Abstract
It is desirable to obtain new antagonists for thyroid hormone (TRs) and other nuclear receptors (NRs). We previously used X-ray structural models of TR ligand binding domains (LBDs) to design compounds, such as NH-3, that impair coactivator binding to activation function 2 (AF-2) and block thyroid hormone (triiodothyronine, T3) actions. However, TRs bind DNA and are transcriptionally active without ligand. Thus, NH-3 could modulate TR activity via effects on other coregulator interaction surfaces, such as activation function (AF-1) and corepressor binding sites. Here, we find that NH-3 blocks TR-LBD interactions with coactivators and corepressors and also inhibits activities of AF-1 and AF-2 in transfections. While NH-3 lacks detectable agonist activity at T3-activated genes in GC pituitary cells it nevertheless activates spot 14 (S14) in HTC liver cells with the latter effect accompanied by enhanced histone H4 acetylation and coactivator recruitment at the S14 promoter. Surprisingly, T3 promotes corepressor recruitment to target promoters. NH-3 effects vary; we observe transient recruitment of N-CoR to S14 in GC cells and dismissal and rebinding of N-CoR to the same promoter in HTC cells. We propose that NH-3 will generally behave as an antagonist by blocking AF-1 and AF-2 but that complex effects on coregulator recruitment may result in partial/mixed agonist effects that are independent of blockade of T3 binding in some contexts. These properties could ultimately be utilized in drug design and development of new selective TR modulators.
There are potentially important applications for thyroid hormone (TH) receptor (TR) antagonists [reviewed in (Baxter, Dillmann et al. 2001; Webb, Nguyen et al. 2002)]. Hyperthyroidism leads to rapid heart rate and arrthymias, muscle weakness, bone loss in post-menopausal women and anxiety. While this condition can be treated by thyroid gland ablation or drugs that block TH production, thyroxine (T4), the major circulating form of TH, has a half-life of 8 days so clinical improvement is slow (Braverman and Utiger 2000). Antagonists that act on TRs should provide more rapid relief and could partially supplant existing therapies (Webb, Nguyen et al. 2002). These drugs could also be useful in amiodarone-induced toxicosis, for which current therapies are inadequate (Conen, Melly et al. 2007), and may reverse cardiac arrhythmias in euthyroid conditions (Webb, Nguyen et al. 2002). Finally, TR antagonists could reduce metabolic rate and prevent tissue damage in anoxic states, such as stroke or injury (Webb, Nguyen et al. 2002; Grover, Dunn et al. 2007).
TRs are nuclear hormone receptors (NRs) (Zhang and Lazar 2000; Yen 2001; Laudet and Gronemeyer 2002). There are three major TR isoforms (TRβ1, TRβ2 and TRα1) that are expressed tissue specifically. Each TR binds to specific DNA sequences (thyroid hormone response elements, TREs) in target gene promoters in the unbound state, often as heterodimers with retinoid X receptors. From this location, TRs recruit coregulators, which, in turn, alter chromatin structure and influence recruitment and processivity of the basal transcription machinery (Lonard and O’Malley 2006). Triiodothyronine (T3), the active form of TH, modulates TR activity by enhancing packing of C-terminal helix 12 against the core of the ligand binding domain (LBD) (Glass and Rosenfeld 2000; Webb, Nguyen et al. 2002). This simultaneously creates a binding surface (activation function 2, AF-2) for coactivators, including steroid receptor coactivators (SRCs) and p300), and blocks binding of corepressors such as nuclear receptor corepressor (N-CoR) (Feng, Ribeiro et al. 1998; Glass and Rosenfeld 2000; Marimuthu, Feng et al. 2002; Webb, Nguyen et al. 2002; Rosenfeld, Lunyak et al. 2006).
Analysis of our X-ray structures of TR-LBDs with agonists suggested a rational strategy for design of TR and NR antagonists (Webb, Nguyen et al. 2002). Since hormone is buried in the core of the domain (Wagner, Apriletti et al. 1995), we proposed that derivatives of agonists with appropriately placed extensions should compete with T3 for binding and inhibit NR activity by displacing H12 (Scanlan, Baxter et al. 1996; Webb, Nguyen et al. 2002). We used this strategy to identify lead compounds that inhibit TRs (Yoshihara, Apriletti et al. 2001; Baxter, Goede et al. 2002; Nguyen, Apriletti et al. 2002; Nguyen, Apriletti et al. 2005). The best, NH-3, is derived from the TRβ selective agonist GC-1 and contains a bulky 5′ nitrophenylethynyl extension (Chiellini, Apriletti et al. 1998; Nguyen, Apriletti et al. 2002; Webb, Nguyen et al. 2002). NH-3 binds TRs with nanomolar affinity, blocks TR AF-2 interactions with coactivators in vitro and in cell culture and antagonizes T3 responses at standard reporters in cell culture. NH-3 also inhibits TH mediated metamorphosis of tadpoles, and antagonizes T3 effects on heart rate, serum cholesterol and plasma thyroid stimulating hormone in rats (Lim, Nguyen et al. 2002; Nguyen, Apriletti et al. 2002; Grover, Dunn et al. 2007).
There are, however, several reasons to suspect that NH-3 will not always act as a pure T3 antagonist. Unliganded TRs, like many other NRs, bind to DNA in the absence of hormone, where they have ligand independent actions. Thus, NH-3 dependent alterations in TR conformation could alter coregulator recruitment and gene expression independently of effects on T3 binding. Indeed, weak partial agonist activity has been observed at high NH-3 doses in rats, but it is not clear whether this involves direct effects on TRs or generation of agonist metabolites (Lim, Nguyen et al. 2002; Grover, Dunn et al. 2007).
How might NH-3 alter coregulator recruitment? We found that NH-3 resembles T3 in its ability to block TR LBD interactions with the corepressors N-CoR and SMRT in mammalian 2-hybrid assays and cell free systems (Nguyen, Apriletti et al. 2002; Moore, Galicia et al. 2004). If NH-3 prevents TR interactions with corepressors, this effect could result in partial agonist or agonist actions (Glass and Rosenfeld 2000; Webb, Nguyen et al. 2002). Moreover, NRs and TRs contain one at least one alternate activation function (AF-1) in their amino terminal domains (NTD), and possibly other coregulator binding loci (Barettino, Vivanco Ruiz et al. 1994; Wilkinson and Towle 1997; Yang, Hong et al. 1999; Oberste-Berghaus, Zanger et al. 2000; Webb, Nguyen et al. 2002; Tian, Mahajan et al. 2006). While it is not obvious how these surfaces would be affected by an antagonist that acts by displacing H12 on the LBD, it is well known that estrogen receptor (ER) antagonists, including the selective ER modulators (SERMs) tamoxifen and raloxifene, activate AF-1 via release of heat shock proteins that prevent the receptors from binding to DNA and indirect effects on AF-1 itself (reviewed in (Webb, Nguyen et al. 2002)). Understanding these issues would be of clear importance for design of antagonists for TRs and other NRs that bind DNA in the absence of ligand.
In this study, we examined NH-3 effects on TR activation functions in transient transfections and upon gene expression and coregulator recruitment in cultured cells. We find that NH-3 blocks both AF-2 and AF-1 and largely lacks agonist activity, but does induce spot 14 (S14) in HTC liver cells. This effect is accompanied by enhanced coactivator recruitment at the S14 promoter, which is not seen in pituitary GC-1 cells where NH-3 fails to enhance S14 expression. NH-3 and T3 exert effects on N-CoR interactions that are counter to current dogma (Glass and Rosenfeld 2000), both promote N-CoR recruitment to TR regulated genes, although NH-3 effects vary with cell type. We suggest that antagonists that interact with DNA bound NRs will mostly block agonist actions, but have the potential to exert limited agonist effects and alter some actions of unliganded receptors. These findings have important ramifications for TR and NR antagonist design.
Results
NH-3 Blocks TR-LBD Interactions with Coactivators and Corepressors
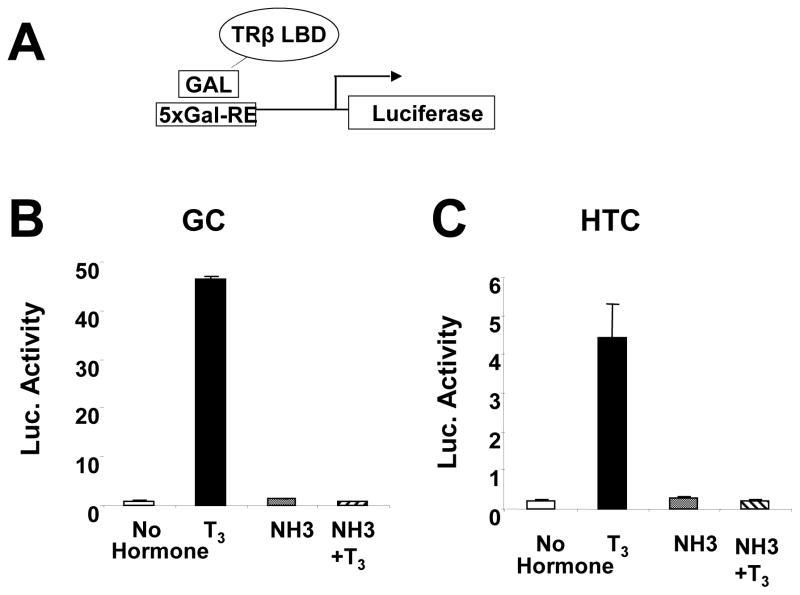
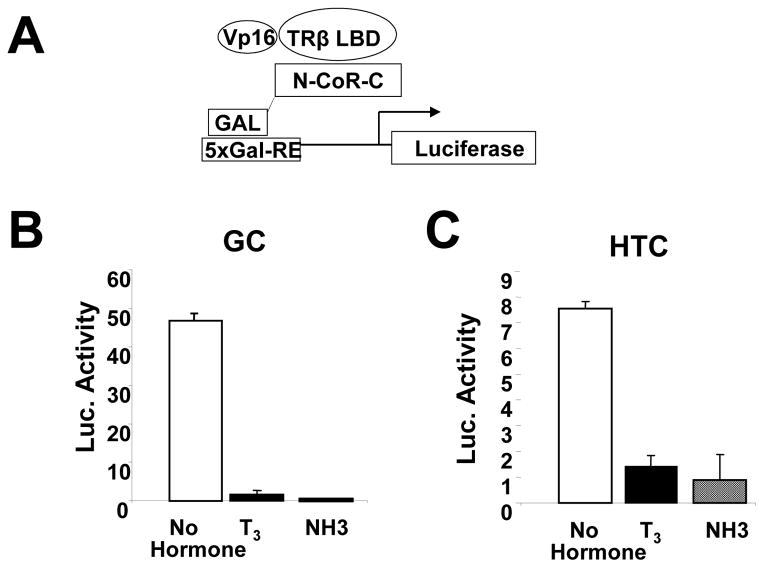
We confirmed that NH-3 blocks TR-LBD interactions with coactivators and corepressors in cells that express endogenous TRs, rat GC pituitary and HTC liver cells (Nguyen, Apriletti et al. 2002). T3 activated a GAL4 reporter in the presence of a GAL-TRβ LBD fusion protein in both cell types, an effect requiring coactivator association with TR AF-2 (Fig. 1). By contrast, NH-3 failed to activate the reporter and blocked T3 induction, confirming that it functions as a TR antagonist. Both ligands also inhibited N-CoR interactions with the TR LBD (Fig. 2); NH-3 and T3 reduced transcriptional readout from the GAL4 responsive reporter in the presence of a GAL4 DBD bait linked to N-CoR NR interacting domains and a VP16-tagged TRβ LBD trap. Similar results were also obtained with a GAL-SMRT fusion (not shown), as previously observed in HeLa cells (Nguyen, Apriletti et al. 2002). Thus, NH-3 blocks TR LBD interactions with coactivators and corepressors.
Fig. 1.
NH-3 does not activate TRβ AF-2 in GC and HTC cells. (A) Transfection components in schematic. (B) Results of transcription readout assays performed in GC cells or (C) HTC cells.
Fig. 2.
NH-3 and T3 promote corepressor release from the TRβ LBD in GC and HTC cells in mammalian 2-hybrid assays. (A) Transfection components in schematic. (B) Results of transcription readout assays performed in GC cells and (C) HTC cells.
TR AF-1 and AF-2 Cooperate in Gene Induction in GC and HTC Cells
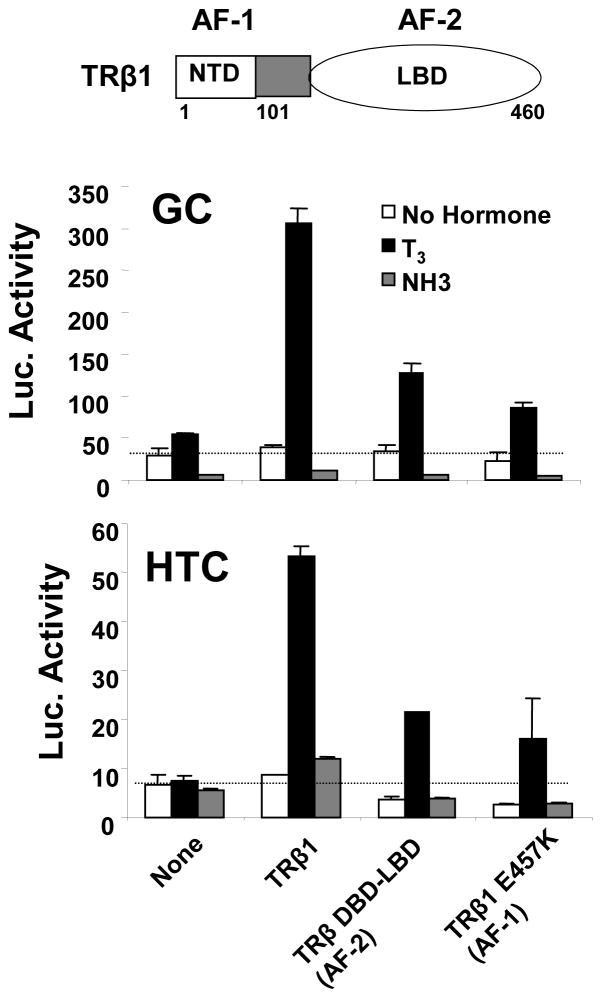
Data in Fig. 3 show effects of NH-3 on activities of full length TR and individual TR transactivation functions (AF-1 and AF-2) at T3-activated TRE-dependent reporter in GC and HTC cells. In GC cells, T3 activated a TRE-dependent reporter (containing an inverted palindrome, IP-6; (Velasco, Togashi et al. 2007)) in the presence of endogenous TRs whereas NH-3 inhibited basal activity (Fig. 3, upper panel). Transfected TRβ1 amplified T3 response, but did not alter the ability of NH-3 to suppress basal reporter activity. In HTC cells; T3 response was only obtained with transfected TRβ1. Here, NH-3 failed to repress basal activity (Fig. 3, lower panel).
Fig. 3.
TR AF-1 and AF-2 cooperate in T3 response at TRE-regulated reporters in GC and HTC cells, but NH-3 lacks agonist activity. The panels shows transfection assays in GC and HTC cells performed with an IP-6 responsive reporter +/− cotransfected expression vectors for TRβ1, TRβ1 DBD-LBD (AF-2 only) or TRβ1 E457K (AF-1 only) and induced with T3 (100nM) or NH-3 (1μM).
Both TR activation functions are active in GC and HTC cells; a TRβ truncation that lacks AF-1 (TRβ1 DBD-LBD) and a TRβ point mutant that lacks AF-2 activity (TRβE457K (Feng, Ribeiro et al. 1998)) exhibit reduced T3-induction relative to TRβ. As expected, NH-3 blocked the activity of the TR truncation that only contained AF-2. However, NH-3 also blocked activity of the TRβE457K mutant that retains AF-1 but lacks AF-2, suggesting that NH-3 must indirectly inhibit TR AF-1 activity in this context.
NH-3 also failed to act as an agonist at standard reporters in a variety of related experimental conditions (not shown). NH-3 did not induce the IP-6 reporter in GC and HTC cells transfected with TRβ2 or TRα1, with any TRs or TR truncations at T3-activated reporters driven by other TREs (Velasco, Togashi et al. 2007), a direct repeat (DR-4) and a palindrome (Pal), and with transiently transfected coactivators that potentiate T3 response, including SRCs and p300. Thus, NH-3 exhibits one cell-specific effect; it suppresses basal TR activity in GC cells but not in HTC cells, but generally lacks agonist activity at standard reporters and can inhibit both AF-1 and AF-2.
NH-3 Is a Cell and Promoter-Specific TR Agonist
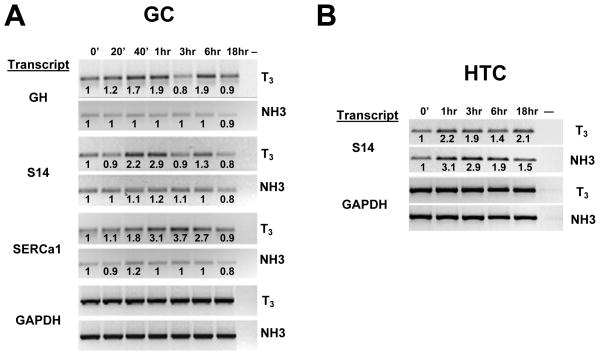
We compared effects of T3 and NH-3 on endogenous T3-responsive genes. Similar to results of others (Sharma and Fondell 2002; Liu, Xia et al. 2006), we observed transient 2–4 fold increases in steady state mRNA levels of growth hormone (GH), spot 14 (S14) and sarcoplasmic endoplasmic reticulum calcium dependent ATPase 1 (SERCa1) in GC cells after T3 treatment (Fig. 4A). There were variations in kinetics of hormone induction; GH and S14 mRNA levels peaked 1 hr after T3 administration, declined at 3 hrs and rose again at 6 hrs, whereas SERCa1 mRNA levels peaked at 3hrs and declined thereafter. T3 also induced S14 mRNA 2–3 fold in HTC cells, with peak induction 1 hr after hormone administration (Fig. 4B; GH and SERCa1 transcripts were not detected in this cell type; not shown). Glyceraldehyde phosphate dehydrogenase (GAPDH) transcripts were unaltered in both cell types, confirming that T3-effects were specific.
Fig. 4.
Effects of T3 and NH-3 on endogenous gene transcription in (A) GC and (B) HTC cells. GC or HTC cells were treated with T3 or NH-3 for different length of times and mRNA expression was analyzed by RT-PCR. The image shows PCR products obtained after amplification of mRNA samples prepared at various times after addition of ligand and separated on a 1% agarose gel. Fold inductions relative to t=0 (estimated by image scanning) are shown at top.
NH-3 exhibited different effects. It failed to induce GH, S14 and SERCa1 transcripts in GC cells at short times and weakly suppressed mRNA levels of these genes at 18hrs (Fig. 4A). By contrast, NH-3 induced S14 mRNA about 3-fold in HTC cells, slightly more efficiently than T3 (Fig. 4B). There were no effects on GAPDH mRNA in either cell type confirming that NH-3 effects were specific. Thus, NH-3 shows cell type specific agonist activity at S14.
NH-3 Induces Histone H4 Acetylation at the S14 Promoter in HTC Cells
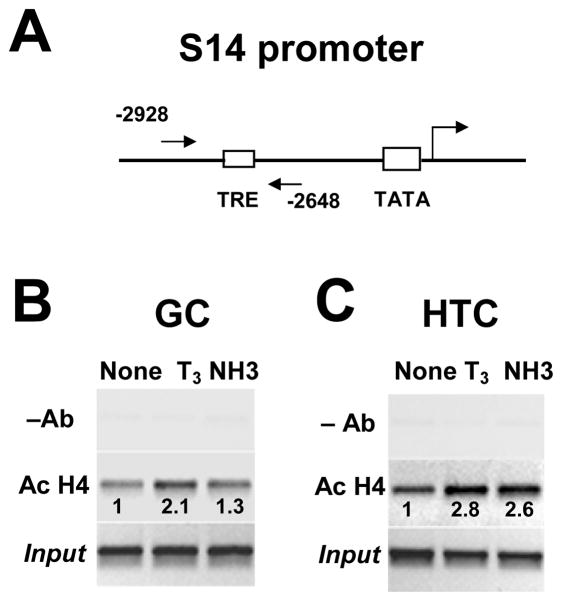
To explore effects of NH-3 on the S14 promoter, we performed ChIP experiments with GC and HTC cell extracts. First, we used an antibody against acetylated histone 4 (H4), a marker for transcriptional activation (Jenuwein and Allis 2001). T3 increased H4 acetylation at the S14 TRE in GC cells whereas NH-3 had a lesser effect (Fig. 5B). Similar results were obtained with the GH and SERCa1 promoters (not shown). By contrast, T3 and NH-3 enhanced H4 acetylation at S14 in HTC cells to similar extents (Fig. 5C). This suggests that NH-3 induces a transcriptionally active state at the S14 promoter in HTC cells.
Fig. 5.
Effect of T3 and NH-3 on histone acetylation at the S14 promoter in GC and HTC cells. (A) Schematic of the S14 promoter with positions of RT primers marked. Immunoprecipitated chromatin from GC (B) and HTC cells (C) was analyzed for the binding of acetylated histone by quantitative PCR in the TRE promoter region of S14 in presence of T3 and NH-3. Products were separated on a 1% agarose gel. Note that expression of S14 transcript in presence of T3 or NH-3 in GC (Fig. 1A) and HTC (Fig. 1B) cells correlates with acetylation of histone. Fold inductions (estimated by image scanning) at top.
NH-3 Enhances TR DNA binding in Cultured Cells
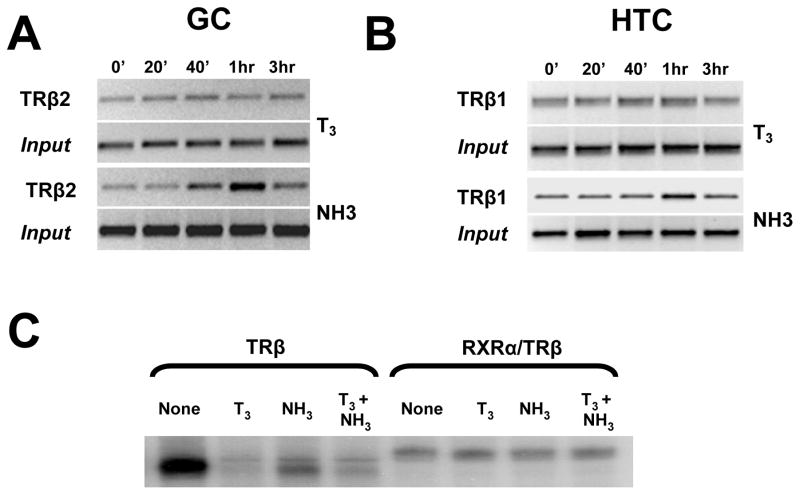
While T3 did not affect TR/DNA interactions, NH-3 enhanced TR binding to target promoters. As reported by others (Yen 2001), ChIP analysis showed that both TRβ2 and TRβ1 occupy the S14 TRE in GC and HTC cells, respectively, and that TR binding is only minimally affected by T3 (Figs. 6A and 6B). By contrast, NH-3 promoted transient increases in TRβ2 at all three target promoters in GC cells (Fig. 6A and not shown) and weaker but qualitatively similar increases in TRβ1 levels at the S14 promoter in HTC cells (Fig. 6B). These effects could not be recapitulated in gel shifts in vitro (Fig. 6C). Like T3, NH-3 promoted TR dissociation from IP-6 (although slightly less than T3, shown) and DR-4 elements (not shown) and failed to affect RXR-TR interactions with either TRE. Thus, NH-3 alters TR DNA binding in cultured cells but not in vitro.
Fig. 6.
Effect of T3 and NH-3 on TR binding to the S14 promoter. (A) ChIP analysis performed on GC cells using anti-TRβ2 antibody. (B) As above, in HTC cells with anti-TRβ1 antibody. (C) Gel shift assay performed with labeled IP-6 oligonucleotide and in vitro translated TRβ1 or a mixture of RXRα and TRβ1.
NH-3 and T3 Promote Different Patterns of Coactivator Recruitment to Target promoters
To explore the idea that NH-3 could induce S14 by altering TR transcription complex formation, we analyzed recruitment of representative TR coactivators to each promoter. We chose three coactivators that are recruited early in transcriptional activation, the p160s SRC2 and SRC1, and the histone acetyl-transferase p300 (Sharma and Fondell 2002; Liu, Xia et al. 2006).
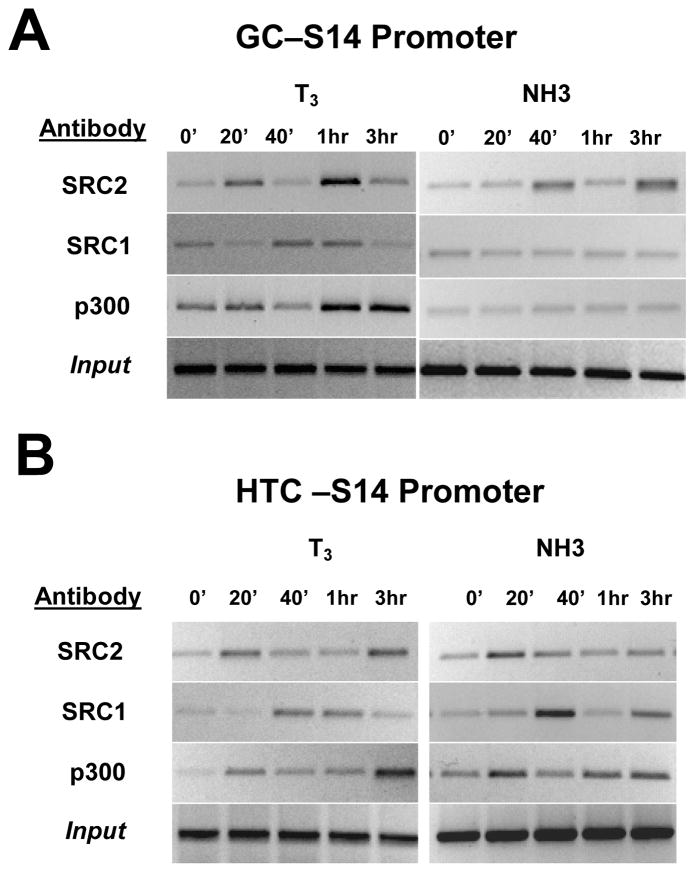
T3 promoted recruitment of all three coactivators to target promoters but the precise pattern varied with cofactor and cell type (Fig. 7). In GC cells (Fig. 7A), SRC2 was rapidly recruited to the S14 promoter, dismissed at 40 minutes and recruited again at 60 minutes. SRC1 was present before T3 induction, dismissed at 20 minutes, re-recruited at 40–60 minutes and dismissed again at 180 minutes. Finally, p300 was also present prior to T3 induction, dismissed at 40 minutes and recruited again at 1–3 hrs. The same profile was obtained with GH and SERCa1 (not shown). Broadly similar results were obtained with the S14 promoter in HTC cells (Fig. 7B); the sole distinction from GC cells is that the second wave of SRC2 and p300 recruitment occurred later in HTC cells.
Fig. 7.
Recruitment of coactivators. (A) Immunoprecipitated chromatin was analyzed for the recruitment of coactivators SRC2, SRC1 and p300 by quantitative PCR in the TRE promoter region of S14 genes in GC cells presence after various times of T3 and NH-3 treatment. (B) As above, using immunoprecipitated chromatin from HTC cells.
NH-3 effects on coactivator recruitment differed from T3. In GC cells, NH-3 promoted cyclical recruitment of SRC2 to the S14 promoter (Fig. 7A, shown) and to the GH and SERCA promoters (not shown). SRC2 recruitment was weaker than obtained with T3 and delayed, peak SRC2 binding occurred 40 minutes and 3hrs after NH-3 treatment, compared to 20 minutes and 1 hour with T3. NH-3 did not promote SRC1 or p300 recruitment to target promoters in this cell type (Fig. 7A and not shown).
By contrast, NH-3 promoted recruitment of all three coactivators to S14 in HTC cells (Fig. 7B). The pattern differed from T3 in several ways. First, NH-3 only promoted one round of SRC2 recruitment during this time frame. Second, NH-3 dependent SRC1 recruitment was stronger than T3 at 40 minutes and p300 recruitment was stronger than with T3 at early times and weaker at later times. Third, recruitment kinetics differed from those of T3; SRC1 was dismissed at 1hr with NH-3 and returned at 3hrs whereas it persisted at 1hr with T3 and was dismissed at 3hrs and the second round of p300 recruitment occurred earlier with NH-3.
Together, our results suggest NH-3 induction of S14 in HTC cells correlates with coactivator recruitment. The fact that NH-3 and T3 induce different patterns of coactivator recruitment may, however, be indicative of different mechanisms of transcriptional activation (see Discussion).
T3-Dependent Recruitment of N-CoR and Cell Type Specific Effects of NH-3
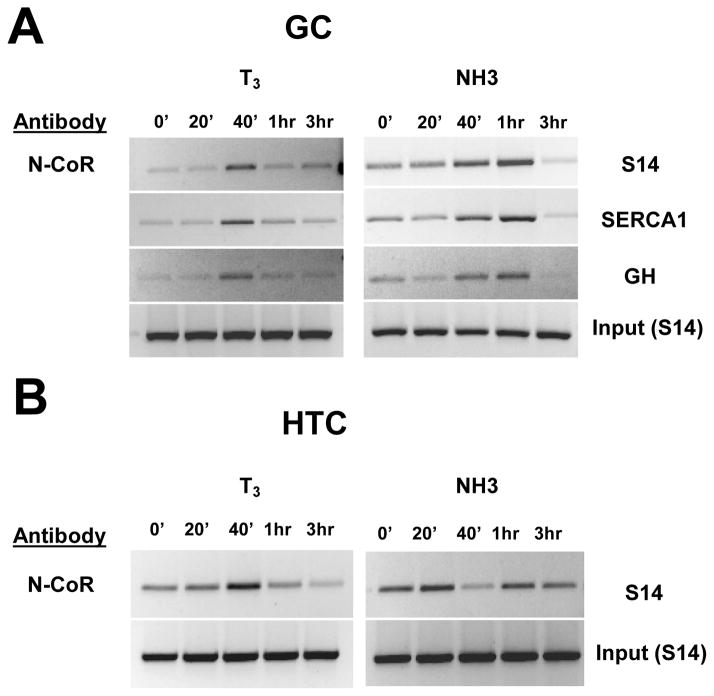
Finally, we examined ligand effects on corepressor recruitment to TR regulated promoters. Unlike current dogma, T3 enhanced N-CoR recruitment in both GC and HTC cells. N-CoR was present at low or undetectable levels at the GH, S14 and SERCA promoters without ligand (Fig. 8A). Administration of T3 led to enhanced N-CoR recruitment, with a peak at 40 minutes that coincided with dismissal of SRC2 and p300 (see Fig. 7A). N-CoR was detectable at the S14 promoter in HTC cells in the absence of ligand (Fig. 8B). Here, T3 promoted a transient increase in N-CoR recruitment at 40 minutes, coinciding again with SRC2 and p300 dismissal (see Fig. 7B).
Fig. 8.
Recruitment of corepressor N-CoR. Immunoprecipitated chromatin was analyzed by quantitative PCR in the TRE promoter region of (A) S14, SERCA1 and GH genes after various times of T3 and NH-3 treatment of GC and (B) HTC cells.
NH-3 effects differed from T3 and varied with cell type. NH-3 promoted weak recruitment of N-CoR to all three promoters in GC cells; this occurred with delayed kinetics relative to T3 and coincided with loss of SRC2 (compare Fig. 8A to Fig. 7A). By contrast, NH-3 led to dismissal of N-CoR at 40 minutes in HTC cells followed by rebinding at later times (Fig. 8B).
Discussion
In this study, we examined actions of NH-3, which blocks T3 action in cell free systems (Nguyen, Apriletti et al. 2002; Moore, Galicia et al. 2004; Nguyen, Apriletti et al. 2005), transfected cells (Nguyen, Apriletti et al. 2002; Nguyen, Apriletti et al. 2005), tadpoles (Lim, Nguyen et al. 2002) and rats (Grover, Dunn et al. 2007). Since unliganded TRs bind DNA and are transcriptionally active we suspected that NH-3 may influence TR conformation and activity in a manner that is independent of effects on T3 binding. This is the case: NH-3 induces S14 in HTC cells, and increases H4 acetylation at the promoter, indicative of a transcriptionally active state. However, it fails to induce this gene, or GH and SERCA, in GC pituitary cells or transfected reporters in either cell type. Thus, NH-3 is a cell-type and promoter-specific agonist.
ChIP analysis confirms that NH-3 alters TR activities. NH-3 induction of S14 in HTC cells is accompanied by enhanced recruitment of three coactivators tested (SRC1, SRC2 and p300) and transient dismissal of N-CoR (opposite to increased N-CoR recruitment with T3). Lack of NH-3 induction of target genes (including S14) in GC cells is associated with large transient increases in TRβ2 binding and delayed recruitment of one coactivator (SRC2) and N-CoR. Thus, NH-3 alters TR transcription complex formation at target promoters, even when it fails to induce the associated gene, and the fact that the drugs tends to push the balance of coregulator recruitment at S14 in towards coactivators in HTC cells but not GC cells probably explains its cell-specific agonist actions.
It is not clear how NH-3 promotes coactivator recruitment. It does not seem likely that preferential expression of TRβ1 vs. TRβ2 in HTC cells explains cell-specific actions of this compound; transfected TRβ1 did not permit NH-3 to activate a T3-inducible S14 reporter in GC (not shown). We also do not know which TR surfaces are involved in coactivator recruitment. NH-3 blocks AF-2 in GC or HTC cells (Figs 1–3), as seen in other cell types (Nguyen, Apriletti et al. 2002; Webb, Nguyen et al. 2002; Moore, Galicia et al. 2004), suggesting that NH-3 must promote coactivator recruitment differently from T3, a potent AF-2 activator. We suspect that NH-3-dependent cofactor recruitment involves AF-1, but have not been able to test this idea directly because TRs paradoxically repress S14 dependent reporters in HTC cells (Ota and Mariash 2003). We note, however, that TR AF-1 mediates T3 response at a reporter with S14 TREs (Wilkinson and Towle 1997) and is required for optimal T3 response at standard reporters in HTC cells, confirming it is active (Fig. 3). We also found that NH-3 allows TR AF-1 dependent interactions with SRC1 and SRC2 in pulldowns in vitro (not shown). These observations are all consistent with the idea that NH-3 could promote coactivator recruitment through AF-1 (Webb, Nguyen et al. 2002).
Surprisingly, T3 enhances corepressor recruitment at all target genes in this study. Whereas current coregulator exchange models predict ligand dependent dismissal of corepressors and replacement with coactivators (Glass and Rosenfeld 2000), we find that T3 promotes transient N-CoR recruitment to S14, GH and SERCA in GC and to S14 in HTC cells. We also expected that NH-3 would act as a partial TR agonist at T3-activated genes by displacing corepressors. Instead, NH-3 failed to induce target genes in GC cells, acts as an agonist at S14 in HTC cells, and, respectively, promotes transient recruitment of N-CoR and cyclical dismissal and rebinding of N-CoR to S14 in these cell types.
We recognize that much available experimental evidence supports each aspect of the coregulator exchange model, including mutational analysis of TRs (Feng, Ribeiro et al. 1998; Hu and Lazar 1999; Nagy, Kao et al. 1999; Jepsen, Hermanson et al. 2000; Marimuthu, Feng et al. 2002), blockade of N-CoR expression with interfering RNAs (Yoon, Chan et al. 2003) or targeted disruption of the N-CoR gene (Jepsen, Hermanson et al. 2000) and ChIP analysis in frogs (Li, Lin et al. 2002; Lim, Nguyen et al. 2002; Buchholz, Hsia et al. 2003; Tomita, Buchholz et al. 2004; Paul, Buchholz et al. 2005; Paul, Fu et al. 2005) and in cultured cells at stably integrated GAL4-responsive reporters regulated by GAL-TR LBD (Sharma and Fondell 2002; Ishizuka and Lazar 2003; Yoon, Choi et al. 2005; Liu, Xia et al. 2006).
How, then, do we reconcile our findings with these studies? Coactivator vs. corepressor recruitment has not been widely explored at native TR-target genes and we propose that the coregulator exchange model applies could only to subsets of T3-induced genes, and none investigated here. This agrees with the fact that unliganded peroxisome proliferator activated receptor gamma recruits corepressors to some promoters and not others (Guan, Ishizuka et al. 2005) and could explain why unliganded TRs only repress a subpopulation of T3 responsive genes in liver in mouse TR knockouts (Yen, Feng et al. 2003). The idea also agrees with observations that steroid receptor agonists sometimes promote corepressor recruitment (Chen, Welsbie et al. 2004; Hodgson, Astapova et al. 2005; Ki, Cho et al. 2005; Wang and Simons 2005; Yoon and Wong 2006; Higgins, Liu et al. 2008). Finally, we note that our findings are exactly consistent with a model of TR action proposed by Lee and coworkers (Sohn, Kim et al. 2003) who envisaged dynamic interplay of coactivator and corepressor complexes at the T3-liganded TR with corepressors serving to restrict T3 response.
It will be interesting to determine the function of ligand-dependent corepressor recruitment. We suspect that N-CoR recruitment serves to limit T3 activation, as proposed by Lee and coworkers (Sohn, Kim et al. 2003), because the timing of this event approximately precedes transient reduction of mRNA levels that are feature of cyclical T3 dependent transcriptional responses (compare Fig. 4 with results of Fig. 8. We must also consider the possibility that NH-3 dependent blockade of AF-1 and AF-2 transactivation functions in GC cells (Fig. 3) may be related to the fact that this ligand promotes corepressor recruitment to target promoters in GC cells while failing to recruit the complete complement of coactivators in these contexts.
We do not know how T3 promotes corepressor binding and why NH-3 effects vary with cell type. TR H12 position could alter in the transcription cycle, respectively favoring coactivator and corepressor binding. Evidence against this argument is that both T3 and NH-3 consistently block TRβ-LBD interactions with N-CoR NR interaction domains in all cell types tested. Alternatively, liganded TRs have been shown to bind a coactivator-like LxxLL peptide in N-CoR (Loinder and Soderstrom 2004; Loinder and Soderstrom 2005), and this may mediate T3-dependent N-CoR recruitment to target genes. This would not explain how NH-3, which blocks TR interactions with LxxLL motifs, promotes N-CoR binding in GC cells. We presently favor the idea that T3-dependent N-CoR recruitment is mediated by alternate interaction surfaces and suggest that this effect could share features with known mechanisms of transcriptional repression (Ogawa, Lozach et al. 2005; Blaschke, Takata et al. 2006; Pascual and Glass 2006; Ghisletti, Huang et al. 2007), where liganded NRs block gene expression by arresting corepressor complexes at target promoters.
Our findings have broad implications for NR selective modulator design. Effects of ligands that inhibit NR AF-2 were established with steroid receptors, where ligands also promote receptor translocation into the nucleus (Webb, Nguyen et al. 2002). This is different from TRs, which bind to nuclear chromatin without ligand. Although NH-3 is a general TR antagonist, it clearly has potential for complex effects on coregulator recruitment and this is also likely to be the case with similar compounds that bind other NRs that interact with chromatin in the absence of ligand. Our results also address potential for TR drugs that inhibit coactivator and corepressor recruitment. Phenotypes of TRα/β-/- knockout mice are mild compared to hypothyroid animals, suggesting that unopposed actions of unliganded TRs mediate harmful effects of this disease and we proposed that TR drugs (such as NH-3) that inhibit corepressor and coactivator recruitment would reverse hypothyroidism without inducing symptoms of thyroid hormone excess (Baxter, Dillmann et al. 2001; Webb, Nguyen et al. 2002). Since TR gene regulation does not always follow the classic coregulator exchange model, we suggest that simple predictions about actions of this class of NR ligand require reevaluation.
Finally, we emphasize that our experiments focus on nuclear TH signaling pathways. It is important to note that TR ligands also elicit rapid responses through binding to the same receptor in the cytoplasm, and that there are alternative TH signaling pathways that do not involve the traditional NRs (Davis, Leonard et al. 2008). It will be interesting to determine how NH-3 affects these alternative signaling pathways and define contributions of classical nuclear actions and alternative actions of such ligands to observed responses in animal models.
Materials and Methods
Cell Culture
HTC cells and GC cells were maintained in DMEM medium containing 10% FBS and streptomycin. Serum was withdrawn 24 hrs prior to the addition of ligand T3 or NH-3 for various periods before harvesting the cells for transcript analysis or ChIP assays. T3 was obtained from Sigma; NH-3 was synthesized as previously described (Nguyen, Apriletti et al. 2002).
Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
GC or HTC cells were treated with T3 or NH-3 (10−7 M) for various times after serum withdrawal. Total RNA was prepared from the cells using TRIZOL (GIBCO-BRL). The cDNA were prepared with Super Script™ first-strand synthesis system for RT-PCR (InVitrogen) from 1μg of total RNA.
The primers were:
| S14: | forward 5′-GTCATGGATCGGTACTCGGC-3′ reverse 5′-CAGCTCCTCCGAAAGCCTGTC-3′ |
| SERCA1: | forward 5′-GTGAGCGAGACCACAGGCCTTACC-3′ reverse 5′-CCTTCAGCGCCTCGATGGCATTCT-3′. |
| GH: | forward 5′-GCCTACATTCCCGAGGGACAGCGC-3′ reverse 5′-GAGCAGAGCGTCATCGCTGCGCAT-3′ |
| GAPDH | forward 5′-GCACAGTCAAGGCTGAGAATGGGA-3′ reverse 5′-CATGGACTGTGGTCATGAGCCCTT-3′. |
Chromatin Immunoprecipitation (ChIP)
Antibodies against TR (06-539 and 06-540), acetyl-Histone H4 (06-866), SRC1 (05-522) were obtained for Upstate Biotechnology, Lake Placid NY. Antibodies against SRC2 (C-20), p300 (C-20), N-CoR (C-20) were obtained from Santa Cruz Biotechnology, Inc.
ChIP assays were performed essentially as previously described in Orlando et al., (Orlando, Strutt et al. 1997) with slight modifications. Approximately 6×106 cells were cross linked with 1% formaldehyde for 15 min at room temperature. Cross linking was stopped by addition of 0.125 M glycine. Cells were washed twice with ice-cold PBS and collected by scraping in 5 ml ice-cold PBS. The pellet was washed with (10 mM Tris, pH 8.0, 10mM EDTA, 0.5mM EGTA, 200 mM NaCl and 1 mM phenylmethylsulfonyl fluoride). Cells were lysed in 0.25% triton buffer (10 mM Tris, pH 8.0, 10mM EDTA, 0.5mM EGTA, 0.25% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride) with rotation at 4°C for 15 minutes. and lysed cell extract resuspended in immunoprecipitation (IP) buffer (20mM Tris HCl, pH 8.0, 0.05% Deoxycholic acid, 0.5% Triton X-100, 0.5% NP-40, 200 mM NaCl, 1mM phenylmethylsulfonyl fluoride) and sonicated 3–5 times for 10 sec to reduce DNA fragment length to approximately 500–2000 bp, as verified by Agarose Gel electrophoresis. Supernatants were collected and immunocleared with salmon sperm DNA/protein A agarose slurry (upstate) for 1 hr at 4°C. One tenth of the supernatant was saved to quantitate the amount of input DNA present in each sample before immunoprecipitation.
Immunoprecipitation was performed overnight with agitation at 4°C with specific antibodies 5μg/500 μg of total lysate. Precipitates then were washed sequentially in low salt IP buffer, IP buffer supplemented with 0.5M NaCl and then final LiCl wash buffer (Orlando, Strutt et al. 1997). Beads were then washed twice in Tris-EDTA buffer and extracted twice with 1% sodium dodecyl sulfate buffer. Pooled eluates as well as saved chromatin solution for quantitating the amount of input DNA from above were heated at 65°C for overnight in 0.2 M NaCl solution to reverse the formaldehyde cross-linking. Diluted sample with TE to lower the concentration of SDS to 0.5% incubated at 45 C for 2 h with 20 μg proteinase K. DNA fragments were purified with phenol/chloroform/isoamyl alcohol (25:24:1) and ethanol precipitation and analyzed by PCR.
All of the ChIP experiments were repeated several times.
| S14: | Forward: 5′-CCAGAGGAACTGGGGTCAAGGGCC-3′ Reverse: 5′-CAGCCCTGACGTAGCGGAGGATAG-3′ |
| SERCA1: | Forward: 5′-GGCCTAAGGGCAAGAGGGCTTACG-3′ Reverse: 5′ CACCTGCCTGTTAACCTGGGCTCC- 3′ |
| GH: | Forward: 5′-CTCCTTGGAGAGGCTCTGTTGCCC-3′ Reverse: 5′-GGCTGGAGCCACTGACAGCTTGTG-3′ |
| GAPDH: | Forward: 5′ GTCAAGCTCCTACCATTCATGCTG-3′ Reverse: 5′CGGTCACCTCACACGGTGGGGTATC-3′ |
PCR was carried out in standard conditions with Taq polymerase. PCR primers used for analysis of ChIP experiments were:
Transfections
The following plasmids have been described previously. Mammalian expression vectors for TRs, GAL4-TRβ LBD, GAL4-N-CoR and Vp16-TRβ LBD (Feng, Ribeiro et al. 1998; Webb, Anderson et al. 2000; Velasco, Togashi et al. 2007). Reporter genes with two copies of IP-6, DR-4 and palindromic TREs upstream of a minimal promoter driving luciferase expression and the GAL responsive reporter comprising five GAL response elements linked upstream of a minimal promoter driving luciferase expression.
Cells were transfected by electroporation with 5μg of the respective reporter gene, 1ug of CMV-β-galactosidase internal control, and 1μg expression vectors for TR or various fusion proteins or empty vector control. Transfected cells were plated and induced overnight with ligand. Luciferase and β-galactosidase assays were performed as previously described (Webb, Anderson et al. 2000; Velasco, Togashi et al. 2007).
Gel Shifts
Binding of TR to TREs were assayed by mixing 20 fmols of 35S labeled TRs produced in a reticulocyte lysate, (TNT T7; Promega) with 10ng oligonucleotide and 1μg of poly(dI-dC) (Amersham Pharmacia Biotech) in a final volume of 20μl 1X binding buffer (c25mM HEPES, 50mM KCl, 1mM DTT, 10μM ZnSO4, 0.1% NP-40, 5% glycerol). After 30′ incubation, the mixture was loaded onto a 5% nondenaturing polyacrylamide gel that was pre-run for 30 min at 200 V and run at 4°C for 120 min at 240 V, in a running buffer of 6.7 mM Tris (pH 7.5), 1 mM EDTA, and 3.3 mM sodium acetate. The gel was then fixed, treated with Amplify (Amersham Pharmacia Biotech), dried and exposed for autoradiography. TRs used in assay were quantified with 125I-T3 binding assay and SDS-PAGE analysis of 35S-TRs.
Acknowledgments
This work was supported by NIH DK61468 and 51281 to JDB and NIH DK52798 to TSS. Dr. Baxter has proprietary interests in and serves as a consultant to Karo Bio AB, which has commercial interests in this area of research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barettino D, Vivanco Ruiz MM, et al. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. Embo J. 1994;13(13):3039–49. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter JD, Dillmann WH, et al. Selective modulation of thyroid hormone receptor action. J Steroid Biochem Mol Biol. 2001;76(1–5):31–42. doi: 10.1016/s0960-0760(01)00052-8. [DOI] [PubMed] [Google Scholar]
- Baxter JD, Goede P, et al. Structure-based design and synthesis of a thyroid hormone receptor (TR) antagonist. Endocrinology. 2002;143(2):517–24. doi: 10.1210/endo.143.2.8617. [DOI] [PubMed] [Google Scholar]
- Blaschke F, Takata Y, et al. A nuclear receptor corepressor-dependent pathway mediates suppression of cytokine-induced C-reactive protein gene expression by liver X receptor. Circ Res. 2006;99(12):e88–99. doi: 10.1161/01.RES.0000252878.34269.06. [DOI] [PubMed] [Google Scholar]
- Braverman LE, Utiger RD, editors. Werner’s and Ingbar’s The Thyroid: A Fundamental and Clinical Text. Philadelphia: Lippincott Williams & Wilkins; 2000. [Google Scholar]
- Buchholz DR, Hsia SC, et al. A dominant-negative thyroid hormone receptor blocks amphibian metamorphosis by retaining corepressors at target genes. Mol Cell Biol. 2003;23(19):6750–8. doi: 10.1128/MCB.23.19.6750-6758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10(1):33–9. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- Chiellini G, Apriletti JW, et al. A high-affinity subtype-selective agonist ligand for the thyroid hormone receptor. Chem Biol. 1998;5(6):299–306. doi: 10.1016/s1074-5521(98)90168-5. [DOI] [PubMed] [Google Scholar]
- Conen D, Melly L, et al. Amiodarone-induced thyrotoxicosis: clinical course and predictors of outcome. J Am Coll Cardiol. 2007;49(24):2350–5. doi: 10.1016/j.jacc.2007.02.054. [DOI] [PubMed] [Google Scholar]
- Davis PJ, Leonard JL, et al. Mechanisms of nongenomic actions of thyroid hormone. Front Neuroendocrinol. 2008;29(2):211–8. doi: 10.1016/j.yfrne.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Feng W, Ribeiro RC, et al. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280(5370):1747–9. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, et al. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25(1):57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14(2):121–41. [PubMed] [Google Scholar]
- Grover GJ, Dunn C, et al. Pharmacological profile of the thyroid hormone receptor antagonist NH3 in rats. J Pharmacol Exp Ther. 2007;322(1):385–90. doi: 10.1124/jpet.106.116152. [DOI] [PubMed] [Google Scholar]
- Guan HP, Ishizuka T, et al. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 2005;19(4):453–61. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins KJ, Liu S, et al. Vascular endothelial growth factor receptor-2 expression is down-regulated by 17beta-estradiol in MCF-7 breast cancer cells by estrogen receptor alpha/Sp proteins. Mol Endocrinol. 2008;22(2):388–402. doi: 10.1210/me.2007-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson MC, Astapova I, et al. The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J Biol Chem. 2005;280(8):6511–9. doi: 10.1074/jbc.M408972200. [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402(6757):93–6. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol. 2003;23(15):5122–31. doi: 10.1128/MCB.23.15.5122-5131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102(6):753–63. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Ki SH, Cho IJ, et al. Glucocorticoid receptor (GR)-associated SMRT binding to C/EBPbeta TAD and Nrf2 Neh4/5: role of SMRT recruited to GR in GSTA2 gene repression. Mol Cell Biol. 2005;25(10):4150–65. doi: 10.1128/MCB.25.10.4150-4165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet V, Gronemeyer H. The Nuclear Receptor Facts Book. London: Academic Press; 2002. [Google Scholar]
- Li J, Lin Q, et al. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 2002;16(6):687–92. doi: 10.1101/gad.962502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W, Nguyen NH, et al. A thyroid hormone antagonist that inhibits thyroid hormone action in vivo. J Biol Chem. 2002;277(38):35664–70. doi: 10.1074/jbc.M205608200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Xia X, et al. Thyroid hormone-regulated target genes have distinct patterns of coactivator recruitment and histone acetylation. Mol Endocrinol. 2006;20(3):483–90. doi: 10.1210/me.2005-0101. [DOI] [PubMed] [Google Scholar]
- Loinder K, Soderstrom M. Functional analyses of an LXXLL motif in nuclear receptor corepressor (N-CoR) J Steroid Biochem Mol Biol. 2004;91(4–5):191–6. doi: 10.1016/j.jsbmb.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Loinder K, Soderstrom M. An LXXLL motif in nuclear receptor corepressor mediates ligand-induced repression of the thyroid stimulating hormone-beta gene. J Steroid Biochem Mol Biol. 2005;97(4):322–7. doi: 10.1016/j.jsbmb.2005.06.031. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125(3):411–4. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Marimuthu A, Feng W, et al. TR surfaces and conformations required to bind nuclear receptor corepressor. Mol Endocrinol. 2002;16(2):271–86. doi: 10.1210/mend.16.2.0777. [DOI] [PubMed] [Google Scholar]
- Moore JM, Galicia SJ, et al. Quantitative proteomics of the thyroid hormone receptor-coregulator interactions. J Biol Chem. 2004;279(26):27584–90. doi: 10.1074/jbc.M403453200. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, et al. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13(24):3209–16. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NH, Apriletti JW, et al. Hammett analysis of selective thyroid hormone receptor modulators reveals structural and electronic requirements for hormone antagonists. J Am Chem Soc. 2005;127(13):4599–608. doi: 10.1021/ja0440093. [DOI] [PubMed] [Google Scholar]
- Nguyen NH, Apriletti JW, et al. Rational design and synthesis of a novel thyroid hormone antagonist that blocks coactivator recruitment. J Med Chem. 2002;45(15):3310–20. doi: 10.1021/jm0201013. [DOI] [PubMed] [Google Scholar]
- Oberste-Berghaus C, Zanger K, et al. Thyroid hormone-independent interaction between the thyroid hormone receptor beta2 amino terminus and coactivators. J Biol Chem. 2000;275(3):1787–92. doi: 10.1074/jbc.275.3.1787. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122(5):707–21. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Strutt H, et al. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11(2):205–14. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Ota Y, Mariash CN. Paradoxical triiodothyronine suppression of S14 transcription in permanent hepatic cell lines. Thyroid. 2003;13(5):437–45. doi: 10.1089/105072503322021098. [DOI] [PubMed] [Google Scholar]
- Pascual G, Glass CK. Nuclear receptors versus inflammation: mechanisms of transrepression. Trends Endocrinol Metab. 2006;17(8):321–7. doi: 10.1016/j.tem.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Paul BD, Buchholz DR, et al. Tissue- and gene-specific recruitment of steroid receptor coactivator-3 by thyroid hormone receptor during development. J Biol Chem. 2005;280(29):27165–72. doi: 10.1074/jbc.M503999200. [DOI] [PubMed] [Google Scholar]
- Paul BD, Fu L, et al. Coactivator recruitment is essential for liganded thyroid hormone receptor to initiate amphibian metamorphosis. Mol Cell Biol. 2005;25(13):5712–24. doi: 10.1128/MCB.25.13.5712-5724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, et al. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405–28. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Scanlan TS, Baxter JD, et al. International Patent. University of California; 1996. Thyroid hormone receptors and ligands. [Google Scholar]
- Sharma D, Fondell JD. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci U S A. 2002;99(12):7934–9. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn YC, Kim SW, et al. Dynamic inhibition of nuclear receptor activation by corepressor binding. Mol Endocrinol. 2003;17(3):366–72. doi: 10.1210/me.2002-0150. [DOI] [PubMed] [Google Scholar]
- Tian H, Mahajan MA, et al. The N-terminal A/B domain of the Thyroid Hormone Receptor-{beta}2 isoform Influences Ligand-dependent Recruitment of Co-activators to the Ligand Binding Domain. Mol Endocrinol. 2006 doi: 10.1210/me.2005-0437. [DOI] [PubMed] [Google Scholar]
- Tomita A, Buchholz DR, et al. Recruitment of N-CoR/SMRT-TBLR1 corepressor complex by unliganded thyroid hormone receptor for gene repression during frog development. Mol Cell Biol. 2004;24(8):3337–46. doi: 10.1128/MCB.24.8.3337-3346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco LF, Togashi M, et al. Thyroid hormone response element organization dictates the composition of active receptor. J Biol Chem. 2007 doi: 10.1074/jbc.M610700200. [DOI] [PubMed] [Google Scholar]
- Wagner RL, Apriletti JW, et al. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378(6558):690–7. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- Wang D, Simons SS., Jr Corepressor binding to progesterone and glucocorticoid receptors involves the activation function-1 domain and is inhibited by molybdate. Mol Endocrinol. 2005;19(6):1483–500. doi: 10.1210/me.2005-0012. [DOI] [PubMed] [Google Scholar]
- Webb P, Anderson CM, et al. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs) Mol Endocrinol. 2000;14(12):1976–85. doi: 10.1210/mend.14.12.0566. [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen NH, et al. Design of thyroid hormone receptor antagonists from first principles. J Steroid Biochem Mol Biol. 2002;83(1–5):59–73. doi: 10.1016/s0960-0760(02)00270-4. [DOI] [PubMed] [Google Scholar]
- Wilkinson JR, Towle HC. Identification and characterization of the AF-1 transactivation domain of thyroid hormone receptor beta1. J Biol Chem. 1997;272(38):23824–32. doi: 10.1074/jbc.272.38.23824. [DOI] [PubMed] [Google Scholar]
- Yang Z, Hong SH, et al. Transcriptional anti-repression. Thyroid hormone receptor beta-2 recruits SMRT corepressor but interferes with subsequent assembly of a functional corepressor complex. J Biol Chem. 1999;274(52):37131–8. doi: 10.1074/jbc.274.52.37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81(3):1097–142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- Yen PM, Feng X, et al. Effects of ligand and thyroid hormone receptor isoforms on hepatic gene expression profiles of thyroid hormone receptor knockout mice. EMBO Rep. 2003;4(6):581–7. doi: 10.1038/sj.embor.embor862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. Embo J. 2003;22(6):1336–46. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HG, Choi Y, et al. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol Cell Biol. 2005;25(1):324–35. doi: 10.1128/MCB.25.1.324-335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HG, Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol. 2006;20(5):1048–60. doi: 10.1210/me.2005-0324. [DOI] [PubMed] [Google Scholar]
- Yoshihara HA, Apriletti JW, et al. A designed antagonist of the thyroid hormone receptor. Bioorg Med Chem Lett. 2001;11(21):2821–5. doi: 10.1016/s0960-894x(01)00521-2. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–66. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]