Abstract
Enzymatic hydrolysis of recalcitrant polysaccharides like cellulose takes place on the solid-liquid interface. Therefore the adsorption of enzymes to the solid surface is a pre-requisite for catalysis. Here we used enzymatic activity measurements with fluorescent model-substrate 4-methyl-umbelliferyl-β-D-lactoside for sensitive monitoring of the binding of cellobiohydrolase TrCel7A from Trichoderma reesei to bacterial cellulose (BC). The binding at low nanomolar free TrCel7A concentrations was exclusively active site mediated and was consistent with Langmuir's one binding site model with K d and A max values of 2.9 nM and 126 nmol/g BC, respectively. This is the strongest binding observed with non-complexed cellulases and apparently represents the productive binding of TrCel7A to cellulose chain ends on the hydrophobic face of BC microfibril. With increasing free TrCel7A concentrations the isotherm gradually deviated from the Langmuir's one binding site model. This was caused by the increasing contribution of lower affinity binding modes that included both active site mediated binding and non-productive binding with active site free from cellulose chain. The binding of TrCel7A to BC was found to be only partially reversible. Furthermore, the isotherm was dependent on the concentration of BC with more efficient binding observed at lower BC concentrations. The phenomenon can be ascribed to the BC concentration dependent aggregation of BC microfibrils with concomitant reduction of specific surface area.
Introduction
Cellulose is the most abundant polysaccharide on Earth and is an appealing raw material for many biotechnological applications. As a structural polysaccharide, cellulose has evolved to heterogeneous structure that makes it recalcitrant towards chemical as well as enzymatic degradation [1]. Individual cellulose chains adhere with each other by hydrogen bonding and van der Waals interactions forming crystalline microfibrils. Enzymatic depolymerization of cellulose chains is carried out by cellulases and it takes place in the solid-liquid interface. Thus the adsorption of cellulases to cellulose surface is a prerequisite for catalysis [2], [3]. To facilitate interactions with substrate many cellulases have modular structure consisting of a catalytic domain (CD) that is connected through a linker peptide with a smaller carbohydrate binding module (CBM) [4]. The best studied cellulase is a processive cellobiohydrolase TrCel7A from Trichoderma reesei (Hypocrea jecorina). The tunnel shaped active site of TrCel7A resides in CD and accommodates 10 consecutive glucose units along cellulose chain. It has been shown that separated CBM and CD of TrCel7A can both bind to cellulose [5]. Furthermore, the linker peptide also contributes to binding [6], [7]. The binding of cellulases is also influenced by the properties of cellulose, which is heterogeneous at different levels: content of different crystalline allomorphs, different crystal faces, amorphous regions, degree of polymerization, content and size of pores, specific surface area etc [3]. In the case of lignocellulosic substrates the binding is further complicated by the presence of lignin and hemicelluloses [8]. Therefore pure model celluloses like bacterial cellulose (BC) and bacterial microcrystalline cellulose (BMCC) are often used in binding studies of cellulases. Modular architecture of the enzymes in combination with heterogeneous substrate is expected to result in a complex binding isotherm. Despite this expected complexity the Langmuiŕs one binding site model has been often found to be sufficient to describe cellulase binding [3], [9]. Among alternative models the Langmuiŕs two independent binding site model [10], [11]. Freundlich model [12], [13], and combined Langmuiŕs-Freundlich model or Hilĺs cooperative binding model [14] have also been used. Although direct measurement of the bound enzyme has been reported [15] the most often applied experimental approach is the depletion method, where the concentration of free enzyme is measured and the amount of cellulose bound enzyme is found as a difference between the concentration of total and free enzyme. The simplest method for the measurement of the concentration of free enzyme is based on the absorbance or the fluorescence measurement of the enzyme protein. However, measurements of cellulase binding are complicated by the possible non-specific adsorption of enzymes to reaction vessels [16]. This may lead to the overestimation of the binding strength, especially at low cellulase concentrations. Therefore blocking agents like bovine serum albumin (BSA) are often used to circumvent the problem. However, in the presence of BSA, the methods relying on direct quantification of protein are not applicable for the measurement of cellulose free cellulase. To measure the binding in the presence of BSA radioactivity [17], [18] or fluorescence labeled cellulases [19] or chimeras of CBMs with fluorescent proteins [14], [20] have been used. Recently we have developed a sensitive method relying on the activity measurement of TrCel7A with low Mw model substrates to monitor the binding of TrCel7A [21], [22]. Since activity measurements reveal the concentration of enzyme with free active site the method can be used to distinguish between different populations of bound enzyme: total bound enzyme, bound through the active site, and bound but with free active site [21], [23]. Here we used fluorescent model substrate, 4-methyl-umbelliferyl-β-D-lactoside (MUL), to study the binding of TrCel7A to BC. Sensitive detection enabled to measure sub-nanomolar concentrations of TrCel7A with free active sites and to reveal the strongest binding observed with non-complexed cellulases.
Results and Discussion
Measuring the binding of TrCel7A to BC
Binding of cellulases to the cellulose surface is a prerequisite step before catalysis and has been in the focus of numerous studies. Despite intensive research there is a gap in our knowledge in many issues like contribution of different binding modes in binding of modular enzymes. This has lead to different and often controversial hypotheses about the role of binding in controlling the overall rate of cellulose degradation [21]–[28]. Beside the nature of enzyme and substrate, the method used for binding measurements along with the range of enzyme and substrate concentrations included in the measurements seems to be important [29]. Binding measurements imply the separation of cellulose bound and free cellulase. Here, the suitability of two methods for this purpose, centrifugation and filtration, were tested. Filtration was found to be more accurate as there was an initial rapid release of TrCel7A from cellulose pellet after centrifugation resulting in the overestimation of the concentration of TrCel7A free from cellulose ([TrCel7A]Free) (Figure S1 in File S1). In this study we used the enzymatic activity of TrCel7A with MUL substrate to measure [TrCel7A]Free. Hydrolysis of MUL by TrCel7A results in the formation of lactose and 4-methylumbelliferone (MU). The latter can easily be detected by the fluorescence at high pH. Advantages of this approach are high sensitivity and applicability in the presence of BSA used to prevent non-specific binding. Control experiments judged that in the presence of BSA (0.1 g/L) there was no non-specific binding of TrCel7A to the reaction vessels and filters. BSA was also found to be necessary to avoid the binding of MU to BC. A certain disadvantage of using activity measurements with MUL is the necessity to remove cellobiose released from cellulose hydrolysis to avoid the cellobiose inhibition of MUL hydrolysis by TrCel7A [21], [22]. Therefore, before the analysis of MUL activity, the filtrates containing TrCel7AFree were treated with β-glucosidase, which hydrolyzes cellobiose into two molecules of glucose. Glucose inhibition of TrCel7A is weak [30] and can be neglected under our experiment conditions. Beside sensitive detection of low enzyme concentrations a virtue of using activity with MUL is the possibility to discriminate between the populations of total bound TrCel7A (TrCel7ABound) and TrCel7A bound through the active site (TrCel7ABound-OA). The latter can be measured by following the inhibition of MUL hydrolysis by cellulose [22], [31]. The subscript OA refers to occupied active site i.e. the active site is occupied by cellulose chain and not available for MUL hydrolysis. Accordingly the subscript FA refers to free active site for MUL hydrolysis. The population of bound TrCel7A with free active sites (TrCel7ABound-FA) is found from the difference between total bound TrCel7A and TrCel7ABound-OA, [TrCel7A]Bound-FA = [TrCel7A]Bound - [TrCel7A]Bound-OA. Beside MUL, other low-Mw model substrates like para-nitrophenyl-β-D-lactoside (pNPL) [21]–[23] have been used to measure the active site mediated binding of TrCel7A.
The measurement of binding kinetics to BC revealed no changes in [TrCel7A]Free within studied incubation times (0.5 – 5 h) (Figure S2 in File S1) and equilibration times between 10 min and 30 min were used in further binding experiments. Although literature reports support fast binding of cellulases to BC and BMCC with equilibrium times in the range of few minutes [10], [25] we used somewhat longer incubation times to reveal possible slow irreversible binding. Constant levels of bound TrCel7A observed over long incubation period (Figure S2 in File S1) indicates that the enzymatic activity of TrCel7A does not interfere with binding. This is in accord with the processive mode of hydrolysis whereby the cellulose crystal is degraded at its surface, “layer by layer”, resulting in thinner crystals with no significant changes in total surface area [11], [32], [33].
Binding of TrCel7A to BC involves multiple binding modes with different affinities
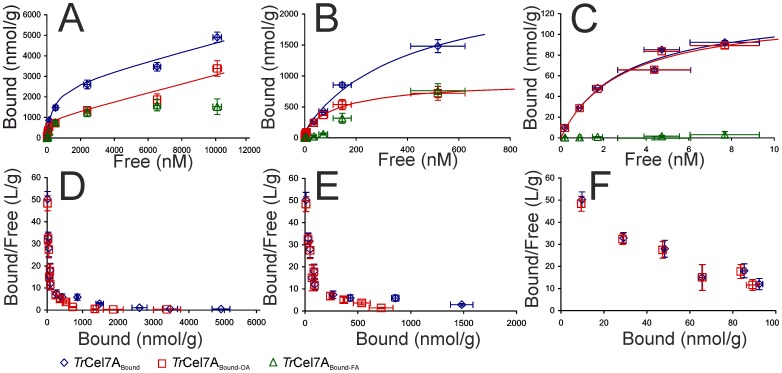
Binding measurements made by varying the concentration of TrCel7A over 4 orders of magnitude (BC was at 1 g/L) revealed complex binding (Figure 1). None of the equations tested provided a very good fit if the full dataset (n = 82) was used in the analysis. Equations tested included Langmuiŕs one (R = 0.9764), two (R = 0.9932), and three independent binding site models (R = 0.9932), Freundlich model (R = 0.9896), Hilĺs cooperative binding model (R = 0.9896), and sum of Langmuiŕs and Hilĺs model (R = 0.9942). Although some models provided a reasonably good fit with R values above 0.99, there was a strong systematic deviation between the experiment and the model in the low-nanomolar range of free TrCel7A concentrations (Figure S3 in File S1). Restricting the dataset with the highest [TrCel7A]Free up to 10 nM resulted in a good accord with Langmuiŕs one binding site model (Figure 1C and F) with K d = 2.9 nM and A max = 126 nmol/g. Next we analyzed the binding in the range of [TrCel7A]Free up to 1.0 µM using the Langmuiŕs two binding sites model (Figure 1B and E). In analysis, the parameters values for the first, high affinity binding mode were fixed (K d = 2.9 nM and A max = 126 nmol/g) and the parameters values for the second binding mode were found by fitting. The K d for the second, medium affinity binding mode was more than two orders of magnitude higher than that for the high affinity binding mode (Table 1). Finally, the full dataset was analyzed using the Langmuiŕs three binding sites model (Figure 1A and D). Here, the parameters values for the first two binding modes were fixed. Because the third, low affinity binding mode is far from saturation, only the A max/K d value can be found for this binding mode (Table 1). It must be noted that although the differences in R values between two and three binding site model does not justify the use of more complex model, the visual inspection of the fits suggests that the presence of at least three binding sites must be assumed to describe the data (Figure S3 in File S1).
Figure 1. Variation of free enzyme concentration over 4 orders of magnitude reveals multi-mode binding of TrCel7A to BC.
[Free] – [Bound] plots (A – C) and Scatchard plots (D – F) of binding of TrCel7A to BC (1 g/L). Full binding isotherm is dissected into three regions with different dominating binding modes. The low affinity binding mode dominates at free TrCel7A concentrations ([TrCel7A]Free) above 1.0 µM (A and D). The medium affinity binding mode dominates in the 0.1 µM – 1.0 µM range of [TrCel7A]Free (B and E). The high affinity binding mode dominates at [TrCel7A]Free up to 10 nM (C and F). Total bound TrCel7A (TrCel7ABound), TrCel7A bound through the active site (TrCel7ABound-OA), and bound TrCel7A with free active site (TrCel7ABound-FA). Solid lines represent best fits of Langmuiŕs one (C), two (B), and three (A) independent binding site model. Error bars are at least from three independent measurements.
Table 1. Binding isotherm parameters for different binding modes of active site mediated and total binding of cellobiohydrolase TrCel7A to bacterial cellulose.
| High affinity binding modeb | Medium affinity binding modec | Low affinity binding moded | ||||
| Parametera | Total bound | Active site bound | Total bound | Active site bound | Total bound | Active site bound |
| A max (nmol/g) | 126±17 | 122±14 | 2440±260 | 790±50 | - | - |
| K d (nM) | 2.9±1.0 | 2.8±0.8 | 406±82 | 156±25 | - | - |
| A max/K d (L/g) | 43.1±14.7 | 43.3±12.7 | 6.0±1.2 | 5.1±0.8 | 0.21±0.01 | 0.22±0.01 |
Parameter values were found by non-linear regression analysis of data in Figure 1. Error limits are the parameter errors from the non-linear regression route and are not primarily statistical in origin.
Dataset was restricted with [TrCel7A]Free up to 10 nM (Figure 1C) and Langmuiŕs one binding site model was used in analysis.
Dataset was restricted with [TrCel7A]Free up to 1.0 µM (Figure 1B) and Langmuiŕs two binding site model was used in analysis. Parameter values for the first, high affinity binding mode were fixed and the parameter values for the second, medium affinity binding mode were found by non-linear regression analysis.
Full dataset (n = 82) was analyzed according Langmuiŕs three binding site model (Figure 1A). Parameter values for the first two binding modes were fixed and the parameter values for the third, low affinity binding mode were found by non-linear regression analysis. Because of the low degree of saturation of this binding mode only the value of A max/K d can be found.
The active site mediated binding of TrCel7A to BC was measured in parallel with the total bound TrCel7A to reveal the contribution of non-productive binding modes, where the enzyme is attached to BC without cellulose chain in the active site. In the range of low-nanomolar [TrCel7A]Free the enzyme was bound exclusively through the active site (Figure 1C and F). However, in the range of [TrCel7A]Free between 0.1 – 1.0 µM the contribution of bound enzyme with free active site (TrCel7ABound-FA) in total bound enzyme was significant (Figure 1B and E). Somewhat surprisingly we found that the low affinity binding that dominates in the high micromolar range of [TrCel7A]Free was active site mediated (Figure 1A and D). Similarly to the total binding none of the tested equations were sufficient to describe the active site mediated binding if the full dataset was used in the analysis. The parameter values for the active site mediated binding were found analogously to those for the total bound enzyme and are listed in Table 1. The presence of different populations of bound enzyme further supports the necessity to include at least three binding modes in the analysis of experiment data. We have two populations of active site bound enzyme with approximately 200 fold different A max/K d values (Table 1) and we also have the population of bound enzyme with free active site that is qualitatively different (Figure 1A and B). Earlier attempts to measure active site mediated binding have shown that the majority of TrCel7A is bound through the active site and the population of TrCel7ABound-FA constitutes usually less than 10% of the total bound enzyme [21], [23]. In contrast, bound enzyme with the free active site has been proposed to be the dominating binding mode of Cel9A from Thermobifida fusca [34]. The data from this study demonstrate that the contribution of active site mediated binding depends primarily on the enzyme to substrate ratio.
Figure 2 shows the proposed mechanistic interpretation of binding modes with different affinity. It has been demonstrated by experiment [35] and also by molecular dynamics simulation [36] that CBM of TrCel7A preferentially binds to the hydrophobic face of cellulose crystal. TrCel7A degrades cellulose crystal also from its hydrophobic face [33]. The estimated surface area of the hydrophobic face is approximately 100 µmoles of cellobiose units per gram BC [10]. Considering the average degree of polymerization of our BC preparation of 825±10 cellobiose units, one can estimate that there is 0.12 µmoles of reducing ends/g BC on the hydrophobic face. This is in good accord with the A max values of the high affinity binding mode for both the total bound and the active site bound TrCel7A (Table 1). Since in this region of the isotherm all of the bound enzyme was bound through the active site (Figure 1C and F) we propose that the high affinity binding mode represents the productive binding to the chain ends on the hydrophobic face, i.e. TrCel7A is attached through both domains, CBM and CD, and the cellulose chain is engaged into the active site tunnel (Figure 2A).
Figure 2. Proposed productive and non-productive interactions between TrCel7A and BC.
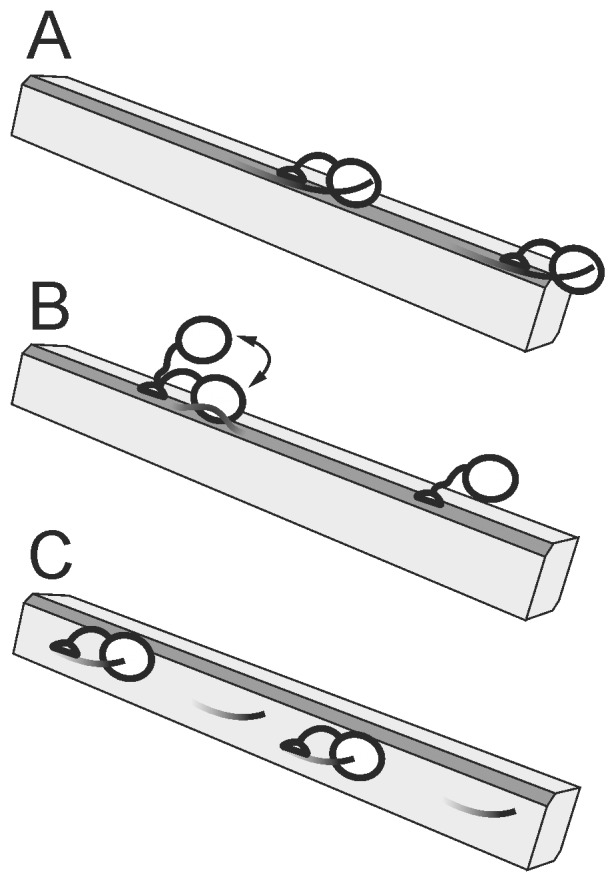
(A) The high affinity binding mode corresponds to the productive binding to cellulose chain ends on the hydrophobic face of BC microcrofibril through both domains, CBM and CD. (B) The medium affinity binding mode includes non-productive binding to the hydrophobic face, where enzyme is attached through CBM only. Latter can become productive binding upon disengagement of cellulose chain into the active site by endo-mode attack. (C) The low affinity binding mode may correspond to the active site mediated binding to cellulose chain ends on the hydrophilic face. The hydrophobic face of cellulose microfibril is shown in dark gray and the cellulose chain ends available for binding through CD are depicted as protruding lines. The A max and K d values for corresponding binding modes are listed in Table 1.
The medium affinity binding dominates in the range of free TrCel7A concentrations between 0.1 µM and 1.0 µM. In this range both populations, the active site bound TrCel7A and the bound TrCel7A with the free active site, were present (Figure 1B and E). CBM of TrCel7A covers 10 cellobiose units [37], whereas CD covers approximately 48 cellobiose units [38]. Thus, for CBM, the binding capacity of the hydrophobic face is expected to be 10 µmoles CBM per gram BC (100 µmoles of cellobiose units per gram BC/10 cellobiose units per CBM). The corresponding figure for CD is approximately 2 µmoles per gram BC. Thus, the binding capacity 2.4 µmol/g found for the total bound enzyme in the medium affinity binding mode (Table 1) is in the same order with the estimated binding capacity of TrCel7A on the hydrophobic face. We propose that in the range of medium affinity binding there is a balance between two populations of bound enzyme. The population of bound TrCel7A with the free active site apparently represents the non-productive binding to the hydrophobic face, where the enzyme is attached through CBM only (Figure 2B). Because TrCel7A can employ an endo-mode initiation on BC [39] the initially non-productive binding through CBM can become productive binding after disengagement of cellulose chain from the crystal lattice to the active site (Figure 2B). The medium affinity active site mediated binding (Table 1) may thus represent the endo-mode complexation of TrCel7A with cellulose chains on the hydrophobic face.
The third, low affinity binding mode dominates in the free TrCel7A concentrations above 1.0 µM (Figure 1A and D). Since its binding capacity is high, it may correspond to the binding of TrCel7A to the hydrophilic face of cellulose crystal (Figure 2C). Because of the large specific area of the hydrophilic face and low affinity we were not able to saturate the low affinity binding mode in our experiments. A recent molecular dynamics study demonstrated that TrCel7A CBM can bind also to the hydrophilic face, although it will diffuse to the hydrophobic face upon vacancy [36]. However, as evidenced by the very similar A max/K d values for the total and the active site mediated binding (Table 1), the low affinity binding mode is active site mediated. One can speculate that the low affinity binding represents the active site mediated binding to the chain ends on the hydrophilic face. Because cellulose chains in the hydrophilic face are more tightly associated with the crystal lattice their disengagement to the enzymes active site is energetically less favorable resulting in a high K d value.
It must be noted that the above mechanistic interpretation was only a simplified view to the binding of TrCel7A to BC. Earlier studies have shown that the analysis of cellulase binding is complicated by the presence of the overlapping binding sites [38] and cooperative effects [14]. This additional complexity may reveal in the insufficiency of Langmuiŕs three independent binding sites model in describing the full dataset. Although the analysis of our binding data in the region of high and medium affinity binding mode using Hill’s plot suggested no cooperative effects within these binding modes (Figure S4 in File S1) more detailed studies are needed to reveal the possible interdependency between different binding modes. The K d value of 2.9 nM measured here for the high affinity binding mode (Figure 1C and F, Figure 2A) is among the strongest bindings observed with non-complexed cellulases. Using radioactivity labeled proteins the A max/K d values of 18 L/g have been reported for the binding of TrCel7A and TrCel6A to BMCC at 22 °C [18]. Using fluorescence measurements of low free enzyme concentrations, Herner et al. [40] reported the K d value of 33±6 nM (at 20°C) for binding of TrCel7A to microcrystalline cellulose. The binding of CBM of exoglucanase Cex from Cellulomonas fimi to BMCC with K d value of 16±3.6 nM (at 30°C) has been measured using isothermal titration calorimetry [41]. Using the measurements of fluorescence recovery after photobleaching Moran-Mirabal et al. [42] have reported K d values in the range of 10 – 50 nM for binding of Thermobifida fusca cellulases to BMCC. However, most of the reported A max and K d values for the binding of different cellulases to BC or BMCC are in the same order with corresponding values found here for the medium affinity binding mode (Figure 1B and E, Figure 2B, Table 1) [7], [10], [43]. The K d values reported for the binding of TrCel7A to other crystalline celluloses like Avicel, algal celluloses, and cellulose III are also in the sub- or low-micromolar range [5], [11], [13], [14], [26], [44]–[49]. The sensitivity of the methods like protein absorbance apparently sets the limits for the lowest observable free enzyme concentration and the possible low capacity strong binding modes may remain undetectable. The results presented here support the conclusions of a recent single molecule tracking study that the performance of TrCel7A measured at low nanomolar enzyme concentrations is strikingly different from that measured at micromolar concentrations [29]. The conclusion from above is that measurements at very low enzyme concentrations have to be included in studies aiming to measure the rate constants for different steps of the hydrolysis of the insoluble substrate like association, processive movement and dissociation.
Reversibility of the binding of TrCel7A to BC
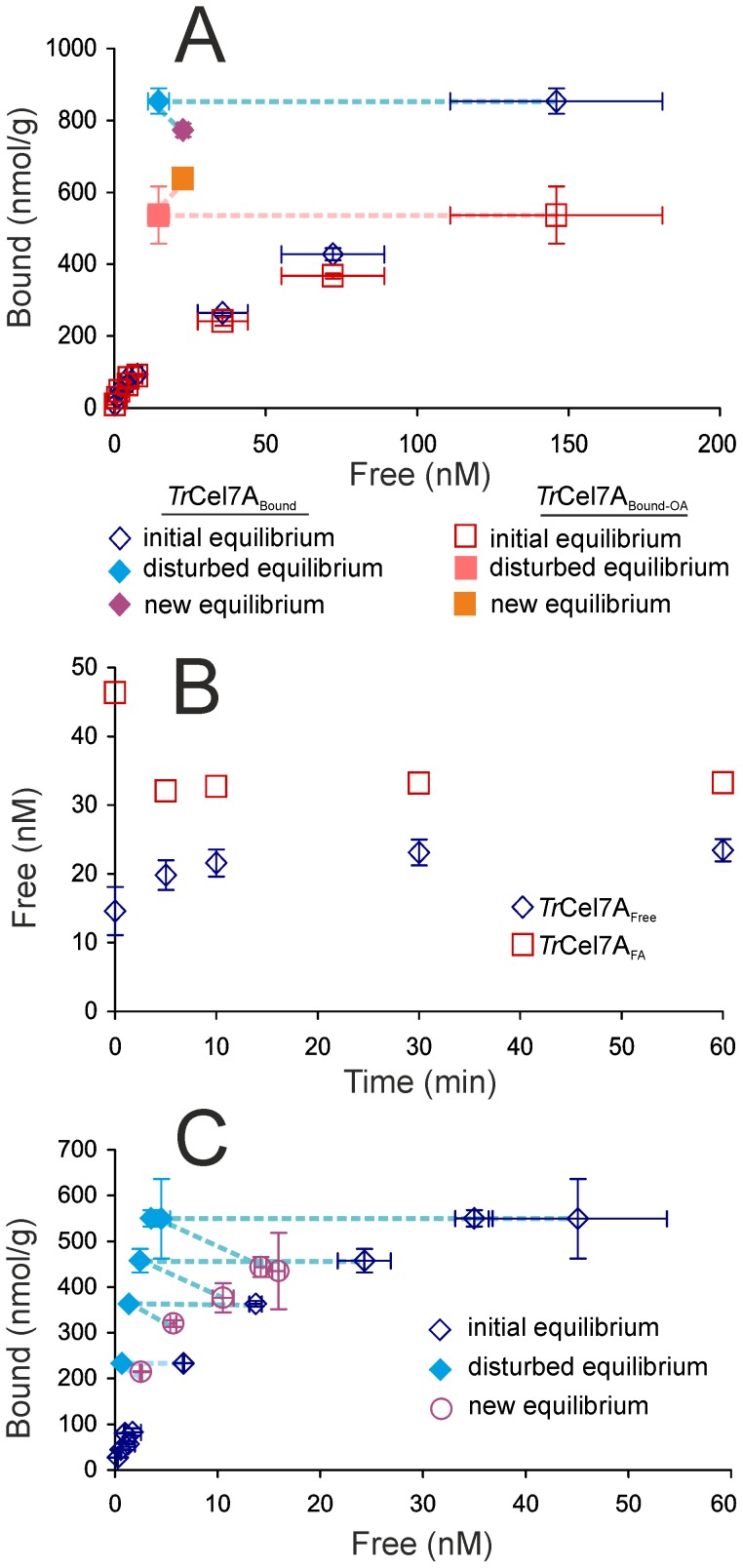
Binding reversibility of cellulases has been a focus of numerous studies but the results have been often controversial. In some studies full reversibility is observed [17], [18], [43] whereas others report significant hysteresis [18], [42], [43], [50]–[54]. Here we studied the reversibility of the binding of TrCel7A using two different approaches, dilution and supernatant replacement experiment. In the dilution experiment BC at 1 g/L was equilibrated with 1.0 µM TrCel7A for 30 min, after which the equilibrium was disturbed by the addition of buffer to bring a tenfold increase in the total volume. Using parallel experiments the binding reversibility was assessed in the basis of both TrCel7ABound and TrCel7ABound-OA (Figure 3A).
Figure 3. Binding of TrCel7A to BC is only partially reversible.
(A and B) Dilution experiment. BC at 1.0 g/L was incubated with 1.0 µM TrCel7A to establish equilibrium. Equilibrium was disturbed by the addition of buffer to bring up tenfold dilution and relaxation to new equilibrium was followed. (A) Binding reversibility was assessed in the basis of both, total bound TrCel7A, TrCel7ABound and active site bound TrCel7A, TrCel7ABound-OA. Dotted lines show the progression from the initial equilibrium to the disturbed equilibrium to the new equilibrium. (B) Change in the concentration of TrCel7A free from cellulose, TrCel7AFree and TrCel7A with free active site TrCel7AFA in time after disturbance of equilibrium by dilution. (C) Free enzyme depletion experiment. BC at 0.1 g/L was incubated with TrCel7A at different concentrations to establish equilibrium. Cellulose with bound enzyme was pelleted by centrifugation, 90% of the supernatant was withdrawn and pellet was resuspended in the same amount of buffer to disturb the equilibrium. The position of the new equilibrium was measured after 30 min from the disturbance. Binding reversibility was assessed in the basis of total bound TrCel7A. Dotted lines show the progression from the initial equilibrium to the disturbed equilibrium to the new equilibrium.
The change in the concentration of TrCel7AFree and TrCel7AFA in time after the disturbance of equilibrium by dilution is shown in Figure 3B. In the case of TrCel7AFree there was a rapid increase in its concentration during first 5 min after the dilution with no further significant changes indicating the relaxation to the new equilibrium. In contrast to TrCel7AFree, there was a decrease in the concentration of TrCel7AFA after dilution resulting in a new equilibrium with lower [TrCel7AFA] (Figure 3B). This indicates an increase in the contribution of the active site mediated binding to the total bound enzyme in the new equilibrium, which is in qualitative agreement with the increased contribution of TrCel7ABound-OA in the region of low [TrCel7A]Free (Figure 1). In the case of reversible binding the new equilibrium position established after dilution is expected to fall into the initial isotherm. However, the position of the new equilibrium remained far from the initial isotherm, indicating strong hysteresis. In the case of TrCel7ABound-OA the amount of bound enzyme even increased upon dilution (Figure 3A). We also assessed the binding reversibility (at the level of total bound TrCel7A) by disturbing the equilibrium without changing the concentration of BC. For that, BC at 0.1 g/L was equilibrated with TrCel7A at different concentrations for 30 min. After equilibration cellulose was pelleted by centrifugation and 90% of the TrCel7AFree containing supernatant was replaced with fresh buffer to disturb the equilibrium. The position of new equilibrium was measured 30 min after the disturbance. The new isotherm did not overlap with the initial isotherm but the hysteresis was less prominent than in the case of the dilution experiment (Figure 3C). The difference between two approaches was that in the case of dilution experiment cellulose was diluted in parallel with TrCel7AFree, whereas in the supernatant exchange experiment cellulose concentration was not changed. This prompted us to measure the binding isotherms at different BC concentrations.
Binding isotherm of TrCel7A depends on the concentration of BC
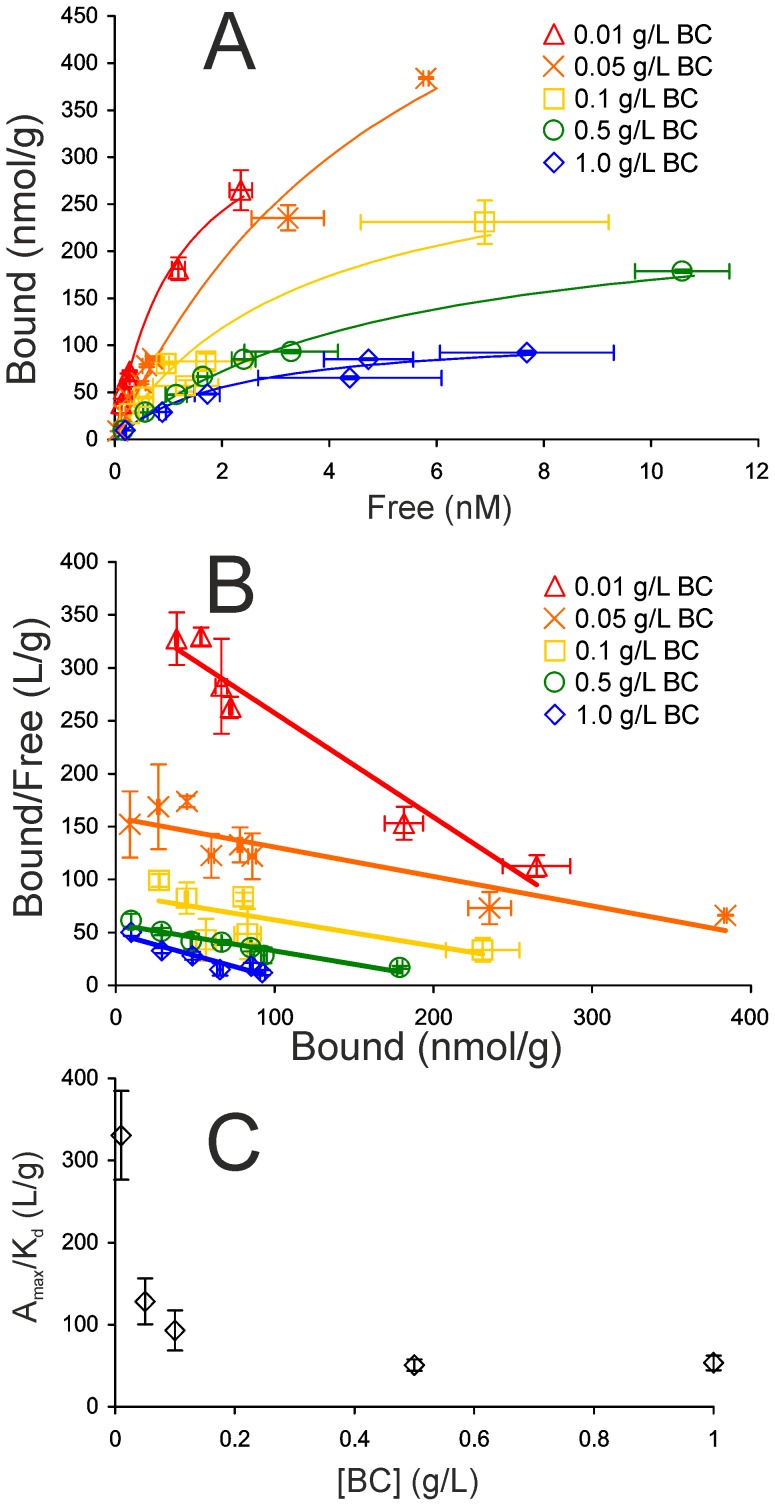
Binding isotherms of TrCel7A (on the total bound enzyme basis) were measured at 5 different BC concentrations between 0.01 – 1.0 g/L. Here we focused only on the high binding affinity region of the isotherm ([TrCel7A]Free up to 10 nM), where the binding was reasonably well consistent with the Langmuiŕs one binding site model (Figure 4A and B). As seen in Figure 4 the isotherms were dependent on the concentration of BC with more efficient binding observed at lower BC concentrations. Plotting the values of A max/K d as a function of BC concentration revealed inverse relationship (Figure 4C). The dependence of A max and K d on BC concentration was less evident than that of A max/K d (Figure S5 in File S1). It must be noted however, that because of the complex binding the determination of the A max and K d values is error prone. Although the Langmuiŕs one binding site model was sufficient to describe the binding at low nanomolar [TrCel7A]Free, further increase in [TrCel7A]Free leads to an increased contribution of the medium affinity binding mode and systematic deviation from the one binding site model (Figure 1). For this reason A max and K d values depend on the highest concentration of TrCel7AFree included into the analysis, which is always somewhat arbitrary. However, A max/K d is given by the initial slope of the isotherm and is less sensitive to the highest concentration of TrCel7AFree included into the analysis.
Figure 4. Binding isotherm of TrCel7A depends on BC concentration.
Binding isotherms (A), and corresponding Scatchard plots (B) of binding of TrCel7A (in the level of total bound TrCel7A) to BC at different concentrations. Solid lines represent best fits of Langmuiŕs one binding site model. Error bars are at least from three independent measurements. (C) A max/K d values at different BC concentrations.
The binding isotherm of cellulases is expected to be independent of the concentration of cellulose. This expectation relies on the assumption, that the total amount of binding sites per gram cellulose as well as binding affinity are constants independent of cellulose concentration. Although the binding isotherms of cellulases have been in the focus of numerous studies, the isotherms are almost exclusively measured at one cellulose concentration. However, Wang et al. studied the binding of crude cellulase to Avicel and found also, that the binding was stronger at lower Avicel concentration [55]. A possible mechanistic interpretation would be that there is a cellulose concentration dependent association of cellulose microfibrils with concomitant decrease in specific surface area available for binding. In recent study Cruys-Bagger et al. proposed that cellulose concentration dependent reduction in the surface area was responsible for lower association rate constants between cellulose and TrCel7A observed at higher cellulose concentrations [28]. Consistent with the binding data reported here a recent scanning electron microscopy study by Kuijk et al. demonstrated the BC concentration dependent formation of large flocks and aggregates in BC suspensions [56]. Association of BC microfibrils should lead to the exclusion of some of the surface area for the binding of cellulases. According to this scenario it is the A max that should decrease with increasing cellulose concentration, whereas K d is expected to remain unaffected. Although the mechanism of the reduced binding efficiency with increasing cellulose concentration remains to be studied, the underlying property of cellulose suspension is expected to depend on cellulose concentration according to that of the A max/K d in Figure 4C. In regard to lignocellulose hydrolysis the data presented in Figure 4 support the suggestion that complete removal of hemicelluloses and lignin during biomass pretreatment may cause the association of cellulose microfibrils with concomitant reduction in surface area available for the binding of cellulases and may thus become a disadvantage [57]. Cellulose concentration dependent association of microfibrils may also contribute in the so called “solids effect” – a decrease in the degree of cellulose conversion with increasing cellulose concentration at constant cellulase to cellulose ratio [58]–[60].
Materials and Methods
Materials
MUL, pNPL and BSA were from Sigma-Aldrich and were used as purchased. TrCel7A was purified from the culture filtrate of Trichoderma reesei QM 9414 as described before [61]. Aspergillus β-glucosidase was purified from Novozyme 188 (Sigma C6105) according to [62]. Enzyme concentration was determined from absorbance at 280 nm using extinction coefficients of 84,400 M−1 cm−1 for TrCel7A and 180,000 M−1 cm−1 for β-glucosidase. BC was prepared by laboratory fermentation of Gluconobacter xylinum strain ATCC 53582 as described before [63]. Degree of polymerization of BC was 1650±20 glucose units as judged by the amount of reducing groups on BC measured using modified bicinchoninic acid method [63], [64].
General conditions
All binding experiments were made in 50 mM sodium acetate buffer pH 5.0 (supplemented with 0.1 g/L BSA) at 25°C. If not stated otherwise the experiments were performed in 1.5 mL polypropylene microcentrifuge tubes without stirring.
Measurement of the concentration of cellulose free TrCel7A
After separation of cellulose bound TrCel7A the concentration of cellulose free TrCel7A was measured by its MUL hydrolyzing activity. Before the activity measurement with MUL 0.4 mL samples were supplemented with β-glucosidase (final concentration 4 nM) and incubated overnight to remove any cellobiose released from the hydrolysis of BC. For MUL-ase activity measurements 0.41 mL of suitably diluted β-glucosidase treated sample was added to 5 µL of 0.5 mM MUL and incubated at 35°C for 0.5 h – 6 h depending on the concentration of TrCel7A. Dilution factors and incubation times were adjusted so that the rate of MUL hydrolysis corresponds to the initial rate. Reactions were quenched by the addition of ammonium hydroxide to the final concentration of 0.1 M and the released MU was quantified by the fluorescence. Excitation and emission wavelengths were set to 360 nm and 450 nm, respectively. Calibration curves were made with known TrCel7A concentrations. TrCel7A used as a reference was treated identically with samples, but the BC was omitted. In the presence of BSA no binding of TrCel7A to reaction vessels and filters was observed.
Measurement of the total bound TrCel7A
BC (0.01 – 1 g/L) was equilibrated with TrCel7A (10 nM – 15 µM) in 0.5 mL total volume for 30 min. The sample was transferred to 2 mL polypropylene syringe and pressed through a glass microfibre filter (Whatman GF-D) to separate cellulose free and bound TrCel7A [21]. The filtrate was centrifuged (2 min 10,000×g) to remove any solids that had passed through the filter and β-glucosidase (final concentration 4 nM) was added to 0.4 mL of the supernatant. After overnight incubation with β-glucosidase [TrCel7A]Free was measured by MUL hydrolyzing activity. [TrCel7A]Bound was found as a difference between the total concentration of TrCel7A and [TrCel7A]Free and was expressed in nanomoles per gram BC.
Measurement of the active site mediated binding of TrCel7A
Here the initial rates of MUL hydrolyzing activity of TrCel7A were measured in the presence of BC. BC (1 g/L) was incubated with TrCel7A (10 nM – 2.0 µM) and β-glucosidase (0.85 µM) in 0.5 mL total volume at 25°C. At selected time 5 µL of 0.5 mM MUL was added, incubated further (at 25°C) for defined time and quenched by the addition of ammonium hydroxide to the final concentration of 0.1 M. BC was separated by centrifugation (2 min 10,000×g) and the released MU was quantified by the fluorescence of the supernatant. Times of the MUL addition and incubation with MUL were selected depending on the concentration of TrCel7A but the total duration of the experiment was always 30 min. [TrCel7A]FA was found from the rate of MUL hydrolysis using standard curves made without BC. [TrCel7ABound-OA] was found as a difference between the total concentration of TrCel7A and [TrCel7A]FA. In the experiments with higher TrCel7A concentrations (1.0 µM – 15 µM) another model substrate, pNPL (0.5 mM final concentration), was used instead of MUL. Here the released para-nitrophenole was measured by the absorbance at 420 nm [21]. At 1.0 µM and 2.0 µM total TrCel7A concentrations [TrCel7A]Bound-OA was measured in parallel experiments using MUL and pNPL model substrates. No differences between using MUL or pNPL were observed within the measurement error limits. In constructing the binding isotherm for TrCel7ABound-OA the [TrCel7A]Free measured in the parallel experiments made under the same total TrCel7A and BC concentrations (see measurement of the total bound TrCel7A) were used instead of [TrCel7A]FA.
Binding reversibility - dilution experiment
Binding reversibility of both TrCel7ABound and TrCel7ABound-OA after dilution was assessed.
For the reversibility on the level of TrCel7ABound, 1 g/L BC was incubated with 1.0 µM TrCel7A and 2.5 µM β-glucosidase in 0.5 mL total volume for 30 min after what 4.5 mL buffer was added. At defined times after dilution 0.5 mL samples were withdrawn, BC was separated by filtration and [TrCel7A]Bound was measured as described in “Measurement of total bound TrCel7A”.
For the reversibility on the level of TrCel7ABound-OA, 1 g/L BC was incubated with 1.0 µM TrCel7A in the presence of 2.5 µM β-glucosidase in 0.5 mL total volume. After 30 min, 4.5 mL MUL in buffer was added (final concentration 10 µM). At defined times after dilution 0.5 mL samples were withdrawn and mixed with 0.5 mL of 0.2 M ammonium hydroxide to quench the MUL hydrolysis. BC was separated by centrifugation and [MU] was quantified by the fluorescence. [TrCel7A]FA was calculated from the rate of MUL hydrolysis in the presence of BC using standard curves made without BC [31]. [TrCel7ABound-OA] was found as a difference between total concentration of TrCel7A and [TrCel7A]FA.
Binding reversibility - free enzyme depletion experiment
BC (0.1 g/L) was incubated with TrCel7A (30 – 100 nM) for 10 min in 0.5 mL total volume. BC was pelleted by centrifugation (2 min, 10,000×g), 0.45 mL of the supernatant was withdrawn and BC pellet was resuspended with 0.45 mL fresh buffer. 10 min after resuspension the cellulose bound and free TrCel7A were separated by filtration and [TrCel7A]Bound was measured as described in “The measurement of total bound TrCel7A”.
Measuring the release of TrCel7A from cellulose pellet
BC (0.1 g/L) was incubated with 30 nM TrCel7A in 0.5 mL total volume for 10 minutes. BC was pelleted by centrifugation (2 min 10,000 × g) and after defined time of standing with cellulose pellet 0.4 mL of supernatant was withdrawn and added to 5 µL of β-glucosidase (final concentration 4 nM). After incubation overnight with β-glucosidase the concentration of free TrCel7A in supernatant was measured by MUL hydrolyzing activity. For the zero time point the cellulose bound and free TrCel7A were separated by filtration through glass microfibre filter (Whatman GF-D) [21]. The filtrate was centrifuged (2 min 10,000×g) and after incubation with β-glucosidase [TrCel7A]Free was measured by MUL-ase activity.
Data treatment
The binding data were analyzed using non-linear regression analysis according to the following equations.
Langmuiŕs independent binding site(s) model
A max is the binding capacity of BC (nmol/g BC) and K d is the equilibrium dissociation constant of BC-TrCel7A complex (nM). In the case of Langmuiŕs two and three independent binding site models the hyperbolas containing the parameters for second (A max(2), K d(2)) and third (A max(3), K d(3)) binding mode were added to the right hand side of the equation.
Freundlich model
where a and 1/m are Freundlich equilibrium constant and power term, respectively.
Hilĺs model
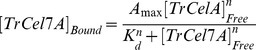 |
where n is cooperativity parameter. In all equations [TrCel7A]Bound and [TrCel7A]Free represent the concentration of cellulose bound (in nmol/g BC) and free (in nM) TrCel7A.
Supporting Information
This file contains following figures: Figure S1. Release of TrCel7A from cellulose pellet after centrifugation. Figure S2. Binding kinetics at different concentrations of TrCel7A and BC. Figure S3. Binding of TrCel7A to BC and best fits of Langmuiŕs two and three binding sites model. Figure S4. Hill's plots for the binding of TrCel7A to BC in the high and medium affinity binding mode. Figure S5. A max and K d values for the binding of TrCel7A in the high affinity binding mode at different BC concentrations.
(PDF)
Acknowledgments
We are grateful to Dr Silja Kuusk from University of Tartu for critical reading of the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Estonian Science Foundation (grant no. 9227), www.etag.ee. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, et al. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315: 804–807. [DOI] [PubMed] [Google Scholar]
- 2. Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66: 506–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang Y-HP, Lynd LR (2004) Toward an aggregated understanding of enzymatic hydrolysis of cellulose: noncomplexed cellulase systems. Biotechnol Bioeng 88: 797–824. [DOI] [PubMed] [Google Scholar]
- 4. Boraston AB, Bolam DN, Gilbert HJ, Davies GJ (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ståhlberg J, Johansson G & Pettersson G (1991) A new model for enzymatic hydrolysis of cellulose based on the two-domain structure of cellobiohydrolase I. Nat Biotechnol. 9: 286–290. [Google Scholar]
- 6. Payne CM, Resch MG, Chen L, Crowley MF, Himmel ME, et al. (2013) Glycosylated linkers in multimodular lignocellulose-degrading enzymes dynamically bind to cellulose. Proc Natl Acad Sci USA 110: 14646–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Srisodsuk M, Reinikainen T, Penttilä M, Teeri TT (1993) Role of the interdomain linker peptide of Trichoderma reesei cellobiohydrolase I in its interaction with crystalline cellulose. J Biol Chem 268: 20756–20761. [PubMed] [Google Scholar]
- 8. Ding S-Y, Liu Y-S, Zeng Y, Himmel ME, Baker JO, et al. (2012) How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science 338: 1055–1060. [DOI] [PubMed] [Google Scholar]
- 9. Bansal P, Hall M, Realff MJ, Lee JH, Bommarius AS (2009) Modeling cellulase kinetics on lignocellulosic substrates. Biotechnol Adv 27: 833–848. [DOI] [PubMed] [Google Scholar]
- 10. Gilkes NR, Jervis E, Henrissat B, Tekant B, Miller RC Jr, et al. (1992) The adsorption of a bacterial cellulase and its two isolated domains to crystalline cellulose. J Biol Chem 267: 6743–6749. [PubMed] [Google Scholar]
- 11. Igarashi K, Uchihashi T, Koivula A, Wada M, Kimura S, et al. (2011) Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333: 1279–1282. [DOI] [PubMed] [Google Scholar]
- 12. Jiang F, Kittle JD, Tan X, Esker AR, Roman M (2013) Effects of sulfate groups on the adsorption and activity of cellulases on cellulose substrates. Langmuir 29: 3280–3291. [DOI] [PubMed] [Google Scholar]
- 13. Medve J, Ståhlberg J, Tjerneld F (1997) Isotherms for adsorption of cellobiohydrolase I and II from Trichoderma reesei on microcrystalline cellulose. Appl Biochem Biotechnol 66: 39–56. [DOI] [PubMed] [Google Scholar]
- 14. Sugimoto N, Igarashi K, Wada M, Samejima M (2012) Adsorption characteristics of fungal family 1 cellulose-binding domain from Trichoderma reesei cellobiohydrolase I on crystalline cellulose: negative cooperative adsorption via a steric exclusion effect. Langmuir 28: 14323–14329. [DOI] [PubMed] [Google Scholar]
- 15. Zhu Z, Sathitsuksanoh N, Percival Zhang Y-H (2009) Direct quantitative determination of adsorbed cellulase on lignocellulosic biomass with its application to study cellulase desorption for potential recycling. Analyst 134: 2267–2272. [DOI] [PubMed] [Google Scholar]
- 16. Bommarius AS, Katona A, Cheben SE, Patel AS, Ragauskas AJ, et al. (2008) Cellulase kinetics as a function of cellulose pretreatment. Metab Eng 10: 370–381. [DOI] [PubMed] [Google Scholar]
- 17. Linder M, Teeri TT (1996) The cellulose-binding domain of the major cellobiohydrolase of Trichoderma reesei exhibits true reversibility and a high exchange rate on crystalline cellulose. Proc Natl Acad Sci USA 93: 12251–12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palonen H, Tenkanen M, Linder M (1999) Dynamic interaction of Trichoderma reesei cellobiohydrolases Cel6A and Cel7A and cellulose at equilibrium and during hydrolysis. Appl Environ Microbiol 65: 5229–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bubner P, Plank H, Nidetzky B (2013) Visualizing cellulase activity. Biotechnol Bioeng 110: 1529–1549. [DOI] [PubMed] [Google Scholar]
- 20. Hong J, Ye X, Zhang Y-HP (2007) Quantitative determination of cellulose accessibility to cellulase based on adsorption of a nonhydrolytic fusion protein containing CBM and GFP with its applications. Langmuir 23: 12535–12540. [DOI] [PubMed] [Google Scholar]
- 21. Jalak J, Väljamäe P (2010) Mechanism of initial rapid rate retardation in cellobiohydrolase catalyzed cellulose hydrolysis. Biotechnol Bioeng 106: 871–883. [DOI] [PubMed] [Google Scholar]
- 22. Jalak J, Kurašin M, Teugjas H, Väljamäe P (2012) Endo-exo synergism in cellulose hydrolysis revisited. J Biol Chem 287: 28802–28815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cruys-Bagger N, Tatsumi H, Borch K, Westh P (2014) A graphene screen-printed carbon electrode for real-time measurements of unoccupied active sites in a cellulase. Anal Biochem 447: 162–168. [DOI] [PubMed] [Google Scholar]
- 24. Fox JM, Levine SE, Clark DS, Blanch HW (2012) Initial- and processive-cut products reveal cellobiohydrolase rate limitations and the role of companion enzymes. Biochemistry 51: 442–452. [DOI] [PubMed] [Google Scholar]
- 25. Cruys-Bagger N, Elmerdahl J, Praestgaard E, Tatsumi H, Spodsberg N, et al. (2012) Pre-steady-state kinetics for hydrolysis of insoluble cellulose by cellobiohydrolase Cel7A. J Biol Chem 287: 18451–18458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao D, Chundawat SPS, Sethi A, Balan V, Gnanakaran S, et al. (2013) Increased enzyme binding to substrate is not necessary for more efficient cellulose hydrolysis. Proc Natl Acad Sci USA 110: 10922–10927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shang BZ, Chang R, Chu J-W (2013) Systems-level modeling with molecular resolution elucidates the rate-limiting mechanisms of cellulose decomposition by cellobiohydrolases. J Biol Chem 288: 29081–29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cruys-Bagger N, Tatsumi H, Ren GR, Borch K, Westh P (2013) Transient kinetics and rate-limiting steps for the processive cellobiohydrolase Cel7A: effects of substrate structure and carbohydrate binding domain. Biochemistry 52: 8938–8948. [DOI] [PubMed] [Google Scholar]
- 29. Shibafuji Y, Nakamura A, Uchihashi T, Sugimoto N, Fukuda S, et al. (2014) Single molecule imaging analysis of elementary reaction steps of Trichoderma reesei cellobiohydrolase I (Cel7A) hydrolyzing crystalline cellulose Iα and IIII . J Biol Chem 289: 14056–14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teugjas H, Väljamäe P (2013) Product inhibition of cellulases studied with 14C-labeled cellulose substrates. Biotechnol Biofuels 6: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horn SJ, Sørlie M, Vårum KM, Väljamäe P, Eijsink VGH (2012) Measuring Processivity. Methods Enzymol 510: 69–95. [DOI] [PubMed] [Google Scholar]
- 32. Boisset C, Fraschini C, Schulein M, Henrissat B, Chanzy H (2000) Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl Environ Microbiol 66: 1444–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y-S, Baker JO, Zeng Y, Himmel ME, Haas T, et al. (2011) Cellobiohydrolase hydrolyzes crystalline cellulose on hydrophobic faces. J Biol Chem 286: 11195–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kostylev M, Moran-Mirabal JM, Walker LP, Wilson DB (2012) Determination of the molecular states of the processive endocellulase Thermobifida fusca Cel9A during crystalline cellulose depolymerization. Biotechnol Bioeng 109: 295–299. [DOI] [PubMed] [Google Scholar]
- 35. Lehtio J, Sugiyama J, Gustavsson M, Fransson L, Linder M, et al. (2003) The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc Natl Acad. Sci USA 100: 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nimlos MR, Beckham GT, Matthews JF, Bu L, Himmel ME, et al. (2012) Binding preferences, surface attachment, diffusivity, and orientation of a family 1 carbohydrate-binding module on cellulose. J Biol Chem 287: 20603–20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo J, Catchmark JM (2013) Binding specificity and thermodynamics of cellulose-binding modules from Trichoderma reesei Cel7A and Cel6A. Biomacromolecules 14: 1268–1277. [DOI] [PubMed] [Google Scholar]
- 38. Sild V, Ståhlberg J, Pettersson G, Johansson G (1996) Effect of potential binding site overlap to binding of cellulose to cellulose: a two-dimensional simulation. FEBS Lett 378: 51–56. [DOI] [PubMed] [Google Scholar]
- 39. Kurašin M, Väljamäe P (2011) Processivity of cellobiohydrolases is limited by the substrate. J Biol Chem 286: 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herner ML, Melnick MS, Rabinovich ML (1999) Enhancement of the affinity of cellobiohydrolase I and its catalytic domain to cellulose in the presence of the reaction product-cellobiose. Biochemistry (Moscow) 64: 1012–1020. [PubMed] [Google Scholar]
- 41. Creagh AL, Ong E, Jervis E, Kilburn DG, Haynes CA (1996) Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc Natl Acad Sci USA 93: 12229–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran-Mirabal JM, Bolewski JC, Walker LP (2011) Reversibility and binding kinetics of Thermobifida fusca cellulases studied through fluorescence recovery after photobleaching microscopy. Biophys Chem 155, 20–28. [DOI] [PubMed]
- 43. Bothwell M, Wilson D, Irwin D, Walker L (1997) Binding reversibility and surface exchange of Thermomonospora fusca E3 and E5 and Trichoderma reesei CBHI. Enzyme Microb Technol 20: 411–417. [Google Scholar]
- 44. Igarashi K, Wada M, Hori R, Samejima M (2006) Surface density of cellobiohydrolase on crystalline celluloses: A critical parameter to evaluate enzymatic kinetics at a solid-liquid interface. FEBS J 273: 2869–2878. [DOI] [PubMed] [Google Scholar]
- 45. Igarashi K, Wada M, Samejima M (2007) Activation of crystalline cellulose to cellulose IIII results in efficient hydrolysis by cellobiohydrolase: hydrolysis of cellulose IIII by cellobiohydrolase. FEBS J 274: 1785–1792. [DOI] [PubMed] [Google Scholar]
- 46. Kamat RK, Ma W, Yang Y, Zhang Y, Wang C, et al. (2013) Adsorption and hydrolytic activity of the polycatalytic cellulase nanocomplex on cellulose. ACS Appl Mater Interfaces 5: 8486–8494. [DOI] [PubMed] [Google Scholar]
- 47. Medve J, Karlsson J, Lee D, Tjerneld F (1998) Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol Bioeng 59: 621–634. [PubMed] [Google Scholar]
- 48. Nidetzky B, Steiner W, Claeyssens M (1994) Cellulose hydrolysis by the cellulases from Trichoderma reesei: adsorptions of two cellobiohydrolases, two endocellulases and their core proteins on filter paper and their relation to hydrolysis. Biochem J 303, 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wahlström R, Rahikainen J, Kruus K, Suurnäkki A (2014) Cellulose hydrolysis and binding with Trichoderma reesei Cel5A and Cel7A and their core domains in ionic liquid solutions. Biotechnol Bioeng 111: 726–733. [DOI] [PubMed] [Google Scholar]
- 50. Kyriacou A, Neufeld RJ, MacKenzie CR (1989) Reversibility and competition in the adsorption of Trichoderma reesei cellulase components. Biotechnol Bioeng 33: 631–637. [DOI] [PubMed] [Google Scholar]
- 51. Ma A, Hu Q, Qu Y, Bai Z, Liu W, et al. (2008) The enzymatic hydrolysis rate of cellulose decreases with irreversible adsorption of cellobiohydrolase I. Enzyme Microb Technol. 42: 543–547. [Google Scholar]
- 52. Maurer SA, Bedbrook CN, Radke CJ (2012) Competitive sorption kinetics of inhibited endo- and exoglucanases on a model cellulose substrate. Langmuir 28: 14598–14608. [DOI] [PubMed] [Google Scholar]
- 53. Maurer SA, Brady NW, Fajardo NP, Radke CJ (2013) Surface kinetics for cooperative fungal cellulase digestion of cellulose from quartz crystal microgravimetry. J Colloid Interface Sci 394: 498–508. [DOI] [PubMed] [Google Scholar]
- 54. McLean BW, Boraston AB, Brouwer D, Sanaie N, Fyfe CA, et al. (2002) Carbohydrate-binding modules recognize fine substructures of cellulose. J Biol Chem 277: 50245–50254. [DOI] [PubMed] [Google Scholar]
- 55. Wang W, Kang L, Wei H, Arora R, Lee YY (2011) Study on the decreased sugar yield in enzymatic hydrolysis of cellulosic substrate at high solid loading. Appl Biochem Biotechnol 164: 1139–1149. [DOI] [PubMed] [Google Scholar]
- 56. Kuijk A, Koppert R, Versluis P, van Dalen G, Remijn C, et al. (2013) Dispersions of attractive semiflexible fiberlike colloidal particles from bacterial cellulose microfibrils. Langmuir 29: 14356–14360. [DOI] [PubMed] [Google Scholar]
- 57. Ishizawa CI, Jeoh T, Adney WS, Himmel ME, Johnson DK, et al. (2009) Can delignification decrease cellulose digestibility in acid pretreated corn stover? Cellulose 16: 677–686. [Google Scholar]
- 58. Kristensen JB, Felby C, Jorgensen H (2009) Yield-determining factors in high-solids enzymatic hydrolysis of lignocelluloses. Biotechnol Biofuels 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roberts KM, Lavenson DM, Tozzi EJ, McCarthy MJ, Jeoh T (2011) The effects of water interactions in cellulose suspensions on mass transfer and saccharification efficiency at high solids loadings Cellulose. 18: 759–773. [Google Scholar]
- 60. Modenbach AA, Nokes SE (2013) Enzymatic hydrolysis of biomass at high-solids loadings – a review. Biomass and Bioenergy 56: 526–544. [Google Scholar]
- 61. Bhikhabhai R, Johansson G, Pettersson G (1984) Isolation of cellulolytic enzymes from Trichoderma reesei QM 9414. J Appl Biochem 6: 336–345. [PubMed] [Google Scholar]
- 62. Sipos B, Benkő Z, Dienes D, Réczey K, Viikari L, et al. (2010) Characterisation of specific activities and hydrolytic properties of cell-wall-degrading enzymes produced by Trichoderma reesei Rut C30 on different carbon sources. Appl Biochem Biotechnol 161: 347–364. [DOI] [PubMed] [Google Scholar]
- 63. Velleste R, Teugjas H, Väljamäe P (2010) Reducing end-specific fluorescence labeled celluloses for cellulase mode of action. Cellulose 17: 125–138. [Google Scholar]
- 64. Zhang Y-HP, Lynd LR (2005) Determination of the number-average degree of polymerization of cellodextrins and cellulose with application to enzymatic hydrolysis. Biomacromolecules 6: 1510–1515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains following figures: Figure S1. Release of TrCel7A from cellulose pellet after centrifugation. Figure S2. Binding kinetics at different concentrations of TrCel7A and BC. Figure S3. Binding of TrCel7A to BC and best fits of Langmuiŕs two and three binding sites model. Figure S4. Hill's plots for the binding of TrCel7A to BC in the high and medium affinity binding mode. Figure S5. A max and K d values for the binding of TrCel7A in the high affinity binding mode at different BC concentrations.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.