Abstract
Rationale
Mice lacking cyclophilin D (CypD−/−), a mitochondrial chaperone protein, have altered cardiac metabolism. As acetylation has been shown to regulate metabolism, we tested whether changes in protein acetylation might play a role in these metabolic changes in CypD−/− hearts.
Objective
To test the hypothesis that loss of CypD alters the cardiac mitochondrial acetylome.
Methods and Results
To identify changes in lysine-acetylated proteins and map acetylation sites following ablation of CypD, we subjected tryptic digests of isolated cardiac mitochondria from WT and CypD−/− mice to immunoprecipitation using agarose beads coupled to anti-acetyl lysine antibodies followed by mass spectrometry. We used label-free analysis for the relative quantification of the 875 common peptides that were acetylated in WT and CypD−/− samples and found 11 peptides (10 proteins) decreased and 96 peptides (48 proteins) increased in the CypD−/− samples. We found increased acetylation of proteins in fatty acid oxidation and branched-chain amino acid metabolism. To evaluate whether this increase in acetylation might play a role in the inhibition of fatty acid oxidation that was previously reported in CypD−/− hearts, we measured the activity of L-3-hydroxyacyl-CoA dehydrogenase (LCHAD), which was acetylated in the CypD−/− hearts. Consistent with the hypothesis, LCHAD activity was inhibited by approximately 50% compared to the WT mitochondria.
Conclusions
These results implicate a role for CypD in modulating protein acetylation. Taken together, these results suggest that ablation of CypD leads to changes in the mitochondrial acetylome, which may contribute to altered mitochondrial metabolism in CypD−/− mice.
Keywords: Cyclophilin D, acetylation, mitochondria, sirtuin 3 (SIRT3), cardiac metabolism, proteomics
INTRODUCTION
Cyclophilin D (CypD) is a peptidyl prolyl cis-trans-isomerase, which functions as a mitochondrial protein chaperone and is therefore likely to have multiple targets. CypD is also known to be an activator of the mitochondrial permeability transition pore (mPTP). The mPTP is a non-selective pore in the inner mitochondrial membrane that is opened by high matrix calcium and/or reactive oxygen species (ROS)1-3. Sustained opening of the mPTP results in loss of membrane potential, uncoupling of oxidative phosphorylation, matrix swelling, ATP depletion, and increased production of reactive oxygen species, ultimately leading to cell death. The mPTP plays a critical role in mediating cell death during ischemia/reperfusion injury and inhibition of mPTP is proposed to be the end-effector of cardioprotective signaling cascades4, 5. In addition to sustained activation of the mPTP, which leads to cell death, the mPTP has been shown to open transiently. It has been proposed that transient opening of the mPTP can serve as a mitochondrial calcium release mechanism1, 6.
Loss or inhibition of CypD has been shown to lead to an increase in mitochondrial calcium, which results in activation of mitochondrial NADH dehydrogenases such as pyruvate dehydrogenase and alterations in the ratio of carbohydrate to fatty acid oxidation6. Using both proteomic and metabolomic approaches, we previously found that mitochondria from CypD−/− hearts have alterations in pyruvate and branched chain amino acid metabolism, as well as changes in levels of mitochondrial histone proteins7. Because acetylation has been shown to regulate mitochondrial metabolism, we consider that the metabolic changes observed with loss of CypD might be due to alterations in the mitochondrial acetylome. Protein acetylation is a reversible post-translational modification (PTM), which adds an acetyl moiety to the epsilon-amino group of lysine residues8-11. Protein acetylation has been shown to play a key role in histone modifications and is well known to influence changes in gene expression12, but much less is known about its role in nonnuclear protein acetylation and cellular regulation. In particular, emerging data indicate that many proteins within the mitochondria are reversibly acetylated13, 14. The acetylation of many metabolic enzymes has been shown in liver and yeast, but there is little comprehensive data in heart. We therefore examined the effect of loss of CypD on cardiac mitochondrial protein acetylation. In the present study, we found that loss of CypD resulted in an increase in acetylation of many mitochondrial proteins in a profile consistent with cardiac metabolic remodeling in CypD−/− mice.
METHODS
Please see the Online Supplement available at http://circres.ahajournals.com for detailed materials and methods related to this study.
Animals
Adult male 12 to 16 week old WT and CypD−/− mice, obtained from Dr. Jeffery Molkentin (Cincinnati Children’s Hospital Medical Center), were studied. All animals were treated and cared for in accordance with the Guide for the Care and Use of Laboratory Animals [National Institutes of Health (NIH), Revised 2011], and protocols were approved by the Institutional Animal Care and Use Committee.
Mitochondria isolation
Mitochondria were isolated by differential centrifugation according to standard procedures15.
Immunopreciptation (IP) for acetylated proteins
Isolated mitochondria (500 μg) were subjected to immunoprecipitation followed by Western blot analyses as previously described16 using anti-GRP75, anti-F1Fo ATP synthase subunit A (ATPA), anti- pyruvate dehydrogenase E1 component subunit alpha (ODPA) antibodies.
Western blot
Equivalent amounts of protein (20-40 μg) from each sample were separated on NuPAGE 4-12% Bis-Tris gels (Invitrogen, Carlsbad, CA) and transferred to nitrocellulose membranes. Gel transfer efficiency and equal loading were verified using reversible Ponceau S staining. The resulting blots were probed with anti-acetylated lysine antibody (Cell Signaling, Danvers, MA), anti-GCN5L1, anti-SIRT3, anti-VDAC-1 (Santa Cruz Biotechnology), anti F1Fo ATP synthase α subunit, or anti Cyclophilin D antibody (Mitosciences, Eugene, Oregon).
Affinity purification of lysine-acetylated peptides for mass spectrometry
Isolated mitochondrial pellets (1 mg) were subjected to immunoprecipitation as previously described 13 to identify lysine-acetylated peptides by mass spectrometry. The LCMS data were searched against the Swiss Prot database, taxonomy Musculus (mouse) using Mascot server (Matrix Science, London, UK; version 2.3). Relative quantification of acetylated peptides were performed using QUOIL (QUantification withOut Isotope Labeling), an in-house software program designed as a label-free approach to peptide quantification by LC-MS/MS17.
SIRT3 activity
Mitochondria were isolated from WT and CypD−/− mouse hearts. SIRT3 activity was measured using SIRT3 Direct Fluorescent Screening Assay Kit (Cayman Chemical, Ann Arbor, MI) in the presence and absence of nicotinamide (NAM).
Langendorff heart perfusion and protocol
Mouse hearts were subjected to Langendorff perfusion as previously described18.
Myocyte isolation
Adult mouse ventricular myocytes were isolated by collagenase digestion as described previously19 and attached to matrigel-coated coverslips for 30 min in a 5% CO2 incubator at 37°C in medium 199 supplemented with 5 mmol/L creatine, 2 mmol/L L-carnitine, 5 mmol/L taurine, 2.5 mmol/L sodium pyruvate, 26 mmol/L NaHCO3, 100 U/mL penicillin, and 100 μg/mL streptomycin.
Mitochondrial NADH measurement
Myocytes were mounted on the stage of a fluorescence microscope (Nikon Diaphot) with a 20× objective and superfused with Tyrode’s solution (25°C) containing (mmol/L): 140 NaCl, 4 KCl, 1 MgCl2, 5 HEPES, and 10 D-glucose (pH 7.4). The endogenouse mitochondrial NADH autofluorescence was excited at λexc = 340 nm (band pass filter) and its emission recorded at λem = 415 nm (long pass filter) into a QuantEM 512 SC electron-multiplying charge-coupled device (CCD) camera (Photometrics, Tucson, AZ). NADH levels were expressed as a percent of the reduced NADH/NAD+ pool, which was calibrated by applying 4 mmol/L NaCN (100%) and 5 μmol/L carbonyl cyanide 3-chlorophenyl hydrazone (CCCP) (0%) for each experiment.
Mitochondrial swelling and calcium retention capacity assays
Mitochondrial permeability transition pore opening in isolated heart mitochondria from WT and CypD−/− mouse hearts was assessed using the calcium retention capacity (CRC) and Ca2+-induced swelling assays. CRC was assessed using 10 μM fluorescent Ca2+ indicator Calcium Green-5N (Molecular Probes, Eugene, OR) with the addition of 10 μM Ca2+ pulses to induce mPTP opening. Ca2+-induced swelling assay was measured spectrophotometrically as a decrease in absorbance at 540 nm after pore opening that was induced by 250 μ M of CaCl2. Both assays were assessed in the presence and absence of 200 nM cyclosporine A (CsA), a known CypD and mPTP inhibitor.
Trifunctional protein enzyme alpha subunit activity measurements
The activity of the L-3-hydroxyacyl-CoA dehydrogenase (LCHAD) was measured in isolated mitochondria from WT and CypD mouse hearts. After mitochondria isolation, the pellet was suspended in 25 mmol/L potassium phosphate, 50 mmole/L MOPS, 0.2 mmol/L EDTA, pH 8.0. The mitochondria suspensions were freeze-thawed 3× and sonicated on ice for 3×10s with a 1-min interval in between. Triton X-100 was then added to the suspensions to give a Triton X-100 to protein ratio of 1:1. After incubation on ice for 30 min, the extracted were centrifuged at 11600g for 10 min. Enzyme activities were then measured in the supernatant at 37°C in the presence of 100 μmole/L NADH and 100 μmole/L acetoacetyl CoA. The LCHAD activity was calculated as the rate of NADH oxidized per min/mg protein.
Statistical analysis
All data were expressed as mean ± standard error. The student’s 2-sample t-test or one-way ANOVA with Bonferroni’s post hoc analyses were used for comparison of differences between groups and a p-value ≤ 0.05 was considered to be significant.
RESULTS
Alterations in the mitochondrial acetylation profile following loss of CypD
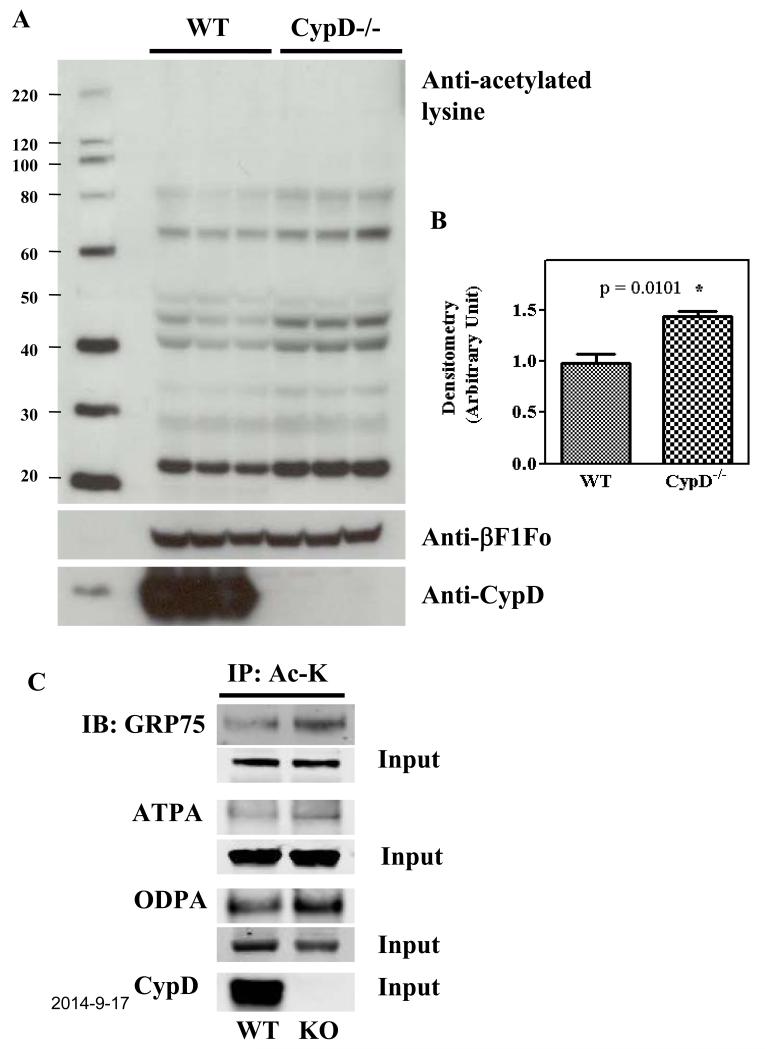
We previously found that CypD−/− hearts have increased pyruvate dehydrogenase (PDH) activity and a reduction in fatty acid oxidation relative to glucose oxidation6. We also found changes in branched-chain amino acid metabolism in CypD−/− hearts7. As mitochondrial acetylation has emerged as a key regulator of mitochondrial fatty acid and branched-chain amino acid metabolisms, we tested whether CypD−/− hearts might have altered mitochondrial acetylation. Acetylation/deacetylation of protein lysine residues has emerged as an important PTM for dynamic regulation of many proteins8-11. We initially examined changes in lysine-acetylated proteins following the ablation of CypD. As shown in Figure 1 (A-B), Western blot analysis shows that acetylation levels were increased by 45% ± 12% in CypD−/− mitochondria.
Figure 1. Increase in acetylation in isolated CypD−/− mitochondria.
A) WT and CypD−/− mouse hearts were perfused for 5 min and mitochondria were isolated as described in the Methods section. Samples were analyzed by Western blot analysis using anti-acetylated lysine, anti-F1Fo ATP synthase β-subunit (as a loading control), and anti-Cyclophilin D. B) mean densitometry data from 3 individual experiments. C) Mitochondria from WT and CypD−/−mouse hearts were subjected to immunoprecipitation using anti-acetyl lysine antibody followed by Western blot using anti-GRP75, anti-F1Fo ATP synthase subunit A (ATPA), anti-pyruvate dehydrogenase E1 component subunit alpha (ODPA) antibodies. The input bands reflect the protein level in the homogenate prior to IP and show no significant change in protein expression of GRP75, ATPA, or ODPA between wild-type and CypD−/− hearts.* p<0.05 compared with WT.
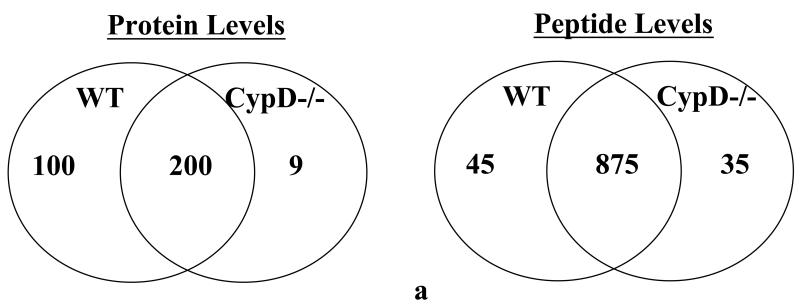
The acetylome has been compiled for yeast, mouse, and rat liver, but the cardiac mitochondrial acetylome has not been well-defined. We therefore decided to define the cardiac mitochondrial acetylome including sites of acetylation at baseline in WT hearts. We accomplished this by subjecting tryptic digests of isolated cardiac mitochondria from WT mice to immunoprecipitation using anti-acetyl lysine antibody coupled to agarose beads followed by mass spectrometry. In WT mitochondria, we found 198 acetylated proteins (with 864 peptides) at baseline (Figure 2A and Online Table I, n=3 biological replicates). As reported in Online Table I, 57 of these proteins were from the electron transport complexes I to V (258 acetylated peptides). More than 50% of the electron transport complexes were found to be acetylated at baseline. We also found that all of the TCA cycle proteins as well as a large number of enzyme involved in fatty acid oxidation were acetylated. We also found 11 mitochondrial carrier proteins acetylated, including regulators (CCDC90A and B) of the recently described mitochondrial Ca2+ uniporter (MCU)20, and mitochondrial pyruvate carrier 2 (Brain protein 44). Of interest, we found CypD is acetylated consistent with previous data21. Thus, most of the enzymes involved in cardiac mitochondrial metabolism and bioenergetics are acetylated at baseline.
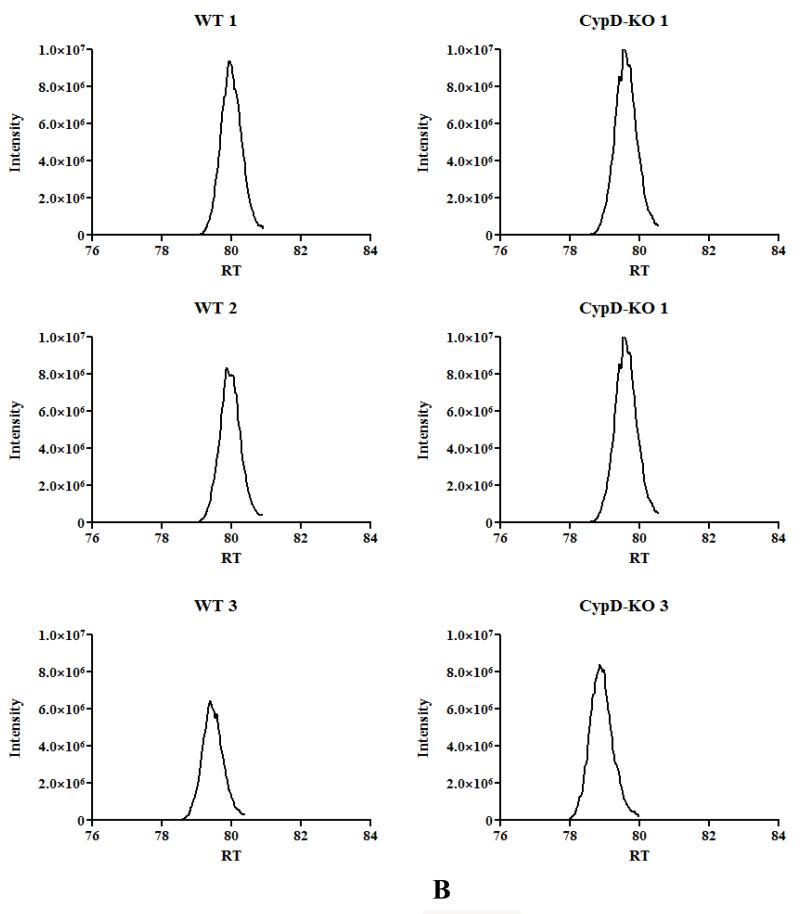
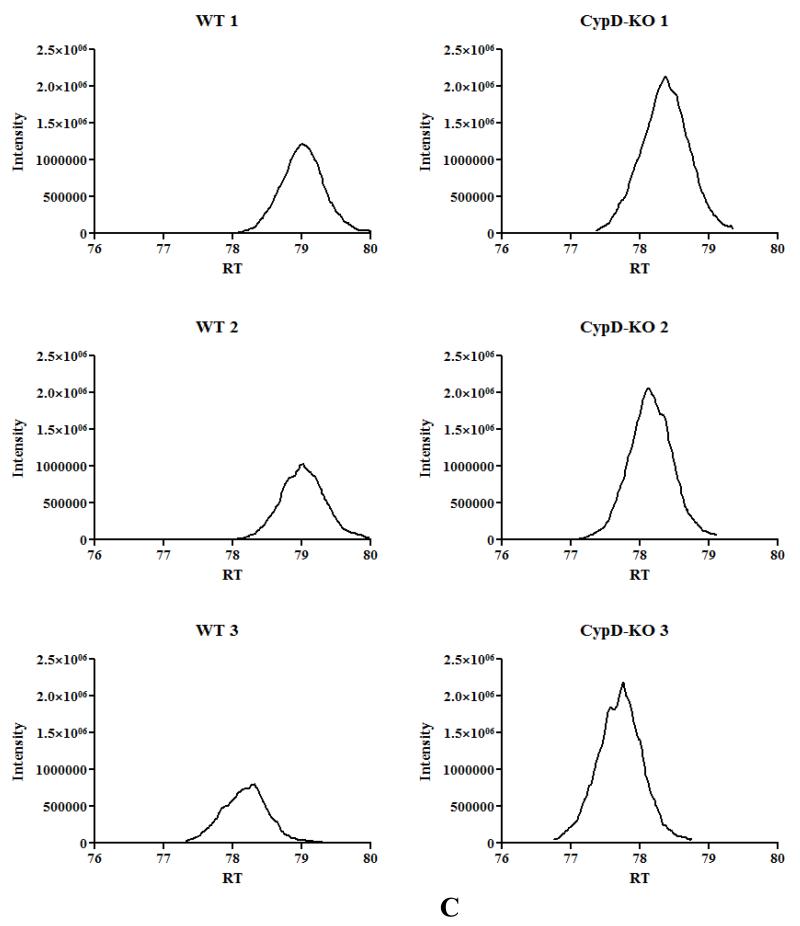
Figure 2. Mitochondrial acetylation following ablation of CypD.
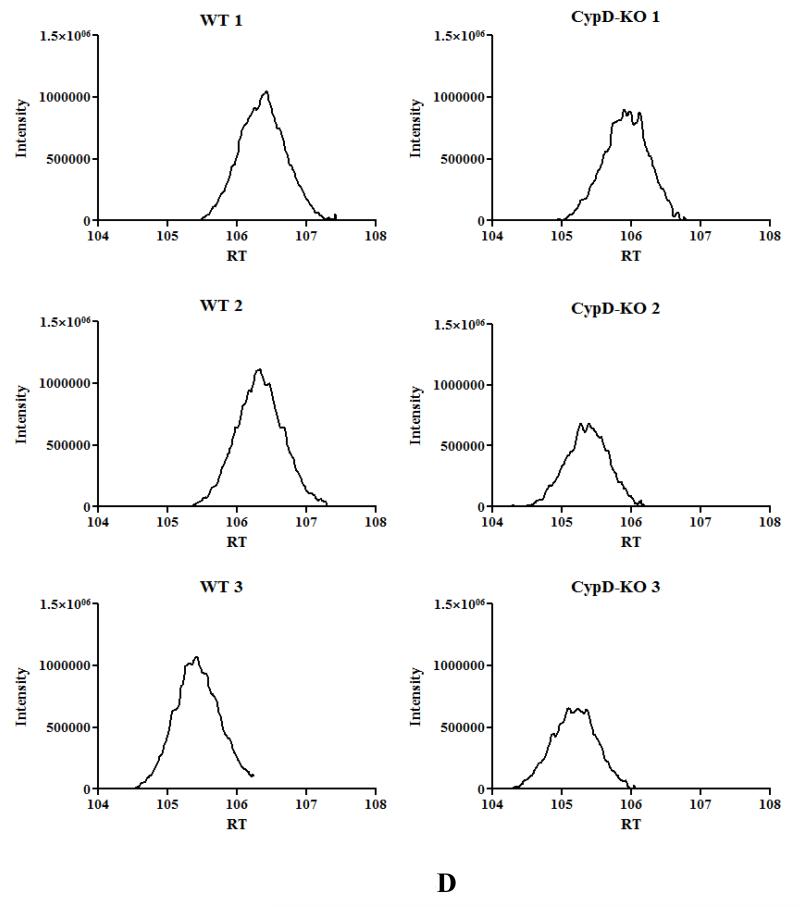
A) acetylated peptides were identified using mass spectrometry as discussed in the Methods section. We manually inspected all the spectraof identified acetylated peptides using Scaffold PTM software in conjunction with the Proteome Discover software. This experiment was performed with 3 biological replicates in each group. To be considered as a common peptide or protein present, it must be present in at least 2 out of 3 replicates in one group and at least 1 out of 3 replicates in the other group (e.g., a peptide is present in 2 out of 3 WT samples, and in 1 out of 3 CypD−/− samples). To be included in the WT only group, a peptide/protein must be present in 2 out of 3 replicates in the WT samples and not present in any of the CypD−/−. To be included in the CypD−/− only group, a peptide/protein must be present in 2 out of 3 replicates in the CypD−/− samples and not present in any of the WT. B) extracted ion chromatography of peptide DDNGkPYVLPSVR from AATM of WT and CypD−/− samples for a peptide that show no change in acetylation between samples. C) extracted ion chromatography of peptide IAkDEGANAFFK from ADT1 (P48962) ADP/ATP translocase 1 of WT and CypD−/− samples for a peptide that show an increase in acetylation following loss of CypD. D) extracted ion chromatography of YkWcEYGLTFTEK from VDAC2_MOUSE (Q60930) Voltage-dependent anion-selective channel protein 2 of WT and CypD−/− samples for a peptide that show a decrease in acetylation following loss of CypD.
As shown in Figure 1A, there is an increase in global acetylation in mitochondria isolated from mice lacking CypD. We therefore compared the acetylome of WT mitochondria to that of CypD−/−. We manually inspected all the spectra of identified acetylated peptides using Scaffold PTM software in conjunction with the Proteome Discover software. As shown in Figure 2A, in the CypD−/− mitochondria we identified 955 acetylated peptides (from 219 proteins) with 875 acetylated peptides common between WT and CypD−/− samples (see Online Table II for a complete list of acetylated proteins and peptides). Using the criteria defined in the legend of Figure 2A, we found 45 peptides that were only acetylated in WT (Online Table III), and 35 peptides that were acetylated only in the CypD−/− mitochondria (Online Table IV). We concentrated our analysis on the 875 acetylated peptides that were detected in both WT and CypD−/− samples. A label-free analysis program for relative quantification of the common acetylated peptides between WT and CypD−/− samples was used and representative ion chromatographs are shown (Figure 2B, C, and D). We found 11 peptides (from 10 proteins) decreased in CypD−/− mitochondria by at least 20% or greater with a p <0.05 (Table 2). Using similar criteria, we found 96 peptides from 48 proteins that showed an increase acetylation in the CypD−/− mitochondria (Online Table V). We confirmed the increase in acetylation of several proteins by subsequent immunoprecipitation of acetylated proteins followed by immunoblot analysis with antibodies recognizing GRP75, F1Fo ATP synthase alpha subunit, and pyruvate dehydrogenase E1 component alpha subunit (Figure 1C). The increase or decrease in acetylation noted in Figure 2A could be due to a change in protein level in the CypD−/− hearts. In a previous study, we examined changes in protein levels in CypD−/− hearts7. We compared the changes in protein levels in CypD−/− heart to changes in acetylation. Two of the 10 proteins (VDAC3 and DHSA) that showed a decrease in acetylation (Table 2) had reduced expression levels in the CypD−/− hearts. Thus, the decrease in acetylation in these proteins is likely due to decreased protein level. Of the 48 proteins that showed an increase in acetylation (Online Table V), only 3 proteins (PRDX5, ODPA and NDUA5) showed an elevated protein level. Therefore, the increase in protein acetylation for the vast majority of proteins cannot be attributed to an increase in protein expression. A change in acetylation has also been suggested to alter protein stability. Only 3 proteins increased in both acetylation and protein levels (PRDX5, ODPA and NDUA5), and these proteins are potential candidates whereby acetylation may increase their stability22. Of the 30 proteins that showed a decrease in protein level in CypD−/− hearts, only 2 showed an increase in acetylation (M2OM and NDUV1). Thus acetylation does not appear to have a dramatic effect on protein expression in the adult myocardium.
Table 2. Acetylated proteins with decreased acetylation in the CypD−/− mitochondria.
| # | Protein | Peptide | WT | SE | KO | SE | KO/WT | p |
|---|---|---|---|---|---|---|---|---|
| 1 | ADT1 MOUSE (P48962) ADP/ATP translocase 1 |
gDQALSFLADFLAGGIAAAVSK | 0.978 | 0.022 | 0.603 | 0.014 | 0.616 | 0.00 |
| 2 | ANXA2_MOUSE (P07356) Annexin A2 | SVcHLQAVFER | 0.910 | 0.092 | 0.565 | 0.021 | 0.621 | 0.02 |
| 3 | CISY_MOUSE (Q9CZU6) Citrate synthase | SMSTDGLMAFVDSK | 0.973 | 0.020 | 0.612 | 0.018 | 0.629 | 0.00 |
| 4 | DHSA_MOUSE (Q8K2B3) Succinate dehydrogenase [ubiquinone] flavoprotein subunit |
AAFGLSEAGFNTAcLTALFPTR | 0.965 | 0.128 | 0.372 | 0.145 | 0.386 | 0.04 |
| 5 | KCRS_MOUSE (Q6P8J7) Creatine kinase S-type | YYALSEMTEQDQQR | 0.950 | 0.116 | 0.480 | 0.018 | 0.505 | 0.02 |
| 6 | SUCA_MOUSE (Q9WUM5) Succinyl-CoA ligase [GDP-forming] subunit alpha |
NTkIIcQGFTGK | 0.853 | 0.073 | 0.530 | 0.048 | 0.621 | 0.02 |
| HLGLPVFNTVAEAK | 0.829 | 0.092 | 0.564 | 0.012 | 0.681 | 0.05 | ||
| 7 | THIL_MOUSE (Q8QZT1) Acetyl-CoA acetyltransferase |
VLkYAGLK | 0.955 | 0.025 | 0.459 | 0.178 | 0.481 | 0.05 |
| 8 | VDAC1_MOUSE (Q60932) Voltage-dependent anion-selective channel protein 1 |
LTFDSSFSPNTGAK | 0.923 | 0.041 | 0.658 | 0.032 | 0.713 | 0.02 |
| 9 | VDAC2_MOUSE (Q60930) Voltage-dependent anion-selective channel protein 2 |
YkWcEYGLTFTEK | 1.035 | 0.021 | 0.717 | 0.080 | 0.693 | 0.02 |
| 10 | VDAC3_MOUSE (Q60931) Voltage-dependent anion-selective channel protein 3 |
cNTPTYcDLGAAAK | 0.992 | 0.068 | 0.715 | 0.053 | 0.720 | 0.03 |
Relative quantification of acetylated peptides were performed using QUOIL (QUantification withOut Isotope Labeling), an in-house software program designed as a label-free approach to peptide quantification by LC-MS/MS. The following criteria were used to determine the effect of chronic or acute inhibition of CypD on acetylated peptides: at least greater than 20% change and p <0.05 vs. WT samples. Accession numbers are from the SWISSPROT/Uniprot database. The acetylated lysine residue is in bold and italicized. SE, standard error; p, p-value.
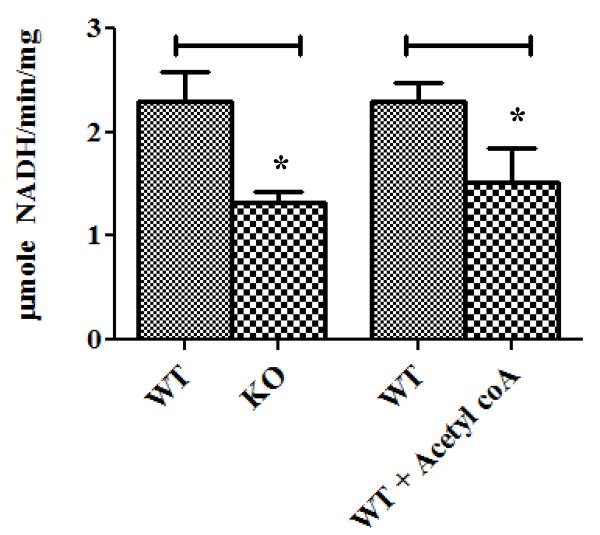
Although acetylation does not have a large impact on protein stability, it could have a major impact on enzyme activity and metabolism. The reduction in fatty acid oxidation observed previously in the CypD−/− hearts6 is entirely consistent with the changes in acetylation observed in the CypD−/− hearts. Increased acetylation has been shown previously to inhibit fatty acid oxidation23, 24. Consistent with the reduction in fatty acid oxidation in the CypD−/− hearts, we saw an increase in the acetylation of key enzymes involved in fatty acid oxidation. We found an increase in acetylation in both subunits of trifunctional enzyme (ECHA and ECHB), long-chain specific acyl-CoA dehydrogenase (ACADL), 2, 4-dienoyl-CoA reductase (DECR), 3-ketoacyl-CoA thiolase (THIM), and enoyl-CoA delta isomerase 1 (ECI1). To test the functional effect of acetylation on enzyme activity, we measured the L-3-hydroxyacyl-CoA dehydrogenase (LCHAD) activity of the trifunctional protein (TFP) alpha subunit, an enzyme involved in fatty acid oxidation, which showed an increase in acetylation in the CypD−/− mitochondria. We found that the LCHAD activity was inhibited by approximately 50% in the CypD−/−mitochondria as compared to the WT mitochondria (Figure 3). To further evaluate whether an increase in acetylation results in a decrease in the LCHAD activity, we preincubated the WT sonicated mitochondria with 100 μM acetyl-CoA (confirming an increase in acetylation of LCHAD by Western blot analysis). Consistent with the hypothesis, with the acetyl CoA treatment, the LCHAD activity was inhibited similarly to the CypD−/− group. To identify the pathways affected by the increase in protein acetylation in the CypD−/− hearts, we performed pathway analysis using Ingenuity software. The acetylated peptides comprise pathways involved in oxidative phosphorylation, mitochondrial dysfunction, butanoate metabolism, pyruvate metabolism, and branched chain amino acid metabolism (Online Figure I).
Figure 3. Decrease in activity of L-3-hydroxyacyl-CoA dehydrogenase (LCHAD) in CypD−/− mitochondria.
Mitochondria were isolated from WT and CypD−/− mouse hearts and LCHAD activity was measured. *p<0.05 vs. WT.
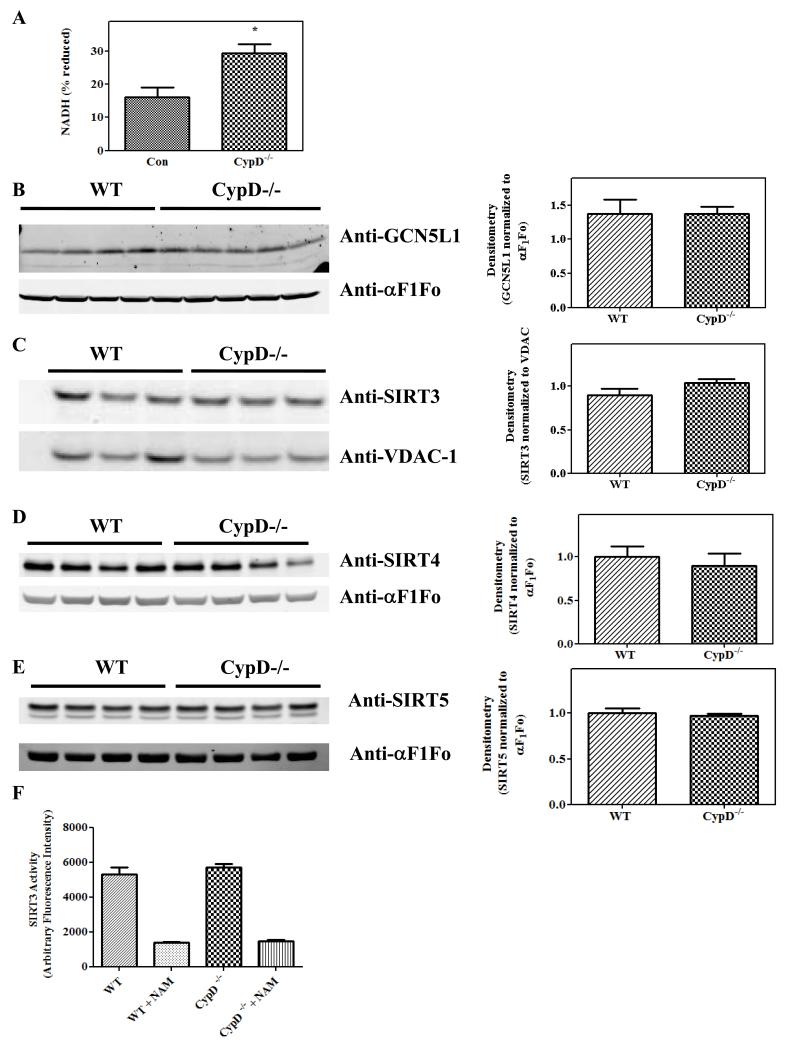
We examined the mechanisms responsible for changes in the mitochondrial acetylome in the CypD−/− mice. A recent study has shown, in an in vitro assay, that increasing NADH while NAD+ is held constant leads to a significant decrease in SIRT3 activity25. As the NADH/NAD+ ratio can change during isolation of mitochondria, we measured the endogenous NADH fluorescence in WT and CypD−/− cardiomyocytes (Figure 3A). Baseline NADH levels were increased by ~80% in CypD−/− hearts relative to WT (29.44 ± 2.63 vs. 16.20 ± 2.86 % of fully-reduced NADH) (Figure 4A). Western blot analysis showed no difference in either the mitochondrial acetyl transferase, GCN5L1 (Figure 4B) or deacetylases, SIRT3 (Figure 4C), SIRT4 (Figure 4D) or SIRT5 (Figure 4E) between WT and CypD−/− mitochondria, which would support a primary role for an increase in NADH in the increase in acetylation. Finally, there was no difference in SIRT3 activity between WT and CypD−/− mitochondria (Figure 3F) measured under conditions where the NAD and NADH were fixed in the assay.
Figure 4. Increased in NADH/NAD+ levels following ablation of CypD.
Measurement of baseline NADH/NAD+ in isolated cardiac myocytes, normalized with NaCN to give the 100% value and CCP to give 0% value (panel A). Mitochondria were isolated from WT and CypD−/− mouse hearts and samples were analyzed by Western blot analysis using anti-GCN5L1 (panel B), anti-SIRT3 (panel C), anti-SIRT4 (panel D) or anti-SIRT5 (panel E). Mean densitometry data from experiments are shown in the right. SIRT3 activity was measured using SIRT3 Direct Fluorescent Screening Assay Kit (Cayman Chemical, Ann Arbor, MI) in the presence and absence of nicotinamide (NAM) (panel F). *p<0.05 vs. WT or Con.
To gain additional insight into the mechanism by which loss of CypD leads to an increase in acetylation, we determined whether the peptides acetylated in CypD−/− hearts were SIRT3 substrates. To accomplish this, we compared the peptides showing an increase in acetylation following ablation of SIRT3 to the list of acetylated peptides increased in hearts lacking CypD. We found 151 peptides that exhibited an increase in acetylation in SIRT3−/− hearts compared to WT littermates; we defined these peptides as potential SIRT3 substrates (Online Table VI). Of interest, CypD itself is a SIRT3 substrate. If we compare these 151 peptides to the 96 peptide that exhibited an increase in acetylation in the CypD−/− hearts, we found only 19 common peptides that are potential SIRT3 substrates (Online Table V, bolded).
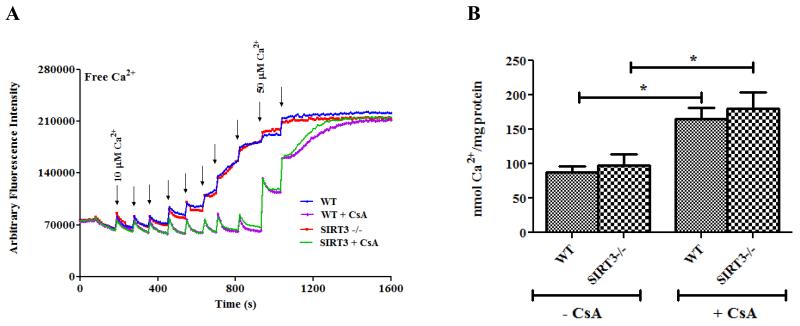
CypD−/− hearts have reduced I/R injury4, and this raises the question as to whether the increase in mitochondrial acetylation plays a role in cardioprotection. Mitochondria isolated from CypD−/− mice have been shown to be more resistant to mPTP opening than WT mice4. To address whether an increase in mitochondrial acetylation reduces mPTP opening, we used SIRT3−/− mice. These mice have been shown (and we confirmed) to have increased protein acetylation26. Consistent with previous studies27, we found no difference in mPTP opening between the WT and SIRT3/- mitochondria isolated from 3 month old mice (Figure 5). The mPTP opening was sensitive to CsA, a known mPTP inhibitor. These data suggest that an increase in mitochondrial acetylation per se is not sufficient to alter mPTP.
Figure 5. Hyperacetylation does not affect mPTP opening in SIRT3−/− mitochondria.
Mitochondria were isolated from WT and SIRT3−/− mouse hearts. Mitochondrial permeability transition pore opening in isolated heart mitochondria from WT and SIRT3−/− mouse hearts was assessed using the calcium retention capacity assay (CRC) (Panel A) and Ca2+-induced swelling assay (Panel B). CRC was assessed using 10 μM fluorescent Ca2+ indicator Calcium Green-5N (Molecular Probes, Eugene, OR) with the addition of Ca2+ pulses to induce mPTP opening in the presence and absence of 200 nM cyclosporine A (CsA), a known mPTP inhibitor. * p<0.05 compared with CsA in each group.
DISCUSSION
A number of studies have shown that acetylation regulates metabolic enzymes by multiple mechanisms, including enzymatic activation10, inhibition28, or protein stability22. A recent proteomics study by Kim et al. revealed that over 20% of liver mitochondrial proteins and enzymes are acetylated and that changes occur in acetylation status in response to acute fasting13. Previous studies have identified several acetylated proteins in cardiac mitochondria29; however, this is the first study to evaluate large scale acetylation in cardiac mitochondria. In this study, we identified over 200 cardiac mitochondrial proteins that are acetylated under basal conditions. We found that more than 50% of the electron transport complex proteins are acetylated at baseline. We also found baseline acetylation of TCA and fatty oxidation enzymes.
This study demonstrates that loss of CypD results in an increase in acetylation. We found an increase in acetylation of many enzymes involved in fatty acid oxidation, and previous studies have reported that increased acetylation of specific fatty acid oxidation enzymes leads to their inhibition23, 24. We provide new data showing that acetylation of LCHAD results in its inhibition.
What is the mechanism responsible for the increase in acetylation in the CypD−/− hearts? Reversible lysine acetylation is regulated by opposing activities of protein acetyltransferases and deacetylases. We found no increase in the protein level of the recently described mitochondrial acetyltransferase (GCN5L1)30. We also found no change in the levels of the three mitochondrial deacytylases, SIRT3, SIRT4 and SIRT5. We also found no change in activity of SIRT3. The data are most consistent with the hypothesis that loss of CypD leads to an increase in NADH/NAD+ resulting in inhibition of deacetylase(s) and an increase in protein acetylation (Online Figure II). Previous studies have suggested that a change in the NADH/NAD+ ratio can alter the activity of sirtuins25, 31-33. A change in NADH/NAD+ would not carry-over in an assay of SIRT from mitochondria, in which the NAD+/NADH level is set as part of the assay. It is likely that this increase in NADH/NAD+ activates all mitochondrial sirtuins. This would be consistent with our data showing that CypD−/− hearts exhibited acetylation of many proteins that are not SIRT3 dependent substrates, as determined either in this study (Online Table V) or previous studies34-37. The current consensus (mostly obtained from data in liver) is that SIRT3 is the primary mitochondrial deacetylase26. SIRT4 and SIRT5 have been shown to exhibit ADP-ribosyltransferase and deacetylase activity, respectively26. However, there are data suggesting that SIRT438 and SIRT525 can also contribute to mitochondrial deacetylation.
It is well established that CypD−/− mitochondria have a decreased susceptibility to mPTP opening. As suggested in online Figure II, we propose that inhibition of transient opening of the mPTP leads to the increase in Ca2+, increased PDH activity, and ultimately, the increase in acetylation. The increase in acetylation per se does not appear to alter mPTP, although acetylation of a specific protein could be involved in regulation. Sinclair et al. reported that mitochondria from SIRT3−/− hearts have an increase in mPTP when measured at 18 months of age, but not at 3 or 6 months of age. They attributed the increase in mPTP at 18 months to an increase in acetylation of CypD27. In SIRT3−/− heart mitochondria, we find a general increase in acetylation and a specific increase in the acetylation of CypD at 3 months of age, but we find no change in mPTP, which is consistent with the study of Sinclair’s group27. The increase in mPTP observed at 18 months in the SIRT3−/− hearts could be due to metabolic changes that are known to occur in these mice rather than a direct effect of acetylation.
In summary, this study provides insights into changes in the cardiac mitochondrial acetylome occurring with loss of CypD and loss of SIRT3. Loss of CypD leads to an increase in protein acetylation which could account for the metabolic changes previously reported in these mice6.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Loss of Cyclophilin D (CypD) leads to a decrease in the ratio of fatty acid oxidation relative to glucose oxidation.
Lysine acetylation has recently emerged as an important post-translational protein modification in the regulation of mitochondrial metabolism.
What New Information Does This Article Contribute?
Identification of the cardiac mitochondrial acetylome.
Loss of CypD results in changes to the cardiac mitochondrial acetylome, with the majority of proteins showing an increase in acetylation.
Acetylation of the mitochondrial trifunctional protein subunit alpha leads to the inhibition of its activity.
We examined whether CypD regulates mitochondrial metabolism by modulating changes in protein acetylation. We used a mass spectrometry-based approach and identified acetylated proteins in cardiac mitochondria under baseline conditions. We then compared differences in protein acetylation profiles in heart mitochondria from WT mice and mice lacking CypD. We found a general increase in mitochondrial protein acetylation in CypD-null hearts including several protein targets involved in fatty acid oxidation. A further examination of one of these protein targets, trifunctional protein subunit alpha, showed that acetylation leads to the inhibition of enzyme activity. This study provides an important link between acetylation and mitochondrial function, and suggests that the mitochondrial acetylome represents a new layer of protein regulation mediating adaptive changes in mitochondrial metabolism.
Table 1. Electron transport chain complex proteins acetylated at baseline.
| # | Accession | Protein ID |
Protein Name | # of acetylated peptides |
|---|---|---|---|---|
| 1 | AT5F1 | Q9CQQ7 | ATP synthase subunit b | 9 |
| 2 | ATP5E | P56382 | ATP synthase subunit epsilon | 4 |
| 3 | ATP5H | Q9DCX2 | ATP synthase subunit d | 15 |
| 4 | ATP5I | Q06185 | ATP synthase subunit e | 1 |
| 5 | ATP5J | P97450 | ATP synthase-coupling factor 6 | 7 |
| 6 | ATP5L | Q9CPQ8 | ATP synthase subunit g | 5 |
| 7 | ATP8 | P03930 | ATP synthase protein 8 | 3 |
| 8 | ATPA | Q03265 | ATP synthase subunit alpha | 16 |
| 9 | ATPB | P56480 | ATP synthase subunit beta | 7 |
| 10 | ATPD | Q9D3D9 | ATP synthase subunit delta | 2 |
| 11 | ATPG | Q91VR2 | ATP synthase subunit gamma | 7 |
| 12 | ATPK | P56135 | ATP synthase subunit f | 4 |
| 13 | ATPO | Q9DB20 | ATP synthase subunit O | 13 |
| 14 | COX17 | P56394 | Cytochrome c oxidase copper chaperone | 2 |
| 15 | COX41 | P19783 | Cytochrome c oxidase subunit 4 isoform 1 | 4 |
| 16 | COX5A | P12787 | Cytochrome c oxidase subunit 5A | 3 |
| 17 | COX5B | P19536 | Cytochrome c oxidase subunit 5B | 5 |
| 18 | COX6C | Q9CPQ1 | Cytochrome c oxidase subunit 6C | 3 |
| 19 | COX7C | P17665 | Cytochrome c oxidase subunit 7C | 2 |
| 20 | CX6B1 | P56391 | Cytochrome c oxidase subunit 6B1 | 1 |
| 21 | CX7A1 | P56392 | Cytochrome c oxidase subunit 7A1 | 1 |
| 22 | CY1 | Q9D0M3 | Cytochrome c1, heme protein | 3 |
| 23 | CYC | P62897 | Cytochrome c, somatic Succinate dehydrogenase [ubiquinone] flavoprotein | 4 |
| 24 | DHSA | Q8K2B3 | subunit Succinate dehydrogenase [ubiquinone] iron-sulfur | 16 |
| 25 | DHSB | Q9CQA3 | subunit | 4 |
| 26 | ETFA | Q99LC5 | Electron transfer flavoprotein subunit alpha | 10 |
| 27 | ETFB | Q9DCW4 | Electron transfer flavoprotein subunit beta Electron transfer flavoprotein-ubiquinone |
10 |
| 28 | ETFD | Q921G7 | oxidoreductase NADH dehydrogenase [ubiquinone] 1 alpha | 6 |
| 29 | NDUA2 | Q9CQ75 | subcomplex subunit 2 NADH dehydrogenase [ubiquinone] 1 alpha | 3 |
| 30 | NDUA4 | Q62425 | subcomplex subunit 4 NADH dehydrogenase [ubiquinone] 1 alpha | 2 |
| 31 | NDUA5 | Q9CPP6 | subcomplex subunit 5 NADH dehydrogenase [ubiquinone] 1 alpha | 6 |
| 32 | NDUA6 | Q9CQZ5 | subcomplex subunit 6 NADH dehydrogenase [ubiquinone] 1 alpha | 1 |
| 33 | NDUA7 | Q9Z1P6 | subcomplex subunit 7 NADH dehydrogenase [ubiquinone] 1 alpha | 2 |
| 34 | NDUA8 | Q9DCJ5 | subcomplex subunit 8 NADH dehydrogenase [ubiquinone] 1 alpha | 3 |
| 35 | NDUA9 | Q9DC69 | subcomplex subunit 9 NADH dehydrogenase [ubiquinone] 1 alpha | 3 |
| 36 | NDUAA | Q99LC3 | subcomplex subunit 10 NADH dehydrogenase [ubiquinone] 1 alpha |
6 |
| 37 | NDUAD | Q9ERS2 | subcomplex subunit 13 NADH dehydrogenase [ubiquinone] 1 beta | 2 |
| 38 | NDUB3 | Q9CQZ6 | subcomplex subunit 3 NADH dehydrogenase [ubiquinone] 1 beta | 2 |
| 39 | NDUB4 | Q9CQC7 | subcomplex subunit 4 NADH dehydrogenase [ubiquinone] 1 beta | 2 |
| 40 | NDUB5 | Q9CQH3 | subcomplex subunit 5 NADH dehydrogenase [ubiquinone] 1 beta | 2 |
| 41 | NDUB6 | Q3UIU2 | subcomplex subunit 6 NADH dehydrogenase [ubiquinone] 1 beta | 1 |
| 42 | NDUB8 | Q9D6J5 | subcomplex subunit 8 NADH dehydrogenase [ubiquinone] 1 beta | 1 |
| 43 | NDUB9 | Q9CQJ8 | subcomplex subunit 9 NADH dehydrogenase [ubiquinone] 1 beta | 2 |
| 44 | NDUBA | Q9DCS9 | subcomplex subunit 10 | 1 |
| 45 | NDUS1 | Q91VD9 | NADH-ubiquinone oxidoreductase 75 kDa subunit NADH dehydrogenase [ubiquinone] iron-sulfur |
8 |
| 46 | NDUS3 | Q9DCT2 | protein 3 NADH dehydrogenase [ubiquinone] iron-sulfur | 1 |
| 47 | NDUS6 | P52503 | protein 6 NADH dehydrogenase [ubiquinone] iron-sulfur | 2 |
| 48 | NDUS7 | Q9DC70 | protein 7 NADH dehydrogenase [ubiquinone] iron-sulfur | 1 |
| 49 | NDUS8 | Q8K3J1) | protein 8 | 2 |
| 50 | NDUV1 | Q91YT0 | NADH dehydrogenase [ubiquinone] flavoprotein 1 | 9 |
| 51 | NDUV2 | Q9D6J6 | NADH dehydrogenase [ubiquinone] flavoprotein 2 | 2 |
| 52 | QCR1 | Q9CZ13 | Cytochrome b-c1 complex subunit 1 | 8 |
| 53 | QCR2 | Q9DB77 | Cytochrome b-c1 complex subunit 2 | 6 |
| 54 | QCR6 | P99028 | Cytochrome b-c1 complex subunit 6 | 2 |
| 55 | QCR7 | Q9D855 | Cytochrome b-c1 complex subunit 7 | 7 |
| 56 | QCR8 | Q9CQ69 | Cytochrome b-c1 complex subunit 8 | 2 |
| 57 | UCRI | Q9CR68 | Cytochrome b-c1 complex subunit Rieske | 2 |
List of electron transport chain complexes from common acetylated proteins in WTs were identified by LC-MS/MS. Accession numbers and Protein ID are from the SWISSPROT/Uniprot database.
Table 3. Acetylated proteins from electron transport chain complexes with increased in acetylation in the CypD-/- mitochondria.
| # | Accession | Protein ID | Protein Name | # of acetylated peptides |
|---|---|---|---|---|
| 1 | AT5F1 | Q9CQQ7 | ATP synthase subunit b | 4 |
| 2 | ATP5H | Q9DCX2 | ATP synthase subunit d | 6 |
| 3 | ATP5J | P97450 | ATP synthase-coupling factor 6 | 4 |
| 4 | ATP5L | Q9CPQ8 | ATP synthase subunit g | 3 |
| 5 | ATP8 | P03930 | ATP synthase protein 8 | 2 |
| 6 | ATPB | P56480 | ATP synthase subunit beta | 3 |
| 7 | ATPG | Q91VR2 | ATP synthase subunit gamma | 1 |
| 8 | ATPK | Q03265 | ATP synthase subunit f | 1 |
| 9 | ATPO | Q9DB20 | ATP synthase subunit O | 4 |
| 10 | COX5B | P19536 | Cytochrome c oxidase subunit 5B | 2 |
| 11 | COX7C | P17665 | Cytochrome c oxidase subunit 7C | 1 |
| 12 | CX7A1 | P56392 | Cytochrome c oxidase subunit 7A1 | 1 |
| 13 | CYC | P62897 | Cytochrome c, somatic | 1 |
| 14 | NDUA5 | Q9CPP6 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | 2 |
| 15 | NDUAA | Q99LC3 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10 | 1 |
| 16 | NDUAD | Q9ERS2 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13 | 1 |
| 17 | NDUV1 | Q91YT0 | NADH dehydrogenase [ubiquinone] flavoprotein 1 | 2 |
| 18 | QCR1 | Q9CZ13 | Cytochrome b-c1 complex subunit 1 | 1 |
| 19 | QCR7 | Q9D855 | Cytochrome b-c1 complex subunit 7 | 3 |
Relative quantification of acetylated peptides were performed using QUOIL (QUantification withOut Isotope Labeling), an in-house software program designed as a label-free approach to peptide quantification by LC-MS/MS. The following criteria were used to determine the effect of ablation of CypD on acetylated peptides: at least greater than 20% change and p <0.05 vs. WT samples. Accession numbers are from the SWISSPROT/Uniprot database.
ACKNOWLEDGEMENTS
We thank Drs. Marcia Haigis and Gaëlle Laurent from Harvard Medical School for kindly providing the SIRT4 antibody.
SOURCES OF FUNDING
This work was supported by the National Heart, Lung, and Blood Institute, National Institutes of Health intramural program.
Nonstandard Abbreviations and Acronyms
- CsA
Cyclosporine A
- CypD
Cyclophilin D
- KO
knockout
- mPTP
mitochondrial permeability transition pore
- PTM
post-translational modification
- Sirtuin 3
SIRT3
- WT
wildtype
Footnotes
DISCLOSURES
None.
REFERENCES
- 1.Bernardi P, Petronilli V. The permeability transition pore as a mitochondrial calcium release channel: A critical appraisal. J Bioenerg Biomembr. 1996;28:131–138. doi: 10.1007/BF02110643. [DOI] [PubMed] [Google Scholar]
- 2.Halestrap AP. What is the mitochondrial permeability transition pore? J Mol Cell Cardiol. 2009;46:821–831. doi: 10.1016/j.yjmcc.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 3.Crompton M. The mitochondrial permeability transition pore and its role in cell death. Biochem J. 1999;341(Pt 2):233–249. [PMC free article] [PubMed] [Google Scholar]
- 4.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, Robbins J, Molkentin JD. Loss of cyclophilin d reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. Cyclophilin d-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- 6.Elrod JW, Wong R, Mishra S, Vagnozzi RJ, Sakthievel B, Goonasekera SA, Karch J, Gabel S, Farber J, Force T, Brown JH, Murphy E, Molkentin JD. Cyclophilin d controls mitochondrial pore-dependent ca(2+) exchange, metabolic flexibility, and propensity for heart failure in mice. J Clin Invest. 2010;120:3680–3687. doi: 10.1172/JCI43171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menazza S, Wong R, Nguyen T, Wang G, Gucek M, Murphy E. Cypd(−/−) hearts have altered levels of proteins involved in krebs cycle, branch chain amino acid degradation and pyruvate metabolism. J Mol Cell Cardiol. 2013;56:81–90. doi: 10.1016/j.yjmcc.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Z, Scott I, Webster BR, Sack MN. The emerging characterization of lysine residue deacetylation on the modulation of mitochondrial function and cardiovascular biology. Circ Res. 2009;105:830–841. doi: 10.1161/CIRCRESAHA.109.204974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 10.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 12.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 13.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Sack MN. Emerging characterization of the role of sirt3-mediated mitochondrial protein deacetylation in the heart. Am J Physiol Heart Circ Physiol. 2011;301:H2191–2197. doi: 10.1152/ajpheart.00199.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholz TD, Balaban RS. Mitochondrial f1-atpase activity of canine myocardium: Effects of hypoxia and stimulation. Am J Physiol. 1994;266:H2396–2403. doi: 10.1152/ajpheart.1994.266.6.H2396. [DOI] [PubMed] [Google Scholar]
- 16.Hirschey MD, Shimazu T, Huang JY, Verdin E. Acetylation of mitochondrial proteins. Methods Enzymol. 2009;457:137–147. doi: 10.1016/S0076-6879(09)05008-3. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using 1c-coupled ion trap or ft mass spectrometry: Reproducibility, linearity, and application with complex proteomes. J Proteome Res. 2006;5:1214–1223. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 18.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in s-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 19.Sun J, Picht E, Ginsburg KS, Bers DM, Steenbergen C, Murphy E. Hypercontractile female hearts exhibit increased s-nitrosylation of the l-type ca2+ channel alpha1 subunit and reduced ischemia/reperfusion injury. Circ Res. 2006;98:403–411. doi: 10.1161/01.RES.0000202707.79018.0a. [DOI] [PubMed] [Google Scholar]
- 20.Mallilankaraman K, Cardenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenar T, Csordas G, Madireddi P, Yang J, Muller M, Miller R, Kolesar JE, Molgo J, Kaufman B, Hajnoczky G, Foskett JK, Madesh M. Mcur1 is an essential component of mitochondrial ca(2+) uptake that regulates cellular metabolism. Nat Cell Biol. 2012:15–123. doi: 10.1038/ncb2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin d induces dissociation of hexokinase ii from the mitochondria. J Cell Sci. 2010;123:894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Caron C, Boyault C, Khochbin S. Regulatory cross-talk between lysine acetylation and ubiquitination: Role in the control of protein stability. Bioessays. 2005;27:408–415. doi: 10.1002/bies.20210. [DOI] [PubMed] [Google Scholar]
- 23.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr., Alt FW, Kahn CR, Verdin E. Sirt3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. Sirt3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl coa synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karamanlidis G, Lee CF, Garcia-Menendez L, Kolwicz SC, Jr., Suthammarak W, Gong G, Sedensky MM, Morgan PG, Wang W, Tian R. Mitochondrial complex i deficiency increases protein acetylation and accelerates heart failure. Cell Metab. 2013;18:239–250. doi: 10.1016/j.cmet.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr., Weissman S, Verdin E, Schwer B. Mammalian sir2 homolog sirt3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, Sinclair DA. Regulation of the mptp by sirt3-mediated deacetylation of cypd at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2010;2:914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by sirt3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinmura K, Tamaki K, Sano M, Nakashima-Kamimura N, Wolf AM, Amo T, Ohta S, Katsumata Y, Fukuda K, Ishiwata K, Suematsu M, Adachi T. Caloric restriction primes mitochondria for ischemic stress by deacetylating specific mitochondrial proteins of the electron transport chain. Circ Res. 2011;109:396–406. doi: 10.1161/CIRCRESAHA.111.243097. [DOI] [PubMed] [Google Scholar]
- 30.Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: A novel role for gcn5l1. Biochem J. 2012;443:655–661. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of nadh. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial nad+levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman JC, He W, Verdin E. Mitochondrial protein acylation and intermediary metabolism: Regulation by sirtuins and implications for metabolic disease. J Biol Chem. 2012;287:42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu W, Dittenhafer-Reed KE, Denu JM. Sirt3 protein deacetylates isocitrate dehydrogenase 2 (idh2) and regulates mitochondrial redox status. J Biol Chem. 2012;287:14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozden O, Park SH, Kim HS, Jiang H, Coleman MC, Spitz DR, Gius D. Acetylation of mnsod directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging (Albany NY) 2011;3:102–107. doi: 10.18632/aging.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of sirt3 in metabolic pathways. Proc Natl Acad Sci U S A. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and sirt3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, Lovitch SB, Sharpe AH, Kurland IJ, Steegborn C, Gygi SP, Muoio DM, Ruderman NB, Haigis MC. Sirt4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl coa decarboxylase. Mol Cell. 2013;50:686–698. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.