Abstract
Background
Current strategies fail to identify most patients with esophageal adenocarcinoma (EAC) before the disease becomes advanced and incurable. Given the dismal prognosis associated with EAC, improvements in detection of early-stage esophageal neoplasia are needed.
Aims
We sought to assess whether differential expression of microRNAs could discriminate between squamous epithelium, Barrett’s esophagus (BE), and EAC.
Methods
We analyzed microRNA expression in a discovery cohort of human endoscopic biopsy samples from 36 patients representing normal squamous esophagus (n=11), BE (n=14), and high-grade dysplasia (HGD)/EAC (n=11). RNA was assessed using microarrays representing 847 human microRNAs followed by qRT-PCR verification of nine microRNAs. In a second cohort (n=18), qRT-PCR validation of five miRNAs was performed. Expression of 59 microRNAs associated with BE/EAC in the literature was assessed in our training cohort. Known esophageal cell lines were used to compare miRNA expression to tissue miRNAs.
Results
After controlling for multiple comparisons, we found 34 miRNAs differentially expressed between squamous esophagus and BE/EAC by microarray analysis. However, miRNA expression did not reliably differentiate non-dysplastic BE from EAC. In the validation cohort, all five microRNAs selected for qRT-PCR validation differentiated between squamous samples and BE/EAC. Microarray results supported 14 of the previously reported microRNAs associated with BE/EAC in the literature. Cell lines did not generally reflect miRNA expression found in vivo.
Conclusions
These data indicate that miRNAs differ between squamous esophageal epithelium and BE/EAC, but do not distinguish between BE and EAC. We suggest prospective evaluation of miRNAs in patients at high risk for EAC.
Keywords: Barrett esophagus, esophageal neoplasms, microarray analysis, microRNAs
Introduction
Risk stratification in esophageal neoplasia is inadequate. More than 90% of patients with esophageal cancer are never diagnosed with a pre-malignant condition[1,2]. Endoscopic screening and surveillance for esophageal neoplasia are expensive and have never been proven to benefit patients in a randomized control trial [3]. Rather than presenting at a stage when intervention is feasible, most patients with esophageal cancer present symptomatically with advanced stage disease, by which time cure is unlikely. Improving the current 17% five-year survival [4] associated with esophageal cancer will require a better strategy for early identification and intervention.
The current clinical approach to Barrett’s esophagus (BE) and esophageal adenocarcinoma (EAC) in the U.S. focuses on BE as the major risk factor for EAC. As suggested by American Gastroenterological Association (AGA) guidelines, patients with risk factors for EAC are screened for BE, and if BE is identified, they are entered into a program of surveillance endoscopy [3]. Patients eligible for screening are currently identified by the following factors: age 50 years or older, male sex, white race, chronic gastroesophageal reflux disease (GERD), hiatal hernia, increased body mass index, increased intra-abdominal body fat [3]. However, these risk factors identify a large proportion of the population, and will identify relatively few cancers. Men who are over 60 with GERD have a risk of BE of only 9.3% [5]. Also, the strategy of targeted screening for BE was based upon a suspected progression rate from BE to EAC of 0.5%/year [6]. However, recent data suggest that the rate of progression is markedly less, perhaps as low as 0.12%/year [2]. This rate of progression is similar to the complication rate traditionally associated with upper endoscopy [7]. Ideally, endoscopy could be reserved for patients who would benefit most from screening, while minimizing harm in the large number of patients who have some risk factors for EAC, but will never actually develop the disease.
MicroRNAs (miRNAs) are small non-coding RNA that modify gene expression within cells [8]. Increasingly, miRNAs are being considered as potential biomarkers because they are relatively stable molecules, less prone to degradation than larger RNA molecules. In addition, miRNAs have been associated with unique patterns of expressions in cancers [9] and identified with several different cancer types as both oncogenes and tumor suppressors [10] [9,11] [12] [13]. We hypothesized that differential expression of miRNAs may be associated with different phases of esophageal neoplasia from squamous mucosa to BE to EAC.
Methods
Discovery set
To evaluate miRNA expression in BE, total RNA was extracted from 36 endoscopically-obtained unique biopsy samples from patients undergoing upper endoscopy for an indication of either clinically-diagnosed GERD or histologically verified BE. While GERD and/or known BE were the clinical indications for endoscopy, patients with erosive esophagitis were excluded from this discovery miRNA analysis to minimize introduction of injury-associated miRNA signal. Patients had been enrolled in the University of North Carolina (UNC) at Chapel Hill esophageal tissue repository, a well-annotated repository that contains samples of squamous esophagus, BE and EAC from adults presenting for upper endoscopy. Each sample had a corresponding pathologic specimen from adjacent tissue interpreted by an expert GI histopathologist. All patients provided informed consent, and the research was approved by Institutional Review Boards at UNC and Duke University.
Samples consisted of three separate phenotypes: squamous esophagus (n=11), BE with no dysplasia or low-grade dysplasia (n=14), and BE with high-grade dysplasia/superficial EAC (n=11).
Clinical data were prospectively collected by a study coordinator using the medical record and questionnaires completed at the time of endoscopy. Body mass index was calculated from measurements obtained in duplicate. Smoking status was dichotomized as an ever/never variable. Prior history of other non-esophageal cancers was elicited. Previous and current endoscopic findings including diagnosis of erosive esophagitis and BE were determined from patient report, the medical record and pathologic reports. BE was defined as any length of endoscopically visible columnar mucosa in the tubular esophagus, with biopsies that demonstrated intestinal metaplasia [14]. Unmatched samples representing different tissue phenotypes were selected from the biorepository for genomic analysis.
RNA extraction
RNA extraction from frozen de-identified samples was performed using mirVana kits (Ambion, Santa Clara, CA). Quality of the RNA was assessed based upon the RNA-integrity number (RIN score) [15].
Inclusion/Exclusion
Acceptable samples were defined as those with a RIN score greater than six. Due to reported changes in systemic (rather than tissue specific) changes in miRNA expression in cancer, patients who carried a known prior diagnosis of a non-esophageal cancer were excluded from this analysis. In addition, while all patients had a diagnosis of GERD or BE as the indication for endoscopy, patients with erosive esophagitis at the time of endoscopy were excluded.
miRNA Microarray
After quantification of total RNA, the RNA from each sample was labeled with a biotinilated signal molecule using FlashTag Biotin HSR RNA labeling. Biotin-labeled samples and hybridization mastermix were incubated and injected into arrays. Then arrays were washed and scanned using standard Affymetrix (Santa Clara, CA) protocol. Affymetrix GeneChip miRNA Arrays representing 847 human miRNA probe sets were used to characterize miRNA expression from each sample. Careful attention was paid to the six essential elements of Minimum Information about a Microarray Experiment (MIAME 2.0) standards in experimental design and data integrity assurance. Details of analysis can be found in the statistical section at the end of this methods section. Briefly, the R statistical environment [R] version 2.15.1 [16] was used for all analysis. Code was written to link clinical annotation to cel files with the original file names and the MD5 Message-Digest algorithm was used to assign a digital fingerprint to the clinical data and cel files. As described later, the Robust Multichip Average (RMA) algorithm from the Bioconductor [17] extension package affy (version 1.34.0) was used to pre-process the CEL files for background correction, normalization, and calculation of expression values [18]. We compared pre-processing using RMA to pre-processing using the miRNA QC tool available from Affymetrix and found comparable results.
Verification of microarray results with qRT-PCR
Quantitative real-time polymerase chain reaction (qRT-PCR) for miRNAs was performed on the same samples used for the microarray-based approach. The primary goal of qRT-verification was to determine if the microarray and qRT-PCR assays would indicate similar patterns of miRNA expression for a subset of probes represented on the microarrays (selection process described in the analysis section). Because qRT-PCR results may be more sensitive and specific than microarrays for miRNAs, it has been suggested that other methods, such as qRT-PCR be used to verify microarray results [19]. In this study, given limitations of availability of qRT-PCR reagents as well as a limiting quantity of original RNA, only a subset of nine miRNAs from the initial microarray analysis was selected for verification by qRT-PCR, and all results are reported. TaqMan miRNA primers and reagents from Ambion (Life Technologies, Carlsbad, CA) were used. Additional details are listed in supplemental materials and primers are listed in Supplemental Table 1. A threshold cycle (CT) value was determined for each sample for analysis of qRT-PCR results. RNU48 was selected as the control probe in all qRT-PCR analyses after comparing its performance to RNU6 (by qRT-PCR) and finding less variability in expression for RNU48. One squamous sample was excluded from qRT-PCR verification analysis due to a technical problem; this resulted in 35 samples total for the PCR analysis portion of the study.
Validation set
A separate validation set was established to assess patterns of expression of miRNAs found to be differentially expressed by microarray and qRT-PCR analysis in the discovery set. Eighteen additional samples from UNC and the Duke Gastrointestinal Tissue Repository were selected for creation of the validation cohort. Phenotypes included: squamous epithelium from patients with reflux without BE (n=7), squamous epithelium from patients with BE (n=4), Barrett’s biopsies (n=4), and EAC (n=3). Samples were removed from this repository under an IRB protocol for miRNA expression studies. RNA extraction was performed as described above, and quality of RNA was assessed with RIN scores.
Analysis/Data Integrity
The microarray CEL files were pre-processed using the Robust Multichip Average (RMA) approach [20]. There were 7815 features assessed for each sample based upon the entire array. All 36 samples from the initial dataset were included in the pre-processing. Further analysis of microarray data was limited to human probes as characterized by “hsa” probe set ids; this limited the set of miRNA features to 847 human probe sets.
In the discovery phase, we evaluated 847 probe sets from the miRNA microarray and generated a list of the 34 miRNAs that demonstrated directional differences in expression across the three ordered phenotypes: squamous, BE and EAC. Given the risk of multiple comparisons and false-discovery associated with microarray experiments, a family-wise error rate was used to adjust p-values and in the discovery phase. Microarray expression values were used to calculate fold change between squamous and BE phenotypes and also squamous and EAC. The goal was to identify miRNAs that demonstrated a step-wise increase or decrease in expression from squamous to BE to HGD/EAC for further validation.
Then, in the next step of the discovery phase, the top 34 miRNAs identified using microarray methods in the discovery set were narrowed to a list of nine miRNAs for reasons of statistical power (avoiding multiple comparisons) and experimental feasibility. The following selection criteria were used to narrow the list. First, miRNAs that represented a complementary strand (as indicated by a “star”) were excluded from further analysis based upon the concern that complementary strands could be unstable and degraded (8 of 34) [21]. A bioinformatics approach was taken as per the National Center for Biotechnology and Informatics (NCBI) and miRBase databases (www. http://www.mirbase.org/_) [22] to determine the location of the miRNAs on the genome, to associate clusters of miRNAs, and to identify related scientific citations. When two miRNAs from the same cluster were available for analysis, one miRNA with strong expression in the microarray data was selected for qRT-PCR verification. Some of the final selection of nine miRNAs for verification was based upon reported association of miRNAs with cancers in the literature, and this was the most subjective part of the selection process. Thus, we present not an exhaustive assessment of all of the “top hits” identified through microarray analysis, but a targeted assessment of nine miRNAs. Finally, a miRNA was excluded that previous experience demonstrated would be difficult to verify with qRT-PCR (miR-30a).
The evaluation of nine miRNAs by qRT-PCR in 35 of the original RNA samples that had been used for microarray analysis were then used to select a set of miRNAs for validation in a separate dataset. The miRNAs selected for phenotypic validation in the separate dataset were chosen based upon demonstration of concordant direction of expression between microarray and qRT-PCR in discovery phase, strength of expression, and relatedness of miRNAs (27a and 27b).
qRT-PCR Analysis
In TaqMan qRT-PCR analysis, a threshold representing a level of fluorescence signal detection is determined. The threshold cycle (CT) values represent the PCR cycle number when reporter fluorescence became greater than the determined threshold. Higher CT values indicate that it took more cycles to detect the particular miRNA of interest, meaning that there is less of the miRNA of interest present in that sample. CT values in these experiments were generated using a manually-determined threshold of 0.4 after visual inspection of the amplification curves (to ensure parallel areas of the amplification curve). Average CT values were determined for each sample based upon a combination of all replicates for that sample and miRNA probe. Delta CT values were defined as the difference between probe and control and were determined based upon the difference between the RNU48 control probe average CT and the average CT of the probe of interest. Simply put, the average delta CT quantitatively indicates the difference in the number of PCR cycles when the probes were detected. The log (base 2) ratio of Delta CT of the probe and control was used to quantify relative expression.
As was performed in the microarray analysis, miRNA probes were tested across the three phenotypes. Concordance [23] was evaluated to quantify the association between expression values for the microarray and qRT-PCR data for each sample.
Based upon discovery cohort results, which indicated more pronounced differences between squamous and BE, and generally similar expression between BE and HGD/EAC, we did not expect that this set of miRNAs would be able to determine a consistent difference between BE and EAC samples in the validation cohort. Thus, the qRT-PCR validation analysis was designed to evaluate expression between squamous samples (n=11) and a combined group of BE and EAC samples (n=7).
Literature Validation
To test the utility of miRNAs previously reported to be associated with BE and EAC, an additional analysis was performed using a list of candidate miRNAs gathered from a systematic review of the literature. MEDLINE was searched and original reports assessing miRNA expression in the BE adenocarcinoma sequence were obtained using combined search terms of “miRNA or microRNAs” and “Barrett’s esophagus or esophageal neoplasia,” yielding a list of 94 reports (search performed in September 2012). Of these, six papers [24–30] were primary reports of overall miRNA expression including either BE or EAC and provided sufficient detail about experiments and miRNA expression results to allow generation of a list of differentially expressed miRNAs. Expression of each miRNA was assessed using a non-parametric rank test.
Cell Line Validation
Finally, to compare miRNA expression from the in vivo experiments to those obtained from culture, four cell lines that are frequently used to study BE and EAC were grown in culture. Two squamous cell lines were included: NES-G2T (from patient without BE) and NES-B3T (from a patient with BE)[31,32]. A Barrett’s cell line, BAR-T, was also included [32–34]. These telomerase-immortalized cell lines (provided courtesy of Rhonda Souza, MD), have been described extensively in the Barrett’s literature. The OE-33 cell line (Sigma-Aldrich, St Louis, MO) is a cell line derived from a BE-related EAC was the fourth cell-type assessed. Cells were grown under standard conditions: OE33 cells were grown in RPMI 1640 with 2mM glutamine (Sigma-Aldrich, St Louis, MO) and 10% fetal bovine serum while the squamous and Barrett’s cell lines were grown per protocols developed by the Souza lab, [32,33] Cells were grown to 80% confluence, and then RNA was extracted from cells using mirVana kits. RNA quality was assessed, and only high-quality RNA was used for microarray experiments (RIN 10). Three biologic replicates were used to generate biological replicates for each of the different cell lines.
As described for the human samples in the training cohort, miRNA expression was assessed for the four cell lines using Affymetrix miRNA arrays. The set of miRNA probes that demonstrated differential expression in our human biopsy samples was specifically evaluated in the cell lines to determine if cells in culture reflected patterns of expression in human tissue.
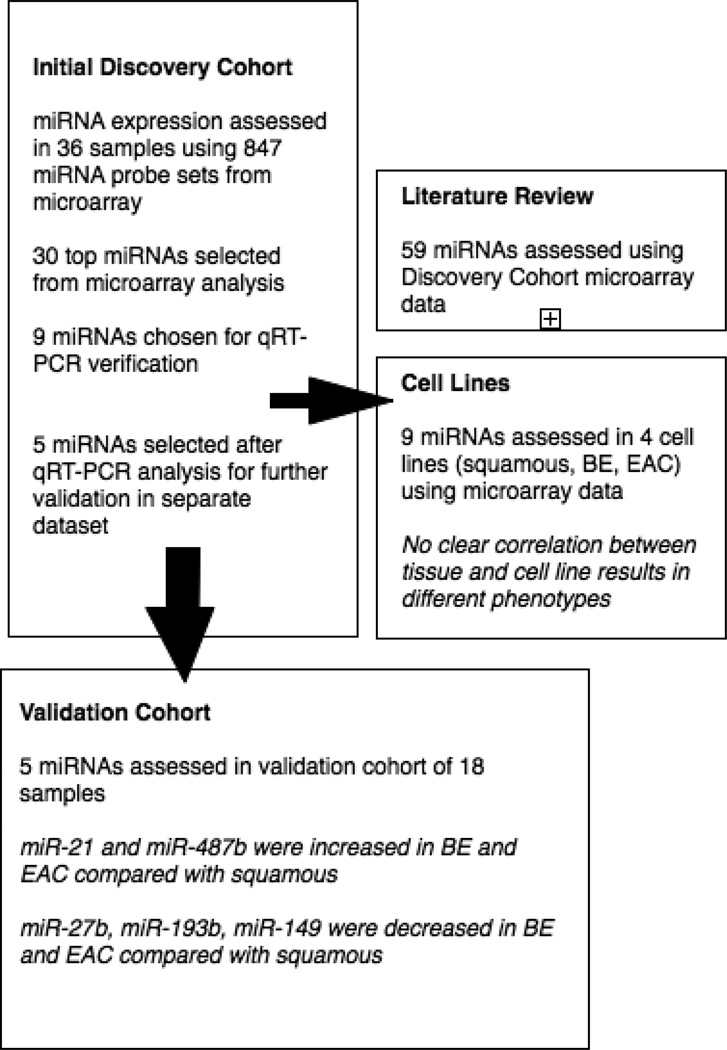
The Flow Diagram in Figure 1 provides a schematic of the analysis performed from discovery set through validation and indicates how the literature review and cell line analysis are related to the discovery cohort.
Figure 1.
This flow diagram represents the steps in the analysis using the discovery cohort and a separate validation cohort. Several steps we involved as the initial set of 847 miRNA probe sets from the microarray was narrowed to a list of five miRNAs for validation.
Statistical Considerations
All statistical analyses were carried out using the R statistical environment [R] version 2.15.1 [16]. The implementation of the Robust Multichip Average (RMA) algorithm from the Bioconductor [17] extension package affy (version 1.34.0) was used to pre-process the CEL files[18]. In the discovery cohort, the Jonckheere-Terpstra test [35] [36] , a non-parametric rank test, was used to generate the list of 34 miRNAs from the microarray analysis, to analyze qRT-PCR results in the discovery cohort, and to analyze the microarray data for literature validation and analysis of cell line results. The implementation of the Jonckheere-Terpstra test from the clinfun extension package version 1.0.1 [37] was used in this analysis. The Kendall Tau [23] measure of concordance was used to assess concordance between microarray expression, and qRT-PCR cycle number. Validation was defined a priori via Wilcoxon rank sum test of the validation cohort; i.e. the corresponding one-sided family-wise error rate (FWER) (to limit type I error) was less than 0.05. The lattice (version 0.20-6) extension package was used for generating figures [38].
Results
Clinical features and demographics of the 36 patients in the initial cohort are represented in Table 1. Patients with HGD/EAC tended to be older, and were more likely to be male than patients from whom squamous or BE samples were obtained. Patients with HGD/EAC also tended to have a higher average body mass index, and were more commonly smokers.
Table 1.
Demographics characterizing the patients in the initial cohort of 36 patients.
| Squamous esophagus samples (n= 11) |
BE samples (n= 14) |
HGD/EAC (n= 11) |
|
|---|---|---|---|
| Age – mean (sdv) | 51.2 (11.2) | 59.5 (13.1) | 63.5 (7.9) |
| Men – n (%) | 6 (55%) | 8 (57%) | 9 (82%) |
| Race (white) – n (%) | 8 (72%) | 14 (100%) | 10 (91%) |
| BMI – mean (sdv) | 27.3 (5.9) | 27.0 (4.2) | 31.2 (5.0) |
| Smoking – n (%) | 6 (55%) | 8 (57%) | 10 (91%) |
The initial microarray analysis on the training cohort yielded the top 34 miRNAs with differential expression when assessing the three tissue types: squamous esophagus, BE, and HGD/EAC. The complete list of top 34 miRNAs with raw and p-values adjusted for multiple comparisons, as well as fold change, is reported in Supplemental Table 2. Also shown in Supplemental Table 2, is the direction of expression in BE/EAC relative to squamous epithelium is reported for the nine miRNAs selected for qRT-PCR verification.
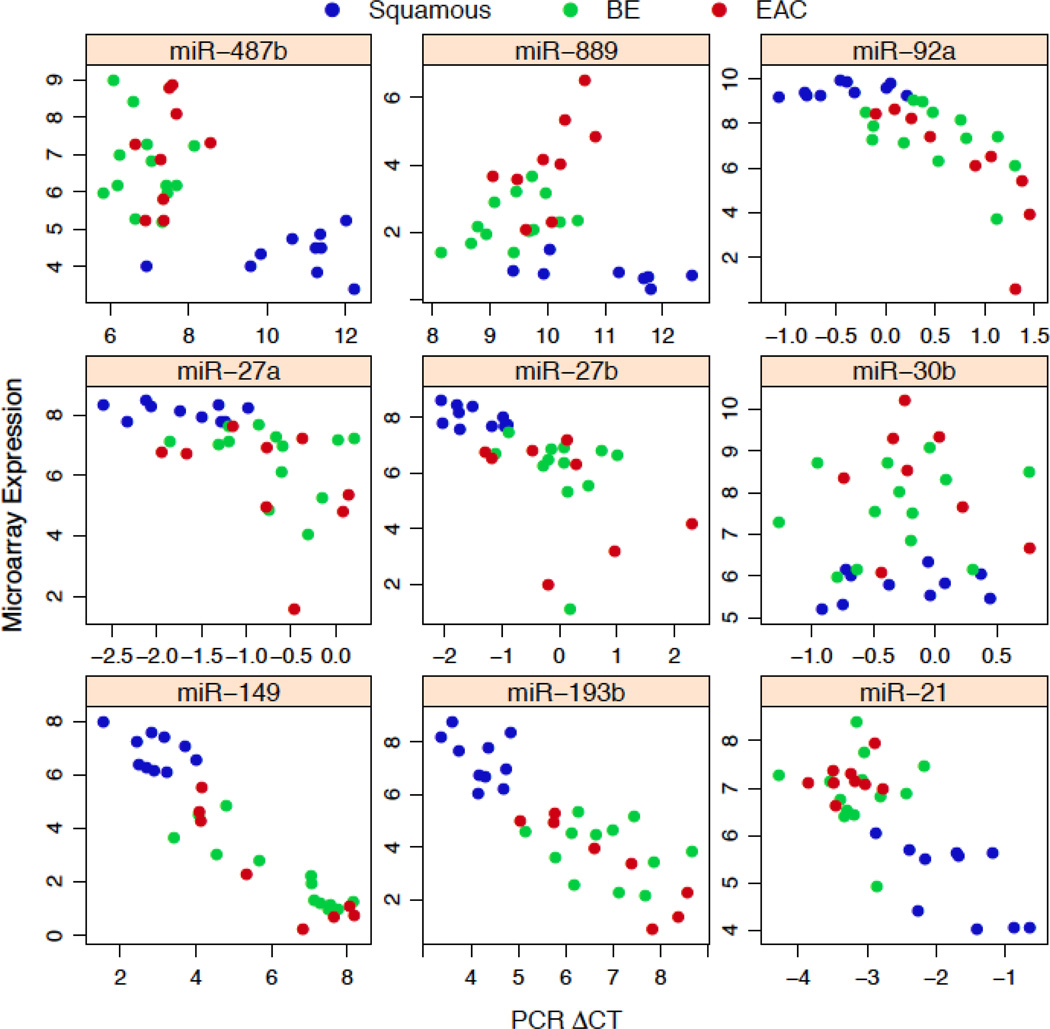
Results of microarray-based miRNA expression plotted against delta CT values from qRT-PCR are shown in Figure 2 for nine miRNAs. This figure demonstrates two important points. First, the direction of the miRNA expression is shown and the direction of expression can be compared between microarray and qRT-PCR results. The predicted inverse relationship between microarray expression and delta CT values exists for most of the miRNAs. The one exception is miR-30b where there is no correlation between microarray and qRT-PCR. Second, it is evident that a clearer distinction exists between squamous samples (shown in blue) and the other two tissue types than between BE and EAC, where there is significant overlap in the BE and HGD/EAC samples (green and red). As seen in these plots, the differences in expression were largely driven by the differences between the squamous and BE/EAC phenotypes.
Figure 2.
In this figure, the vertical axis represents expression by microarray analysis and the horizontal access represents the delta CT values for each of the nine miRNA selected for qRT-PCR verification of microarray results. This figure provides information on direction of expression by tissue type for both the microarray and PCR platforms. Blue represents squamous samples, green represent BE, while red represents EAC. miR-889, miR-487b, miR-30b and miR-21 increased in BE compared to squamous. miR-27a, miR-92a, miR-27b, miR-193b and miR-149 decreased in BE compared to squamous. The figure also demonstrates concordance between microarray and qRT-PCR results (high concordance appears as a diagonal line). In general, the miRNA expression differentiated squamous tissue from BE/EAC, but not BE from EAC.
On the basis of qRT-PCR analysis, seven of nine miRNAs assessed in the discovery cohort were of sufficient quality to warrant phenotypic validation. Specific p-values for the qRT-PCR analysis for all of the nine miRNAs are shown in Table 2. In addition, Table 2 reports the concordance between expression values from microarray and the qRT-PCR miRNA. As with the microarray results, differences in expression were much more pronounced between squamous esophagus and BE/EAC than between BE and EAC.
Table 2.
Significance test results of the discovery cohort for both microarray data and delta CT values for each qRT-PCR probe. Concordance between microarray and qRT-PCR results is presented in the fourth column; a negative tau is expected when there is a true relationship between microarray and qRT-PCR results given the inverse relationship between microarray expression and CT values.
| miRNA | p-value Microarray |
p-value qRT-PCR |
tau | p-value for concordance |
|---|---|---|---|---|
| miR-149 | 2.82e-05 | 2.83e-04 | −0.720 | 1.86e-10 |
| miR-193b | 4.65e-06 | 7.21e-05 | −0.660 | 2.46e-08 |
| miR-21 | 3.29e-05 | 6.95e-05 | −0.431 | 3.83e-04 |
| miR-27a | 1.71e-07 | 2.86e-03 | −0.452 | 1.89e-04 |
| miR-27b | 2.27e-06 | 7.19e-04 | −0.563 | 4.04e-06 |
| miR-30b | 7.78e-06 | 7.87e-01 | 0.0538 | 6.86e-01 |
| miR-487b | 1.30e-06 | 6.56e-03 | −0.343 | 5.47e-03 |
| miR-889 | 1.18e-10 | 3.82e-01 | −5.75e-02 | 6.71e-01 |
| miR-92a | 1.30e-06 | 4.87e-05 | −6.01e-01 | 2.42e-07 |
In general, we found significant concordance between microarray results and qRT-PCR results. In the verification of microarray results, if a true relationship existed between microarray and qRT-PCR results, an inverse relationship between expression values and log delta CT should be observed. Of the nine miRNAs assessed by microarray and qRT-PCR, only miR-30b did not reveal the expected inverse relationship and neither miR-889 nor miR-30b showed significant differential expression among the phenotype groups and were consequently excluded from further phenotypic validation.
For miR-889 and miR-487b, miRNA expression in squamous samples was extremely low, making the qRT-PCR results difficult to interpret as there was little separation between samples, and cycle number was high, indicating very low levels (estimated at 0–2 copies) of miRNA expression.
Validation
Five miRNAs: miR-21, miR-487b (both increased in BE and EAC) and miR-27b, miR-149, miR-193b (decreased in BE and EAC), were selected for further validation in a separate cohort. Discovery results guided the design of the validation study in terms of the phenotype comparison and the selection of five miRNAs for validation in a separate cohort. Both miR-30b and miR-889 were excluded based upon their performance in the qRT-PCR verification. Given very low levels of expression on verification qRT-PCR, miR-92a (part of the miR-106a cluster) was also excluded from the validation. Finally, miR-27b was selected for validation and miR-27a was excluded as miR-27b demonstrated higher overall expression.
In comparing the five miRNAs in esophageal squamous epithelium to samples with BE or EAC, all five of the miRNAs were significantly differentially expressed between squamous and BE. Table 3 provides the p-values for the comparison of the five probes in squamous versus BE/EAC samples.
Table 3.
Results of qRT-PCR Validation in Validation Cohort (n=18)
| miRNA | one-sided test p-value |
Direction in BE/EAC vs squamous |
Location on Genome |
|---|---|---|---|
| miR-21 | 0.00022 | increased | Chromosome 17q23.1 |
| miR-487b | 0.0077 | increased | Chromosome 14q32.31 |
| miR-27b | 0.0023 | decreased | Chromosome 9q22.32 |
| miR-149 | 0.00060 | decreased | Chromosome 2q37.3 |
| miR-193b | 0.00060 | decreased | Chromosome 16p13.12 |
All five miRNA probes performed in the hypothesized directions in the qRT-PCR validation. Based upon a priori determination of significance, (p-value < 0.01 for each individual probe in the one-sided Wilcoxon rank-sum test), miR-21, miR-27b, miR-149, and miR-193b all validated in this cohort, with significant differences in expression between squamous and BE/EAC samples.
Evaluation of miRNAs from the Literature
After performing a literature search for reports of miRNA expression in BE and/or EAC, we identified a list of 59 miRNA probes. Then, we assessed the association of those miRNA probes with BE and EAC in our discovery cohort. In the discovery dataset, 14 of the miRNAs were expressed in a direction consistent with that reported in the literature; Table 4 lists these 14 miRNA probe sets, for which the marginal p value is less than 0.0008 (= 0.05 / 59), indicating significance. (Results for all 59 miRNAs are reported in Supplemental Table 3.) Again, the differences in expression were observed primarily in the squamous to BE/EAC comparison.
Table 4.
Microarray Data for the miRNAs identified through Literature Review: In this table, we list all of the miRNA probes for which the marginal p-value is less than 0.0008 (=0.05 / 59).
| miRNA | Direction of Expression | Validation p-value |
|---|---|---|
| hsa-miR-143_st | Increased in BE/EAC [25,30] | 0.00038 |
| hsa-miR-145_st | Increased in BE/EAC [25,30] | 0.000323 |
| hsa-miR-21_st | Increased in BE/EAC [24,29,30] | 4.1 × 10−05* |
| hsa-miR-192_st | Increased in BE/EAC [24,26,28,29] | 1.39 × 10−05 |
| hsa-miR-194_st | Increased in BE/EAC [24,28,30] | 9.78 × 10−06 |
| hsa-miR-215_st | Increased in BE/EAC [28–30] | 7.16 × 10−05 |
| hsa-miR-203_st | Decreased in BE/EAC [24,25,29,30] | 7.54 × 10−05 |
| hsa-miR-205_st | Decreased in BE/EAC [24,25,29,30] | 1.17 × 10−05 |
| hsa-miR-27b_st | Decreased in EAC [24,25] | 1.22 × 10−05* |
| hsa-miR-99a_st | Decreased in EAC [25,28] | 3.53 × 10−05* |
| hsa-miR-149_st | Decreased in EAC [25] | 1.69 × 10−05* |
| hsa-miR-210_st | Decreased in EAC [25] | 0.000246 |
| hsa-miR-221_st | Decreased in EAC [25] | 1.89 × 10−05 |
| hsa-miR-617_st | Decreased in EAC [25] | 4.25 × 10−05* |
An asterix identifies miRNAs included in our list of 30 miRNAs identified by microarray in the discovery cohort.
When a miRNA had been reported in the BE and EAC literature multiple times, it was more likely to be associated with BE/EAC in our training set. The 14 miRNAs from the literature that met our significance threshold, controlling for multiple comparisons, were identified in an average of 2.5 references compared to 1.1 references for the group that did not validate.
Cell Lines
Of the nine miRNAs included in our microarray analysis of miRNA expression in cell lines, miR-92a expression by the cell lines most closely resembled the pattern of miRNA expression found in our set of human samples with lower expression in BE/EAC, yet these results differed miR-92 expression reported in the literature [29]. We found that the squamous cells (NES-G2T and NES-B3T) expressed higher levels of miR-92a than the BE cells with lower expression of miR-92a in the EAC-derived cell line OE33 cells (p-value 0.00592). Expression of miR21 also differed among cell lines: the adenocarcinoma cell line (OE33) had the highest level of expression and slightly lower expression was found in NES-B3T and BAR-T cells, however, the other squamous cell line (G2T) demonstrated miR-21 expression similar to the OE33 cells. Overall, the range of miRNA expression appeared to be much more restricted in the cell lines than in the human esophageal tissue, resulting in far smaller differences in microRNA expression across the spectrum of esophageal cell lines than between benign versus neoplastic tissue samples from patients.
Discussion
We assessed whether differential expression of miRNAs could discriminate between squamous epithelium, BE, and EAC. Based upon our analysis, we conclude that miR-21 and miR-487b (increased in BE and EAC) as well as miR-27b, miR-193b, miR-149 (all decreased in BE and EAC) demonstrate differential expression in squamous epithelium compared to BE and EAC. However, we did not find miRNAs that consistently differed between BE and EAC. These findings highlight the overall similarity in miRNA expression between BE and EAC [24,39] and suggest that miRNAs can be used to distinguish between squamous esophagus and BE, but not between BE, BE with dysplasia, and EAC.
Several of the miRNAs reported here are novel: miR-193b has not been previously reported as decreased in BE and EAC. Neither miR-889 nor miR-487b had previously been noted as increased in BE or EAC. Our qRT-PCR results revealed high CT values for miR-889 and miR-487b, indicating very low levels of expression in squamous epithelium compared to the increased expression in BE and EAC, making these potentially useful biomarkers.
The approach to select for miRNAs that increase or decrease expression from squamous to BE to EAC does not capture all biology (as some miRNAs may increase in the metaplasia from squamous to BE, but decrease in the progression to cancer). Several of the miRNAs identified as highly expressed in BE are linked to the intestinal phenotype, and are expressed in the mouse small intestine [40]. Logically, this similarity between BE and intestinal phenotype makes sense, and recently, investigators have begun to assess the role of specific miRNAs (such as miR-145) in the development of BE [41]. Some of the miRNAs that increase in BE then decrease in some EACs as the cancer phenotype develops. Because our goal was to advance the field of biomarker development, we focused on miRNAs with the step-wise increase across phenotypes, a reasonable strategy for possible biomarker identification that has been recently described by other groups [39].
Previous studies have described miR-203 and miR-205 as highly expressed in squamous epithelium compared to BE or EAC, while miR-21 and miR-194 have demonstrated increased expression in BE, gastric epithelium, and EAC compared to squamous esophageal epithelium [24],[28–30]. Most recently, Bansal and colleagues identified miRNAs that were differentially expressed in patients with Barrett’s esophagus with and without dysplasia [26]. However, these attempts have not resulted in a consensus list of miRNAs to be used in prospective studies to aid in risk-stratification. In an effort to derive such a list of likely biomarker candidates, we provided an additional analysis of these reported miRNAs in our own cohort, supporting some, but not all, of the previous findings.
We were able to reproduce results from the literature for 14 miRNAs previously reported as associated with BE/EAC using our microarray dataset. In our analysis, miR143, miR145, miR-21, miR-192, miR-194, miR-215 were all increased in BE and EAC while miR-203, miR-205, miR-27b, miR-99c, miR-149, miR-210, miR-221, and miR-617 were all decreased in BE/EAC. Six of these miRNAs were identified by our initial original microarray analysis. However, these miRNAs did not reliably discern non-dysplasic BE from HGD/EAC.
As noted, none of the miRNAs tested reliably distinguished non-dysplastic BE from HGD/EAC. This has implications for how such a biomarker might be used. Because endoscopists do not generally have difficulty discerning between squamous and Barrett’s mucosa, assay of these miRNAs in endoscopic biopsies may be of limited utility. On the other hand, a biomarker that differentiates squamous from BE tissue might be of great use in the ongoing development of non-endoscopic methods for BE screening in a targeted population at risk for EAC based on clinical risk factors. An initial, relatively inexpensive, non-endoscopic test such as the recently described swallowed sponge[42] might act as a gateway to endoscopy for those who are likely to have BE based upon a miRNA-based biomarker, resulting in cost savings with little loss in diagnostic sensitivity.
There are several limitations to the work presented here. This is a cross-sectional study and the temporal appearance of the biomarkers is difficult to ascertain from such a study. To better understand the potential utility of miRNAs in risk stratification in BE, future studies should include a prospective cohort study of high-risk patients. Assessment of miRNAs from esophageal specimens, as well as miRNA expression from peripheral blood in these patients when they underwent upper endoscopies over years would be ideal. Another limitation is our reliance upon paired biopsies used to classify samples histologically. Using histologic classification as a gold-standard is not ideal, although in cross-sectional studies, there are no alternatives. It is well-known that pathologists experience considerable intra- and inter-rater disagreement in their classification of BE [43]. For this reason, a separate category of LGD was not included. Similarly, HGD and EAC were also grouped together in our cohort, representing advanced neoplasia. In general, pathologists agree on the general diagnosis of BE versus squamous tissue, and EAC versus BE. Cellularity of different tissue samples (squamous, BE, EAC) may not be equal, and this could account for some of our findings. We acknowledge that our validation sample-size was small, and as such, we limited the number of miRNAs selected for validation to only five. It is also worth noting that microarrays are limited as a miRNA detection technology by reliance upon uniform hybridization conditions and as such, do not always correlate with qRT-PCR microRNA results [19]. For the probes we assessed by qRT-PCR, correlation with microarray was good. While verification of several miRNAs identified through microarray was performed, future work could continue qRT-PCR verification of microarray findings. Finally, at the time that this study was designed and initiated, RNA-Seq, which is now rapidly becoming the gold-standard method for assessing RNA expression, was not readily available [44]. Even now, small RNAs may be difficult when miRNAs with low copy number are of interest [19].
There are also several strengths to this study. Our study design included separate discovery and validation cohorts. We approached the question of miRNA expression using an original analysis as well as an assessment of the reproducibility of results from miRNA studies published in the literature. The cell line experiments here add depth to this report, contrasting miRNA expression in human cell lines with concurrent in vivo work. The results of the in vitro experiments suggest a cautionary approach in the interpretation of cell culture studies of miRNA in the development of EAC, as the cell culture studies do not reflect findings found in vivo.
There are several potential explanations for differences between human tissue miRNA expression and that found in cell culture. Changes in miRNA expression could be related to the immortalization process, the cell culture media conditions, or a non-epithelial esophageal cell type may be needed to express or support expression of miRNAs in culture.
In our study, miR-21 differentiated squamous from BE tissue. More than any other microRNA, increased expression of miR-21 has been associated with many cancers [13]. In a transgenic animal model, miR-21 has been clearly linked to carcinogenesis. In the animals with over-expression of miR-21 in lymph and brain tissues, lymphoid tumors developed that then regressed after inactivation of miR-21. As for its gene targets, miR-21 targets the 3’-UTR of programmed cell death 4 (PDCD4) and miR-21 is a negative regulator of PDCD4 [45]. Fassan et al showed that the basal layer of normal squamous epithelium and the non-intestinal columnar metaplasia demonstrated PDCD4, but this target was lost in dysplasia and EAC as predicted [46]. Thus, miR-21 is an especially interesting miRNA because of the current data and given its putative function as a cancer-causing miRNA.
Our study suggests distinct miRNA expression in a comparison of squamous and BE/EAC tissue, with notable similarities between miRNA expression in BE and EAC. In this cross-sectional study, miRNAs did not distinguish between grades of dysplasia. While the current study cannot support the use of miRNAs as biomarkers for BE dysplasia, moving forward, these results may have the greatest clinical value in non-endoscopic screening for BE in patients who are at risk for EAC. This might advance care by targeting endoscopy to patients who are most likely to both have BE (based upon non-invasive miRNA biomarkers). Even the population of patients who are at increased risk of EAC based upon clinical risks such as male sex, white race, age, GERD history, family history or smoking status still has a relatively low risk of developing EAC. A non-invasive screening test for BE would help narrow the field of patients most likely to benefit from upper endoscopy.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by an American College of Gastroenterology Junior Faculty Development Award (KG) and by the Emeline Brown Cancer Genomics Research Fund.
References
- 1.Dulai GS, Guha S, Kahn KL, Gornbein J, Weinstein WM. Preoperative prevalence of Barrett's esophagus in esophageal adenocarcinoma: a systematic review. Gastroenterology. 2002;122:26–33. doi: 10.1053/gast.2002.30297. [DOI] [PubMed] [Google Scholar]
- 2.Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett's esophagus. N Engl J Med. 2011;365:1375–1383. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- 3.Spechler SJ, Sharma P, Souza RF, Inadomi JM, Shaheen NJ. American Gastroenterological Association medical position statement on the management of Barrett's esophagus. Gastroenterology. 2011;140:1084–1091. doi: 10.1053/j.gastro.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 4.National Cancer Institute. [Accessed March 14, 2012, 2012];SEER Cancer Statistics Review. 2011 Available at: http://seer.cancer.gov/csr/1975_2008/.
- 5.Rubenstein JH, Mattek N, Eisen G. Age- and sex-specific yield of Barrett's esophagus by endoscopy indication. Gastrointest Endosc. 2010;71:21–27. doi: 10.1016/j.gie.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaheen NJ, Crosby MA, Bozymski EM, Sandler RS. Is there publication bias in the reporting of cancer risk in Barrett's esophagus? Gastroenterology. 2000;119:333–338. doi: 10.1053/gast.2000.9302. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Menachem T, Decker GA, Early DS, et al. Adverse events of upper GI endoscopy. Gastrointest Endosc. 2012;76:707–718. doi: 10.1016/j.gie.2012.03.252. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Croce CM. miRNAs in the spotlight: Understanding cancer gene dependency. Nat Med. 2011;17:935–936. doi: 10.1038/nm0811-935. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spechler SJ, Sharma P, Souza RF, Inadomi JM, Shaheen NJ. American Gastroenterological Association technical review on the management of Barrett's esophagus. Gastroenterology. 2011;140:e18–e52. doi: 10.1053/j.gastro.2011.01.031. quiz e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Opitz L, Salinas-Riester G, Grade M, et al. Impact of RNA degradation on gene expression profiling. BMC Med Genomics. 2010;3:36. doi: 10.1186/1755-8794-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R_Core_Team. R: A language and environment for statistical computing. City: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 19.Koshiol J, Wang E, Zhao Y, Marincola F, Landi MT. Strengths and limitations of laboratory procedures for microRNA detection. Cancer Epidemiol Biomarkers Prev. 2010;19:907–911. doi: 10.1158/1055-9965.EPI-10-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 21.Chen DT, Hernandez JM, Shibata D, et al. Complementary strand microRNAs mediate acquisition of metastatic potential in colonic adenocarcinoma. J Gastrointest Surg. 2012;16:905–912. doi: 10.1007/s11605-011-1815-0. discussion 912–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kendall M. A New Measure of Rank Correlation. Biometrika. 1938;30:81–89. [Google Scholar]
- 24.Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–260. doi: 10.1016/j.jtcvs.2007.08.055. discussion 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Gu J, Wang KK, et al. MicroRNA expression signatures in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:5744–5752. doi: 10.1158/1078-0432.CCR-09-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bansal A, Lee IH, Hong X, et al. Feasibility of MicroRNAs as Biomarkers for Barrett's Esophagus Progression: A Pilot Cross-Sectional, Phase 2 Biomarker Study. Am J Gastroenterol. 2011;106:1055–1063. doi: 10.1038/ajg.2011.37. [DOI] [PubMed] [Google Scholar]
- 27.Leidner RS, Ravi L, Leahy P, et al. The microRNAs, MiR-31 and MiR-375, as candidate markers in Barrett's esophageal carcinogenesis. Genes Chromosomes Cancer. 2012;51:473–479. doi: 10.1002/gcc.21934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fassan M, Volinia S, Palatini J, et al. MicroRNA expression profiling in human Barrett's carcinogenesis. Int J Cancer. 2010 doi: 10.1002/ijc.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathe EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853–861. doi: 10.1002/bjs.7000. [DOI] [PubMed] [Google Scholar]
- 31.Huo X, Zhang HY, Zhang XI, et al. Acid and bile salt-induced CDX2 expression differs in esophageal squamous cells from patients with and without Barrett's esophagus. Gastroenterology. 2010;139:194–203. doi: 10.1053/j.gastro.2010.03.035. e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang HY, Zhang X, Chen X, et al. Differences in activity and phosphorylation of MAPK enzymes in esophageal squamous cells of GERD patients with and without Barrett's esophagus. Am J Physiol Gastrointest Liver Physiol. 2008;295:G470–G478. doi: 10.1152/ajpgi.90262.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaiswal KR, Morales CP, Feagins LA, et al. Characterization of telomerase-immortalized, non-neoplastic, human Barrett's cell line (BAR-T) Dis Esophagus. 2007;20:256–264. doi: 10.1111/j.1442-2050.2007.00683.x. [DOI] [PubMed] [Google Scholar]
- 34.Hormi-Carver K, Zhang X, Zhang HY, et al. Unlike esophageal squamous cells, Barrett's epithelial cells resist apoptosis by activating the nuclear factor-kappaB pathway. Cancer Res. 2009;69:672–677. doi: 10.1158/0008-5472.CAN-08-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonckheere A. A distribution-free k-sample test again ordered alternatives. Biometrika. 1954;41:133–145. [Google Scholar]
- 36.Terpstra T. The asymptotic normality and consistency of Kendall's test against trend, when ties are present in one ranking Indagationes. Mathematicae. 1952;14:327–333. [Google Scholar]
- 37.clinfun. [Accessed Oct 8, 2012];Clinical Trial Design and Data Analysis Functions. R Package version 1.0.3. 2012 Available at: http://CRAN.R-project.org/package=clinfun.
- 38.Sarkar D. Lattice: Multivariate Data Visualization with R. City: Springer; 2008. [Google Scholar]
- 39.Wu X, Ajani JA, Gu J, et al. MicroRNA Expression Signatures during Malignant Progression from Barrett's Esophagus to Esophageal Adenocarcinoma. Cancer Prev Res (Phila) 2013;6:196–205. doi: 10.1158/1940-6207.CAPR-12-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenna LB, Schug J, Vourekas A, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654–1664. doi: 10.1053/j.gastro.2010.07.040. 1664 e1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Baal JW, Verbeek RE, Bus P, et al. microRNA-145 in Barrett's oesophagus: regulating BMP4 signalling via GATA6. Gut. 2013;62:664–675. doi: 10.1136/gutjnl-2011-301061. [DOI] [PubMed] [Google Scholar]
- 42.Kadri SR, Lao-Sirieix P, O'Donovan M, et al. Acceptability and accuracy of a non-endoscopic screening test for Barrett's oesophagus in primary care: cohort study. BMJ. 2010;341:c4372. doi: 10.1136/bmj.c4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montgomery E, Bronner MP, Goldblum JR, et al. Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum Pathol. 2001;32:368–378. doi: 10.1053/hupa.2001.23510. [DOI] [PubMed] [Google Scholar]
- 44.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fassan M, Pizzi M, Battaglia G, et al. Programmed cell death 4 (PDCD4) expression during multistep Barrett's carcinogenesis. J Clin Pathol. 2010;63:692–696. doi: 10.1136/jcp.2010.078253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.