Abstract
Objective:
The purpose of this study was to develop an algorithm incorporating MRI metrics to classify patients with mild traumatic brain injury (mTBI) and controls.
Methods:
This was an institutional review board–approved, Health Insurance Portability and Accountability Act–compliant prospective study. We recruited patients with mTBI and healthy controls through the emergency department and general population. We acquired data on a 3.0T Siemens Trio magnet including conventional brain imaging, resting-state fMRI, diffusion-weighted imaging, and magnetic field correlation (MFC), and performed multifeature analysis using the following MRI metrics: mean kurtosis (MK) of thalamus, MFC of thalamus and frontal white matter, thalamocortical resting-state networks, and 5 regional gray matter and white matter volumes including the anterior cingulum and left frontal and temporal poles. Feature selection was performed using minimal-redundancy maximal-relevance. We used classifiers including support vector machine, naive Bayesian, Bayesian network, radial basis network, and multilayer perceptron to test maximal accuracy.
Results:
We studied 24 patients with mTBI and 26 controls. Best single-feature classification uses thalamic MK yielding 74% accuracy. Multifeature analysis yields 80% accuracy using the full feature set, and up to 86% accuracy using minimal-redundancy maximal-relevance feature selection (MK thalamus, right anterior cingulate volume, thalamic thickness, thalamocortical resting-state network, thalamic microscopic MFC, and sex).
Conclusion:
Multifeature analysis using diffusion-weighted imaging, MFC, fMRI, and volumetrics may aid in the classification of patients with mTBI compared with controls based on optimal feature selection and classification methods.
Classification of evidence:
This study provides Class III evidence that classification algorithms using multiple MRI features accurately identifies patients with mTBI as defined by American Congress of Rehabilitation Medicine criteria compared with healthy controls.
Mild traumatic brain injury (mTBI) is a growing public health problem.1 Twenty to thirty percent of patients have persistent symptoms after injury resulting in substantial disability.2 One of the major obstacles in development of appropriate treatment strategies is the lack of an accurate and objective means to establish diagnosis.
Currently, several different definitions of mTBI exist (World Health Organization,3 American Congress of Rehabilitation Medicine [ACRM],4 Centers for Disease Control and Prevention,5 Department of Defense, and Department of Veteran Affairs6). There is universal agreement that a unified, objective definition is needed.7,8 Furthermore, most classification schemes rely on Glasgow Coma Scale score,9 which was recently deemed insufficient as a single classifier for traumatic brain injury by the National Institute for Neurological Disorders and Stroke, which proposed that neuroimaging have a larger role in the classification scheme for mTBI.10 Recent work using MRI revealed that there are areas of subtle brain injury after mTBI11−23; however, no single imaging metric has thus far been shown to be useful as an independent biomarker.
In the machine learning community, it is well known that using multiple features can improve classification performance compared with a single feature alone. The purpose of this work is to develop a computational tool for the classification of mTBI using minimal-redundancy maximal-relevance (mRMR) feature selection24 from a larger set of features that includes imaging metrics we have previously investigated and shown to be different between subject and control groups, and use 10-fold cross-validation to test the performance of classification algorithms in this cohort.
METHODS
Methodology informing classification of evidence.
We seek to answer the following research question: Can we develop a computational tool that accurately classifies subjects with mTBI as defined by ACRM criteria compared with healthy controls? Class III level of evidence is assigned to this question.
Standard protocol approvals, registrations, and patient consents.
This work is part of an institutional review board–approved and Health Insurance Portability and Accountability Act–compliant prospective study, and written informed consent was obtained from all participants.
Participants.
We recruited subjects with mTBI fulfilling ACRM criteria,4 which were used as a gold standard for classification. Mean interval between MRI and trauma was 23 days (3–56 days). Mechanism of injury varied, including 7 motor vehicle accidents, 5 falls, 4 assaults, 3 sports-related, 2 bicycle collisions, and 2 other. Patients were excluded if there was prior head injury, known neurologic or psychiatric disorder, and if there was a contraindication to MRI. Healthy control subjects matched for age, sex, and educational level were also recruited from the general population.
MRI.
The methodologic details for MRI and analysis techniques, including structural magnetization-prepared rapid-acquisition gradient echo,25,26 resting-state fMRI,19,27 magnetic field correlation (MFC),28,29 and diffusion techniques,16,17,19,30 have been previously described. In summary, experiments used 3T Trio MRI scanner (Siemens Medical Solutions, Erlangen, Germany), body coil for transmission, and 12-element SENSE receive head coil. We selected MRI metrics based on their ability to detect subtle differences between subjects with mTBI and controls, some of which have been previously published in an overlapping cohort.16–19,25
Feature selection.
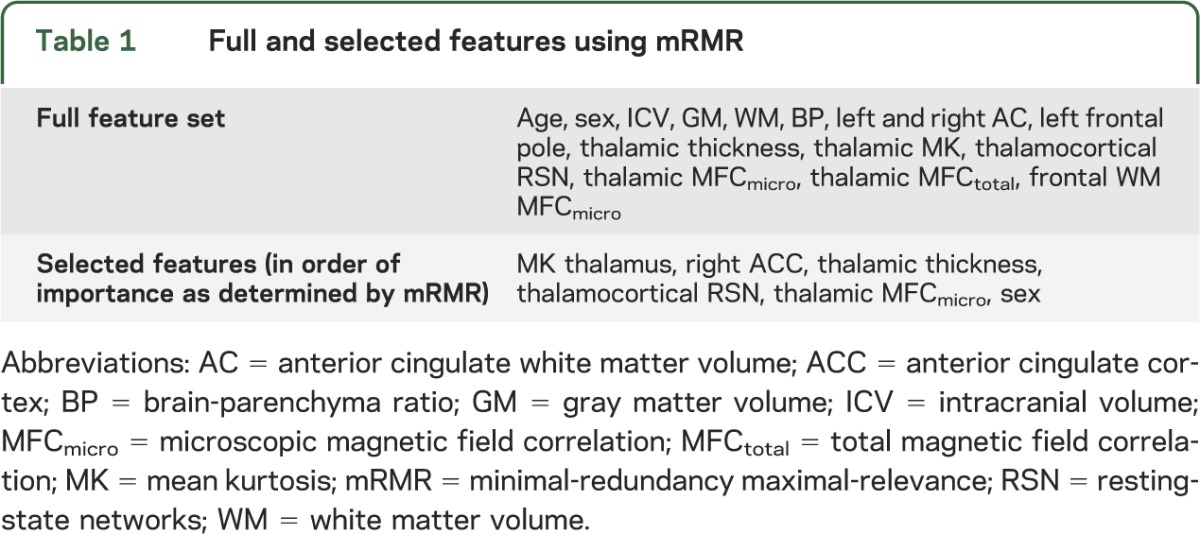
We selected features from 15 imaging metrics (table 1): 2 general demographic features, 3 global brain volumetric features, and 10 regional brain MRI metrics based on previously demonstrated differences between mTBI and control cohorts.16–19,25 All original features are normalized by removing the mean of each feature and dividing by its SD. We used the feature selection procedure, mRMR,24 to incrementally choose the most representative subset of imaging features, to increase relevance, and decrease redundancy. The mRMR algorithm was chosen because it selects a subset of features that are not only significant, but unique from one another. This ensures that the feature space is maximally informative with the smallest dimensionality. The mRMR algorithm chooses features from the full set one at a time. At each iteration, the mRMR evaluates the mutual information of a candidate feature from the pool of remaining features with the desired output and the average mutual information with the chosen features, and selects the candidate feature that yields maximal difference between the 2 mutual information measures. In our implementation, the mRMR iteration stops when this mutual information difference is ≤0. The mRMR algorithm is based on established methodology24 and implemented using a MATLAB Toolbox function (The MathWorks Inc., Natick, MA).
Table 1.
Full and selected features using mRMR
Classification algorithms.
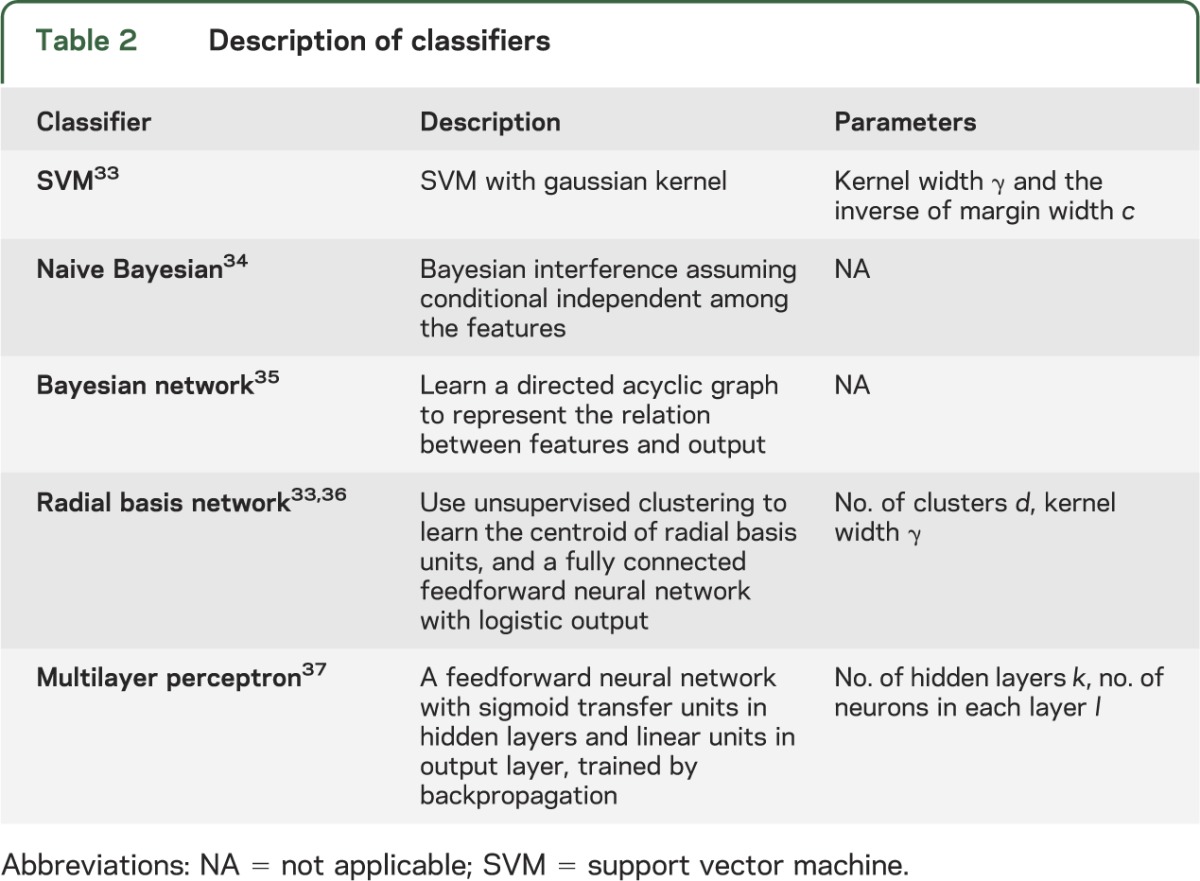
We used 5 types of mainstream classifiers on the features chosen by mRMR: support vector machine (SVM), naive Bayesian, Bayesian network, radial basis network, and multilayer perceptron. A detailed description of each classifier and its parameters is found in table 2. To prevent overfitting, given the limited training set available, and to maximize generalizability, we trained classifiers using 10-fold cross-validation. The entire dataset is randomly divided into 10 equally numbered, nonoverlapping subsets, each called a fold. Nine of 10 folds are then used as the training set and the remaining tenth as the validation set. Each classifier is first trained using a training set, and then tested on the validation set. The above procedure is repeated 10 times using each of the 10 folds as a validation set. Finally, this computational method averages recorded error rates for each trial to arrive at an average cross-validation error rate, used to assess the classification algorithm. For each classifier, we found the optimal parameters by evaluating the average cross-validation error associated with each possible parameter value over a chosen search space, and using the ones leading to the minimal cross-validation error.
Table 2.
Description of classifiers
We also applied the above methodology to evaluate the achievable performance of different classifiers using the single best feature alone and for mRMR selected features.
Note that the cross-validation procedure is to provide a performance measure that is expected for unseen data. To apply the best performing classifier identified from this research in future nontraining data, one should retrain the classifier using all available training data, and use the resulting classifier weights as the final classifier.
Of note, using the classification algorithms described, there are no specific thresholds for individual features. An iterative process arrives at an algorithm that gives each of the features differing weights to optimize accuracy.
RESULTS
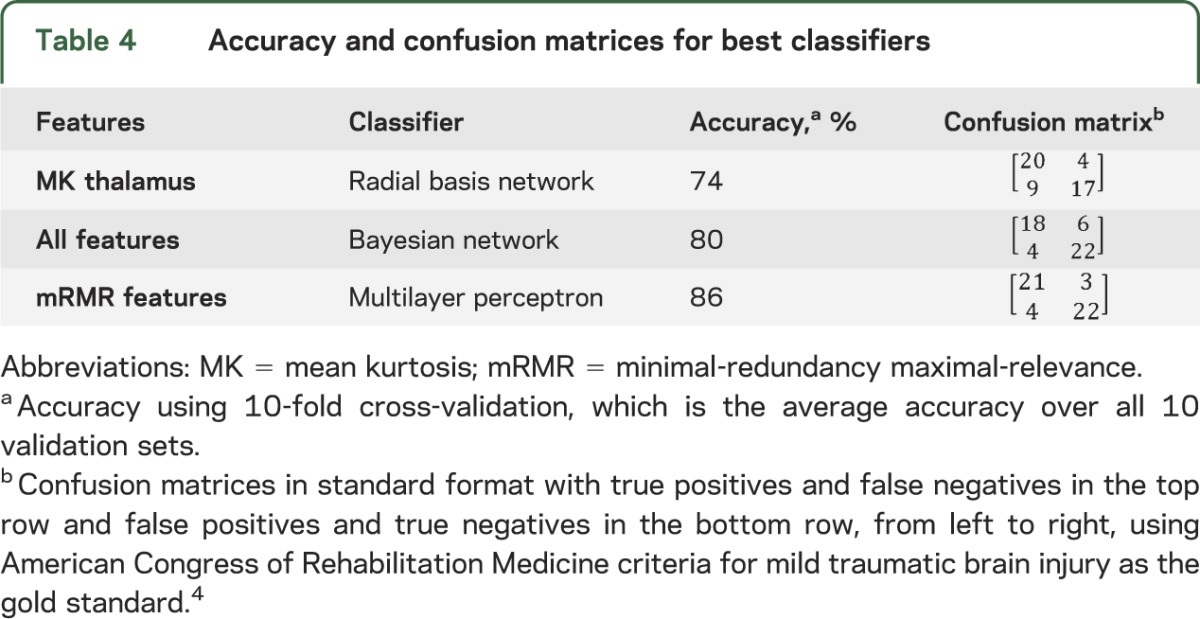
We studied 23 patients (mean age 33.65 ± 11.21 years; 6 female and 17 male) and 25 healthy controls matched for age, sex, and education (mean age 36.68 ± 11.5 years; 13 female and 12 male) recruited prospectively from the emergency department of a level I trauma center. All patients had data on all features. Regarding feature selection, the subset of nonredundant, informative features identified using mRMR are listed in table 1 in order of importance. The first feature chosen by mRMR is thalamic mean kurtosis. This feature is considered the most informative among all features considered, because it has the highest mutual information with the target classification outcome. The results for classification using the different standard classifiers are shown in table 3 based on highest performance and consistency with all parameters for the classifiers indicated. Using the single best (most informative) feature, thalamic mean kurtosis, the highest classification accuracy was achieved using the radial basis network classification algorithm. Classification using all features achieved highest accuracy using the Bayesian net classification algorithm. Using mRMR selected features, the highest accuracy was achieved using the multilayer perceptron classification algorithm (table 4).
Table 3.
Summary of classification algorithm accuracy (averaged over all validation sets in 10-fold cross-validation)
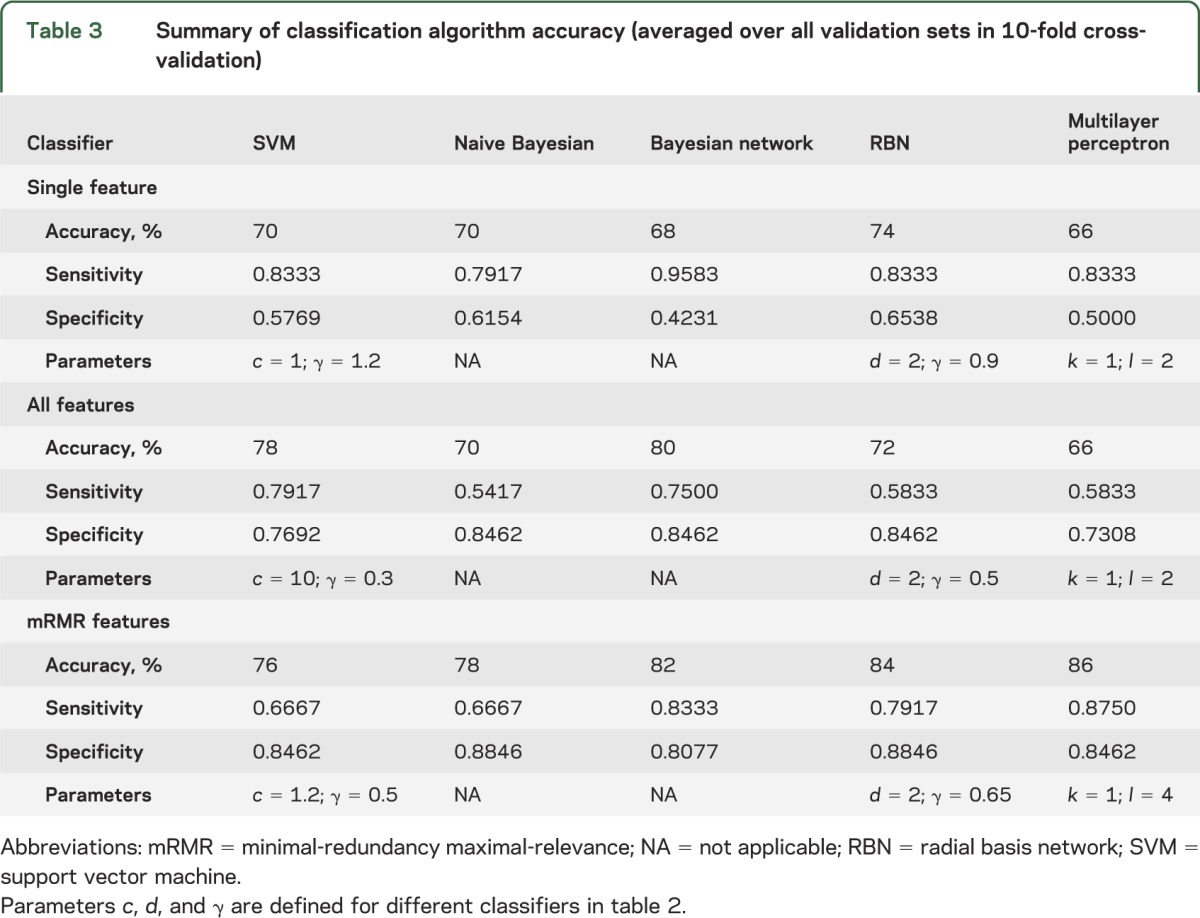
Table 4.
Accuracy and confusion matrices for best classifiers
DISCUSSION
In this study, we developed a classification algorithm using multifeature analysis for identifying subjects with mTBI by incorporating multiple features including demographic, global, and regional imaging MRI metrics. Multifeature classification improved accuracy using a subset of most relevant features obtained via mRMR feature selection by 12% over using the single most significant metric alone and 6% over using all metrics.
Throughout the medical community, there is growing interest in using machine learning techniques to understand and use medical data in an informative way including as applied to mTBI.31 Our results provide a preliminary step toward developing a diagnostic tool for classification of patients with mTBI based on objective metrics that could complement qualitative clinical assessment, the current standard of care.
The features selected by mRMR represent a truly distinct sample of the full feature set and, when reviewed, are certainly unique from one another. Global brain morphologic features, such as intracranial volume, gray matter, and white matter, are eschewed for more targeted regional brain volumes, such as right anterior cingulate cortex, an area implicated in depression. Furthermore, it is not surprising that thalamic microscopic MFC was chosen at the exclusion of total MFC, despite both metrics showing differences between the 2 populations as these 2 metrics are mathematically related to one another: microscopic MFC quantifies magnetic field inhomogeneities at a subvoxel level, reflecting what is believed to be important cellular-level iron content, whereas total MFC also contains contributions from macroscopic field inhomogeneities, which may relate to artifact. Across classifiers, mRMR feature selection improves overall classification performance compared with using one feature alone and also improves classification performance in all classifiers except SVM when compared with using all features. This supports the use of mRMR feature selection.
We tackled potential overfitting for this relatively small dataset in the following 3 ways: (1) by selecting features, we effectively narrow the number of parameters used to classify, (2) cross-validation repeatedly trains and tests on nonoverlapped subsets to evaluate the classifier performance on unseen data, and (3) for each classifier, we impose the constraint that the number of trainable parameters be strictly below the number of training samples, which is considered a threshold to avoid overfitting. Note that when searching for the optimal configurations for a given classifier type (e.g., the number of layers and the number of nodes per layer in the neural net classifier), we also restrict our search range reflecting the above consideration.32 In addition, the best classifiers radial basis network and multilayer perceptron performed well using 10-fold cross-validation, confirming no gross overfitting to be present.
Using all features, no performance improvement is achieved over using one feature with the radial basis network and multilayer perceptron classification algorithms. This is likely attributable to insufficient training, because the number of trainable parameters in both classifiers is proportional to the number of features. With limited training data available for evaluation using all features, we were not able to derive reliable parameters from 9 of 10 of the total available training data, that can classify accurately remaining 1 of 10 of the available training data, in our 10-fold cross-validation study. We observed that in this case, SVM yielded improved accuracy with all features compared with mRMR selected features, a unique finding among the classifiers. This can be attributed to the fact that the number of parameters in SVM is determined by the number of training samples and independent of the number of features. Increasing the number of features does not increase the dimensionality using this particular classifier. On the contrary, having a higher dimensional feature space seems to provide a better separating hyperplane for this problem. We also point out that the optimal performance for SVM is obtained with a c value significantly larger than other cases, effectively restricting the allowable support vectors.
Overall, our study is limited by a relatively small training dataset. The optimal feature set and classification algorithm need to be validated over a larger dataset. Instead of using the mRMR feature selection method, future research could compare all possible feature combinations for a given classifier using an exhaustive search, and it is possible that an even better classification performance could be achieved. Another limitation is that the original feature set studied included primarily MRI metrics selected from our previous work based on differences we observed between study and control populations. It would be instructive to use additional features, such as fractional anisotropy and susceptibility-weighted imaging findings, as well as clinical characteristics, to enrich the feature selection and classification algorithms. We did not incorporate clinical and cognitive data here in order to avoid complications with higher dimensionality. Future work incorporating clinical features and imaging such as diffusion tensor imaging and susceptibility-weighted imaging could potentially achieve even greater classification accuracy. In addition, automated extraction of features using data mining techniques could be considered with a larger dataset. The results of this pilot study should be considered provisional, and correlation with clinical characteristics is needed to validate the approach. Our future goal is to recruit a unique cohort to test validity and reproducibility of classifiers.
This work serves as a pilot study showing that a combination of features including MRI metrics can classify patients with mTBI and controls with 86% accuracy, up from 74% for the best single feature alone. Furthermore, mRMR feature selection optimizes this process by selecting relevant and nonredundant features. These results show that there is promise for use of multifeature classification as a viable tool to aid in the objective diagnosis of mTBI.
Supplementary Material
GLOSSARY
- ACRM
American Congress of Rehabilitation Medicine
- MFC
magnetic field correlation
- MK
mean kurtosis
- mRMR
minimal-redundancy maximal-relevance
- mTBI
mild traumatic brain injury
- SVM
support vector machine
Footnotes
Editorial, page 1226
AUTHOR CONTRIBUTIONS
Yvonne W. Lui: drafting/revising manuscript for content, including medical writing for content, analysis or interpretation of data, and study concept or design. Yuanyi Xue: drafting/revising manuscript for content, including medical writing for content, and analysis or interpretation of data. Damon Kenul: drafting/revising manuscript for content, including medical writing for content and analysis or interpretation of data, acquisition of data. Yulin Ge: drafting/revising manuscript for content, including medical writing for content, analysis or interpretation of data, and study concept or design. Robert I. Grossman: study concept or design. Yao Wang: drafting/revising manuscript for content, including medical writing for content, analysis or interpretation of data, and study concept or design.
STUDY FUNDING
Supported in part by NIH grants UL1 TR000038 and 2 RO1 NS039135-11.
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Faul MLW, Wald MM, Coronado VG. Traumatic brain injury in the United States: emergency department visits, hospitalizations and deaths 2002–2006. Available at: http://www.cdc.gov/traumaticbraininjury/pdf/blue_book.pdf. Accessed May 02, 2014
- 2.McMahon P, Hricik A, Yue JK, et al. Symptomatology and functional outcome in mild traumatic brain injury: results from the prospective TRACK-TBI study. J Neurotrauma 2014;31:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll LJ, Cassidy JD, Peloso PM, et al. Prognosis for mild traumatic brain injury: results of the WHO Collaborating Centre Task Force on Mild Traumatic Brain Injury. J Rehabil Med 2004;(43 suppl):84–105 [DOI] [PubMed] [Google Scholar]
- 4.Kay T, Harrington D, Adams R, et al. Definition of mild traumatic brain injury. J Head Trauma Rehabil 1993;8:86–87 [Google Scholar]
- 5.Marr A, Corronado V, editors. Central Nervous System Injury Surveillance Data Submission Standards—2002. Atlanta: Centers for Disease Control and Prevention, National Center for Injury Prevention and Control; 2004 [Google Scholar]
- 6.Veterans Administration Department of Defense Clinical Practice Guideline for Mangement of Concussion/Mild Traumatic Brain Injury [online]. Available at: http://www.dcoe.mil/content/navigation/documents/VA%20DoD%20Management%20of%20Concussion%20mild%20Traumatic%20Brain%20Injury.pdf. Accessed August 22, 2014 [Google Scholar]
- 7.Arciniegas DB, Silver JM. Regarding the search for a unified definition of mild traumatic brain injury. Brain Inj 2001;15:649–652 [DOI] [PubMed] [Google Scholar]
- 8.Ruff RM, Jurica P. In search of a unified definition for mild traumatic brain injury. Brain Inj 1999;13:943–952 [DOI] [PubMed] [Google Scholar]
- 9.Teasdale G, Jennett B. Assessment of coma and impaired consciousness: a practical scale. Lancet 1974;2:81–84 [DOI] [PubMed] [Google Scholar]
- 10.Voller B, Auff E, Schnider P, Aichner F. To do or not to do? Magnetic resonance imaging in mild traumatic brain injury. Brain Inj 2001;15:107–115 [DOI] [PubMed] [Google Scholar]
- 11.Kraus MF, Susmaras T, Caughlin BP, Walker CJ, Sweeney JA, Little DM. White matter integrity and cognition in chronic traumatic brain injury: a diffusion tensor imaging study. Brain 2007;130:2508–2519 [DOI] [PubMed] [Google Scholar]
- 12.Bogner J, Corrigan JD. Reliability and predictive validity of the Ohio State University TBI identification method with prisoners. J Head Trauma Rehabil 2009;24:279–291 [DOI] [PubMed] [Google Scholar]
- 13.Mayer AR, Ling J, Mannell MV, et al. A prospective diffusion tensor imaging study in mild traumatic brain injury. Neurology 2010;74:643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson CV, Wood DM, Bigler ED, Blatter DD. Lesion volume, injury severity, and thalamic integrity following head injury. J Neurotrauma 1996;13:35–40 [DOI] [PubMed] [Google Scholar]
- 15.Ge Y, Patel MB, Chen Q, et al. Assessment of thalamic perfusion in patients with mild traumatic brain injury by true FISP arterial spin labelling MR imaging at 3T. Brain Inj 2009;23:666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossman EJ, Ge Y, Jensen JH, et al. Thalamus and cognitive impairment in mild traumatic brain injury: a diffusional kurtosis imaging study. J Neurotrauma 2014;29:2318–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miles L, Grossman RI, Johnson G, Babb JS, Diller L, Inglese M. Short-term DTI predictors of cognitive dysfunction in mild traumatic brain injury. Brain Inj 2008;22:115–122 [DOI] [PubMed] [Google Scholar]
- 18.Raz E, Jensen JH, Ge Y, et al. Brain iron quantification in mild traumatic brain injury: a magnetic field correlation study. AJNR Am J Neuroradiol 2011;32:1851–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang L, Ge Y, Sodickson DK, et al. Thalamic resting-state functional networks: disruption in patients with mild traumatic brain injury. Radiology 2011;260:831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood DM, Bigler ED. Diencephalic changes in traumatic brain injury: relationship to sensory perceptual function. Brain Res Bull 1995;38:545–549 [DOI] [PubMed] [Google Scholar]
- 21.Hulkower MB, Poliak DB, Rosenbaum SB, Zimmerman ME, Lipton ML. A decade of DTI in traumatic brain injury: 10 years and 100 articles later. AJNR Am J Neuroradiol 2013;34:2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luther N, Niogi S, Kutner K, et al. Diffusion tensor and susceptibility-weighted imaging in concussion assessment of National Football League Players. Neurosurgery 2012;71:E558 [Google Scholar]
- 23.Yuh EL, Mukherjee P, Lingsma HF, et al. Magnetic resonance imaging improves 3-month outcome prediction in mild traumatic brain injury. Ann Neurol 2013;73:224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Kierans A, Kenul D, et al. Longitudinal regional brain volume changes in mild traumatic brain injury patients. Radiology 2013;267:880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischl B, van der Kouwe A, Destrieux C, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex 2004;14:11–22 [DOI] [PubMed] [Google Scholar]
- 26.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 2012;59:2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen JH, Chandra R, Ramani A, et al. Magnetic field correlation imaging. Magn Reson Med 2006;55:1350–1361 [DOI] [PubMed] [Google Scholar]
- 28.Jensen JH, Szulc K, Hu C, et al. Magnetic field correlation as a measure of iron-generated magnetic field inhomogeneities in the brain. Magn Reson Med 2009;61:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossman EJ, Jensen JH, Babb JS, et al. Cognitive impairment in mild traumatic brain injury: a longitudinal diffusional kurtosis and perfusion imaging study. AJNR Am J Neuroradiol 2013;34:951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng HC, Long FH, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 2005;27:1226–1238 [DOI] [PubMed] [Google Scholar]
- 31.Bianchi A, Bhanu B, Donovan V, Obenaus A. Visual and contextual modeling for the detection of repeated mild traumatic brain injury. IEEE Trans Med Imaging Epub 2013 Jun 18 [DOI] [PubMed]
- 32.Battiti R. Using mutual information for selecting features in supervised neural-net learning. IEEE Trans Neural Netw 1994;5:537–550 [DOI] [PubMed] [Google Scholar]
- 33.Schölkopf B, Burges CJC, Smola AJ. Advances in Kernel Methods: Support Vector Learning. Cambridge, MA: MIT Press; 1999 [Google Scholar]
- 34.Manning CD, Raghavan P, Schütze H. Introduction to Information Retrieval. New York: Cambridge University Press; 2008 [Google Scholar]
- 35.Heckerman D. A Tutorial on Learning with Bayesian Networks. Redmond, WA: Microsoft Research; 1996 [Google Scholar]
- 36.Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed New York: Springer; 2009 [Google Scholar]
- 37.Rumelhart DE, Hinton GE, Williams RJ. Learning internal representations by error propagation. In: Rumelhart DE, McClelland JL, editors. Parallel Distributed Processing: Explorations in the Microstructure of Cognition, vol 1 Cambridge, MA: MIT Press; 1986:318–362 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


