Abstract
Recent advances in our structural understanding of telomerase and telomere-associated proteins have contributed significantly to elucidating the molecular mechanisms of telomere maintenance. The structures of telomerase TERT domains have provided valuable insights into how experimentally identified conserved motifs contribute to the telomerase reverse transcriptase reaction. Additionally, structures of telomere-associated proteins in a variety of organisms have revealed that, across evolution, telomere-maintenance mechanisms employ common structural elements. For example, the single-stranded 3′ overhang of telomeric DNA is specifically and tightly bound by an OB-fold in nearly all species, including ciliates (TEBP and Pot1a), fission yeast (SpPot1), budding yeast (Cdc13), and humans (hPOT1). Structures of the yeast Cdc13, Stn1, and Ten1 proteins demonstrated that telomere maintenance is regulated by a complex that bears significant similarity to the RPA heterotrimer. Similarly, proteins that specifically bind double-stranded telomeric DNA in divergent species use homeodomains to execute their functions (human TRF1 and TRF2 and budding yeast ScRap1). Likewise, the conserved protein Rap1, found in budding yeast, fission yeast, and humans, contains a structural motif that is known to be critical for protein-protein interaction. In addition to revealing the common underlying themes of telomere maintenance, structures have also elucidated the specific mechanisms by which many of these proteins function, including identifying a telomere-specific domain in Stn1 and how the human TRF proteins avoid heterodimerization. In this review, we summarize the high-resolution structures of telomerase and telomere-associated proteins and discuss the emergent common structural themes among these proteins. We also address how these high-resolution structures complement biochemical and cellular studies to enhance our understanding of telomere maintenance and function.
Keywords: telomere, telomerase, shelterin, tRPA, OB fold, TPP1, POT1, RAP1, TRF1, TRF2, TRFH, TERT, TRBD, TEN, RCT, TIN2, Apollo, Sir3, Taz1, Cdc13, Stn1, Ten1, TEBP
Introduction
Telomeres are nucleoprotein complexes required for chromosomal stability. They shield the ends of linear chromosomes from recognition by DNA damage machinery and provide a solution to the end-replication problem through the action of the reverse transcriptase telomerase (de Lange, 2009; Greider and Blackburn, 1987). Protection of the chromosomal end is conferred by essential protein complexes that prevent the severe and lethal consequences of a cellular response to exposed DNA ends, including chromosomal end-to-end fusions and nucleolytic processing (de Lange, 2009). Additionally, the limitations of semiconservative DNA replication result in gradual telomere shortening, limiting the number of cell divisions as short telomeres trigger cellular senescence (Harley et al., 1990; Hayflick, 1979; Lundblad and Szostak, 1989). This limitation can be circumvented by the telomerase-mediated extension of telomeric DNA, as observed in unicellular eukaryotic organisms and proliferative metazoan cells (Bodnar et al., 1998; Lundblad and Szostak, 1989; Yu et al., 1990). Both the end-protection and telomerase activities are tightly controlled (de Lange, 2009), and their dysregulation is associated with several human diseases (Armanios, 2009; Calado and Young, 2009; Garcia et al., 2007).
The sequences of telomere-associated proteins diverge rapidly (Linger and Price, 2009), confounding our ability to identify the unifying themes underlying telomere maintenance. Fortunately, the high-resolution structures of telomere-associated factors have revealed the repeated use of common structural elements with some intriguing elaborations. In particular, some apparently divergent telomere-associated proteins in distantly related species share folds, while others are similar to well-characterized non-telomeric proteins, suggesting the evolution of telomere-specific function. These structures, both through their similarities and their differences, provide direction for biochemical and cellular studies that aim to define the mechanisms of action of telomere-associated factors. In this review, we will discuss the emergent common structural themes in telomere-associated proteins and describe how these high-resolution structures have enhanced our understanding of telomere maintenance and function.
Telomerase reverse transcriptase
The ends of linear chromosomes terminate in G-rich single-stranded 3′ overhangs (Klobutcher et al., 1981; Larrivee et al., 2004; McElligott and Wellinger, 1997; Moyzis et al., 1988; Shampay et al., 1984; Wright et al., 1997). In vitro, telomeric single-stranded DNA (ssDNA) readily forms higher order G-quadruplex structures amenable to high-resolution characterization (Burge et al., 2006; Neidle and Parkinson, 2003), although these structures have only been reported in vivo at ciliate telomeres (Lipps and Rhodes, 2009; Paeschke et al., 2005). Chromosome ends also form large DNA duplex “t-loops”, where the ssDNA overhang invades the duplex region to form a structure similar to a recombination intermediate, which may represent the higher-order structure in vivo (de Lange, 2004; Griffith et al., 1999; Munoz-Jordan et al., 2001), but they have not yet been amenable to high-resolution structural studies in vitro.
Telomerase catalyzes the addition of telomeric repeats onto this 3′ overhang using an integrated ribonucleoprotein complex that consists of a reverse transcriptase protein (TERT) and a large RNA component (TR) which provides the template sequence for the telomeric repeat (Greider and Blackburn, 1989; Lingner et al., 1997). Within the family of reverse transcriptases, telomerase is anomalous in that its RNA component is a constitutive component of the transcriptase with multiple functions (Autexier and Lue, 2006). TERT and TR associate with additional proteins in vivo to form a functional holoenzyme (Lendvay et al., 1996; Lingner and Cech, 1996; Venteicher et al., 2009; Witkin and Collins, 2004; Witkin et al., 2007). Although the structure of an intact telomerase holoenzyme has yet to be solved, structures of individual telomerase and TR subdomains have informed our understanding of its organization and the mechanisms of its unique reverse transcriptase activity (Kelleher et al., 2002). Here, we limit our discussion to structures of TERT, as the structures of domains within telomerase RNA have been expertly reviewed elsewhere (Zhang et al., 2011a).
TERTs contain a reverse transcriptase domain (RT) that possesses the canonical RT motifs 1, 2, A, B′, C, D, and E, including the three invariant catalytic aspartate residues (Lingner et al., 1997). Three additional domains fully define a TERT protein: a “telomerase essential N-terminal” domain (TEN); a telomerase RNA binding domain (TRBD); and a C-terminal extension (CTE) (Fig. 1A) (Blackburn, 2005; Bryan and Cech, 1999; Wyatt et al., 2010). Much of our current structural understanding of TERT has come from the Tetrahymena thermophila TRBD and TEN domain structures and the structure of the putative TERT from the flour beetle Tribolium castaneum (TcTERT). TcTERT is somewhat removed from other known telomerases; it lacks the TEN domain that is present in all other known telomerases, the RNA component has not been identified, and as yet there is no in vivo evidence for telomerase-mediated telomere maintenance in this organism (Osanai et al., 2006). However, TcTERT is clearly a reverse transcriptase that contains telomerase-specific sequence motifs (see below), which suggests that, if not an active telomerase, it may be an evolutionary intermediate. TcTERT thus serves as a judicious starting point for understanding the structural framework of telomerase action while we await a high-resolution structure of an intact TERT+TR holoenzyme.
Figure 1. Telomerase enzymes are telomere-specific reverse transcriptases.
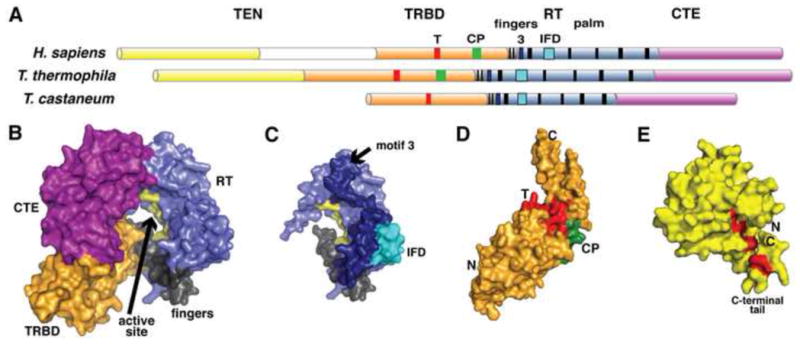
(A) Telomerase domain topology (TEN, yellow; TRBD, orange; fingers, gray; RT, blue; CTE, magenta) with conserved motifs (T, green; CP, red; IFD, cyan; and left to right in black, motifs 1, 2, 3, and the canonical RT motifs A, B′, C, D, E, and F, black). (B) Surface representation of T. castaneum TERT. Domains are labeled and colored as in (A), with the active site residues in yellow (PDB: 3DU6). (C) The T. castaneum RT domain is shown in light blue, showing the locations of the telomerase-specific motifs 3 (dark blue) and IFD (cyan). As in (B), the active site residues are in yellow. (D) The T. thermophila TRBD domain is shown as a surface representation, with the T motif (red) and CP motif (green) highlighted (PDB: 2R4G). (E) The T. thermophila TEN domain is also shown as a surface (PDB: 2B2A). Mutations that alter nucleotide binding are shown as red sticks and red surface lining the pocket formed by the C-terminal tail, which is labeled. All structures were modeled using the PyMol Molecular Graphics System, Version 1.3 Schrödinger, LLC (Schrodinger, 2010).
TcTERT forms a ring-like structure with the TRBD making considerable contact with the CTE even though they are separated in primary sequence by the RT domain (Gillis et al., 2008) (Fig. 1A,B). The RT folds into a palm-and-fingers organization reminiscent of other reverse transcriptases (Das and Georgiadis, 2004; Rodgers et al., 1995). The active site is in the palm of the RT, and contains universally catalytic aspartates and a Mg2+ ion (Gillis et al., 2008; Mitchell et al., 2010a) (Fig. 1B). The CTE curls around to contact the TRBD and complete the ring (Fig. 1B). A co-crystal of TcTERT and a hybrid RNA/DNA hairpin shows that, analogous to retroviral reverse transcriptases, the nucleic acid docks in the center of the ring, contacting elements from the TRBD, RT, and CTE (Mitchell et al., 2010a). The TcTERT structure also reveals the context of two previously characterized telomerase-specific motifs in the RT palm that affect activity: motif 3 and IFD (Fig. 1A,C). Motif 3 is located between motifs 2 and A in the primary sequence (Xie et al., 2010) (Fig. 1A). In TcTERT, motif 3 forms two helices on the RT surface adjacent to the active site (Fig. 1C). IFD is an insertion between the A and B′ motifs, originally named the “insertion in the fingers domain” based on mapping to the HIV-1 structure (Lue et al., 2003) (Fig. 1A,C). The TcTERT IFD is part of the solvent-exposed surface on the outside of the RT domain (Fig. 1C), where it likely affects the active site through a helix that interacts with the IFD on one side and with the RNA substrate in the active site on the other (Lue et al., 2003).
The specialized RNA-binding activity of TERT is conferred by the essential and highly conserved TRBD, which uses the telomerase-specific T and CP motifs to recognize essential elements of TR (Bryan et al., 1998; Nakamura et al., 1997; O’Connor et al., 2005). TRBD structures are available in isolation from T. thermophila and within the full-length T. castaneum TERT. These structures are very similar; both consist of two asymmetrical helical lobes connected by a β-hairpin hinge region (Gillis et al., 2008; Rouda and Skordalakes, 2007) (Fig. 1D). This novel topology places the phylogenetically conserved T and CP motifs adjacent on the hinge surface (Lai et al., 2001; Rouda and Skordalakes, 2007) (Fig. 1D). The details of RNA binding will require a co-crystal structure, although mutagenesis suggests that RNA binding employs hydrophobic interactions within the conserved T and CP motifs. Full understanding of nucleic acid recognition will also require analysis of the structurally uncharacterized N-terminal region of the T. thermophila TRBD (residues 195–253), which was found to be required for biochemical activity (Lai et al., 2001) but was not contained in the structurally characterized construct (residues 254–519) (Rouda and Skordalakes, 2007).
Telomerases are additionally distinguished from canonical reverse transcriptases by the TEN domain, which is essential for telomerase activity in vivo and in vitro (Bryan et al., 2000; Friedman and Cech, 1999; O’Connor et al., 2005; Xia et al., 2000). The TEN domain binds both TR and the telomeric ssDNA substrate, and is critical for the telomerase-specific repeat-addition processivity (RAP) activity (Moriarty et al., 2004; Zaug et al., 2008). The T. thermophila TEN domain adopts a novel protein fold, consisting of N-terminal and C-terminal subdomains ending in a C-terminal tail (Jacobs et al., 2006) (Fig. 1E, key tail residues highlighted in red). The structure was used to direct mutagenesis experiments that identified the C-terminal tail (residues 177–191) as essential for RNA binding and weak binding of telomeric ssDNA primers (Jacobs et al., 2006) (Fig. 1E). Protein flexibility may contribute to nucleic acid binding, as deletion of the disordered C-terminal tail of TEN compromises its ability bind RNA (Jacobs et al., 2006).
Telomere-Associated Proteins
Our knowledge of telomere organization is being built from the ground up, focusing on the structural characterization of individual proteins and subcomplexes from a range of organisms (Fig. 2). This review focuses on the structurally characterized protein domains and their complexes of factors whose primary cellular role is linked to telomere maintenance. Metazoan telomeres contain the six-member shelterin complex (Palm and de Lange, 2008) (Fig. 2), comprised of the double-stranded binding proteins TRF1 and TRF2, which bind telomeric dsDNA as homodimers and interact with RAP1 and TIN2. TIN2 interacts with hTPP1, which in turn binds the telomeric ssDNA binding protein, hPOT1 (Fig. 2). Fission yeast also employ a shelterin-like complex, comprised of a single TRF1/2 homolog, Taz1, which interacts with a Rap1 homolog. This Rap1 also interacts with Poz1, which then binds the hTPP1-hPOT1 homologs SpTpz1-SpPot1 (de Lange, 2009; Miyoshi et al., 2008) (Fig. 2). Budding yeast appear to use a distinct mechanism of telomere maintenance through the Rap1/Rif1/Rif2 complex, which binds telomeric dsDNA, and a telomere-specific RPA-like complex containing Cdc13/Stn1/Ten1 (t-RPA), which binds telomeric ssDNA (Shore and Bianchi, 2009) (Fig. 2). Fission yeast and metazoan telomeres also employ an RPA-like complex (Martín et al., 2007; Miyake et al., 2009; Song et al., 2008). While the double-stranded DNA-binding factors in ciliates have not been reported, O. nova amitotic macronuclear telomeres employ the heterodimeric complex TEBP (Gottschling and Zakian, 1986), while T. thermophila contains a Pot1/TPP1-like complex (Jacob et al., 2007; Linger et al., 2011).
Figure 2. Telomere-associated protein complexes in different species.
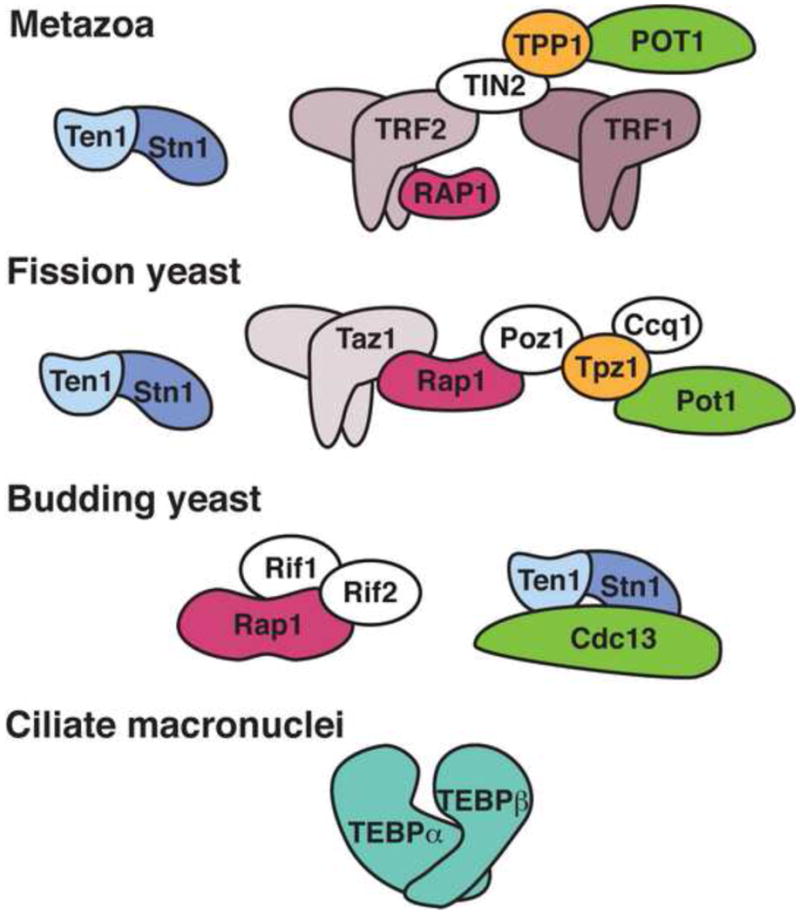
The proteins that are shown in color indicate that high-resolution structural data are available for either that protein or a close homolog. Metazoans: shelterin complex of TRF1, TRF2, RAP1, TIN2, hTPP1, and hPOT1; RPA-like proteins Stn1 and Ten1. Fission yeast: shelterin-like complex of Taz1, Rap1, Poz1, Tpz1, Ccq1, and Pot1; also present are Stn1 and Ten1. Budding yeast: dsDNA-binding complex of Rap1, Rif1, and Rif2; ssDNA-binding complex of Cdc13, Stn1, and Ten1. Ciliate macronuclei: TEBPα/β.
Single-stranded DNA binders and their complexes
High-resolution structures have revealed similarities among the telomere end-protection (TEP) proteins that were not predicted from their primary sequences. TEP proteins universally bind the ssDNA overhang using the oligosaccharide/oligonucleotide/oligopeptide binding (OB) fold, a common Greek key motif in ssDNA and RNA binding proteins (Theobald et al., 2003). Since ssDNA is present throughout the genome during replication, TEP proteins must discriminate between telomere and non-telomere sequence. Some TEPs execute this exquisite specificity while also accommodating degeneracy or variable spacer sequences within the telomeric repeats. All of this is accompanied by very high binding affinities, with unusually tight KD values often in the low pico- to nanomolar range (Croy and Wuttke, 2006). The high-resolution structures of several TEP domains have provided insight into how OB folds perform these myriad duties simultaneously.
OnTEBP
The first high-resolution TEP structures were of the heterodimeric telomere end-binding protein complex (TEBP) from the hypotrichous ciliate Oxytricha nova (“On”, now called Sterkiella nova (Foissner and Berger, 1999)). TEBP consists of two subunits, α and β, which bind tenaciously to O. nova macronuclear chromosomal termini as a heterodimeric complex specific for T4G4 repeats (Gottschling and Zakian, 1986; Gray et al., 1991; Horvath et al., 1998). TEBPα consists of two OB folds in an N-terminal domain (OB1 and OB2) and a third OB fold as a C-terminal domain (OB3) (Horvath et al., 1998), while TEBPβ is comprised of a single N-terminal OB fold (OB4), a central globular domain, and a lysine-rich unstructured tail (CTD) (Buczek and Horvath, 2006; Horvath et al., 1998) (Fig. 3A,B). This first TEP structure unveiled new mechanisms for OB fold binding and set the benchmark for how this protein family functions. Notably, OB1 and OB2 were found to form a single extended ssDNA-binding surface that cooperates with OB4 to form the complete binding pocket. Additionally, the canonical ligand-binding site in OB3 interacts with the globular domain of TEBPβ (Horvath et al., 1998) (Fig. 3B). This discovery expanded the set of known OB fold ligands to include oligopeptides as well as oligonucleotides and oligosaccharides (Horvath et al., 1998).
Figure 3. OnTEBP proteins bind ciliate telomeric ssDNA.
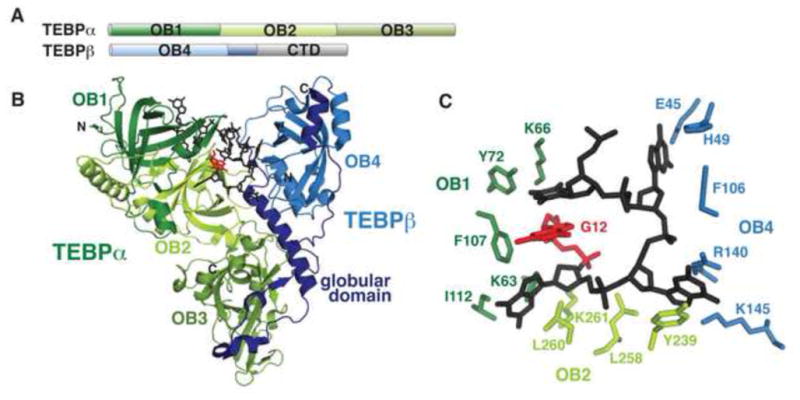
(A) Domain topology of O. nova TEBP proteins. Four OB folds are present, as well as a structurally uncharacterized C-terminal domain (CTD) in TEBPβ. (B) TEBPα OB1-3 (green) forms a complex with TEBPβ (blue) to bind ssDNA ligand (black; PDB: 2I0Q). The TEBPβ globular domain that is bound by OB3 is in dark blue. The 3′ base, G12, is colored red and is fully buried in the groove between the protein subunits. (C) OB1, OB2, and OB4 make critical contacts with the 3′ loop of the ssDNA ligand to sequester the bases from solvent.
The TEBPαβ heterodimer and ssDNA co-fold into a stable complex, with the ssDNA forming a loop within a groove formed by the N-terminal OB folds of TEBPα and the globular domain of TEBPβ (Fig. 3B). The nucleotide bases are generally buried, with the 3′ end completely solvent inaccessible, and make extensive contacts with amino acid side chains through a chemically diverse range of interactions, including aromatic stacking, hydrophobic interactions, hydrogen-bonding, and electrostatic interactions, with electrostatics contributing little to the thermodynamics of binding (Horvath et al., 1998) (Fig. 3C). The occlusion of the bases and the 3′ end established a physical basis for sequence-specific binding and end protection (Horvath et al., 1998). The number and diversity of nucleotide-protein contacts provides a mechanism for the exquisite specificity of TEP proteins for their cognate ligands. As TEBPα can also bind ssDNA independently, a model emerged in which TEBPα coats ssDNA with a terminal TEBPβ binding event to form a stable complex that caps the very end of the chromosome (Classen et al., 2001; Peersen et al., 2002).
Structures of TEBPαβ bound to a panel of noncognate ligands showed that OnTEBPαβ accommodates sequence variation using modest changes in side chain conformation and dramatic shifts in nucleic acid binding register to retain key specificity contacts (Theobald and Schultz, 2003). The ligand rearrangement explains why profound nucleotide sequence alterations caused less than a 10-fold change in affinity (Theobald and Schultz, 2003). Such nucleotide shuffling may be a primary mechanism by which TEP proteins bind variable 3′ overhangs; evidence for similar ligand accommodation is present in fission and budding yeast, although those mechanisms are currently structurally undefined.
Pot1 and hTPP1
Weak sequence identity between TEBPα OB1 and an uncharacterized S. pombe protein led to the identification of Pot1 (protection of telomere-1) in fission yeast and humans (Baumann and Cech, 2001). Pot1 binds telomeric ssDNA as part of shelterin, and is conserved in eukaryotes from S. pombe (SpPot1) to humans (hPOT1) (Croy and Wuttke, 2006). Deletion of Pot1 from either S. pombe or vertebrate cells is catastrophic, resulting in telomeric instability, chromosomal end-to-end fusions, and cell death (Baumann and Price, 2010). Like TEBPα, Pot1 is comprised of an N-terminal DNA-binding domain (DBD) and a C-terminal protein-protein interaction domain (Fig. 4A). Also like TEBPα, Pot1 binds an OB-fold containing partner protein (hTPP1/SpTpz1) (Liu et al., 2004; Miyoshi et al., 2008; Ye et al., 2004).
Figure 4. Pot1 and hTPP1.
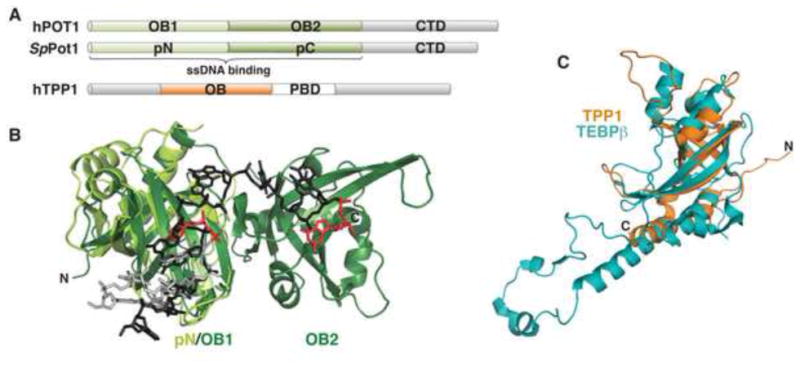
(A) Domain topology of hPOT1, SpPot1, and hTPP1 proteins. Following the tandem OB folds that comprise the DNA-binding domain, Pot1 contains a C-terminal protein-protein interaction domain (CTD), and hTPP1 contains a central Pot1-binding domain (PBD). (B) Superposition of SpPot1pN (bright green; PDB: 1QZH) on hPOT1-DBD (dark green; rmsd = 1.09 Å; PDB: 3KJP). ssDNA ligands are modeled as sticks, with 6mer bound to SpPot1pN (light gray) and 12mer bound to hPOT1-DBD (dark gray). The 3′ bases of both ligands are shown in red. (C) hTPP1-OB (orange; PDB: 2I46) superimposed on TEBPβ (teal; PDB: 2I0Q; rmsd =2.0 Å).
Human hPOT1 and hTPP1
The hPOT1-DBD comprises residues 1–340, and has been co-crystallized with ssDNA (Lei et al., 2004; Nandakumar et al., 2010) (Fig. 4B, dark green and dark gray, respectively). The DBD is comprised of tandem OB folds (OB1 and OB2). OB1 was predicted by a variety of sequence alignments, but OB2 was only weakly predicted using a profile-based sequence analysis (Theobald and Wuttke, 2004). The DNA adopts an extended conformation and makes specific contacts with both OB folds, where hydrogen bonds between amino acid side chains and nucleotide bases provide a structural basis for the biochemically observed nucleotide specificity preference for the 5′ end of the ligand and the terminal 3′ guanine (Lei et al., 2004) (Fig. 4B).
hPOT1 is localized to the larger multimeric shelterin complex through the interaction of its C-terminal domain with a central domain in hTPP1 (Hockemeyer et al., 2007; Xin et al., 2007; Ye et al., 2004). hTPP1 also contains an N-terminal OB fold that bears striking resemblance to that of TEBPβ (residues 90–250) and an as-yet uncharacterized C-terminal hPOT1-interaction domain (Wang et al., 2007). hTPP1 additionally enhances hPOT1 affinity by 10-fold (Gray et al., 1991; Wang et al., 2007), and also refines hPOT1 discrimination against ribonucleic acids (Nandakumar et al., 2010). Although the isolated domains of hPOT1/hTPP1 closely correspond structurally to those of TEBPαβ, the two complexes exhibit somewhat different biochemical behavior (Fig. 4C). The DNA-binding domains of TEBPα and hPOT1 alone exhibit modest preference for the nature of the 3′ base (Classen et al., 2003; Lei et al., 2004), and the structures reveal a partially buried 3′OH. In contrast, TEBPαβ completely buries the 3′G (Horvath et al., 1998). While no structure is yet available of the hPOT1/hTPP1 complex, biochemically the 3′ end requirement is relaxed (Wang et al., 2007). Both the TEBPα and TEBPαβ complexes inhibit telomerase activity, presumably through steric occlusion of the 3′ end (Froelich-Ammon et al., 1998). Similarly, when localized to the 3′ end, hPOT1 alone inhibits human telomerase in vitro (Lei et al., 2005). However, when bound coincidentally with hPOT1, hTPP1 triggers recovery of telomerase activity, behaving as a telomerase processivity factor by decreasing the rate of primer dissociation (Abreu et al., 2010; Latrick and Cech, 2010; Tejera et al., 2010; Wang et al., 2007; Xin et al., 2007). The structural similarity and functional divergence of hTPP1 and TEBPβ is a prime example of the species-specific adaptations that are characteristic of telomere maintenance complexes throughout phylogeny.
S. pombe Pot1
Like hPOT1, S. pombe Pot1 is also comprised of a DBD (1–389) and a C-terminal domain (Croy et al., 2006) (Fig. 4A). Although sequence homology is limited to the N-terminal OB fold, protein threading models and biochemical data strongly suggest that the DBD, like hPOT1, contains two tandem OB folds, SpPot1pN and SpPot1pC (Croy et al., 2006; Croy et al., 2009). Only SpPot1pN has been structurally characterized to date (Fig. 4B), and as the first Pot1 structure to be elucidated, it provided initial insight into how Pot1 specifically recognizes ssDNA (Lei et al., 2003). SpPot1pN superpositions well on hPOT1-OB1, with the central ligand bases overlaying tightly with more divergence at the 3′ and 5′ ends. Stacking interactions between aromatic amino acid on the protein and bases of the ssDNA are largely conserved (Fig. 4B). Solution dynamics analysis of the free and bound states of SpPot1pN suggests that specificity is achieved by a conformational selection mechanism where residues involved in forming the specific contacts experience dynamics that are quenched upon binding (Croy and Wuttke, 2009; Croy et al., 2008).
In contrast to TEBPα and hPOT1, the SpPot1 OB folds exhibit independent DNA-binding activities, allowing for characterization of how these domains work in concert to perform sequence-specific recognition of DNA. SpPot1pN binds a single S. pombe telomeric repeat, d(GGTTAC), while SpPot1pC minimally binds to one and a half repeats (Croy et al., 2009; Lei et al., 2002). These binding activities are decoupled in the intact SpPot1-DBD, which binds the combined two-and-a-half repeat ssDNA with low picomolar affinity (Croy et al., 2009). Interestingly, SpPot1-DBD can also bind ssDNA of two repeats with identical affinity but with different specificities and tolerance for substitution as the longer ligand (Altschuler et al., 2011). These biochemical observations suggest that SpPot1 is capable of remarkable conformational plasticity and ligand accommodation. Supporting this hypothesis, structures of SpPot1pN complexed with non-cognate ligands identified novel ligand conformations that nonetheless bound with similar thermodynamic parameters (Croy et al., 2008). As the core S. pombe telomere sequence repeats are often separated by a variable number of nucleotides (Trujillo et al., 2005), the ability to accommodate alternate telomeric sequences with minimal thermodynamic impact may be an essential element of SpPot1 function.
RPA-like complexes
An additional widely conserved complex also contributes to telomere function. This complex, first discovered in S. cerevisiae, is composed of three essential proteins Cdc13, Stn1, and Ten1 and impacts several aspects of telomere maintenance (Grandin et al., 2001; Grandin et al., 1997; Nugent et al., 1996) (Fig. 5). Cdc13 positively regulates telomere length by recruiting telomerase (Bianchi et al., 2004; Nugent et al., 1996; Pennock et al., 2001), and all three proteins are also genetically implicated in negative length regulation (Chandra et al., 2001; Grandin et al., 2001; Grandin et al., 1997; Qi and Zakian, 2000). Cdc13 specifically binds yeast telomeric ssDNA with 300 pM affinity through its DBD, which is a single OB fold with no sequence similarity to TEBP or Pot1 proteins (Anderson et al., 2002; Nugent et al., 1996) (Fig. 5A). The isolated DBD binds more tightly, with 3 pM affinity, using an unusually long, structured loop between β2 and β3 (L2–3) to extend the binding interface (Eldridge and Wuttke, 2008; Mitton-Fry et al., 2002; Mitton-Fry et al., 2004). As seen in TEBP and Pot1, aromatic stacking and hydrophobic interactions mediate recognition of nucleotide bases and electrostatics stabilizing the backbone phosphates (Anderson et al., 2003; Mitton-Fry et al., 2002). As observed in the Pot1 proteins, the most critical protein-DNA contacts are located in the 5′ end of the ligand, where these contacts define the highly sequence-specific binding behavior of Cdc13 (Anderson et al., 2003; Eldridge et al., 2006) (Fig. 5A).
Figure 5. Budding yeast Cdc13, Stn1, and Ten1.
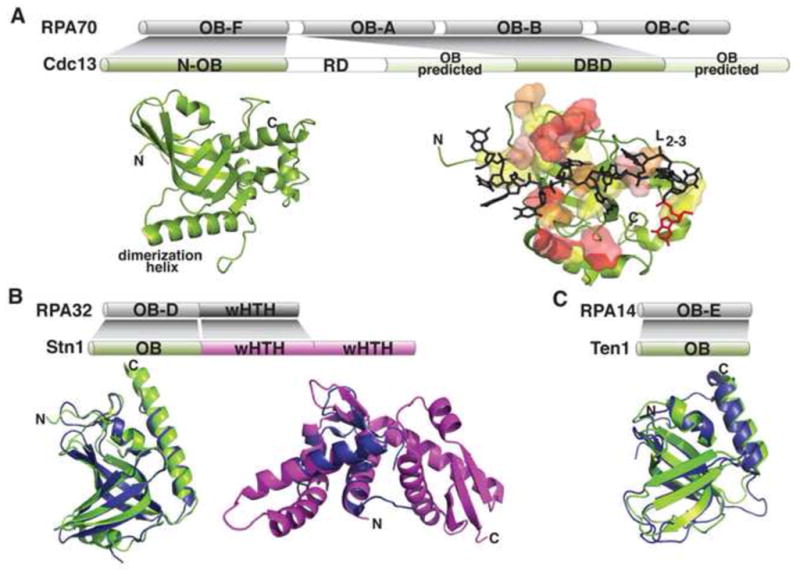
(A) Cdc13 domain topology and subdomain structures compared to RPA70. N-OB is in green, with the dimerization helix labeled (PDB: 3NWS). The DBD is shown as a cartoon, with superimposed transparent surfaces of the residues involved in DNA binding in increasing thermodynamic contribution from yellow to red (PDB: 1S40). The 11mer ligand is shown as sticks (gray) with the 3′ base in red. (B) Stn1 domain topology and structures compared to RPA32. Left, the S. pombe Stn1-N (green) is superimposed on RPA32-N (blue; rmsd = 1.6Å; PDB: 3KF6 and 1QUQ, respectively); right, the S. cerevisiae Stn1-C (magenta) is superimposed on RPA32-C (blue; rmsd = 1.9Å; PDB: 3K10 and 1DPU, respectively). (C) Ten1 domain topology and structure compared to RPA14. The S. pombe Ten1 (green) is superimposed on RPA14 (blue; rmsd =1.96Å; PDB 3K0X and 1QUQ, respectively).
Exciting insights into the function of this complex came from the proposal that Cdc13, Stn1, and Ten1 form a telomere-specific RPA-like heterotrimer (t-RPA) (Gao et al., 2007) (Fig. 5), based on sequence analysis of Cdc13, Stn1, and Ten1 and in vivo domain swapping (Gao et al., 2007; Theobald and Wuttke, 2004). This was a paradigm-shifting idea, as RPA nonspecifically binds to ssDNA throughout the genome but Cdc13 specifically recognizes telomeric ssDNA. Structural data have been central to refining this hypothesis (Fig. 5). Like RPA70, Cdc13 has an OB fold in the extreme N-terminus (N-OB), but unlike RPA70, this domain contains a long helix that mediates dimerization (Mitchell et al., 2010b; Sun et al., 2011) (Fig. 5A). The N-OB has been alternately proposed to bind either ssDNA or DNA polymerase α (Mitchell et al., 2010b; Sun et al., 2011). In addition to the N-OB and DBD domains, two additional OB folds flanking the DBD have been predicted (Sun et al., 2011; Theobald and Wuttke, 2004). The demonstrated presence of four OB folds in Cdc13 would conclusively establish a domain organization analogous to RPA70 (Fig. 5A). However, several elaborations confer the unique telomere functions of Cdc13, namely that the DBD is a single OB fold with specificity for telomeric DNA, the observation of dimerization, and the presence of a telomerase regulatory domain (RD) within the N-terminus (Fig. 5A).
Structures of Stn1 and Ten1 domains revealed that they closely mimic their RPA counterparts (Fig. 5B,C). The N-terminal domains of Stn1 (Stn1-N) from the divergent yeasts S. pombe and Candida tropicalis are OB folds that superimpose on the RPA32 OB fold (Sun et al., 2009) (Fig. 5B). The C-terminal domain (Stn1-C) from S. cerevisiae is comprised of tandem winged helix-turn-helix (wHTH) motifs (Gelinas et al., 2009; Sun et al., 2009). The N-terminal wHTH superimposes on the lone wHTH of RPA32 (Fig. 5B), while in vivo studies identified a telomere-specific function for the C-terminal wHTH in negative telomere length regulation (Gelinas et al., 2009). Like RPA14, both the S. pombe and C. tropicalis Ten1proteins are single OB folds (Gelinas et al., 2009; Sun et al., 2009) (Fig. 5C). Furthermore, RPA32/14 and Stn1-N/Ten1 form stable heterodimers in vitro (Bochkarev et al., 1999; Gao et al., 2007; Sun et al., 2009). The presence of multiple OB folds in Cdc13 and the remarkable structural identity between Stn1/Ten1 and RPA32/14 strongly support the hypothesis that Cdc13, Stn1, and Ten1 are RPA-like proteins (Fig. 5).
Although the RPA-like telomere proteins have been studied most extensively in budding yeast, a heterotrimeric complex composed of the well-conserved Stn1 and Ten1 proteins and a less conserved DNA-binding large subunit contributes to telomere function in a diverse set of species, including fission yeast, plants, and mammals (Casteel et al., 2009; Martín et al., 2007; Miyake et al., 2009; Surovtseva et al., 2009). In fission yeast, stn1-Δ and ten1-Δ strains exhibit the same phenotype as a Pot1 knockout, with rapid telomere degradation accompanied by high levels of inviability (Martín et al., 2007). Knockout of a Stn1 homolog in Arabidopsis, AtStn1, also exhibits a telomere shortening phenotype and severe morphological defects (Song et al., 2008). However, the rapid divergence of the large subunit has confounded the analysis of this complex. Even Cdc13 proteins from other yeast do not retain the level of telomere-specific DNA binding observed for S. cerevisiae Cdc13 (Mandell et al., 2011). Structural studies will clearly play a major role in the refinement of these models, including high-resolution structures of multimeric complexes and structure-directed in vivo mutagenesis.
Double-stranded DNA binders and their complexes
A separate set of factors specifically binds double-stranded telomeric DNA, where they function in chromosomal protection and telomere-length regulation (de Lange, 2009; Shore and Bianchi, 2009) (Fig. 2). These proteins share common folds despite considerable sequence variability and differences in domain topology. As observed in the ssDNA-binding proteins, dsDNA-protein interactions and protein-protein interactions at the telomere in evolutionarily divergent species employ similar structural solutions to execute telomere-specific functions.
ScRap1
S. cerevisiae telomeric dsDNA is bound by ScRap1, which interacts with the proteins Rif1 and Rif2 to regulate telomere length (Shore and Bianchi, 2009). ScRap1 also mediates gene silencing both at the telomere and at mating-type loci through interactions with Sir3 and Sir4 (Moretti and Shore, 2001). The DNA-binding domain (DBD; residues 361–596), the BRCT domain (residues 6–102), and the C-terminal protein-protein interaction domain (CTD; residues 672–827) have been structurally characterized, revealing the physical bases for these interactions and providing functional insights (Fig. 6A).
Figure 6. Domains of telomeric dsDNA binding proteins.
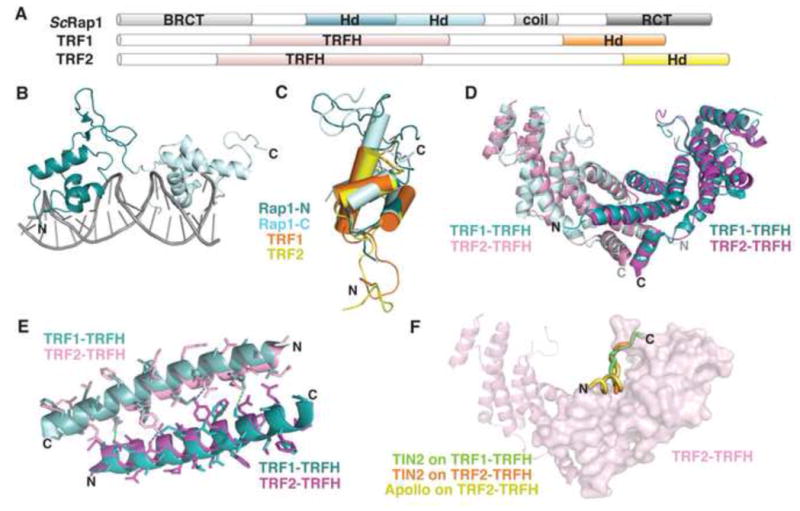
(A) Domain topology of TRF1, TRF2, and ScRap1. (B) ScRap1 binds to double-stranded telomeric DNA with two homeodomains (N-terminal, teal; C-terminal, light blue; PDB: 1IGN). (C) Overlay of the homeodomains from ScRap1 (ScRap1-N, teal; ScRap1-C, light blue; PDB: 1IGN), TRF1-DBD (orange; PDB: 1ITY), and TRF2-DBD (yellow; PDB: 1VF9). (D) Superposition of the TRF1-TRFH homodimer (aqua and light blue; PDB: 1H6O) with the TRF2-TRFH homodimer (pink and magenta; PDB: 1H6P; rmsd =1.23Å). (E) Superposition of the α1 helices that comprise part of the homodimerization interface. TRF1, teal and light blue; TRF2, pink and magenta. Dashes indicate hydrogen bonding between monomers (TRF1, green; TRF2, blue). (F) Superposition of three peptide ligands on the structure of the TRF2-TRFH homodimer. TIN2 peptide bound to TRF1, green (PDB: 3BQO); TIN2 peptide bound to TRF2, orange (PDB: 3BU8); Apollo peptide bound to TRF2, yellow (PDB: 3BUA). For simplicity, only one peptide binding site is shown on the surface of one monomer in the homodimer.
The structure of ScRap1-DBD was seminal for the field, and demonstrated that DNA binding activity is carried out by two homeodomains, each comprised of an N-terminal arm and a Myb-like three-helical bundle that includes a recognition helix, followed by a C-terminal tail (Konig et al., 1996) (Fig. 6B). The N-terminal arms of each homeodomain make specific contacts within the minor groove and nonspecific contacts on the backbone of the dsDNA, and the recognition helix lies within the major groove. The tandem homeodomains wrap around the dsDNA with a separation of 8 basepairs (Fig. 6B). The C-terminal tail contacts major groove bases and the recognition helix of the first homeodomain as it wraps back around the DNA to fully enclose the substrate (Konig et al., 1996). Specificity is mediated through direct and water-mediated hydrogen-bond contacts between the recognition helix and nucleotide bases of both strands.
The C-terminal protein-protein interaction domain of ScRap1 (RCT, for Rap1 C-terminus) adopts an entirely novel helical topology, in which the N-terminal helices form a highly hydrophobic cleft (Feeser and Wolberger, 2008) (Fig. 7A). A recent crystal structure of ScRap1-RCT complexed with a Sir3 peptide showed the peptide buried in the hydrophobic cleft (Chen et al., 2011). Recently, the structure of the ScRap1 BRCT domain was found to contain less secondary structure and more flexible loops than observed in canonical BRCT domains, but the physiological relevance of the more flexible structure has yet to be elucidated (Zhang et al., 2011b).
Figure 7. Protein-protein interactions.
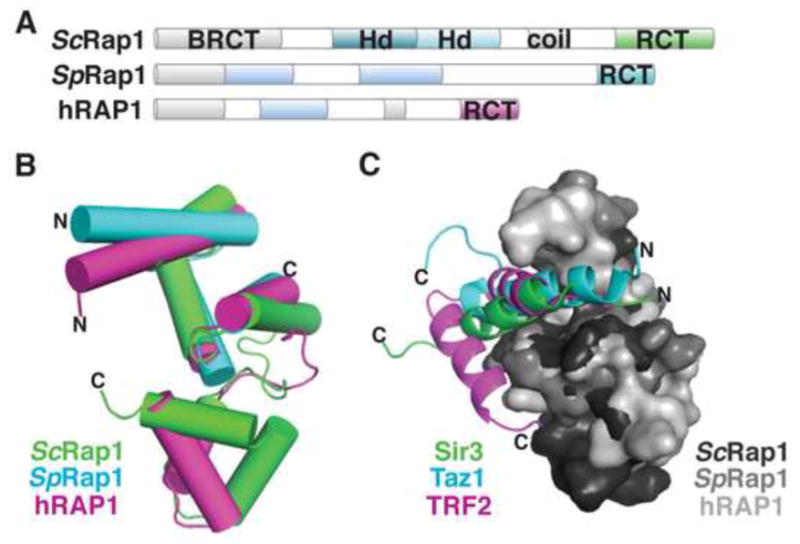
(A) Domain topology of Rap1 proteins. (B) Cartoon overlay of the Rap1 C-terminal domains (RCT) from S. cerevisiae (green; PDB: 3OWT), S. pombe (cyan; PDB: 2L3N), and human (magenta; PDB: 3K6G). (C) Surface overlay of the RCTs from S. cerevisiae (dark gray), S. pombe (medium gray), and human (light gray), shown with their respective peptide ligands shown as cartoons: Sir3 (green), Taz1 (cyan), and TRF2 (magenta).
TRF1, TRF2 and human Rap1
Human TRF1 and TRF2 bind to telomeric dsDNA as homodimers, with each monomer containing a single dsDNA-binding homeodomain that is similar to those that comprise ScRap1-DBD (Bianchi et al., 1999) (Fig. 6A,C). The free and bound forms of TRF1-DBD and TRF2-DBD were independently solved by both solution NMR and x-ray crystallography (Court et al., 2005; Hanaoka et al., 2005; Nishikawa et al., 1998; Nishikawa et al., 2001). Specificity of binding is achieved primarily by the DNA recognition helix, H3, which lies in the major groove of the DNA where it makes numerous sequence-specific contacts with the telomeric dsDNA. Additionally, the N-terminal arms become more rigid in the bound state as they make specific contacts in the minor groove, similar to the specific contacts made between the ScRap1 loops and its cognate DNA (Court et al., 2005; Nishikawa et al., 2001) (Fig. 6C).
Homodimerization of the TRF proteins is mediated by their TRF homology domains (TRFH) (Fig. 6A). TRF1-TRFH and TRF2-TRFH share 27% sequence identity, and the crystal structures showed highly similar tertiary structures between the two homodimers (Fairall et al., 2001) (Fig. 6D). Despite their structural similarity, TRF1 and TRF2 do not heterodimerize, due to incompatible hydrophobic networks that form the dimerization interface (Fairall et al., 2001) (Fig. 6E). A superposition of the α1 helices from the TRF1 and TRF2 homodimers shows the incompatibility between sidechains (Fig. 6E).
While the high-resolution structure of the full shelterin complex has yet to be achieved, insights into higher order protein assembly have been obtained from structures of shelterin domains complexed with peptides derived from binding partners. For example, the TRFH homodimers harbor interfaces for protein-protein interactions that stabilize shelterin and bind shelterin-interacting factors (Chen et al., 2008). Both TRF1 and TRF2 bind a TIN2-derived peptide; TRF2 also binds a peptide from the nuclease Apollo, which is associated with telomere protection during S phase (van Overbeek and de Lange, 2006). These peptides share a common binding interface, in a groove at the base of the homodimer horseshoe, and interact through a conserved hydrophobic surface (Chen et al., 2008) (Fig. 6F). This peptide-based approach has also been employed to understand the mechanism of interaction between Rap1 proteins and their potential binding partners (Fig. 2). Although the Rap1 DBD is not conserved, the C-terminal domains of the S. pombe and human Rap1 proteins share significant structural homology with the C-terminal domain of S. cerevisiae Rap1 (Fig. 7A, B), and also bind α-helices from their respective interaction partners in a helical cleft (Chen et al., 2011) (Fig. 7C). The hRAP1 C-terminal domain (RAP1-RCT) (residues 303–399) binds to a peptide derived from the intervening sequence between the TRFH and DBD domains of TRF2. SpRap1-RCT (residues 639–693) binds a peptide derived from the S. pombe TRF-like protein Taz1 (Fig. 8C). As the shelterin complex is reconstituted, it will be exciting to see how many more structural and mechanistic similarities exist between species that were not detectable by sequence homology.
Conclusion
High-resolution structures have been instrumental in informing telomere function and guiding biological studies aimed at elucidating the underlying mechanisms of telomere maintenance. The three-dimensional structures of several sets of telomerase and telomere-associated proteins revealed that they often display similar topologies in the absence of discernible sequence relationship, while variations on these shared protein folds highlight the unique ways in which organisms address their species-specific functions. With the structural picture of telomere function coming into focus, the next challenges will be the study of larger and larger complexes, as well as to probe the alteration in their state as a function of outside triggers, such as phosphorylation. The next frontier is to understand how regulation of the telomere is achieved, and how interactions between DNA, RNA, and proteins work dynamically together to perform telomere biogenesis, regulation, and maintenance.
Acknowledgments
We gratefully acknowledge research funding from the NIH (GM059414) and the NSF (MCB0617956) to D.S.W., and a postdoctoral fellowship from the NIH (GM093528) to K.A.L. We thank Tom Cech, Vicki Lundblad, Sarah Altschuler, Thayne Dickey, Robert Hom, Jayakrishnan Nandakumar, and Danielle Pfaff for helpful discussions and thoughtful comments on the manuscript. We apologize to all those whose work we have been unable to include due to space restrictions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler SE, Dickey TH, Wuttke DS. Schizosaccharomyces pombe Protection of Telomeres 1 Utilizes Alternate Binding Modes To Accommodate Different Telomeric Sequences. Biochemistry. 2011 doi: 10.1021/bi200826a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EM, Halsey W, Wuttke D. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EM, Halsey W, Wuttke D. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry. 2003;42:3751–3758. doi: 10.1021/bi027047c. [DOI] [PubMed] [Google Scholar]
- Armanios M. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Baumann P, Price C. Pot1 and telomere maintenance. FEBS Lett. 2010;584:3779–3784. doi: 10.1016/j.febslet.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol Cell. 2000;6:493–499. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Sperger JM, Chapman KB, Cech TR. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczek P, Horvath MP. Structural reorganization and the cooperative binding of single-stranded telomere DNA in Sterkiella nova. J Biol Chem. 2006;281:40124–40134. doi: 10.1074/jbc.M607749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado RT, Young NS. Telomere diseases. New Engl J Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. A DNA polymerase-alpha-primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284:5807–5818. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, et al. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol. 2011;18:213–221. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- Classen S, Lyons D, Cech TR, Schultz SC. Sequence-specific and 3′-end selective single-strand DNA binding by the Oxytricha nova telomere end binding protein alpha subunit. Biochemistry. 2003;42:9269–9277. doi: 10.1021/bi0273718. [DOI] [PubMed] [Google Scholar]
- Classen S, Ruggles JA, Schultz SC. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein alpha subunit both uncomplexed and complexed with telomeric ssDNA. J Mol Biol. 2001;314:1113–1125. doi: 10.1006/jmbi.2000.5191. [DOI] [PubMed] [Google Scholar]
- Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croy J, Podell E, Wuttke D. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11–389. J Mol Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Croy J, Wuttke D. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Croy J, Wuttke D. Insights into the dynamics of specific telomeric single-stranded DNA recognition by Pot1pN. J Mol Biol. 2009;387:935–948. doi: 10.1016/j.jmb.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croy JE, Altschuler SE, Grimm NE, Wuttke DS. Nonadditivity in the recognition of single-stranded DNA by the Schizosaccharomyces pombe protection of telomeres 1 DNA-binding domain, Pot1-DBD. Biochemistry. 2009;48:6864–6875. doi: 10.1021/bi900307x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croy JE, Fast JL, Grimm NE, Wuttke DS. Deciphering the mechanism of thermodynamic accommodation of telomeric oligonucleotide sequences by the Schizosaccharomyces pombe protection of telomeres 1 (Pot1pN) protein. Biochemistry. 2008;47:4345–4358. doi: 10.1021/bi701778x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Georgiadis MM. The crystal structure of the monomeric reverse transcriptase from Moloney murine leukemia virus. Structure. 2004;12:819–829. doi: 10.1016/j.str.2004.02.032. [DOI] [PubMed] [Google Scholar]
- de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge A, Halsey W, Wuttke D. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry. 2006;45:871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge A, Wuttke D. Probing the mechanism of recognition of ssDNA by the Cdc13-DBD. Nucleic Acids Res. 2008;36:1624–1633. doi: 10.1093/nar/gkn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L, Chapman L, Moss H, de Lange T, Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- Feeser EA, Wolberger C. Structural and functional studies of the Rap1 C-terminus reveal novel separation-of-function mutants. J Mol Biol. 2008;380:520–531. doi: 10.1016/j.jmb.2008.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner W, Berger H. Identification and ontogenesis of the nomen nudum Hypotrichs (Protozoa: Ciliophora) Oxytricha nova (= Sterkiella nova sp n.) and O. trifallax (= S. histriomuscorum) Acta Protozool. 1999;38:215–248. [Google Scholar]
- Friedman KL, Cech TR. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich-Ammon SJ, Dickinson BA, Bevilacqua JM, Schultz SC, Cech TR. Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev. 1998;12:1504–1514. doi: 10.1101/gad.12.10.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- Garcia C, Wright W, Shay J. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas AD, Paschini M, Reyes FE, Héroux A, Batey RT, Lundblad V, Wuttke DS. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc Natl Acad Sci USA. 2009;106:19298–19303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Zakian VA. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- Gray JT, Celander DW, Price CM, Cech TR. Cloning and expression of genes for the Oxytricha telomere-binding protein: specific subunit interactions in the telomeric complex. Cell. 1991;67:807–814. doi: 10.1016/0092-8674(91)90075-a. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Hanaoka S, Nagadoi A, Nishimura Y. Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 2005;14:119–130. doi: 10.1110/ps.04983705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Hayflick L. The cell biology of aging. J Invest Dermatol. 1979;73:8–14. doi: 10.1111/1523-1747.ep12532752. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- Jacob NK, Lescasse R, Linger BR, Price CM. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol Cell Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Kelleher C, Teixeira MT, Forstemann K, Lingner J. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem Sci. 2002;27:572–579. doi: 10.1016/s0968-0004(02)02206-5. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Swanton MT, Donini P, Prescott DM. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci USA. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger BR, Morin GB, Price CM. The Pot1a-associated proteins Tpt1 and Pat1 coordinate telomere protection and length regulation in Tetrahymena. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech T. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O’Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- Lue NF, Lin YC, Mian IS. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol Cell Biol. 2003;23:8440–8449. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Szostak J. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Mandell EK, Gelinas AD, Wuttke DS, Lundblad V. Sequence-specific binding to telomeric DNA is not a conserved property of the cdc13 DNA binding domain. Biochemistry. 2011;50:6289–6291. doi: 10.1021/bi2005448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA. 2007;104:14038–14043. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010a;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- Mitchell MT, Smith JS, Mason M, Harper S, Speicher DW, Johnson FB, Skordalakes E. Cdc13 N-terminal dimerization, DNA binding and telomere length regulation. Mol Cell Biol. 2010b;30:5325–5334. doi: 10.1128/MCB.00515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke D. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J Mol Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- Moretti P, Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty TJ, Marie-Egyptienne DT, Autexier C. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol Cell Biol. 2004;24:3720–3733. doi: 10.1128/MCB.24.9.3720-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Cross GA, de Lange T, Griffith JD. t-loops at trypanosome telomeres. EMBO J. 2001;20:579–588. doi: 10.1093/emboj/20.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Morin G, Chapman K, Weinrich S, Andrews W, Lingner J, Harley C, Cech T. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc Natl Acad Sci USA. 2010;107:651–656. doi: 10.1073/pnas.0911099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle S, Parkinson GN. The structure of telomeric DNA. Curr Opin Struct Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- Nishikawa T, Nagadoi A, Yoshimura S, Aimoto S, Nishimura Y. Solution structure of the DNA-binding domain of human telomeric protein, hTRF1. Structure. 1998;6:1057–1065. doi: 10.1016/s0969-2126(98)00106-3. [DOI] [PubMed] [Google Scholar]
- Nishikawa T, Okamura H, Nagadoi A, König P, Rhodes D, Nishimura Y. Solution structure of a telomeric DNA complex of human TRF1. Structure. 2001;9:1237–1251. doi: 10.1016/s0969-2126(01)00688-8. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- O’Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanai M, Kojima KK, Futahashi R, Yaguchi S, Fujiwara H. Identification and characterization of the telomerase reverse transcriptase of Bombyx mori (silkworm) and Tribolium castaneum (flour beetle) Gene. 2006;376:281–289. doi: 10.1016/j.gene.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Peersen OB, Ruggles JA, Schultz SC. Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA. Nat Struct Biol. 2002;9:182–187. doi: 10.1038/nsb761. [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated Est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- Shampay J, Szostak JW, Blackburn EH. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- Shore D, Bianchi A. Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J. 2009;28:2309–2322. doi: 10.1038/emboj.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsisthaliana. Proc Natl Acad Sci U S A. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, et al. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase α. Cell Res. 2011;21:258–274. doi: 10.1038/cr.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera AM, Stagno d’Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell. 2010;18:775–789. doi: 10.1016/j.devcel.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Schultz SC. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 2003;22:4314–4324. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Bunch JT, Baumann P. Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of heterogeneous fission yeast telomeres. J Biol Chem. 2005;280:9119–9128. doi: 10.1074/jbc.M414511200. [DOI] [PubMed] [Google Scholar]
- van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin KL, Prathapam R, Collins K. Positive and negative regulation of Tetrahymena telomerase holoenzyme. Mol Cell Biol. 2007;27:2074–2083. doi: 10.1128/MCB.02105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–5622. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Peng Y, Mian IS, Lue NF. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol Cell Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 2010;38:1982–1996. doi: 10.1093/nar/gkp1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O’Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GL, Bradley JD, Attardi LD, Blackburn EH. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kim NK, Feigon J. Telomerase and Retrotransposons: Reverse Transcriptases That Shaped Genomes Special Feature Sackler Colloquium: Architecture of human telomerase RNA. Proceedings of the National Academy of Sciences of the United States of America; 2011a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang J, Zhang X, Xu C, Tu X. Solution structure of Rap1 BRCT domain from Saccharomyces cerevisiae reveals a novel fold. Biochem Biophys Res Commun. 2011b;404:1055–1059. doi: 10.1016/j.bbrc.2010.12.109. [DOI] [PubMed] [Google Scholar]


