Abstract
Injection drug use has been responsible for most HIV-1 transmission in Asia and has led to the generation and/or spreading of circulating recombinant CRF01_AE, CRF07_BC, and CRF08_BC, which are currently the predominant HIV-1 strains in China. Monitoring HIV-1 transmissions among injection drug users (IDUs) still provides critical information on HIV-1 prevalence and control. In the present study, we detected three HIV-1 sequences, BH048/057/066, from IDUs in Beihai city, Guangxi province, all of which share an extremely complex mosaic recombination pattern. CRF01_AE and CRF08_BC contributed to the rise of new recombinants, which contain CRF08_BC-derived env fragments with evidence of further evolution. Our data, combined with those of other studies, suggest that the BH048/057/066 recombinants represent early strains of a circulating recombinant form of HIV-1 in southern China. Information concerning such newly appearing variants is critical for both HIV-1 control and ongoing HIV-1 vaccine trials in Guangxi, China.
The routes of transmission of human immunodeficiency virus type 1 (HIV-1) in China have been expanding since it was first discovered there.1 Several early outbreaks of HIV-1 infection occurred in southern China, which is located on the trafficking routes for heroin and other drugs from southern/southeastern Asia into China.2,3 Therefore, HIV-1 infections in the early outbreaks occurred mainly among injection drug users (IDUs).2–5 Although sexual activity has recently become the major pathway for HIV-1 transmission, transmission via IDUs still requires attention. New HIV-1 infections continue to be reported among IDUs, especially in southern China,6 which may lead to the generation of novel circulating recombinant forms (CRFs) of HIV-1. These CRFs should be beneficial to the virus, because they combine different gene fragments from different subtypes, making it easier for the virus to adapt to local cell tropisms, counteract host defense mechanisms, and/or spread under evolutionary selection pressure.6,7 Here, we report that we have identified a novel recombination form among three HIV-1 sequences retrieved from IDUs in Beihai city in Guangxi province, China.
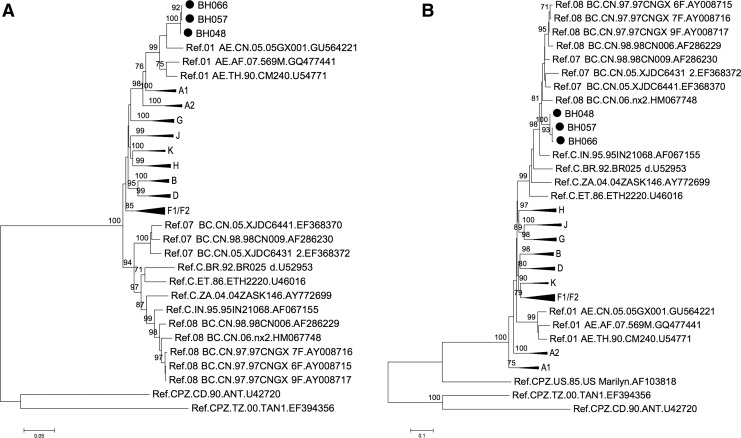
Previously, in Guangxi province, we successfully retrieved a total of 65 HIV-1 sequences from patients who were infected with HIV-1 through injection drug use.6 Analysis of partial gag/pol and env fragments with the help of MEGA58 suggested that these strains belonged to existing subtypes such as CRF01_AE, CRF07_BC, and CRF08_BC,6 with CRF08_BC and CRF01_AE being the predominant forms in southern China as seen in previous studies.9–11 However, three additional strains, termed BH048, BH057, and BH066, presented inconsistent subtyping results in the gag/pol and env regions (Fig. 1), suggesting the possible existence of HIV-1 recombination forms of CRF01_AE and CRF08_BC among these samples from IDUs in Beihai.
FIG. 1.
Phylogenetic analyses of HIV-1 sequences retrieved from Beihai city in China reveal possible recombination events. (A) Phylogenetic analysis performed with retrieved gag/pol sequences from Beihai city suggests that BH048/057/066 belong to CRF01_AE. (B) Phylogenetic analysis performed with retrieved env sequences from Beihai city suggests BH048/057/066 likely evolved from subtype CRF08_BC, based on the locally predominant HIV-1 subtypes. The reference sequences were retrieved from the HIV database at the Los Alamos National Laboratory (www.hiv.lanl.gov). The sequences used correspond to positions 1,850–2,961 in HXB2 for gag/pol and 6883–7642 for env. Nucleotide sequence alignment was performed with the help of MEGA5 software. Phylogenetic trees were constructed with the neighbor-joining statistical method and Kimura two-parameter as the model. The phylogenetic tree was determined for 1,000 replicates with random seeds. Only strong bootstrap values (>70%) are shown. The scale bar shows nucleotide substitutions per site.
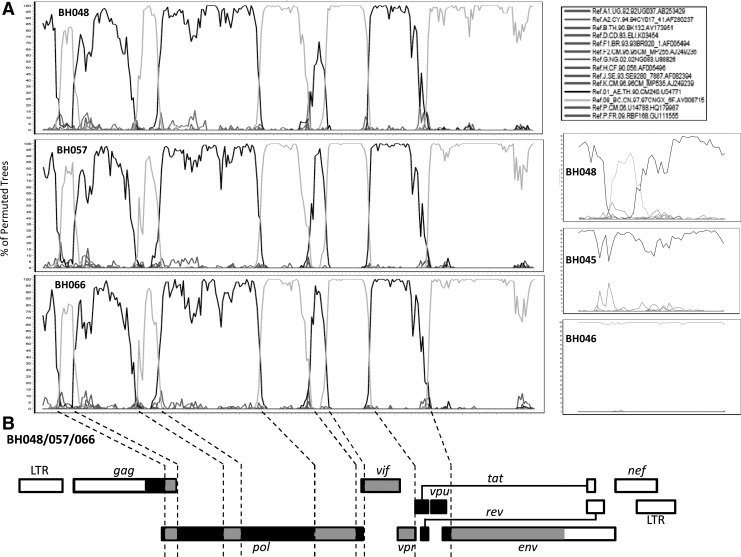
To further confirm our recombination events, we performed single nested polymerase chain reaction (PCR) with the help of ExTaq DNA polymerase (TaKaRa, Japan) and successfully retrieved the 4-kb fragments between known gag/pol and env sequences of these HIV-1 strains. SimPlot (version 3.5.1)12 was used to reveal the existence of recombination, and similarity tests (data not shown) were later done to confirm the recombination. The combined sequence of BH048 (covering HXB2 1850–7642) revealed a complex recombination pattern. Recombination between the parental subtypes (i.e., CRF01_AE and CRF08_BC) occurred several times along the sequence (Fig. 2A). Indeed, a mosaic pattern was observed in both gag and pol, which is different from bootscanning results of BH045 and BH046 that were sampled at the same time as BH048 but were previously determined to be CRF01_AE and CRF08_BC, respectively.6 CRF08_BC contributed to vif and vpr, while CRF01_AE was the source of vpu and at least part of tat and rev (Fig. 2B). Interestingly, although it was genetically related to CRF08_BC, the env fragment 6392–7642 of BH048 still contained on average 167 nucleotide (or 54 amino acids among Env fragment 58–473) differences compared to the CRF08_BC strains circulating in China; the magnitude of the differences was similar to what appeared between subtypes C, CRF07_BC, and CRF08_BC (Table 1), suggesting that the BH048 env sequence might represent a significant variant of the CRF08_BC env fragment. Surprisingly, despite the evolved env fragments, bootscanning results for BH048/057/066 demonstrated an extremely similar recombination pattern among all three HIV-1 sequences (Fig. 2A).
FIG. 2.
A novel recombination form of HIV-1 was detected in Beihai city. (A) Bootscanning analysis of BH048/057/066. The results indicate that these sequences present a similar, yet mosaic, pattern of CRF01_AE/CRF08_BC recombination. The sequences used correspond to positions 1,850–7,642 in HXB2. BH048/057/066 were first aligned with HIV-1 reference sequences retrieved from the LANL HIV-1 database (www.hiv.lanl.gov). BH048, 057, or 066 was set as the query sequence and the bootscanning was performed with a window size of 200 nt and steps of 20 nt, with the help of SimPlot software. Bootscanning results among the gag/pol fragment (HXB2 1,850–2,961) of BH048, BH045 (CRF01_AE), and BH046 (CRF08_BC) were shown on the right for direct comparison of the recombination events. (B) A schematic cartoon showing recombination in the BH048 genome. Black indicates CRF08_BC, light gray indicates CRF01_AE, and white indicates unsequenced regions. Dashed lines were used to indicate possible break points determined by bootscanning.
Table 1.
Number of Different Nucleotides/Amino Acids Among env Fragments Between Subtypes C/CRF07_BC/CRF08_BC and BH048
| Nucleotide | ||||
|---|---|---|---|---|
| Subtypes | BH048 | C | CRF07_BC | CRF08_BC |
| BH048 (n=3) | 17±5.2 | |||
| C (n=6) | 186±19 | 131±26 | ||
| CRF07_BC (n=27) | 192±13 | 176±23 | 110±30 | |
| CRF08_BC (n=16) | 167±18 | 135±24 | 179±25 | 95±35 |
| Amino acid | ||||
|---|---|---|---|---|
| BH048 (n=3) | 9±1 | |||
| C (n=6) | 67±2 | 61±8 | ||
| CRF07_BC (n=27) | 66±13 | 63±16 | 56±24 | |
| CRF08_BC (n=16) | 54±8 | 64±8 | 62±17 | 49±17 |
Sequences corresponding to HXB2 6392–7642 in the RNA genome (or Env 58–473 in protein sequence). The analysis was performed with MEGA5. Data are shown as means±SD. Subtype C/CRF07_BC/CRF08_BC sequences circulating in China were retrieved from the HIV database at the Los Alamos National Laboratory (www.hiv.lanl.gov).
Patients with BH048/057/066 (hereafter designated BH048) were all male and were in their 30s when the samples were obtained in 2008, but they shared no other common demographic characteristics, such as marriage status, occupation, or period of IDU. The only apparent connection among these people was their use of injection drugs. Therefore, we believed we might have discovered a new CRF of HIV-1 in Guangxi province, and we therefore checked the circulating status of BH048. Literature screening revealed that HIV-1 strain GXDY129913 and strain 226110 share an extremely similar recombination pattern with BH048, yet GXDY1299 and 2261 are transmitted through sexual contact, not injection drug use. Only 11 different nucleotides were detected between BH048 and 2261 in overlapping fragments 1850–5096. Thus, strains of BH048 are now circulating among IDUs and sexual contacts, two high-risk groups for HIV-1 transmission in China.
Recently, new HIV-1 recombination patterns have been reported in China10,13; most of them, however, have been determined to be unique recombinant forms (URFs) that may not actually circulate. URFs are more easily detected among IDUs because of these individuals' high-risk activities, such as sharing needles. Here we report that we have also detected a novel recombinant form of HIV-1 among IDUs in the Guangxi province of China. CRF01_AE and CRF08_BC entered Guangxi province through different drug trafficking routes,2 became locally dominant HIV-1 subtypes,6 and have now merged again among IDUs to generate recombinant strains of BH048/057/066.
Alerts should be raised in response to the generation of this new recombinant form of BH048, which may be circulating. Evidence from the sequencing of partial or nearly full-length HIV-1 genomes indicates that the 2261 and GXDY1299 strains detected after 2009 share high-level similarity in terms of both nucleotide sequence and recombination pattern with BH048. Thus, our sequences represent an early form of this new HIV-1 CRF. More importantly, both GXDY1299 and 2261 were transmitted through heterosexual activity, indicating that BH048, which was generated among IDUs, has already started to be transmitted through sexual contact, currently a predominant pathway for HIV-1 transmission. Therefore, although the criteria for a novel CRF designation14 have not been met, the extremely complex recombination pattern identically observed among all five sequences retrieved by three independent studies suggests that HIV-1BH048 has continued to spread during the past several years, has moved from one high-risk group to another, and is still responsible for novel infection events.
In addition, with the help of the HIV Drug Resistance Database at Stanford University (http://hivdb.stanford.edu),15 we detected antiviral drug-resistant mutations in two of these three strains (V179D in the reverse transcriptase of strain BH048 and possibly E138K in the integrase of BH057). Therefore, uncontrolled transmission of this new HIV-1 CRF may be coupled with the spread of drug-resistant mutations. It should also be noted that all the strains of this novel CRF were detected in Guangxi province, where a phase II HIV-1 vaccine trial is currently being conducted. The evolved env sequences detected in BH048 suggest an adaptation of this new CRF to local cell tropism, which points to a serious need for further attention and possible adjustments to the process of anti-HIV-1 vaccine development.
Sequence Data
All HIV-1 sequences retrieved from this study have been submitted to GenBank, with accession numbers KF803577 to KF803579.
Acknowledgments
We thank the staff at the Centers for Disease Control and Prevention in Beihai city for sample collection, Drs. Haiyan Qi, Linzhang Li, and Fei Xu for technical assistance, and Deborah McClellan for editorial assistance.
Written consent was obtained from all participants involved in the study. This study was reviewed and approved by the institutional review board of Guangxi Medical University in Nanning, Guangxi, China.
This work was supported in part by funding from the Chinese Ministry of Science and Technology (2012CB911100 and 2013ZX0001-005) and the Chinese Ministry of Education (IRT1016); the Key Laboratory of Molecular Virology, Jilin Province (20102209), China; and a grant (2R56AI62644-6) from the NIAID.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Teng T. and Shao Y: Scientific approaches to AIDS prevention and control in China. Adv Dent Res 2011;23(1):10–12 [DOI] [PubMed] [Google Scholar]
- 2.Beyrer C, Razak MH, Lisam K, et al. : Overland heroin trafficking routes and HIV-1 spread in south and south-east Asia. AIDS 2000;14(1):75–83 [DOI] [PubMed] [Google Scholar]
- 3.Chu TX, Levy JA. Injection drug use and HIV/AIDS transmission in China. Cell Res 2005;15(11–12):865–869 [DOI] [PubMed] [Google Scholar]
- 4.Piyasirisilp S, McCutchan FE, Carr JK, et al. : A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J Virol 2000;74(23):11286–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu XF, Liu W, Chen J, et al. : Maintaining low HIV type 1 env genetic diversity among injection drug users infected with a B/C recombinant and CRF01_AE HIV type 1 in southern China. AIDS Res Hum Retroviruses 2002;18(2):167–170 [DOI] [PubMed] [Google Scholar]
- 6.Qi H, Zhao K, Xu F, et al. : HIV-1 diversity, drug-resistant mutations, and viral evolution among high-risk individuals in phase II HIV vaccine trial sites in southern China. PLoS One 2013;8(7):e68656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Chen L, Yang S, et al. : Recombination form and epidemiology of HIV-1 unique recombinant strains identified in Yunnan, China. PLoS One 2012;7(10):e46777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura K, Peterson D, Peterson N, et al. : MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28(10):2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laeyendecker O, Zhang GW, Quinn TC, et al. : Molecular epidemiology of HIV-1 subtypes in southern China. J Acquir Immune Defic Syndr 2005;38(3):356–362 [PubMed] [Google Scholar]
- 10.Li L, Chen L, Liang S, et al. : Subtype CRF01_AE dominates the sexually transmitted human immunodeficiency virus type 1 epidemic in Guangxi, China. J Med Virol 2013;85(3):388–395 [DOI] [PubMed] [Google Scholar]
- 11.Li L, Sun G, Liang S, et al. : Different distribution of HIV-1 subtype and drug resistance were found among treatment naive individuals in Henan, Guangxi, and Yunnan province of China. PLoS One 2013;8(10):e75777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lole KS, Bollinger RC, Paranjape RS, et al. : Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999;73(1):152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang N, Wei H, Xiong R, et al. : Near full-length genome characterization of a new CRF01_AE/CRF08_BC recombinant transmitted between a heterosexual couple in Guangxi, China. AIDS Res Hum Retroviruses 2014;30(5):484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson DL, Anderson JP, Bradac JA, et al. : HIV-1 nomenclature proposal. Science 2000;288(5463):55–56 [DOI] [PubMed] [Google Scholar]
- 15.Liu TF. and Shafer RW: Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis 2006;42(11):1608–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]