SUMMARY
During cell division cells form the microtubule-based mitotic spindle, a highly specialized and dynamic structure that mediates proper chromosome transmission to daughter cells. Cancer cells can show perturbed mitotic spindles and an approach in cancer treatment has been to trigger cell killing by targeting microtubule dynamics or spindle assembly. To identify and characterize proteins necessary for spindle assembly, and potential antimitotic targets, we performed a proteomic and genetic analysis of 592 mitotic microtubule co-purifying proteins (MMCPs). Screening for regulators that affect both mitosis and apoptosis, we report the identification and characterization of STARD9, a kinesin-3 family member, which localizes to centrosomes and stabilizes the pericentriolar material (PCM). STARD9-depleted cells have fragmented PCM, form multipolar spindles, activate the spindle assembly checkpoint (SAC), arrest in mitosis, and undergo apoptosis. Interestingly, STARD9-depletion synergizes with the chemotherapeutic agent taxol to increase mitotic death, demonstrating that STARD9 is a mitotic kinesin and a potential anti-mitotic target.
INTRODUCTION
Mitotic spindle assembly is a highly complex and orchestrated event that organizes cell division. Mitosis relies on a multitude of protein complexes, protein-protein interactions, and regulatory mechanisms (Walczak and Heald, 2008). To date, many proteins that associate with microtubules and function in mitotic spindle assembly have been identified and characterized (Loughlin et al., 2008; Manning and Compton, 2008a, b; Walczak and Heald, 2008). Non-motor proteins, for example, function in microtubule nucleation, crosslinking, and stability, and can influence the activities of motor proteins (Manning and Compton, 2008b). Motor proteins, in addition to their transport roles, influence microtubule dynamics, kinetochore microtubule attachment, and centrosome separation (Walczak and Heald, 2008).
A strategy in the treatment of cancer has been to inhibit cell division with antimitotic drugs, a set of natural and synthetic small molecules that characteristically arrest cells in mitosis, and induce programmed cell death (Gascoigne and Taylor, 2008; Shi et al., 2008). The spindle is the major target of antimitotics and three major microtubule spindle targets and associated inhibitors have been explored in the clinic: microtubule inhibitors including taxanes and epothilones; inhibitors of the Polo-like kinase Plk1, a regulator of spindle assembly, including BI 2536; and inhibitors of the mitotic kinesin-5, including monastrol and ispinesib (Kapoor et al., 2000; Lansing et al., 2007). Recent clinical trials have questioned the long-term efficacy of current antimitotic drugs. Although taxol remains the most widely-used and efficacious chemotherapeutic agent, it shows dose-limiting toxicities, including neutropenia and severe neuropathies, driving a need to identify alternative antimitotic drug targets that can be targeted and combined with lower doses of taxol to reduce the toxicity associated with high-dose taxol.
To identify proteins involved in mitotic spindle assembly, the linkage to cell death, and thus uncover potential targets for cancer therapeutics, we performed a proteomic analysis to identify mitotic microtubule co-purifying proteins (MMCPs) and genetic RNAi screening to test the contribution of these proteins to mitotic progression and induction of apoptosis. We report the results of these screens, which we exemplify by the identification and characterization of STARD9, a mitotic kinesin. STARD9 is necessary for PCM cohesion during the establishment of spindle bipolarity. The absence of STARD9 causes the pericentriolar material to fragment and dissociate from the centrioles, along with a failure to congress chromosomes, multipolar spindle formation, mitotic arrest, and apoptotic cell death. Most importantly, depletion of STARD9 synergizes with taxol treatment, making STARD9 a candidate target to extend current cancer therapeutics.
RESULTS
Identification of Mitotic Microtubule Co-purifying Proteins (MMCPs)
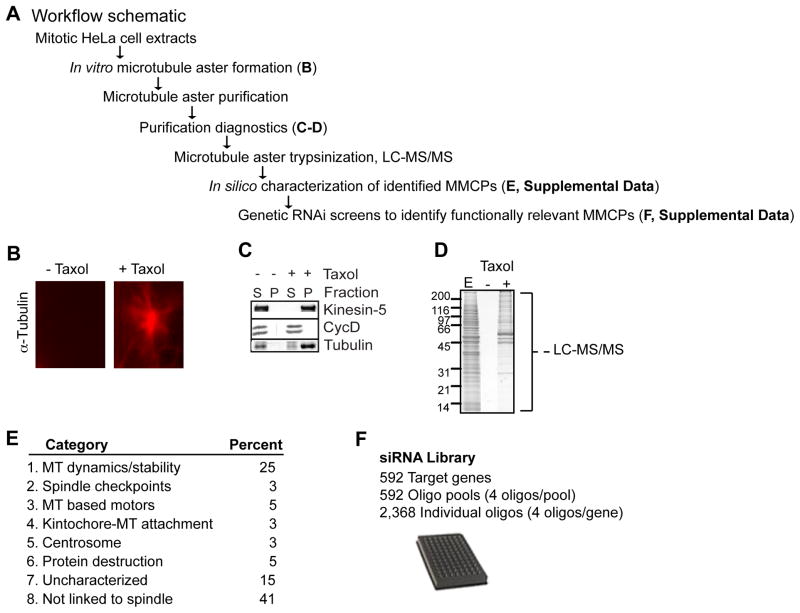
To identify MMCPs that contribute to mitotic spindle formation, we performed a proteomic analysis of microtubule aster co-purifying proteins (Figure 1A–D). Mitotic HeLa cell extracts were induced to undergo in vitro microtubule polymerization in the presence or absence of the microtubule-stabilizing drug, taxol (see (Mack and Compton, 2001) (Figure 1B). Polymerized microtubules and associated proteins were purified by sedimentation through a sucrose cushion. The selectivity of the purification was assessed by immunoblotting protein samples from the supernatant (S) and the pelleted microtubule aster (P) fractions for Kinesin-5 and cyclin D (Figure 1C). Kinesin-5 associated with the taxol stabilized microtubule pellet, whereas cyclin D remained in the supernatant (Figure 1C). Neither protein pelleted in the absence of taxol, demonstrating minimal non-specific pelleting (Figure 1C). Purified microtubule asters were trypsinized in solution and 592 MMCPs were identified by mass spectrometry (LC-MS/MS) (Figure 1D, Table S1, and Supplemental Information).
Figure 1. Proteomic and In Silico Analysis of MMCPs.
(A) Workflow for purification and identification of MMCPs. (B) In vitro mitotic aster microtubule polymerization reactions ± taxol, visualized with anti α–tubulin antibodies. (C) Immunoblot analysis of supernatant (S) and pellet (P) fractions from microtubule polymerization reactions. Blots were probed with anti Kinesin-5, CycD, and α–tubulin antibodies. (D) Purified microtubule pellets and associated proteins were analyzed by SDS-PAGE and stained with Coomassie blue. E = input extract. Microtubule pellets were digested in solution and proteins identified by LC-MS/MS. (E) In silico analysis of 592 genes and encoded proteins, for details see text and Supplemental Information. (F) Generation of a siRNA library targeting 592 genes corresponding to the MMCP set. (A–F) See also Supplemental Experimental Procedures and Tables S1 and S2.
To understand the potential roles of these proteins in cell division, the 592 MMCPs were systematically queried in PubMed and one of eight functional annotations was assigned to each protein based on existing literature (Figure 1E and Table S1). 25% of the proteins had a previously validated role in microtubule dynamics and stability, including TPX2, NuMA, and Astrin. Another 14% were involved in spindle-associated activities, including spindle checkpoints, kinetochore-microtubule attachment, centrosome homeostasis, regulation of mitosis, or were microtubule-based molecular motors including kinesins. Interestingly, over half of the proteins were either uncharacterized or had not been linked to mitotic spindle assembly (Figure 1E and Table S1).
The domain compositions of the 592 MMCPs were analyzed by querying the Unison (http://unison-db.org) (Hart and Mukhyala, 2009) database, which calculates the presence of domains from the Pfam (Finn et al., 2008) and Prosite (Hulo et al., 2006) databases, as well as from signal sequence and transmembrane prediction algorithms (see Supplemental Experimental Procedures). Proteins and domains were hierarchically clustered (Eisen et al., 1998) using the Pearson correlations of the numbers of each domain appearing in each protein (Table S1 and S2). A notable cluster of 21 MMCPs contained kinesin motor domains, including an uncharacterized protein STARD9, a family of ATPases whose members are important for spindle bipolarity, spindle pole focusing, kinetochore function, and regulation of microtubule dynamics.
Functional Characterization of MMCPs
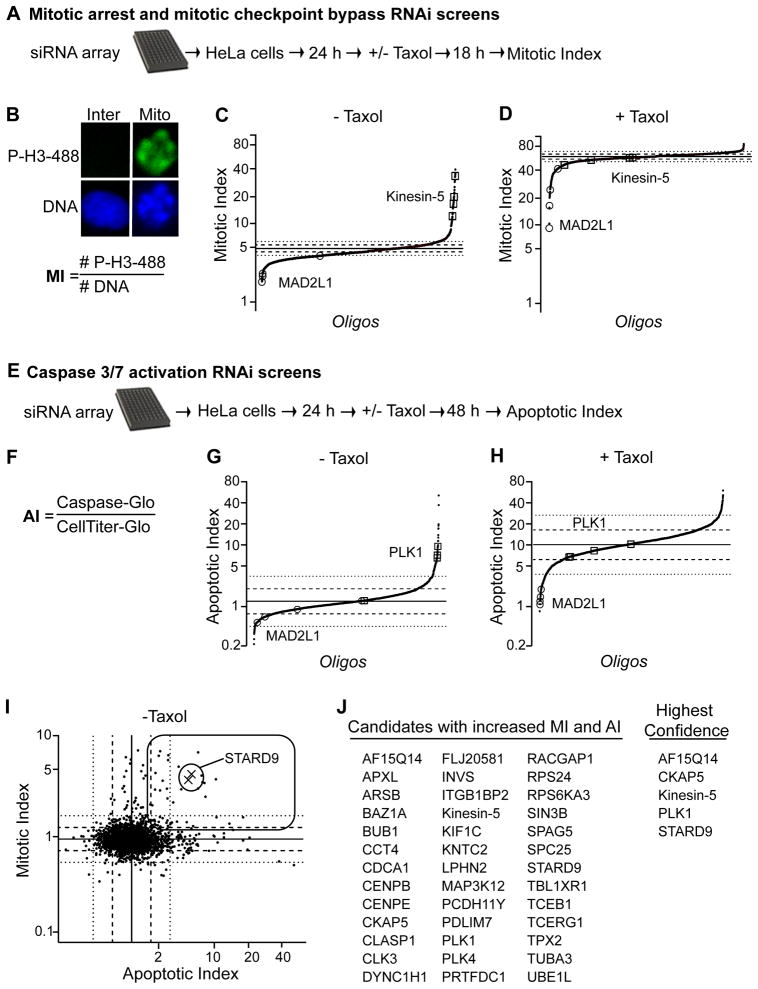
To assess the contribution of MMCPs to mitotic progression, we developed a siRNA library targeting the knockdown of all MMCP-encoding genes (Figure 1F). The library was used to perform four functional screens: mitotic arrest; mitotic spindle assembly checkpoint (SAC) bypass; induction of apoptosis; and synergy with taxol to induce apoptosis (Figure 2A–H, Table S3, Supplemental Information). Due to space constraints we focus our discussion on two screens. First, we sought to identify gene depletions leading to mitotic arrest. HeLa cells were reverse transfected with siRNAs for 48 hr prior to fixation and staining with Hoechst (DNA dye) and anti p-H3 antibodies, which recognize phosphorylated histone H3 (a marker of mitotic cells). Data was collected on 1,000 Hoechst positive cells per well using an image-based, high-content screening microscope. The mitotic index (MI) per well was derived from the number of mitotic nuclei (p-H3 positive) divided by the total nuclei (Hoechst positive) (Figure 2B). The results were graphed as scatter plots, with normalized MI on the Y-axis (in log scale) and individual oligonucleotides spread across the X-axis. 194 siRNAs, corresponding to 138 genes showed an increased MI greater than two standard deviations from the mean, and within this set 32 genes had two or more oligonucleotides score as hits (Figure 2C and Table S3). The majority of these gene products had a validated role in spindle assembly like TOGp, TPX2, TACC3, and Kinein-5. Additionally, this set also included multiple proteins with potential roles in spindle assembly, like STARD9.
Figure 2. Functional Characterization of MMCPs.
(A) Schematic of mitotic arrest and mitotic checkpoint bypass RNAi screens. (B) The mitotic index (MI= # cells phospho-histone H3 positive/total # Hoechst stained nuclei) was quantified per well. (C–D) RNAi screen summary scatter plots for individual oligonucleotides ± taxol. Individual oligonucleotides are spread across the X-axis and MI along the Y-axis. Solid line represents mean and dashed and dotted lines indicate one and two standard deviations from the mean of control oligonucleotides, respectively. (E) Schematic of RNAi screens for detection of increased apoptosis and synergy with taxol to induce apoptosis. (F) The apoptotic index (AI) was quantified as the total caspase 3/7 activities (Caspase-Glo) divided by the total ATP levels (CellTiter-Glo) per well. (G–H) Apoptosis screens summary scatter plots for individual oligonucleotides ±taxol. X-axis indicates oligonucleotides and Y-axis indicates normalized AI. Graphs are as in D. (I) Correlation analysis of individual oligonucleotides in - taxol screens, based on normalized MI (over control oligonucleotides) (Y-axis) versus normalized AI (over control oligonucleotides) (X-axis). Rectangle represents quadrant containing gene depletions with increased MI and AI. Xs represents STARD9 RNAi. (J) List of top hits with increased MI and AI. (A–J) See also Supplemental Experimental Procedures and Table S3.
Next, we screened for gene depletions leading to apoptosis (Figure 2E–H). Apoptosis relies on caspase 3/7 activity (Chowdhury et al., 2008). Therefore, we used a luciferase-based assay (caspase substrate linked to luciferin, which when cleaved is converted to light by luciferase and can be measured by a luminometer) to measure total caspase 3/7 activities (Caspase-Glo assay) per well. Additionally, the CellTiter-Glo assay, which measures total ATP levels (indicative of metabolically active cells) was used to measure the number of viable cells per well. HeLa cells were transfected with siRNA and the apoptotic index (AI, caspase activity per cell/ATP levels as described in (Peterson et al., 2010) was measured 72 hr post transfection (Figure 2F). Depletion of 45 genes (with ≥ 2 oligonucleotides scored as hits) showed an increased AI one standard deviation above the mean (Figure 2G). Among this set were Plk1, NuMA, AF15Q14, and STARD9.
Identification of STARD9
When inhibited, antimitotic targets arrest cells in mitosis and induce apoptosis (Gascoigne and Taylor, 2008; Shi et al., 2008). To assess the potential of MMCPs as antimitotic drug targets, we performed a correlation analysis of the normalized MI and normalized AI for each oligonucleotide from our screens (Figure 2I). This separated data into 4 quadrants, including a quadrant with 39 genes whose depletion caused an increase in both MI and AI (Figure 2I–J). This quadrant included Plk1 and Kinesin-5 whose depletion have been previously demonstrated to both induce mitotic arrest and activate the apoptotic response (Petronczki et al., 2008; Shi et al., 2008) (Figure 2J). Within this set we identified STARD9 (steroidgenic acute regulatory protein-related lipid transfer (START) domain containing 9), a previously uncharacterized kinesin-like protein (Halama et al., 2006) (Figure 2J). STARD9 was selected for further analysis, based on its co-purification with mitotic microtubule asters, requirement for mitotic progression, role in suppressing apoptosis, and novelty as a mitotic kinesin.
STARD9 is a Kinesin-3 Family Member
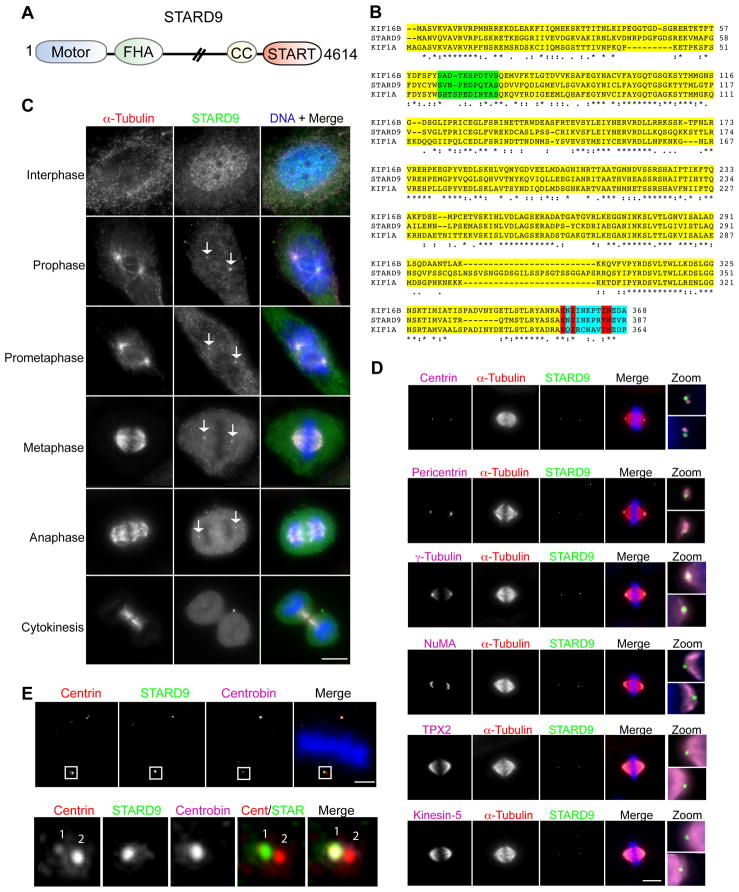
Database searches predicted three STARD9 isoforms; the longest encoding a 4,614 amino acid protein with an apparent molecular mass of 506.7 kDa (Figures S1A–B, and 3A). STARD9 is a member of the Kinesin-3 family, and shares 52% and 48% sequence identity within the motor domain (amino acids 1-374) with its closest human paralogues KIF16B and KIF1A respectively (Lawrence et al., 2004; Wickstead and Gull, 2006) (Figure 3B). The Kinesin-3 family (composed of the KIF1, KIF13, KIF14, KIF16, and KIF28 subfamilies) is implicated in transporting vesicles and organelles (Hoepfner et al., 2005; Miki et al., 2005; Nangaku et al., 1994). The STARD9 motor and FHA domains are conserved among all family members (Vale, 2003) (Figure 3B). STARD9 also has a C-terminal START domain predicted to bind phospholipids and/or sterols, analogous to the KIF1A/KIF1B PH domain and the KIF16B PX domain that bind phospholipids (Alpy and Tomasetto, 2005). Interestingly, STARD9 has a unique 26 amino acid residue insertion in loop 12, a microtubule interaction region of kinesins (Figure 3B). STARD9 is ubiquitously expressed at low levels in all tissues analyzed, including skin, brain, and heart (Figure S1C). Additionally, STARD9 orthologs are only found in vertebrates (Figure S2).
Figure 3. STARD9 is a Centrosomal Protein Enriched at Daughter Centrioles.
(A) Schematic of STARD9 protein composition, including N-terminal kinesin motor domain, FHA domain, predicted coiled coil, and C-terminal START domain. (B) STARD9 motor domain alignment. The STARD9 motor domain shares 49% identity to KIF1A and 52% identity to KIF16B. A conserved insertion within loop 3 of Kinesin-3 family members is highlighted in green. Sequence C-terminal to motor domain, highlighted in blue, contains polarity determining amino acids residues and key residues predicting plus end directionality are in red. (C) Cell cycle subcellular localization of STARD9. HeLa cells were co-stained for STARD9, α-tubulin, and DNA. STARD9 accumulates at the centrosomes from prophase to late anaphase, see arrows (Bar = 5 μm). (D) Co-staining of STARD9, α-tubulin, DNA, and either centrin, pericentrin, γ-tubulin, NuMA, TPX2, or Kinesin-5. Zoom depicts magnified view of two centrosomes in one cell (Bar = 5 μm). (E) Co-staining of STARD9, α-tubulin, DNA, and centrobin (Bar = 2 μm). Bottom panel is magnified view of one centrosome showing STARD9 and centrobin colocalization. (A–E) See also Supplemental Experimental Procedures and Figures S1, S2 and S3.
STARD9 Localizes to Daughter Centrioles During Mitosis
To assess the role of STARD9 in mitosis, we analyzed its sub-cellular localization through the cell cycle. HeLa cells were fixed and co-stained for DNA, STARD9, and α–tubulin. STARD9 was distributed throughout the nucleus and cytoplasm in interphase, localized to the centrosomes from prophase to late anaphase, and disappeared at cytokinesis (Figure 3C, see arrows). To define the localization of STARD9, we performed co-localization studies with centrosome and spindle pole markers (Figure 3D). Centrin staining detects two centrioles per centrosome and STARD9 consistently co-localized to one of the two centrioles (Figure 3D, panel 1 zoom). STARD9 also co-localized with pericentrin and γ–tubulin and localized to the outer periphery of the spindle pole as defined by NUMA, TPX2, and Kinesin-5 staining (Figure 3D, panels 2–6 zoom). Interestingly, STARD9 co-localized with centrobin, a notable daughter centriole-associated protein (Figure 3E, see zoom) (Zou et al., 2005). Thus, the localization of STARD9 to daughter centrioles is cell cycle specific. Additionally, in cells containing multiple centrosomes, STARD9 only localized to the daughter centrioles of two centrosomes (Figure S3A), suggesting that it is under a restrictive control that blocks its appearance at supernumerary centrioles.
The STARD9 Motor Domain Contains Microtubule-binding and ATPase Activities
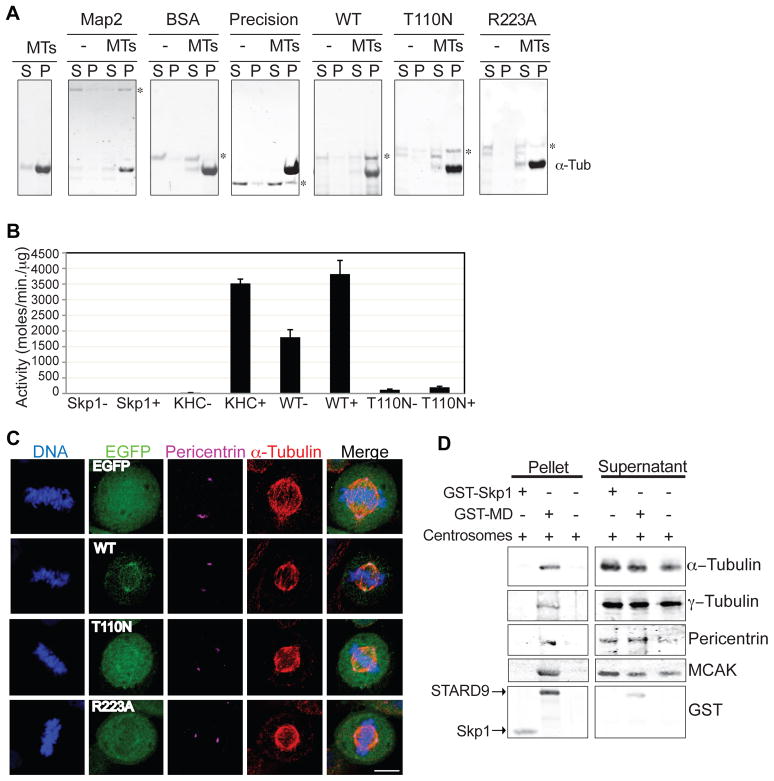
STARD9 is a kinesin-like protein with no previous molecular dissection, thus we analyzed whether GST-tagged STARD9 motor domain (MD) fusion proteins, wild type (WT) and mutants could bind microtubules and undergo an ATPase cycle, two defining properties of kinesins. Mutant T110N is a threonine to asparagine substitution at position 110 within the P-loop of the ATP binding site, predicted to show rigor binding to microtubules with no ATP hydrolysis or movement (Blangy et al., 1998). Mutant R223A is an arginine to alanine substitution within the Switch I region, predicted to reduce its affinity for microtubules (Yun et al., 2001). For microtubule binding assays (as described in (Goode and Feinstein, 1994), MD-WT and MD-mutants were incubated with or without polymerized microtubules and their ability to bind microtubules was determined by pelleting microtubules at 100,000 × g and analyzing samples from the pellet and supernatant by SDS-PAGE and Coomassie blue staining (Figure 4A). Similar to microtubule associated protein 2 (Map2, positive control) MD-WT and MD-T110N pelleted with microtubules. However, MD-R223A showed a significantly reduced ability to pellet with microtubules (Figure 4A). Next, the ATP-hydrolyzing activity of STARD9 was analyzed by performing ATPase assays (enzyme linked inorganic phosphate assay, Cytoskeleton Inc.) in the presence or absence of microtubules (Figure 4B). In this assay ATP hydrolysis releases inorganic phosphate (Pi), which is converted into an absorbance shift from 330 to 360 nm that can be measured by a spectrophotometer (Webb, 1992). Similar to kinesin heavy chain, MD-WT displayed microtubule-stimulated ATPase activity. However, MD-T110N and Skp1 (negative control) showed minimal ATPase activity (Figure 4B). Therefore, the motor domain of STARD9 possesses microtubule-binding and ATP hydrolyzing activities comparable to that of other kinesins.
Figure 4. STARD9 Microtubule-Binding and ATPase Activities are Required for its Centrosomal Localization.
(A) Map2, BSA, GST-Precision, or GST-tagged motor domain (MD)-WT, T110N and R223A were incubated with or without microtubules and their ability to bind microtubules determined by analyzing the supernatant and pellet fractions by SDS-PAGE and Coomassie blue staining. Asterisks indicate protein bands of interest. (B) The ATPase activity of kinesin heavy chain (KHC), GST-Skp1, GST-tagged WT or T110N was assessed by an ATPase end point assay. Activity is in moles/minute/μg. Error bars indicate ±STD. (C) HeLa cells were transfected with eGFP, or eGFP-tagged WT, T110N or R223A for 24 hr. Cells were fixed and co-stained for α-tubulin, DNA, and eGFP (Bar = 5 μm). (D) Recombinant GST-Skp1 or GST-WT were immobilized on a bead matrix and incubated with centrosome preparations. Samples from pelleted beads and supernatants were resolved by SDS-PAGE and immunoblotted for α-tubulin, γ-tubulin, pericentrin, and MCAK. (A–D) See also Supplemental Experimental Procedures.
Kinesins rely on their microtubule-binding and ATP hydrolyzing activities to localize to specific subcellular structures. Thus, we analyzed whether STARD9s localization to centrioles was dependent on these activities. EGFP-tagged MD-WT, MD-T110N, and MD-R223A were expressed in HeLa cells and their localization was monitored by immunofluorescence. EFGP-MD-WT localized to the daughter centriole, consistent with endogenous STARD9 localization, along with the mother centriole and the surrounding PCM, indicating that this truncated fragment lacked determinants that selectively target it to the daughter centriole. Consistently, GST-MD-WT, but not a control GST-Skp1, was able to bind α–tubulin and the centrosomal proteins γ–tubulin, pericentrin, and MCAK in capture assays with purified centrosomes (Figure 4D). However, eGFP-MD-T110N and -MD-R223A were unable to localize to centrosomes (Figure 4C). These data indicate that the STARD9 microtubule-binding and ATP hydrolyzing activities are required for its localization to centrosomes.
Depletion of STARD9 Induces PCM Fragmentation in Varied Cancer Cell Lines
To understand the role of STARD9 in mitosis, we analyzed the cellular consequences of depleting STARD9. First, we identified two siRNA oligonucleotides, which reduced STARD9 mRNA levels to less than 10% (oligonucleotides 1 (herein referred to as siSTARD9) and 4) (Table S4). HeLa cells were transfected with siSTARD9 for 48 hr and the effect on mitosis was analyzed by co-staining for DNA, centromeres, and α-tubulin (Figure 5A). STARD9-depleted cells failed to congress mitotic chromosomes, which remained scattered along a defective multipolar spindle (Figure 5A). Consistently, no STARD9 daughter centriole staining was observed in siSTARD9 cells, showing the specificity of these antibodies for STARD9 (Figure S3B). Similar defects were observed with STARD9 siRNA oligonucleotide 4 (Figure S3C–D).
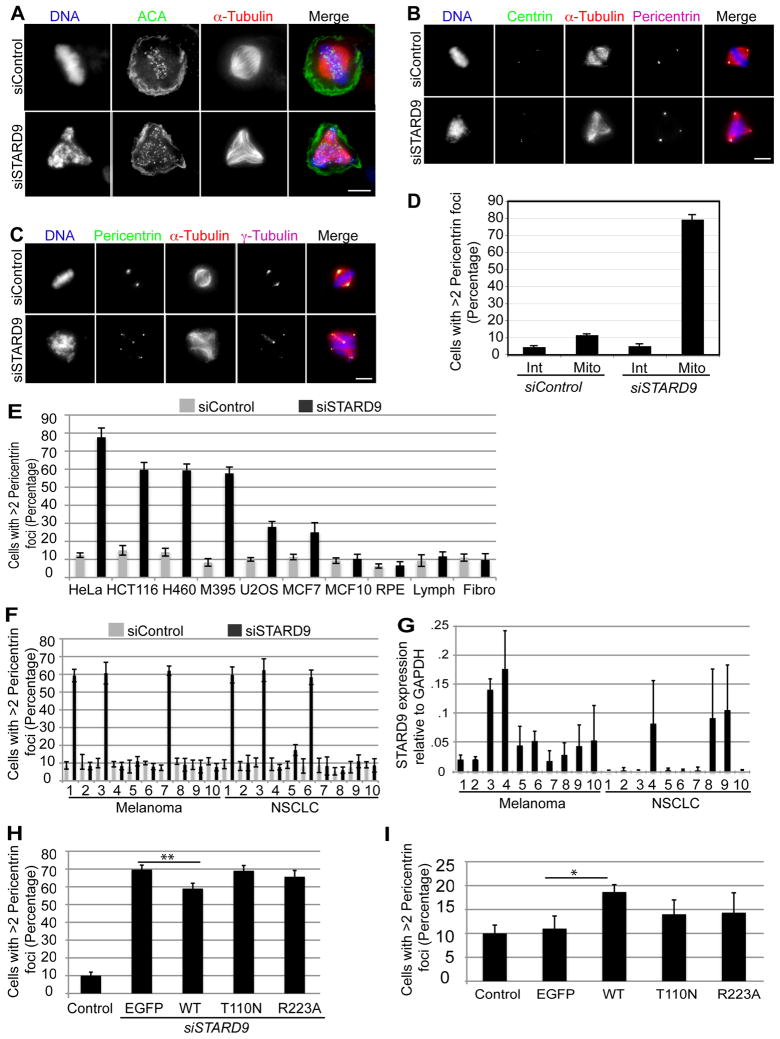
Figure 5. STARD9 Depletion Induces Fragmentation and Dissociation of the PCM from Centrosomes in Multiple Cancer Types.
(A–C) Control or STARD9 siRNA treated HeLa cells were fixed and co-stained for α-tubulin, DNA, and ACA (centromere marker) (A), centrin and pericentrin (B), pericentrin and γ-tubulin (C). (D) Quantitation of the percentage of interphase (Int) and mitotic (Mito) cells with greater than two pericentrin foci. (E) HeLa, HCT116, H460, M395, U2OS, MCF7, MCF10a, hTERT-RPE-1, lymphoblast, and fibroblast cells were treated with siControl or siSTARD9 for 48 hr. Cells were fixed and co-stained for DNA, α-tubulin and pericentrin and the percentage of mitotic cells with greater than two pericentrin foci was quantified. (F) The same analysis in E was carried out with panels of melanoma and non-small-cell lung carcinoma cell lines (NSCLC). (G) STARD9 mRNA expression levels relative to GAPDH in melanoma and NSCLC cell lines. (H) Overexpression of STARD9-MD partially rescues PCM fragmentation. Cells were co-transfected with siSTARD9 and vectors expressing eGFP, or siRNA resistant eGFP-tagged STARD9 motor domain WT, T110N, or R223A. Data represent the average ± SD of 3 independent experiments, 100 cells counted for each. **p < 0.005. (I) Overexpression of the STARD9 motor domain leads to a dominant negative phenotype. EGFP-tagged STARD9 motor domain WT, T110N, or R223A were expressed for 48 hr in HeLa cells and the percentage of cells with greater than two pericentrin foci was quantified as in H *p < 0.05. (D–I) Error bars indicate ±STD. (A–E) See also Supplemental Experimental Procedures, Figures S3, S4, S5 and Tables S4 and S5.
Multiple pathways exist for generating multipolar spindles, including centrosome amplification, centriole fragmentation, centriole splitting, and pericentriolar material (PCM) fragmentation (Fukasawa, 2007; Oshimori et al., 2006). To define the mechanism(s) by which siSTARD9 cells generate multipolar spindles, we visualized the status of the centrosomes, centrioles (centrin) and PCM (pericentrin). Similar to siControl cells, mitotic siSTARD9 cells had two centrosomes with no centriole splitting or fragmentation (Figure 5B). In interphase, > 90 % of siControl and siSTARD9 cells had one or two pericentrin or γ-tubulin foci, indicating that STARD9 depletion did not trigger centrosome over-duplication (Figure 5C–D). In mitosis, ~82 % of siSTARD9 cells had more than two pericentrin foci, which correspond to sites of ectopic microtubule nucleation, compared to ~12 % of siControl cells (Figure 5C–D). These results indicate that siSTARD9 cells enter mitosis with a normal number of centrosomes, and also suggest that multipolar spindles do not arise from centriole splitting or centriole fragmentation, but instead from fragmentation and dissociation of the PCM from centrosomes.
Our analysis of STARD9 centered on HeLa (cervical adenocarcinoma) cells, but we sought to assess the importance of STARD9 for cell division in other types of cancers. Thus, we analyzed the consequences of depleting STARD9 in a panel of cancer cell lines. Similar to the defects observed in HeLa cells, HCT116 (colorectal carcinoma), H460 (non small cell lung carcinoma), and M-395 (melanoma) cells showed an increase in the number of mitotic cells with greater than 2 pericentrin foci (Figures 5E and S4). In contrast, only a small increase in PCM fragmentation was observed in U2OS (bone osteosarcoma) and MCF-7 (breast adenocarcinoma) cells and no significant change was seen in hTERT-RPE-1 (retinal pigment epithelial cells, which have properties of more normal cells, and show strongly reduced responses to other anti-mitotic agents), MCF-10a (normal breast cells), lymphoblasts, and fibroblasts (Figures 5E and S4). These results indicate that various cancer cells are sensitive to STARD9 depletion, but not normal cells. However, cancer subtypes tend to have varied genetic backgrounds, checkpoint competencies, and sensitivities to drug treatments. Therefore, we analyzed the response to STARD9 depletion in two panels of cancer cell lines, one containing 10 melanoma cell lines and the other 10 non small cell lung carcinoma (NSCLC) cell lines. Surprisingly, only 3 out of 10 melanoma and 3 out of 10 NSCLC cell lines displayed PCM fragmentation above 55% (Figure 5F). Thus, although many types of cancer cells are sensitive to STARD9 depletion, there is a need to identify the specific subtypes.
Deregulating the levels of microtubule-associated proteins can contribute to tumor formation, for example, TOGp, NuMA, and TPX2, which nucleate, crosslink, and stabilize microtubules in mitosis are over-expressed in cancer and correlate with a high incidence of chromosomal instability (Carter et al., 2006). Additionally, RASSF1A and MTUS1, which control mitotic progression, are underexpressed in specific cancers and act as tumor suppressors (Rodrigues-Ferreira et al., 2009; Song et al., 2004). Thus, we asked if proteins within the MMCP set (including STARD9) were deregulated in cancer by analyzing the differential gene expression levels of the MMCP set across 27 cancer tissues versus normal samples from the same tissue (Figure S5 and Table S5). Consistent with previous studies, the spindle checkpoint regulators Mad2L1, Bub1, and Aurora kinase B (AurKB) clustered within a large group showing over-expression across multiple cancer types. This set included genes involved in centrosome (AurKA and Plk4) and spindle pole (Astrin) homeostasis, suggesting that spindle poles show altered regulation in cancer. Interestingly, STARD9 clustered with a large group down-regulated across multiple cancer types that included the centriolar and ciliary protein Alms1 (Figure S5 and Table S5). Consistently, qRT-PCR analysis showed that STARD9 was expressed at low levels in most cancers (Figure 5G). However, we found no simple correlation between the basal STARD9 expression levels and the extent of PCM fragmentation induced by STARD9 depletion.
Overexpression of the STARD9 Motor Domain Partially Rescues PCM Fragmentation in STARD9-depleted Cells
Possibly due to its large size, our attempts to express full length STARD9 were unsuccessful. As the STARD9 motor domain has microtubule-binding and ATPase activities required for its localization to centrosomes, we asked if expression of the motor domain rescued the PCM fragmentation phenotype observed in STARD9-depleted cells. HeLa cells were co-transfected with siSTARD9 and siRNA resistant eGFP-tagged MD-WT, MD-T110N, and MD-R223A for 48 hr and the extent of PCM fragmentation was determined. Overexpression of MD-WT, but not MD-T110N or MD-R223A led to a modest but significant rescue (~10%, p-value .001; Figure 5H), indicating that the STARD9 motor domain not only localizes STARD9 to centrioles, but also functions to maintain PCM cohesion. Interestingly, overexpression of MD-WT in control HeLa cells led to modest increase (~9%, p-value .0491) in PCM fragmentation and mitotic spindle defects, indicating a dominant negative effect (Figure 5I).
STARD9 is not Required for Recruitment of Spindle Pole Focusing Activities
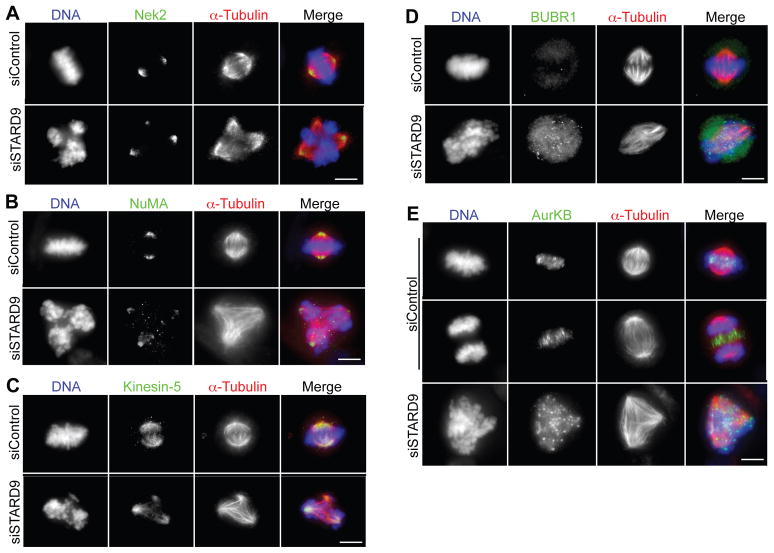
Kinesins are often involved in the transport of cargo along microtubules, therefore it was possible that STARD9s role in PCM cohesion involved the transport of factors required for centrosome homeostasis. Thus, we analyzed whether depletion of STARD9 had an effect on the localization of proteins involved in centrosome maturation (NEK2, AurKA, Plk1), centrosome separation (Kinesin-5), microtubule nucleation (TPX2) and spindle organization (NuMA) by immunofluorescence microscopy. NEK2, AurKA, Plk1, Kinesin-5, NuMA, and TPX2 were each localized to spindle poles and ectopic microtubule nucleating sites (fragmented PCM) that formed multipolar spindles in siSTARD9 cells (Figures 6A–C, S6A–C). These results indicate that STARD9 does not recruit spindle pole focusing activities and motors necessary for spindle bipolarity.
Figure 6. STARD9 is not Required for Recruitment of Spindle Pole Focusing Activities and its Depletion Activates the SAC.
(A–E) Control or STARD9 siRNA treated HeLa cells were fixed and co-stained for STARD9, α-tubulin, DNA, and NEK2 (A), NuMA (B), Kinesin-5 (C), BUBR1 (D) and AurKB (E). Bar = 2 μm. (A–E) See also Supplemental Experimental Procedures and Figure S6.
Depletion of STARD9 Arrests Cells in Mitosis With an Active SAC
Depletion of STARD9 led to defects in chromosome congression and arrested cells in early mitosis, indicative of an activated spindle assembly checkpoint (SAC), which monitors microtubule-kinetochore attachment and inter-kinetochore tension. When active, SAC proteins like BubR1 and Bub1 reside at the kinetochore region and disperse upon proper microtubule-kinetochore attachment (Musacchio and Salmon, 2007). Consistently, siControl cells aligned their chromosomes at the metaphase plate with minimal BubR1 and Bub1 staining (Figures 6D and S6D). Conversely, siSTARD9 cells arrested in prometaphase, with BubR1 and Bub1 prominently localized to kinetochores (Figures 6D and S6D). The chromosomal passenger complex (CPC, composed of AurKB, INCENP, Survivin, and Borealin) is a SAC complex required to arrest cells in response to lack of inter-kinetochore tension (Ruchaud et al., 2007). At the onset of anaphase, in siControl cells, the CPC was dynamically delocalized from the kinetochores and transitioned to the central spindle (Figure 6E). In contrast, in siSTARD9 cells, AurKB and INCENP remained at the kinetochore region and never redistributed to the central spindle (Figures 6E and S6E). These results indicate that STARD9-depleted cells lack proper microtubule-kinetochore attachments and/or inter-kinetochore tension, which activates the SAC and arrests cells in early mitosis.
Chemical Inhibition of Cdk1, Plk1, or Kinesin-5 Function Prevents PCM Fragmentation in STARD9-depleted Cells
To define the timing and order of events leading to PCM fragmentation and mitotic arrest in siSTARD9 cells, we combined siSTARD9 treatment with inhibition of mitotic regulators. Synchronized siControl and siSTARD9 cells were treated with small molecule inhibitors of Plk1 (centrosome maturation), Kinesin-5 (centrosome separation), Cdk1 (G2/M transition), and microtubule dynamics (taxol and nocodazole) prior to mitotic entry. Two hours post mitotic entry, cells were fixed and co-stained for DNA, α-tubulin, pericentrin or centrin, and STARD9. Inhibition of Plk1 with the ATP-competitive inhibitor compound 1 (Lansing et al., 2007) arrested siControl and siSTARD9 cells with monopolar spindles and a focused PCM around the two centriole pairs (Figure S7A, top panels). Similarly, inhibition of Kinesin-5 with monastrol (Kapoor et al., 2000) arrested siControl and siSTARD9 cells with monopolar spindles and a matured PCM around the two centriole pairs (Figure S7B, top panels). Thus, both treatments blocked siSTARD9 cells from transitioning to PCM fragmentation, indicating that STARD9 functions to maintain PCM cohesion downstream of centrosome maturation and separation. Consistent with this possibility, inhibition of Cdk1 with RO-3306 (Vassilev et al., 2006) arrested cells at the G2/M transition and prevented PCM fragmentation in siSTARD9 cells (Figure S7C, top panels) (Vassilev et al., 2006). Additionally, STARD9 did not localize to daughter centrioles in G2/M arrested cells (Figure S7C, lower panel), confirming our previous results showing STARD9 localization to daughter centrioles only from prophase to late anaphase (Figure 3C).
In early mitosis, microtubule-dependent forces are applied to the centrosome by proteins that organize and form the bipolar spindle (Abal et al., 2005). When PCM cohesion is compromised, these forces fragment the PCM (Oshimori et al., 2006). Thus, it was possible that microtubule-dependent forces played a role in the PCM fragmentation observed in siSTARD9 cells. Under normal conditions ~80% of siSTARD9 cells exhibit PCM fragmentation compared to ~10% of siControl cells (Figure 5D). However, depolymerization of microtubules with nocodazole treatment drastically reduced PCM fragmentation in siSTARD9 cells to undetectable levels (Figure S7D, top panels). Interestingly, stabilization of microtubules in siSTARD9 cells with taxol treatment (interferes with microtubule-dependent forces) also modestly reduced PCM fragmentation (to 51%; Figure S7E, top panels). These results indicate that microtubule-dependent forces drive PCM fragmentation in siSTARD9 cells. In addition, STARD9 was weakly localized to centrioles in taxol treated cells and absent in nocodazole treated cells (Figure S7D lower panels and S7E, right panels), indicating that the localization of STARD9 to the centrioles requires microtubules.
STARD9 Depletion Induces Mitotic Apoptosis
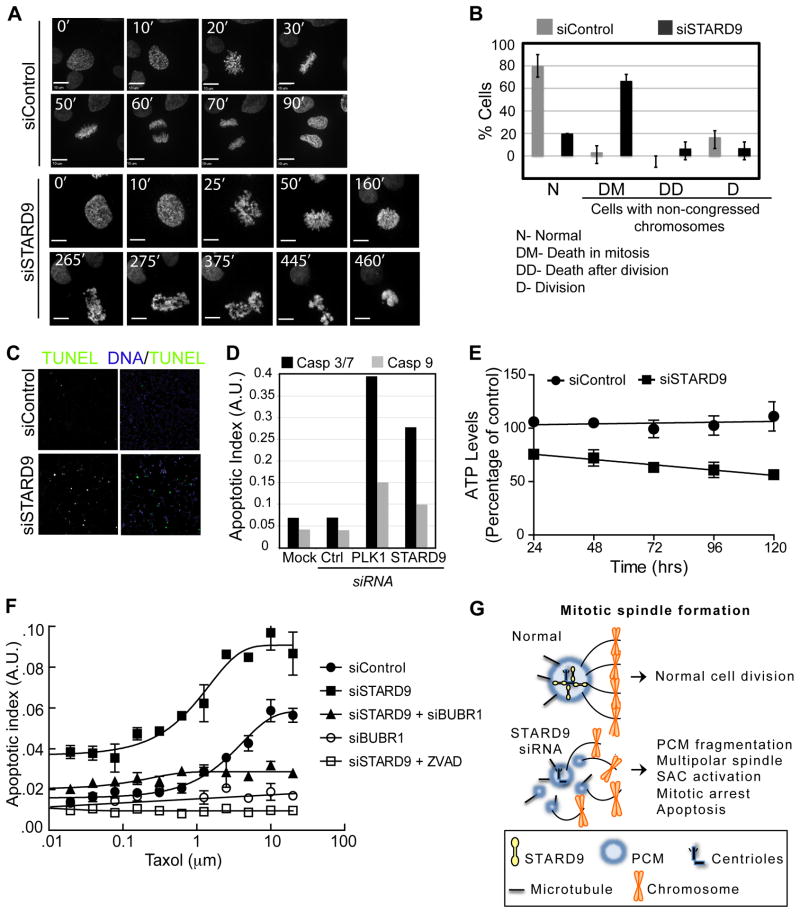
Since siSTARD9 cells exhibited elevated levels of mitotic arrest, SAC activation, and apoptosis, we hypothesized that they were apoptosing in mitosis. To test this, we imaged siControl and siSTARD9 cells by time-lapse microscopy using a HeLa cell line expressing eGFP-histone 2B to mark chromosomes. In control cultures, 90 % of cells transitioned through mitosis with normal kinetics (~80 minutes) and 10 % had defects in chromosome congression. In contrast, 75 % of siSTARD9 cells had defects in chromosome congression and arrested in mitosis for an average of 3.5 hr, at which time their chromatin became fragmented and highly condensed, indicative of apoptosis (Figure 7A–B, Movies S1 and S2).
Figure 7. STARD9 Depletion Induces Mitotic Apoptosis and Synergizes with Taxol.
(A–B) Time-lapse microscopy of HeLa-GFP-H2B cells treated with control or STARD9 siRNA, frames are at 5-minute intervals (see Movies S1 and S2). (B) Twenty siControl or siSTARD9 cells were visualized by time-lapse microscopy and chromosome congression and mitotic outcome were quantified. (C) Visualization of DNA fragmentation in siControl or siSTARD9 HeLa cells with the DeadEnd Fluorometric TUNEL assay. Cells were fixed and imaged 5 hr post mitotic entry. (D) Caspase 3/7 and 9 activity was determined for siControl, siSTARD9, or siPlk1 HeLa cells. The apoptotic index (AI, Caspase-Glo/CellTiter-Glo) was quantified and plotted on a log scale as arbitrary units. (E) STARD9 depletion decreases cell viability. Total ATP levels (CellTiter-Glo assay) were quantified in siControl and siSTARD9 cells at indicated time points post siRNA transfection. (F) Depletion of STARD9 synergizes with taxol treatment and is dependent on the spindle assembly checkpoint. Cells treated with control, STARD9, BUBR1, or STARD9 + BUBR1 siRNA were incubated with increasing concentrations (0–10 μm) of taxol and the apoptotic index was quantified as in D. Additionally, the small molecule ZVAD was added to STARD9 siRNA-treated cells. (G) Model: STARD9 localizes to daughter centrioles in prophase and stabilizes the PCM during bipolar spindle assembly. (B, E, F) Error bars indicate ±STD. (A–G) See also Supplemental Experimental Procedures, Figure S7, and Movies S1 and S2.
To substantiate our in vivo findings, we tested siSTARD9 cells for biochemical hallmarks of apoptosis, namely DNA fragmentation, apoptosis, and a decrease in cell viability. Synchronized siControl and siSTARD9 cells were fixed 5 hr post mitotic entry and analyzed by the DeadEnd fluorometric TUNEL assay (Promega) for DNA fragmentation (see Supplemental Information). In this assay DNA breaks are labeled with fluorescein-12-dUTP and can be visualized and quantified by fluorescence microscopy. In contrast to siControl cells, siSTARD9 cells had a marked increase in TUNEL-positive cells (Figure 7C). Apoptosis relies on the activation of initiator (caspase 9) and effector (caspase 3/7) caspases (Chowdhury et al., 2008). Thus, we analyzed whether these caspases were activated in siSTARD9 cells. Seventy-two hours post transfection, asynchronous siControl, siPlk1, or siSTARD9 cells were fixed and the AI was measured as described in our apoptotic screen. Depletion of STARD9 or Plk1 led to an increased AI, confirming our screen results and published data (Petronczki et al., 2008) (Figure 7D). We then measured the cell viability of siControl and siSTARD9 cells using the CellTiter-Glo assay that measures total ATP levels (indicative of metabolically active cells) at 24, 48, 72, 96, and 120 hr post siRNA transfection. As expected, the viability of siSTARD9 cells decreased over time compared to siControl cells (Figure 7E). These results indicate that STARD9 depletion leads to mitotic arrest, activation of apoptosis and mitotic cell death.
STARD9 Depletion Synergizes with Taxol Treatment
Although paclitaxel (Taxol) remains one of the most widely used anticancer agents, its efficacy is limited by neutropenia and neurotoxicities. If a combination treatment were to shift the dose-efficacy curve, it might be possible to improve the therapeutic index and efficacy of taxol. Thus, we asked if depletion of STARD9 could synergize with taxol to induce apoptosis. siControl and siSTARD9 cells were treated with increasing concentrations of taxol 48 hr post-transfection and their AI was measured 72 hr post-transfection. Indeed, depletion of STARD9 increased the apoptotic response with increasing concentrations of taxol, with an IC50 of 1.15 μM for siSTARD9 and IC50 of 3.28 μM for siControl (Figure 7F). siSTARD9 treatment also increased the peak AI by ~1.7×, consistent with a higher percentage of cells undergoing apoptosis at this time point. These results indicate that depletion of STARD9 synergizes with taxol treatment.
The SAC including Mad2 and BUBR1 plays a critical role in the induction of mitotic apoptosis. Thus, we asked if siSTARD9-induced apoptosis was dependent on the SAC by depleting BUBR1 and assessing its effect on the siSTARD9-taxol combination treatment. In comparison to siSTARD9, co-depletion of STARD9 and BUBR1 led to a marked decrease in AI (Figure 7F), indicating that the SAC plays a critical role in the induction of mitotic apoptosis in siSTARD9 cells. Furthermore, addition of ZVAD, caspase inhibitor, led to a complete inhibition of siSTARD9-induced apoptosis (Figure 7F).
DISCUSSION
The importance of mitotic spindle assembly to cell division and cancer biology has fueled intensive study of this structure. Our interest in identifying targets for cancer therapeutics prompted us to conduct a focused screen for regulators of mitosis. We report the identification of 592 mitotic microtubule co-purifying proteins by mass spectrometry and analysis of their contribution to mitosis by functional RNAi screens for mitotic arrest, checkpoint bypass, and induction of apoptosis. Interestingly, we identified a kinesin, STARD9, with a critical role in cancer cell division. Several insights have emerged from our studies. First, we have uncovered a previously undescribed mitotic kinesin whose localization and function is unique and significantly different from other kinesins. Second, based on STARD9s function and enzymatic activity, it appears to be a potential cancer target. Third, the triggers that induce mitotic apoptosis are poorly-defined and our study indicates that compromising the integrity of the PCM may have a role in inducing apoptosis. Finally, our study highlights the utility of performing focused siRNA screens to understand cellular processes, as many of the genes we have identified as regulating mitosis and apoptosis were not identified in previous genome-wide screens for mitotic regulators (Kittler et al., 2007).
Role of STARD9 in Mitosis
STARD9-depletion shares many phenotypic similarities to depletion of Kizuna, a centrosomal protein critical for centrosome integrity and spindle bipolarity, which forms a bond that tethers the expanded PCM to the centriole (Oshimori et al., 2006). Kizuna is also a Plk1 substrate and functions downstream of Plk1-mediated PCM organization in early mitosis (Oshimori et al., 2006). Similar to Kizuna, we hypothesize that STARD9 stabilizes the PCM under microtubule-mediated tension during bipolar spindle assembly. However, the localization of STARD9 to centrioles indicates that unlike Kizuna, which forms a general PCM bond, STARD9 may form a link between the daughter centriole and the PCM. Interestingly, both Kizuna and STARD9 are conserved among vertebrates, but not invertebrates or other lower eukaryotes (Figure S2). It is intriguing to consider whether higher eukaryotes may have more sophisticated centrosome compositions that allow them to accommodate the microtubule-mediated tension that occurs during the formation of a bipolar spindle.
Identification of STARD9 interacting proteins will be critical to understanding its role in mitosis. A recent genome-wide yeast two-hybrid analysis identified an interaction between STARD9 and CDK5RAP2 (the human homolog of D. melanogaster Centrosomin) (Stelzl et al., 2005). CDK5RAP2 localizes to centrosomes during mitosis and has been implicated in centriole engagement, PCM cohesion, and microtubule nucleation (Barrera et al., 2010; Choi et al., 2010). However, a detailed analysis will be necessary to determine if the STARD9-CDK5RAP2 interaction is important for PCM cohesion. Future efforts to define the mechanism of STARD9s’ function will benefit from biochemical purifications and mass spectrometry analyses aimed at identifying STARD9 interacting proteins, which may cooperate with STARD9 or regulate STARD9 activity.
The localization of STARD9 to daughter centrioles is unique, as centrobin is the only protein known to predominantly localize to daughter centrioles (Zou et al., 2005). However, unlike centrobin, STARD9 centriolar localization is cell cycle-regulated, appearing only at the onset of prophase and disappearing by cytokinesis (Figure 3C). Treatment with nocodazole abolishes this localization indicating that it is microtubule-dependent (Figure S7D). Interestingly, the STARD9 motor domain displays microtubule-binding and ATPase activities, both of which are required for its centriolar localization (Figure 4C).
STARD9 Depletion and Apoptosis
The triggers that activate the apoptotic pathway in mitosis are poorly understood. Interestingly, Plk1 and Kinesin-5 inhibitors tend to arrest the cells in mitosis for prolonged lengths of time (up to 18 hr) before the induction of apoptosis (Petronczki et al., 2008; Shi et al., 2008). However, the induction of apoptosis observed in STARD9-depleted cells appears to show rapid kinetics, implying that siSTARD9 prometaphase arrest differs from that observed in Plk1 and Kinesin-5 inhibited cells. It is tempting to consider that PCM fragmentation, defective microtubule dynamics, or DNA tearing observed in STARD9-depleted cells (Figures 5 and 7) are important apoptotic triggers and should be explored further.
STARD9 as a Cancer Target
Several of our findings indicate that inhibition of STARD9s enzymatic activity may be a viable approach to inhibiting cancer cell division. First, the PCM fragmentation and spindle assembly defects caused by STARD9-depletion appear to be conserved among multiple cancer cell lines and not in normal cell lines (Figure 5E and S4). Second, siSTARD9 cells apoptose during mitosis and with faster kinetics than other antimitotic treatments like Kinesin-5 or Plk1 inhibition. Third, the STARD9 microtubule-binding and ATPase activities are critical to its localization to centrioles and function in PCM cohesion (Figure 4C and 5H). Fourth, depletion of STARD9 synergizes with taxol treatment (Figure 7F), opening the possibility for combined therapies. Many details of STARD9 function will need to be explored to fully assess its potential as a cancer target, including a structure-function analysis. In addition, as the basal expression levels of STARD9 do not correlate with the level of STARD9-depletion sensitivity, future studies should be aimed at identifying the variables that lend specific types of cancers responsive to STARD9 inhibition. Finally, it is possible that the low STARD9 levels observed in many cancers could be a contributing factor to cancer progression and should be analyzed further.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa cells were grown in F12:DMEM 50:50 medium (GIBCO) with 10% FBS, 2 mM L-glutamine and antibiotics, in 5% CO2 at 37° C. To obtain synchronized HeLa cells in mitosis, cells were treated with 300 nM nocodazole (Sigma-Aldrich) for 18 hrs. To synchronize HeLa cells in G1/S, cells were treated with 2 mM thymidine (Sigma-Aldrich) for 18 hrs. For in vivo small molecule inhibition, cells were incubated with 100 μM Monastrol (Sigma-Aldrich) (Kinesin-5 inhibition), 200 nM compound 1 (GlaxoSmithKline) (Plk1 inhibition), 10 μM R0-3306 (EMD Chemicals) (Cdk1 inhibition), 5 μM taxol, or 300 nM nocodazole 6 hrs post release, fixed, and stained 4 hrs post small molecule addition. For gene knockdowns, cells were transfected with indicated Dharmacon ON-TARGETplus siRNA at 50 nM using Lipofectamine RNAiMAX (Invitrogen) for 48 hrs.
Microtubule aster co-purifying proteins
Mitotic aster microtubule co-pelleting assays were performed essentially as described in (Mack and Compton, 2001) Sixteen 15cm plates of nocodazole (300 nM for 18 hours) arrested mitotic HeLa cells were harvested by mitotic shake-off, washed in PBS plus 20 μg/ml cytochalasin B (Sigma-Aldrich) twice, washed with KHMD (78 mM KCL, 50 mM Hepes pH 7.0, 4 mM MgCl2, 2 mM EGTA, 1 mM DTT, 20 μg/ml cytochalasin B) once and resuspended in KHMDL (KHMD plus protease inhibitors leupeptin-pepstatin-chymostatin 1 μg/ml). Cells were dounce homogenized and extracts were cleared by ultracentrifugation at 38,000 RPM for 15 minutes. All steps were carried out at 4° C unless otherwise noted. Cleared lysates were supplemented with 5 μg/ml latrunculin B (Sigma-Aldrich). Microtubule polymerization reactions were carried out in the presence of control vehicle DMSO or 10 μM taxol (Sigma-Aldrich) at 33° C for 30 minutes. Polymerization reactions were layered onto a 50% W/V sucrose/KHM cushion supplemented with 10 μM taxol for reactions with taxol-stabilized microtubules. Layered reactions were centrifuged for 2 hours at 39,000 RPM in a TLS-55 (Beckman) swinging bucket ultracentrifuge rotor. The microtubule co-pelleting fraction was washed three times with KHM buffer and processed for LC-MS/MS analysis (See Supplemental Experimental Procedures).
RNAi Screen Conditions
Dharmacon ON-TARGETplus siRNA smart pools and individual oligos targeting the MMCP gene set were arrayed on 96 well plates. For transfection, Oligofectamine (Invitrogen) in OPTI-MEM (Invitrogen) was added to each well of a 96 well plate (poly-D-Lysine coated, Beckton Dickinson Labware) containing the prespotted siRNA library. The mixture was incubated for 40 minutes and HeLa cells, 10,000 cells/well, were plated onto the siRNA mixture using a multidrop combi (Thermo Scientific) to ensure even distribution. Cells were incubated at 37° C for indicated times. For + taxol screens, taxol (100 nM final concentration) was added to each well 24 hours post transfection using a RapidPlate liquid handler (Caliper Life Sciences). For mitotic arrest screens, 48 hours post transfection cells were fixed by paraformaldehyde (4% final concentration) perfusion for 15 minutes at room temperature. Following fixation, cells were permeabilized with .2% Triton X-100/PBS for 5 minutes at room temperature. Plates were washed with PBS one time and stained for phospho-histone H3 and DNA (1:50 Alexa 488 phospho-histone H3 (Ser10) (Cell Signaling) and 1 μg/ml Hoechst 33342 in 5% fish gelatin (Invitrogen) for 1 hour. After staining plates were washed two times with .1% Triton X-100/TBS using a BioTek microplate washer ELx405 (BioTek Instruments). Screen plates were imaged on a Cellomics ArrayScanVTI HCS Reader (Cellomics). The ArrayScan/vHCS:Scan software was used for image acquisition using the following parameters: objective: 10× .5 NA, bio application algorithm: Spot Detector, number of channels: two Hoechst and FITC, focus channel: Hoechst, scan limits: 1000 objects (nuclei), with background correction and object segmentation on. Data was analyzed using ArrayScan/vHCS:View software and exported to Excel spreadsheets. For apoptotic screens, 72 hours post transfection cells were fixed and subjected to the Caspase-Glo assay (Promega) or the CellTiter-Glo assay (Promega) and the apoptotic index (AI, caspase activity per cell/ATP levels as described in (Peterson et al., 2010) was measured using a luminometer. For statistical analysis, see Supplemental Experimental Procedures.
Motor Domain Analysis
For STARD9 motor domain expression (modified from (Moyer et al., 1996), microtubule-binding assays (as described in (Goode and Feinstein, 1994), centrosome binding assays (as described in (Mitchison and Kirschner, 1986), ATPase assays (as described in(Funk et al., 2004), localization studies, and rescue experiments, the first 387 amino acids of STARD9 (WT, T110N, and R223A) were fused to the C-terminus of either GST (pGEX-6P vector) or eGFP (pGLAP1 vector, (Torres et al., 2009). For localization, overexpression, and rescue experiments, eGFP, or eGFP-tagged STARD9-MD-WT, T110N, or R223A were transfected in to HeLa cells for the indicated times. Cells were fixed and stained for DNA, α-tubulin, eGFP, and pericentrin. For centrosome binding experiments, 100 μg of GST-STARD9-MD or GST-Skp1, were pre-bound to glutathione agarose beads, and incubated with 500 μg of mitotic centrosome preps, after 1 hr beads were pelleted, washed three times with 110mM NaCl, 5mM ATP, 80mM PIPES pH 6.9, 5mM MgCl2, 1mM EGTA, and samples from the first supernatant and the pellet fraction were resolved by SDS-PAGE and analyzed by immunoblot with indicated antibodies.
Immunofluorescence
Immunofluorescence was carried out essentially as described in (Torres et al., 2010). HeLa cells were transfected with control or indicated siRNA (ON-TARGETplus siRNA, Dharmacon) for 48 h, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100/PBS, and co-stained with 0.5 μg/ml Hoechst 33342 and indicated antibodies. Images were captured with a Zeiss Axio Imager.Z1 microscope and processed using SlideBook 4.2 (Intelligent Imaging).
Supplementary Material
Acknowledgments
We thank; B. Brinkley, D. Compton, Q. Gao, W. Jiang, J. Salisbury, and H. Yu for antibodies; A. Ribas, F. Mortazavi, G. Miranda-Carboni, C. Miceli, R. Gatti, and B. Gomperts for cell lines; M. White for sharing data prior to publication; members of the Torres and Jackson labs for helpful discussions. This work was supported by Genentech Inc, NIH grants RO1 GM054811 and RO1 GM063023 (P.K.J.), Stanford Cancer Biology Postdoctoral Fellowship (J.Z.T.), The Leukemia and Lymphoma Society Postdoctoral Fellowship (J.Z.T.), and The V Foundation for Cancer Research V Scholar Award (J.Z.T.).
Footnotes
Supplemental Information includes 7 Supplemental Figures, 5 Supplemental Tables, 2 Supplemental Movies, Supplemental Experimental Procedures, and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abal M, Keryer G, Bornens M. Centrioles resist forces applied on centrosomes during G2/M transition. Biol Cell. 2005;97:425–434. doi: 10.1042/BC20040112. [DOI] [PubMed] [Google Scholar]
- Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- Barrera JA, Kao LR, Hammer RE, Seemann J, Fuchs JL, Megraw TL. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell. 2010;18:913–926. doi: 10.1016/j.devcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Chaussepied P, Nigg EA. Rigor-type mutation in the kinesin-related protein HsEg5 changes its subcellular localization and induces microtubule bundling. Cell Motil Cytoskeleton. 1998;40:174–182. doi: 10.1002/(SICI)1097-0169(1998)40:2<174::AID-CM6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol. 2010;191:1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury I, Tharakan B, Bhat GK. Caspases - an update. Comp Biochem Physiol B Biochem Mol Biol. 2008;151:10–27. doi: 10.1016/j.cbpb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- Funk CJ, Davis AS, Hopkins JA, Middleton KM. Development of high-throughput screens for discovery of kinesin adenosine triphosphatase modulators. Anal Biochem. 2004;329:68–76. doi: 10.1016/j.ab.2004.02.032. [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Goode BL, Feinstein SC. Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J Cell Biol. 1994;124:769–782. doi: 10.1083/jcb.124.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halama N, Grauling-Halama SA, Jager D. Identification and characterization of the human StARD9 gene in the LGMD2A-region on chromosome 15q15 by in silico methods. Int J Mol Med. 2006;18:653–656. [PubMed] [Google Scholar]
- Hart RK, Mukhyala K. Unison: an integrated platform for computational biology discovery. Pac Symp Biocomput. 2009:403–414. [PubMed] [Google Scholar]
- Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. The PROSITE database. Nucleic Acids Res. 2006;34:D227–230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, Poser I, Lawo S, Grabner H, Kozak K, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- Lansing TJ, McConnell RT, Duckett DR, Spehar GM, Knick VB, Hassler DF, Noro N, Furuta M, Emmitte KA, Gilmer TM, et al. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol Cancer Ther. 2007;6:450–459. doi: 10.1158/1535-7163.MCT-06-0543. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin R, Riggs B, Heald R. SnapShot: motor proteins in spindle assembly. Cell. 2008;134:548–548. e541. doi: 10.1016/j.cell.2008.07.038. [DOI] [PubMed] [Google Scholar]
- Mack GJ, Compton DA. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc Natl Acad Sci U S A. 2001;98:14434–14439. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Compton DA. SnapShot: Nonmotor proteins in spindle assembly. Cell. 2008a;134:694. doi: 10.1016/j.cell.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Compton DA. Structural and regulatory roles of nonmotor spindle proteins. Curr Opin Cell Biol. 2008b;20:101–106. doi: 10.1016/j.ceb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Isolation of mammalian centrosomes. Methods Enzymol. 1986;134:261–268. doi: 10.1016/0076-6879(86)34094-1. [DOI] [PubMed] [Google Scholar]
- Moyer ML, Gilbert SP, Johnson KA. Purification and characterization of two monomeric kinesin constructs. Biochemistry. 1996;35:6321–6329. doi: 10.1021/bi960017n. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Oshimori N, Ohsugi M, Yamamoto T. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol. 2006;8:1095–1101. doi: 10.1038/ncb1474. [DOI] [PubMed] [Google Scholar]
- Peterson D, Lee J, Lei XC, Forrest WF, Davis DP, Jackson PK, Belmont LD. A chemosensitization screen identifies TP53RK, a kinase that restrains apoptosis after mitotic stress. Cancer Res. 2010;70:6325–6335. doi: 10.1158/0008-5472.CAN-10-0015. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Ferreira S, Di Tommaso A, Dimitrov A, Cazaubon S, Gruel N, Colasson H, Nicolas A, Chaverot N, Molinie V, Reyal F, et al. 8p22 MTUS1 gene product ATIP3 is a novel anti-mitotic protein underexpressed in invasive breast carcinoma of poor prognosis. PLoS One. 2009;4:e7239. doi: 10.1371/journal.pone.0007239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Shi J, Orth JD, Mitchison T. Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res. 2008;68:3269–3276. doi: 10.1158/0008-5472.CAN-07-6699. [DOI] [PubMed] [Google Scholar]
- Song MS, Song SJ, Ayad NG, Chang JS, Lee JH, Hong HK, Lee H, Choi N, Kim J, Kim H, et al. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat Cell Biol. 2004;6:129–137. doi: 10.1038/ncb1091. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Torres JZ, Ban KH, Jackson PK. A Specific Form of Phospho Protein Phosphatase 2 Regulates Anaphase-promoting Complex/Cyclosome Association with Spindle Poles. Mol Biol Cell. 2010;21:897–904. doi: 10.1091/mbc.E09-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JZ, Miller JJ, Jackson PK. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics. 2009;9:2888–2891. doi: 10.1002/pmic.200800873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci U S A. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Zhang X, Park CG, Park HW, Endow SA. A structural pathway for activation of the kinesin motor ATPase. Embo J. 2001;20:2611–2618. doi: 10.1093/emboj/20.11.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, Gao Q. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol. 2005;171:437–445. doi: 10.1083/jcb.200506185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.