIQ peptide in subsequent experiments. The intrinsic tryptophan fluorescence of NCS-1 was used to measure interactions with the PQIQ peptide (Figure 4), using an approach similar to that described for the interactions with the D2 dopamine receptor peptide.27 NCS-1 has two tryptophan residues; addition of the PQIQ peptide resulted in a decrease in fluorescence and allowed titration of the peptide over a range of concentrations (Figure 2A). The data for the change in fluorescence versus concentration were analyzed using a logistic fit. The dose response upon titration with the PQIQ indicated half-maximal binding at 1.036 ± 0.07 μM with a Hill coefficient of 1.7. Hill coefficients greater than 1 would be consistent with a 2:1 stoichiometry of peptide binding to NCS-1 because NCS-1 itself was monomeric.27 ITC shows that the interaction is entropically driven. Because of the tendency of the protein complex to aggregate at the high peptide (100 μM) and protein concentrations (1 mM) required to observe binding by this method, it was not possible to obtain reliable equilibrium binding constants using ITC. The affinity obtained for the interaction of NCS-1 with the PQIQ peptide is almost 1 order of magnitude lower than the reported affinities involving the calmodulin lobes. From ITC experiments, Kd values of 51 ± 20 and 4.32 ± 0.39 nM were obtained for binding of the Ca2+/N and Ca2+/C lobes, respectively, to the IQ domain of the PQ channel.31 The two lobes in NCS-1 are intimately linked, and attempts to express the two NCS-1 lobes as independently folded domains soluble at micomolar concentrations have met with little success so far. Therefore, experiments that aimed to disentangle the relative affinities of the two NCS-1 domains for the PQIQ peptide have not been possible.
Figure 4.
Binding of the PQIQ peptide to NCS-1 monitored using tryptophan fluorescence. Changes in tryptophan fluorescence following sequential additions of the PQIQ peptide to the final indicated concentrations. Recombinant NCS-1 at a concentration of 1 μM in a buffer [50 mM Tris (pH 7.5), 50 mM NaCl, and 5 mM CaCl2] was titrated with PQIQ at 50 μM to 1 mM under the same buffer conditions. The data were fit using a logistic equation and nonlinear curve fitting. Data are plotted as means ± SEM for three independent experiments.
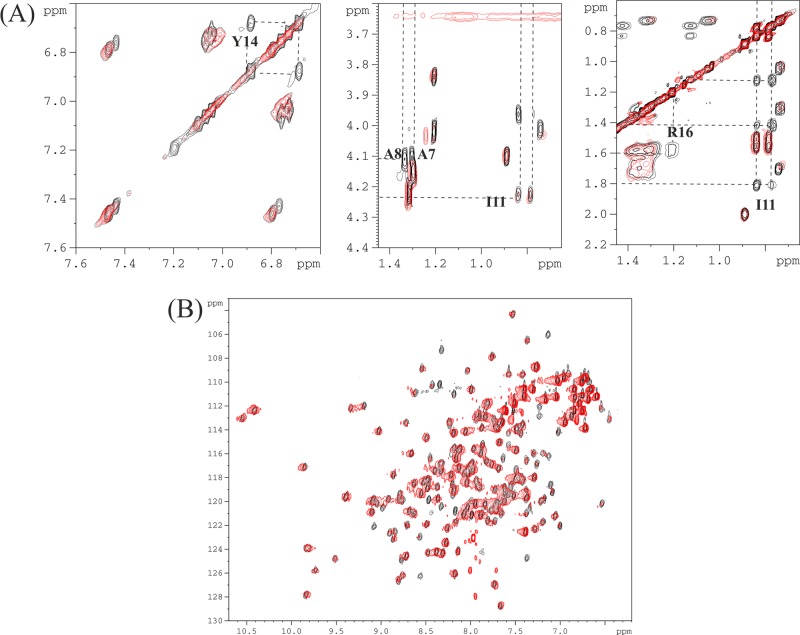
To determine which region of the PQIQ peptide forms the NCS-1-binding site, we again used NMR. In these experiments, [13C,15N]NCS-1 was titrated into a sample of the unlabeled PQIQ peptide and 13C- and 15N-filtered TOCSY and NOESY data were acquired. Analysis of the side chain resonances, in particular the methyl and tyrosine resonances, reveals selective line broadening for 14 residues, YAAMMIMEYYRQSK (Figure 5A). Hence, a subset of the 27 amino acids in the polypeptide sequence forms the main interaction site for NCS-1 binding, with the residues beyond this core peptide region further enhancing the interaction. The length of this motif correlates well with the recently reported structure of NCS-1 in complex with the C-terminal peptide from the human D2 dopamine receptor (Protein Data Bank entry 2YOU). The 14-amino acid motif is short enough for it to bind NCS-1 with a stoichiometry of 2:1 (peptide:NCS-1) and supports the fluorescence binding data.
Figure 5.
1H TOCSY and HSQC spectra showing PQIQ–NCS-1 interactions. (A) The TOCSY spectrum of the PQIQ peptide delineates the NCS-1-binding region. Line broadening was observed for selected resonances in the presence of a 2-fold excess of NCS-1. Line-broadened resolved resonances from Y4, A7, A8, I11, and R16 are highlighted: aromatic resonances (left) and aliphatic resonances (middle and right). (B) 1H–15N HSQC spectra of [15N]NCS-1 (0.8 mM, black) in the presence of PQIQ (final concentration of 0.4 mM, red) in 50 mM Tris buffer (pH 6.5), 50 mM NaCl, and 5 mM CaCl2 at 300 K.
NMR 1H–15N HSQC of NCS-1 in the presence of a 2-fold excess of PQIQ shows resonances that are universally broadened although still discernible to confirm that the protein remains folded (Figure 5B). Addition of equimolar amounts of unlabeled CaM/Ca2+ to the preformed 15N-labeled NCS-1/PQIQ complex led to a complete reversion of the 1H–15N HSQC C spectrum to that of free NCS-1. This is further confirmation that NCS-1 and CaM bind to similar sites on the PQ channel, with, as expected, CaM binding the peptide sufficiently strongly to displace it from NCS-1.
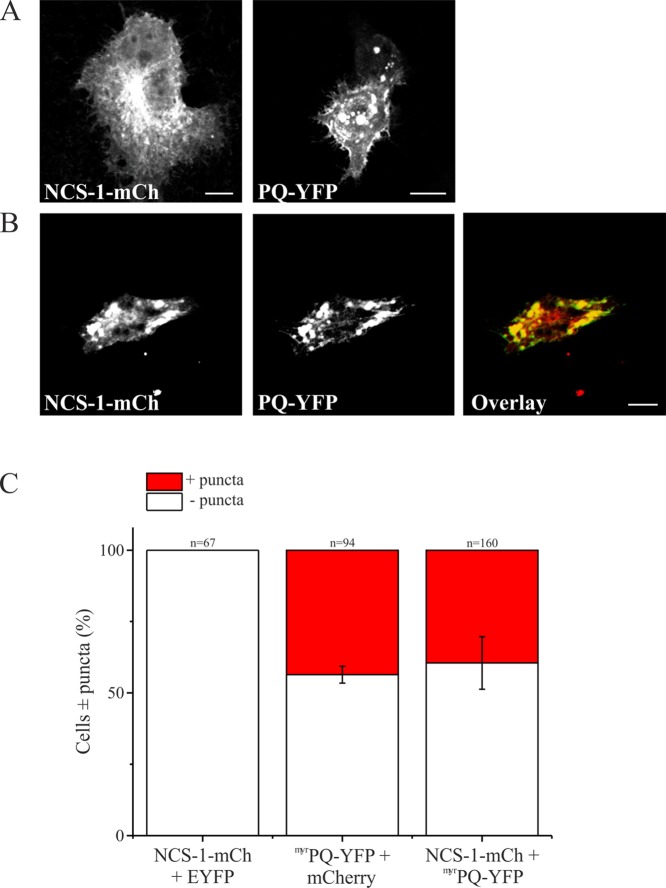
The experiments described above show that a direct interaction of NCS-1 with the P/Q C-terminus could be detected using three distinct in vitro biochemical approaches. To provide evidence of the interaction of NCS-1 with the P/Q channel regulatory domain in a more physiological setting, we studied the localization of these proteins ex vivo in cultured HeLa cells. For these experiments, we returned to the use of a longer recombinant construct encompassing both the IQ and CBD domains of the P/Q C-terminus. We first examined the cellular localization of both NCS-1-mCherry18 and a C-terminally YFP-tagged variant of P/Q 1898–2035 to which an N-terminal myristoylation consensus sequence had been attached (myrPQ-YFP). In previous studies, this approach has been used to successfully direct isolated soluble domains of plasma membrane proteins to the plasma membrane for functional analyses.25 NCS-1-mCherry exhibited a predominantly perinuclear localization consistent with its association with the TGN in addition to some plasma membrane localization (Figure 6A).32myrPQ-YFP was observed to target the plasma membrane and/or large cytosolic puncta (Figure 6A). Plasma membrane association of myrPQ-YFP suggested that myristoylation of the peptide was accurately targeting a fraction of PQ to the expected location. Interestingly, in cells exhibiting myrPQ-YFP puncta, co-expressed NCS-1-mCherry failed to localize to the TGN or plasma membrane and instead colocalized with myrPQ-YFP on the enlarged punctate structures (Figure 6B). We quantified these phenomena and were able to demonstrate that in 39.5 ± 9.2% of cells cotransfected with NCS-1-mCherry and myrPQ-YFP puncta were observed and that in 100% of this subpopulation NCS-1 protein was associated with large myrPQ-YFP positive punctate structures (Figure 6C, NCS-1-mCh + myrPQ-YFP). This was similar to the number of cells exhibiting puncta in myrPQ-YFP/mCherry controls [43.6 ± 3% (Figure 6C, myrPQ-YFP + mCherry)]. In contrast, control cells expressing NCS-1-mCherry along with EYFP protein exhibited no observable formation of puncta (Figure 6C, NCS-1-mCh + EYFP). It should be noted that colocalization of NCS-1-mCherry with myrPQ-YFP in punctate structures was specific for NCS-1 as no colocalization was observed with the soluble mCherry protein in control cotransfections (data not shown). These observations are consistent with a direct interaction of NCS-1-mCherry with myrPQ-YFP in a cellular context.
Figure 6.
Expression of NCS-1 and PQ in HeLa cells. (A) HeLa cells were cotransfected with NCS-1-mCherry (red) and EYFP (green) or mCherry (red) or (B) NCS-1-mCherry (red) and myrPQ-YFP (green). Regions of colocalization are colored yellow in overlay images. Scale bars are 10 μm. (C) Quantification of data from panels A and B. Cells were counted and scored for the presence of puncta {three independent experiments from each control condition [NCS-1-mCh + EYFP and myrPQ-YFP + mCherry (A)]} or four independent experiments [NCS-1-mCh + myrPQ-YFP (B)]. The total number of cells counted for all experiments and for each condition is shown above the corresponding bar on the histogram. All data are plotted as means ± SEM.
Discussion
In this paper, we set out to examine whether NCS-1 is able to interact directly with the C-terminus of the α1 subunit of the P/Q-type channel. To avoid a potentially misleading indication of a positive interaction, we tested this using three separate in vitro biochemical/biophysical assays using either a long construct from the P/Q C-terminus or a synthetic IQ domain peptide as well as a test through cellular colocalization. The use of multiple techniques gives a more rigorous approach to addressing the question of whether a significant direct interaction does exist. In all cases, we were able to detect a direct interaction between NCS-1 and P/Q-type Ca2+ channels.
P/Q-type channel regulation by small Ca2+-binding proteins is essential in the mammalian CNS for the processes of CDI and CDF that drive short-term presynaptic plasticity.6 Alterations in synaptic plasticity of this form in turn influence information processing and neural network behavior that underpin the normal functioning of the CNS.33 A large body of experimental evidence that describes at the molecular level how CaM is able to regulate all Cav-type channels, including those of the P/Q subtype, has accumulated. CaM is constitutively associated with the P/Q α subunit and is considered an auxiliary subunit of the channel. Apo-CaM is believed to bind to the P/Q α subunit at a site outside of, but in the proximity of, the Ca2+-dependent IQ-binding site, as determined using a Förster resonance energy transfer approach in cultured mammalian cells.34 It has been suggested that this arrangement locates CaM within striking distance of the regulatory IQ motif to ensure fast responses to Ca2+ influx on channel opening.34 Ca2+-dependent binding of CaM to the IQ motif is then able to modulate both CDI and CDF depending on the precise nature of the Ca2+ signal. Mechanistically, this system has been resolved in elegant experiments showing that Ca2+ entry through individual P/Qα channels selectively activates the CaM C-lobe to mediate CDF. Larger Ca2+ signals emanating from multiple local channel opening events conversely activate the CaM N-lobe to drive CDI.3,7,35 Our data confirming a Ca2+-dependent interaction of CaM with the P/Q containing the IQ motif are consistent with previous biochemical7 and structural analyses.31 A second CaM-binding site, the CaM-binding domain (CBD), has been reported in related studies of P/Q–channel interactions.5,13 In this study, although we have been unable to replicate binding of CaM to this region of the rat α1A CBD using pull-down experiments, the NMR data suggest that CaM, but not NCS-1, binds to the proposed CBD, albeit rather weakly. The weak nature of this interaction might explain why there are discrepancies in the literature regarding the significance of the CBD. Pull-down-type experiments select for high-affinity interactions, whereas NMR approaches are able to detect lower-affinity transient binding events consistent with these results. Our data help reconcile apparently conflicting experimental data as some studies do observe a CaM–CBD interaction5,13 whereas others do not.7
NCS-1, a multifunctional Ca2+ sensor conserved from yeast to humans, has documented roles in the regulation of membrane trafficking,36,37 ion channel activity,38,39 dopamine receptor signaling,27,40 long-term depression,41 autism,18,42 and memory acquisition.43,44 NCS-1 exhibits enriched expression in mammalian brain tissue,45 and many of the aforementioned functions are related to neuronal activity. NCS-1 interacts with a number of targets in common with CaM,24 and a previous functional study identified links between NCS-1 and CDF of P/Q-type channel activity in rat primary neurons.20 Other studies have additionally indicated a cellular connection between NCS-1 and P/Q-type channel activity in bovine adrenal chromaffin cells21,22 and Drosophila.23 With this information in mind, we have tested the possibility that NCS-1 can directly bind to P/Q regulatory elements in common with CaM and other EF-hand-containing Ca2+ sensors.10,14 Our data show that NCS-1 can associate with the IQ motif of rat P/Qα1A in a Ca2+-dependent manner and that this binding site must overlap to some extent with that of CaM because CaM can compete for binding to the same region leading to NCS-1 displacement. This is the first example of a CaM-related Ca2+-sensing protein behaving in a manner almost identical to that of CaM with respect to IQ binding. Both CaBP110 and VILIP-214 interact with the CBD or require this domain for functional activity. CaM binds predominantly to the IQ domain with a possible weak interaction with the CBD, and NCS-1 binds to only the IQ domain in the presence of Ca2+. The weak CaM–CBD interaction that we have observed in this study might also help to explain why CaBP1 and VILIP-2 are able to effectively compete with CaM at this site.10,14
The precise details of the NCS-1 interaction are still to be elucidated. In rat primary neurons, NCS-1 appears to play a role during CDF,20 and in Drosophila, loss of the NCS-1 orthologue Frequenin leads to a reduced level of entry of Ca2+ into neurons and defective synaptic transmission.23 These data suggest that at least one function of NCS-1 is in positively regulating P/Q channel opening. Our data now provide an attractive biochemical explanation for these cellular studies and act as a platform for further cellular and structural investigations studying the interplay between CaM and NCS-1 during regulation of P/Q channel activity. CaM is expressed in all neurons, and one possibility is that specific neuronal populations that also express NCS-1 are able to utilize this Ca2+ sensor as an additional modulator of P/Q activity. NCS-1 has a Ca2+ affinity higher than that of CaM15,46 and therefore could potentially interact with the IQ motif over a different range of local Ca2+ concentrations to modulate activity in a manner independent of CaM. As Ca2+ concentrations increase, CaM could then displace NCS-1 to exert its documented roles in CDF and CDI. The N- and C-lobes of CaM, as described, exert distinct modes of P/Q channel regulation. NCS-1, unlike CaM, does not have distinct lobes but rather a compact globular structure.47 The orientations of the N- and C-lobes with respect to one another differ in the two proteins, giving rise to different conformations of the ligand-binding site. Residues from both lobes in NCS-1 together form a large, contiguous solvent-exposed hydrophobic ligand-binding crevice, resembling the palm of a partially open hand. In the Ca2+/CaM–PQIQ complex, the hydrophobic binding pocket is more enclosed, resembling a closed hand that envelops the PQIQ peptide. It is, therefore, not possible to model a structure of NCS-1 in complex with the PQIQ peptide based on the Ca2+/CaM complex structure. It is likely that binding the PQIQ peptide would require a significant conformational change to the overall structure of NCS-1 to accommodate the peptide.
Using a combination of in vitro biochemical, NMR and ITC biophysical, and cellular approaches, we provide the first evidence of a direct interaction of NCS-1 with P/Q-type voltage-gated Ca2+ channels. Voltage-gated Ca2+ channel modulation has direct implications for synaptic plasticity and complex neural processing in higher organisms. Our work expands the number of small Ca2+-sensing proteins that can interact with the essential P/Q-type neuronal channel and provides further insights into the complexity of the regulation of voltage-gated Ca2+ channels in the mammalian CNS. Extensive further work will be required to determine the full structural basis for the interaction of NCS-1 with the P/Q IQ domain.
Glossary
Abbreviations
- Ca2+
calcium
- CaM
calmodulin
- NCS-1
neuronal calcium sensor-1
- CaBP
calcium-binding protein
- Cav
voltage-gated Ca2+ channel
- CDI
Ca2+-dependent inactivation
- CDF
Ca2+-dependent facilitation
- CNS
central nervous system
- PBS
phosphate-buffered saline
- NTA
nitrilotriacetic acid
- EGTA
ethylene glycol tetraacetic acid
- EDTA
ethylenediaminetetraacetic acid
- SDS
sodium dodecyl sulfate
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid
- DTT
dithiothreitol
- HRP
horseradish peroxidase
- ECL
enhanced chemiluminescence
- ITC
isothermal titration calorimetry
- NMR
nuclear magnetic resonance
- COSY
correlation spectroscopy
- HSQC
heteronuclear single-quantum correlation
- NOESY
nuclear Overhauser spectroscopy
- TOCSY
total correlation spectroscopy
- SEM
standard error of the mean.
This work was supported by Wellcome Trust Prize Ph.D. studentships awarded to V.M.M. and P.A.C.T. and a University of Liverpool Studentship awarded to S.R.P.
The authors declare no competing financial interest.
References
- Olivera B. M.; Miljanich G. P.; Ramachandran J.; Adams M. E. (1994) Calcium channel diversity and neurotransmitter release: The ω-conotoxins and ω-agatoxins. Annu. Rev. Biochem. 63, 823–867. [DOI] [PubMed] [Google Scholar]
- Rajakulendran S.; Kaski D.; Hanna M. G. (2012) Neuronal P/Q-type calcium channel dysfunction in inherited disorders of the CNS. Nat. Rev. Neurol. 8, 86–96. [DOI] [PubMed] [Google Scholar]
- Liang H.; DeMaria C. D.; Erickson M. G.; Mori M. X.; Alseikhan B. A.; Yue D. T. (2003) Unified mechanisms of Ca2+ regulation across the Ca2+ channel family. Neuron 39, 951–960. [DOI] [PubMed] [Google Scholar]
- Budde T.; Meuth S.; Pape H. C. (2002) Calcium-dependent inactivation of neuronal calcium channels. Nat. Rev. Neurosci. 3, 873–883. [DOI] [PubMed] [Google Scholar]
- Lee A.; Scheuer T.; Catterall W. A. (2000) Ca2+/calmodulin-dependent facilitation and inactivation of P/Q-type Ca2+ channels. J. Neurosci. 20, 6830–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W. A.; Few A. P. (2008) Calcium channel regulation and presynaptic plasticity. Neuron 59, 882–901. [DOI] [PubMed] [Google Scholar]
- DeMaria C. D.; Soong T. W.; Alseikhan B. A.; Alvania R. S.; Yue D. T. (2001) Calmodulin bifurcates the local Ca2+ signal that modulates P/Q-type Ca2+ channels. Nature 411, 484–489. [DOI] [PubMed] [Google Scholar]
- Lee A.; Zhou H.; Scheuer T.; Catterall W. A. (2003) Molecular determinants of Ca2+/calmodulin-dependent regulation of Cav2.1 channels. Proc. Natl. Acad. Sci. U.S.A. 100, 16059–16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeisen F.; Minor D. L. Jr. (2010) Structural basis for the differential effects of CaBP1 and calmodulin on CaV1.2 calcium-dependent inactivation. Structure 18, 1617–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.; Westenbroek R. E.; Haeseleer F.; Palczewski K.; Scheuer T.; Catterall W. A. (2002) Differential modulation of Cav2.1 channels by calmodulin and Ca2+-binding protein 1. Nat. Neurosci. 5, 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate P.; Mochca-Morales J.; Wu Y.; Zhang J. Z.; Rodney G. G.; Serysheva I. I.; Williams B. Y.; Anderson M. E.; Hamilton S. L. (2000) Determinants for calmodulin binding on voltage-dependent Ca2+ channels. J. Biol. Chem. 275, 39786–39792. [DOI] [PubMed] [Google Scholar]
- Minor D. L. Jr.; Findeisen F. (2010) Progress in the structural understanding of voltage-gated calcium channel (CaV) function and modulation. Channels 4, 459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.; Wong S. T.; Gallagher D.; Li B.; Storm D. R.; Scheuer T.; Catterall W. A. (1999) Ca2+/calmodulin binds to and modulates P/Q-type calcium channels. Nature 399, 155–159. [DOI] [PubMed] [Google Scholar]
- Lautermilch N. J.; Few A. P.; Scheuer T.; Catterall W. A. (2005) Modulation of CaV2.1 channels by the neuronal calcium-binding protein visinin-like protein-2. J. Neurosci. 25, 7062–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R. D.; Weiss J. L. (2001) The neuronal calcium sensor family of Ca2+-binding proteins. Biochem. J. 353, 1–12. [PMC free article] [PubMed] [Google Scholar]
- Weiss J. L.; Hui H.; Burgoyne R. D. (2010) Neuronal calcium sensor-1 regulation of calcium channels, secretion, and neuronal outgrowth. Cell. Mol. Neurobiol. 30, 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dason J. S.; Romero-Pozuelo J.; Atwood H. L.; Ferrus A. (2012) Multiple roles for frequenin/NCS-1 in synaptic function and development. Mol. Neurobiol. 45, 388–402. [DOI] [PubMed] [Google Scholar]
- Handley M. T.; Lian L. Y.; Haynes L. P.; Burgoyne R. D. (2010) Structural and functional deficits in a neuronal calcium sensor-1 mutant identified in a case of autistic spectrum disorder. PLoS One 5, e10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlowsky A.; Gianfelice A.; Pallotto M.; Zanchi A.; Vara H.; Khelfaoui M.; Valnegri P.; Rezai X.; Bassani S.; Brambilla D.; Kumpost J.; Blahos J.; Roux M. J.; Humeau Y.; Chelly J.; Passafaro M.; Giustetto M.; Billuart P.; Sala C. (2010) A postsynaptic signaling pathway that may account for the cognitive defect due to IL1RAPL1 mutation. Curr. Biol. 20, 103–115. [DOI] [PubMed] [Google Scholar]
- Tsujimoto T.; Jeromin A.; Saitoh N.; Roder J. C.; Takahashi T. (2002) Neuronal calcium sensor 1 and activity-dependent facilitation of P/Q-type calcium currents at presynaptic nerve terminals. Science 295, 2276–2279. [DOI] [PubMed] [Google Scholar]
- Weiss J. L.; Archer D. A.; Burgoyne R. D. (2000) Neuronal Ca2+ sensor-1/frequenin functions in an autocrine pathway regulating Ca2+ channels in bovine adrenal chromaffin cells. J. Biol. Chem. 275, 40082–40087. [DOI] [PubMed] [Google Scholar]
- Weiss J. L.; Burgoyne R. D. (2001) Voltage-independent inhibition of P/Q-type Ca2+ channels in adrenal chromaffin cells via a neuronal Ca2+ sensor-1-dependent pathway involves Src family tyrosine kinase. J. Biol. Chem. 276, 44804–44811. [DOI] [PubMed] [Google Scholar]
- Dason J. S.; Romero-Pozuelo J.; Marin L.; Iyengar B. G.; Klose M. K.; Ferrus A.; Atwood H. L. (2009) Frequenin/NCS-1 and the Ca2+-channel α1-subunit co-regulate synaptic transmission and nerve-terminal growth. J. Cell Sci. 122, 4109–4121. [DOI] [PubMed] [Google Scholar]
- Haynes L. P.; Fitzgerald D. J.; Wareing B.; O’Callaghan D. W.; Morgan A.; Burgoyne R. D. (2006) Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin delta. Proteomics 6, 1822–1832. [DOI] [PubMed] [Google Scholar]
- Oz S.; Benmocha A.; Sasson Y.; Sachyani D.; Almagor L.; Lee A.; Hirsch J. A.; Dascal N. (2013) Competitive and non-competitive regulation of calcium-dependent inactivation in CaV1.2 L-type Ca2+ channels by calmodulin and Ca2+-binding protein 1. J. Biol. Chem. 288, 12680–12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFerran B. W.; Graham M. E.; Burgoyne R. D. (1998) Neuronal Ca2+ sensor 1, the mammalian homologue of frequenin, is expressed in chromaffin and PC12 cells and regulates neurosecretion from dense-core granules. J. Biol. Chem. 273, 22768–22772. [DOI] [PubMed] [Google Scholar]
- Lian L. Y.; Pandalaneni S. R.; Patel P.; McCue H. V.; Haynes L. P.; Burgoyne R. D. (2011) Characterisation of the interaction of the C-terminus of the dopamine d2 receptor with neuronal calcium sensor-1. PLoS One 6, e27779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFerran B. W.; Weiss J. L.; Burgoyne R. D. (1999) Neuronal Ca2+ sensor 1. Characterization of the myristoylated protein, its cellular effects in permeabilized adrenal chromaffin cells, Ca2+-independent membrane association, and interaction with binding proteins, suggesting a role in rapid Ca2+ signal transduction. J. Biol. Chem. 274, 30258–30265. [DOI] [PubMed] [Google Scholar]
- Haynes L. P.; Tepikin A. V.; Burgoyne R. D. (2004) Calcium-binding protein 1 is an inhibitor of agonist-evoked, inositol 1,4,5-trisphosphate-mediated calcium signaling. J. Biol. Chem. 279, 547–555. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. (2005) The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696. [DOI] [PubMed] [Google Scholar]
- Kim E. Y.; Rumpf C. H.; Fujiwara Y.; Cooley E. S.; Van Petegem F.; Minor D. L. Jr. (2008) Structures of CaV2 Ca2+/CaM-IQ domain complexes reveal binding modes that underlie calcium-dependent inactivation and facilitation. Structure 16, 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes L. P.; Sherwood M. W.; Dolman N. J.; Burgoyne R. D. (2007) Specificity, promiscuity and localization of ARF protein interactions with NCS-1 and phosphatidylinositol-4 kinase-III β. Traffic 8, 1080–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott L. F.; Regehr W. G. (2004) Synaptic computation. Nature 431, 796–803. [DOI] [PubMed] [Google Scholar]
- Erickson M. G.; Alseikhan B. A.; Peterson B. Z.; Yue D. T. (2001) Preassociation of calmodulin with voltage-gated Ca2+ channels revealed by FRET in single living cells. Neuron 31, 973–985. [DOI] [PubMed] [Google Scholar]
- Park H. Y.; Kim S. A.; Korlach J.; Rhoades E.; Kwok L. W.; Zipfel W. R.; Waxham M. N.; Webb W. W.; Pollack L. (2008) Conformational changes of calmodulin upon Ca2+ binding studied with a microfluidic mixer. Proc. Natl. Acad. Sci. U.S.A. 105, 542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.; Varnai P.; Tuymetova G.; Balla A.; Toth Z. E.; Oker-Blom C.; Roder J.; Jeromin A.; Balla T. (2001) Interaction of neuronal calcium sensor-1 (NCS-1) with phosphatidylinositol 4-kinase β stimulates lipid kinase activity and affects membrane trafficking in COS-7 cells. J. Biol. Chem. 276, 40183–40189. [DOI] [PubMed] [Google Scholar]
- Haynes L. P.; Thomas G. M.; Burgoyne R. D. (2005) Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase β and trans-Golgi network-plasma membrane traffic. J. Biol. Chem. 280, 6047–6054. [DOI] [PubMed] [Google Scholar]
- Hui H.; McHugh D.; Hannan M.; Zeng F.; Xu S. Z.; Khan S. U.; Levenson R.; Beech D. J.; Weiss J. L. (2006) Calcium-sensing mechanism in TRPC5 channels contributing to retardation of neurite outgrowth. J. Physiol. 572, 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.; Malin S. A.; Johns D. C.; Jeromin A.; Nerbonne J. M. (2002) Modulation of Kv4-encoded K+ currents in the mammalian myocardium by neuronal calcium sensor-1. J. Biol. Chem. 277, 26436–26443. [DOI] [PubMed] [Google Scholar]
- Kabbani N.; Negyessy L.; Lin R.; Goldman-Rakic P.; Levenson R. (2002) Interaction with neuronal calcium sensor NCS-1 mediates desensitization of the D2 dopamine receptor. J. Neurosci. 22, 8476–8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J.; Heon S.; Kim M. J.; Son G. H.; Park Y.; Henley J. M.; Weiss J. L.; Sheng M.; Collingridge G. L.; Cho K. (2008) Metabotropic glutamate receptor-mediated LTD involves two interacting Ca2+ sensors, NCS-1 and PICK1. Neuron 60, 1095–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A.; Michaud J. L.; Peng H.; Aradhya S.; Gauthier J.; Mottron L.; Champagne N.; Lafreniere R. G.; Hamdan F. F.; S2D team; Joober R.; Fombonne E.; Marineau C.; Cossette P.; Dube M. P.; Haghighi P.; Drapeau P.; Barker P. A.; Carbonetto S.; Rouleau G. A. (2008) Mutations in the calcium-related gene IL1RAPL1 are associated with autism. Hum. Mol. Genet. 17, 3965–3974. [DOI] [PubMed] [Google Scholar]
- Gomez M.; De Castro E.; Guarin E.; Sasakura H.; Kuhara A.; Mori I.; Bartfai T.; Bargmann C. I.; Nef P. (2001) Ca2+ signaling via the neuronal calcium sensor-1 regulates associative learning and memory in C. elegans. Neuron 30, 241–248. [DOI] [PubMed] [Google Scholar]
- Saab B. J.; Georgiou J.; Nath A.; Lee F. J.; Wang M.; Michalon A.; Liu F.; Mansuy I. M.; Roder J. C. (2009) NCS-1 in the dentate gyrus promotes exploration, synaptic plasticity, and rapid acquisition of spatial memory. Neuron 63, 643–656. [DOI] [PubMed] [Google Scholar]
- Paterlini M.; Revilla V.; Grant A. L.; Wisden W. (2000) Expression of the neuronal calcium sensor protein family in the rat brain. Neuroscience 99, 205–216. [DOI] [PubMed] [Google Scholar]
- Burgoyne R. D. (2007) Neuronal calcium sensor proteins: Generating diversity in neuronal Ca2+ signalling. Nat. Rev. Neurosci. 8, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y.; Dannenberg J.; Pollmann V.; Marchot P.; Pongs O. (2001) Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1). J. Biol. Chem. 276, 11949–11955. [DOI] [PubMed] [Google Scholar]