Abstract
We describe a multiplexed aberration measurement method that modulates the intensity or phase of light rays at multiple pupil segments in parallel to determine their phase gradients. Applicable to fluorescent-protein-labeled structures of arbitrary complexity, it allows us to obtain diffraction-limited resolution in various samples in vivo. For the strongly scattering mouse brain, a single aberration correction improves structural and functional imaging of fine neuronal processes over a large imaging volume.
Tissue refractive index heterogeneity limits the imaging quality of two-photon fluorescence microscopy in vivo by distorting its excitation wavefront and resulting in an enlarged focal volume and reduced focal intensity. To image fine structures at depth, it is essential to measure and correct these sample-induced aberrations through adaptive optics (AO)1. An ideal aberration-measurement method should work with signal from structures intrinsic to the sample, under any labeling density (e.g., from sparse to dense), with any labeling strategy (e.g., from bright synthetic dye to dim fluorescent proteins), in any sample (e.g., from transparent to strongly scattering), and, if the sample permits, generate a corrective pattern that improves image quality over a large volume. Here, we describe a multiplexed aberration measurement method that satisfies all of the above criteria.
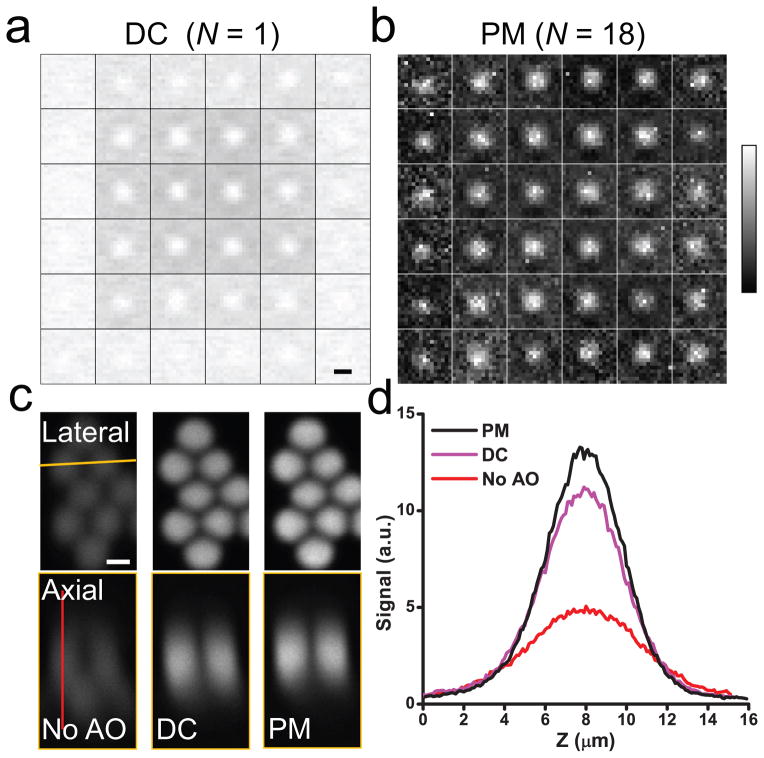
To form a perfect focus, all light rays have to intersect at the same point and interfere with the same phase. Aberration modifies the directions and phases of these rays, which can be measured to obtain the corrective wavefront needed for restoring diffraction-limited focus. Consider the wavefront of an excitation beam illuminating the back pupil of a microscope objective: if we apply local phase ramps to a spatial light modulator (SLM) to raster scan the ray at one pupil segment while keeping all other rays fixed as reference, the varying interference between them modulates the focal intensity and consequently signal strength. Plotting the signal relative to the displacement of the ray in the focal plane as a 2D “signal vs. displacement” map, the local wavefront tilt for this particular ray can be determined by measuring the extra phase ramp needed to generate maximal interference2. Repeating the procedure for all light rays (Fig. 1a), we get the local wavefront gradients, from which the entire corrective wavefront can be obtained and applied to the SLM for aberration correction3, 4. This method, however, is slow: the interference between rays at a small pupil segment and the rest leads to only small signal variations (~1–2% for edge segments in Fig. 1a), whose detection requires long integration time; Sequential measurement of each ray further increases the time needed. An aberration measurement using a moderate number (e.g., 36 in Fig. 1a) of pupil segments may take longer than 20 minutes, making it impractical for in vivo applications due to sample photobleaching and motion. To speed it up, we measure rays at multiple pupil segments in parallel through multiplexing5: tagging each ray with a characteristic frequency ω by modulating either its intensity or phase using a digital micromirror device (DMD) or a segmented deformable mirror (SDM) conjugated to the SLM (Supplementary Fig. 1), we separate the contribution of each ray in the final interference signal by Fourier-transforming the signal trace and plotting the Fourier magnitude for each ray at its distinct modulation frequency ω in the 2D map (Fig. 1b, intensity modulation with DMD) (Supplementary Fig. 2 and Online Methods). Besides speeding up the aberration measurement (by 18× in Fig. 1b, see Supplementary Table 1 for measurement time and laser power), moving the measurement away from DC also leads to better signal-to-noise ratio, because noise such as that associated with excitation power fluctuations is much smaller at higher frequency, resulting in a better corrective performance (Fig. 1c,d). This multiplexed aberration measurement method works in both sparsely and densely labeled samples, and allowed us to correct sample-induced aberration using fluorescent protein signal from a wide variety of biological samples in vivo (see Online Methods and Supplementary Figs. 3 and 4 for examples on zebrafish and C. elegans). Below we demonstrated its application to in vivo imaging in the mouse brain.
Figure 1.
Multiplexed aberration measurement with parallel modulation (PM) of light ray intensity at multiple pupil segments. (a) and (b) “Signal vs. displacement” maps for 36 pupil segments obtained from the DC signal (“DC (N=1)”) and from the Fourier magnitudes (“PM (N=18)”, 18 pupil segments modulated simultaneously with DMD), respectively. Each map is normalized between 0 and its maximum. (c) Lateral and axial (along the yellow line) images of 2-μm-diameter beads obtained without AO, with the corrective wavefront from the DC method (“DC”), and with the corrective wavefront from the multiplexed method (“PM”), respectively. (d) Signal profiles along the red line in c. Scale bar: 2 μm.
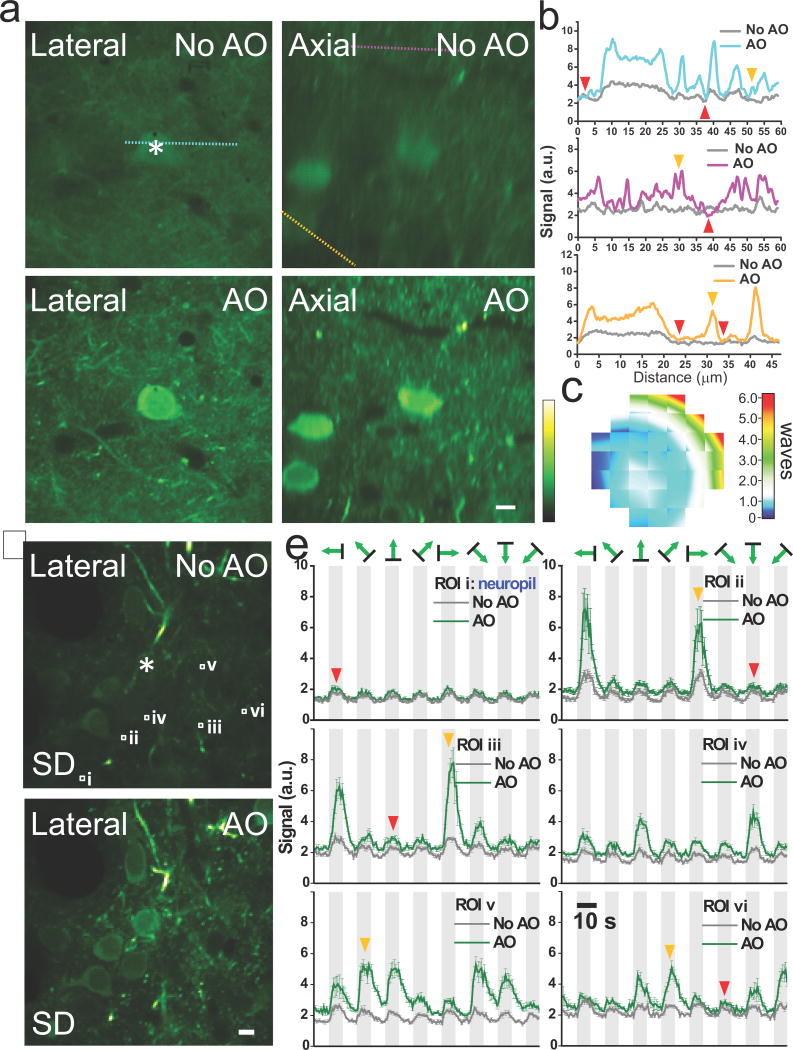
At layer 2/3 in the primary visual cortex (V1) of a mouse transfected with the genetically encoded calcium sensor GCaMP6s6, a single corrective wavefront (Fig. 2c) obtained at the cell body marked with an asterisk (Fig. 2a, intensity modulation with DMD) improves both the signal and resolution of layer 2/3 neurons, with many more fine neuronal processes resolvable after AO correction (Fig. 2a,b). High label density leads to a diffuse fluorescence background (e.g., red arrowheads, Fig. 2b), which makes it difficult to find isolated structures to quantify the signal improvement before and after AO, but the line intensity profile comparisons show that many previously invisible neuronal processes become easily detectable after AO correction (e.g., orange arrowheads, Fig. 2b). In contrast, the fluorescence background signal changes little, because whereas the signal from smaller features is improved due to the higher excitation intensity of an AO-corrected focus, an aberrated and enlarged focus can compensate for the loss of focal intensity by exciting more fluorophores in spatially extended structures7. This has important implications for in vivo calcium imaging experiments, where the detected fluorescence change caused by neural activity may come from two sources: the individual neuronal structure of interest (e.g., soma, axons, and dendrites) and its surrounding diffusely-stained neuropil, with the latter signal often uncorrelated with stimulus feature and considered a contamination8. With AO improving the strength of the desired signal (e.g., orange arrowheads, Fig. 2e) much more than neuropil contamination (e.g., ROI i, red arrowheads, Fig. 2e), more calcium events can be detected (represented by pixels with large standard deviation in Fig. 2d, also see Supplementary Videos 1) and, with the increase of signal-to-neuropil ratio, the orientation-selectivity of dendrites can now be determined with confidence (e.g., ROIs ii–vi, Fig. 2e).
Figure 2.
AO correction with multiplexed aberration measurement at layer 2/3 of a GCaMP6s-expressing mouse brain in vivo. (a) Lateral and axial images at 115 μm below dura without and with AO correction. (b) Signal profiles along the cyan, purple, and orange lines in a. Orange arrowheads label the structures rendered invisible by the diffusely-labeled background (red arrowheads). (c) Corrective wavefront in units of waves. (d) Lateral images at 150 μm depth without and with AO correction. Pixel brightness reflects the standard deviation (SD) of pixel values across 800 calcium imaging frames, with larger SD values indicating higher neuronal activity. (e) Fluorescent signals reflecting calcium transients measured without (dark gray curves) and with (green curves) AO correction at 6 regions of interest (ROIs) (white squares i–vi in d) under visual stimulations of gratings moving in eight different directions. Orange arrowheads mark the example responses from dendrites at the preferred directions of grating motion, while the red arrowheads label example responses dominated by neuropil contamination. Light gray denotes the duration with stimuli on. Signals are averages of five trials of calcium imaging with the error bars indicating the standard errors of the mean. Signals at the asterisks were used to measure aberration through multiplexed intensity modulation via DMD. Greenhot lookup table in ImageJ is used for all images. Scale bar: 10 μm.
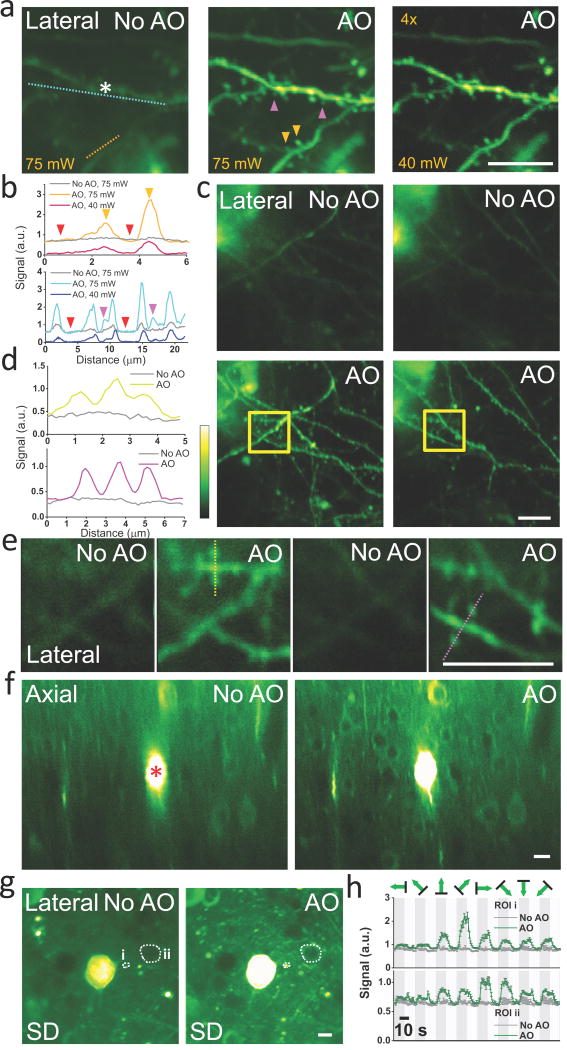
With increasing imaging depth, the excitation laser power has to be increased to compensate for the scattering loss of ballistic photons at the focus. Eventually, the laser intensity at the surface of the brain is high enough to excite superficially labeled structures without requiring the spatial confinement of a focus, resulting in a background signal even in sparsely labeled brains (e.g., Fig. 3a, post-objective power at 75 mW, at 385 μm depth inside a thy1-YFP-H mouse brain), which would ultimately overwhelm the signal generated at the focus and limit the imaging depth9. This out-of-focus signal is not affected by the presence of aberration10, and therefore not increased by AO correction (e.g., Fig. 3b, red arrowheads). The in-focus signal, however, strongly depends on the focus quality. After AO correction (phase modulation with SDM at the asterisk in Fig. 3a), dendritic spines previously rendered invisible by the background in the uncorrected image become clearly visible (e.g., orange and purple arrowheads, Fig. 3a,b). The improvement in both resolution and signal allows us to reduce excitation power (e.g., from 75 mW to 40 mW) yet still obtain a higher quality image with much suppressed out-of-focus background.
Figure 3.
AO correction with multiplexed aberration measurement at layers 4 and 5 of the mouse brain in vivo. (a) Maximal intensity projections of dendrites at 376–395 μm below dura measured without and with AO at excitation powers 75 mW and 40 mW, respectively. The gain of the image taken at 40 mW is increased 4×. (b) Line intensity profiles along the orange and cyan lines in a. Purple and orange arrowheads label dendritic spines invisible before AO correction. Red arrowheads indicate the out-of-focus background level at 75 mW excitation. (c) Maximal intensity projections of basal dendrites at 473–481 μm (left column) and 481–490 μm (right column) measured without and with AO correction. (d) Line intensity profiles along the yellow and violet lines in e. (e) Images taken inside the yellow squares in c at 480 μm (left) and 484 μm (right) depths without and with AO correction. (f) Axial images at 427–547 μm depth measured without and with AO. (g) From f, lateral images at 490 μm depth without and with AO correction. Pixel brightness reflects the SD of pixel values across 800 calcium imaging frames. (h) Calcium transients measured without (dark gray curves) and with (green curves) AO correction at the two ROIs in g (ROI1: i, dendrite; ROI2: ii, soma). a,f, corrective wavefront was measured at the asterisk; c, correction was at 30 μm above the imaging volume. a,c, phase modulation with SDM; f, intensity modulation with SDM. a–e, thy1-YFP-H mice; f–h, wild-type mouse expressing GCaMP6s. Greenhot lookup table in ImageJ is used for all images. Scale bar: 10 μm.
With the synaptic-level resolution provided by AO, we can now resolve the sub-micron-sized spines on the basal dendrites of layer 5 pyramidal neurons at depth. Without AO correction, at 458–500 μm depth, only the crude profiles of these dendrites can be detected. AO correction elevates the signal from dendritic spines to rise above background noise, making them easily identifiable in the corrected images (Fig. 3c–e, Supplementary Videos 2 and 3, thy1-YFP-H mouse, phase modulation with SDM). For more densely labeled brain, fine neuronal processes and even the much larger somata go from invisible to clearly resolvable after AO correction (Fig. 3f, 427–547 μm depth, wild-type mouse with GCaMP6s expression, intensity modulation with SDM), where the nuclei of neurons appear dark due to the nucleus-exclusion of GCaMP6s in healthy neurons. Finally, AO also drastically improves calcium imaging of fine neuronal processes and somata at depth: at 490 μm depth, structures whose calcium activity are undetectable without aberration correction show clear visually-evoked calcium transients after correction (e.g., Fig. 3g,h, Supplementary Video 4).
Besides aberration, light scattering by minuscule scatterers such as cellular organelles also limits image quality in vivo. These much higher spatial-frequency wavefront perturbations may be partially corrected via wavefront shaping11, 12. However, because tissue scattering profile is highly spatially variable, a corrective pattern only improves signal in a small volume (e.g., 6×6×7 μm3 in fixed brain tissues13). In contrast, because aberration correction removes the lower-order wavefront errors, in the case of mouse brain in vivo, one correction pattern improves signal over a much larger volume (e.g., 168×168×120 μm3 in Fig. 3f). Furthermore, for all the in vivo samples tested by us, optical aberration is temporally stable over the experimental duration (e.g., data in Figs. 3g,h was obtained 3 hours after aberration measurement). In comparison, because of its susceptibility to microscopic motions, tissue scattering profile may vary rapidly in time14. Therefore, for applications requiring large field of view and extended imaging period, correcting optical aberration may be a more generally applicable approach for deep tissue imaging.
With current wavefront shaping hardware and fluorescent protein brightness, we obtain a corrective wavefront within minutes. Although it is still slow compared to direct wavefront sensing methods15, which only work in transparent samples, our method is applicable to strongly scattering tissues. Since, in mouse brain, a single correction improves ensuing experiments for hours, the extra time required for aberration measurement does not constitute a substantial burden while allowing us to image the major input and output layers of mouse cortex in vivo with synaptic-level resolution.
ONLINE METHODS
All experiments were performed according to methods approved by the Janelia Farm Institutional Animal Care and Use Committee.
Adaptive-optical (AO) two photon fluorescence microscope
A simplified schematic of our microscope is shown (Supplementary Fig. 1a). Femtosecond pulses at 900 nm from a Titanium:Sapphire laser (Ti:Sapphire, Chameleon Ultra II, Coherent Inc.) are reflected off either a segmented deformable mirror (SDM, Iris AO Inc.) or a high-speed digital micromirror device (DMD) (V4100, ViALUX Messtechnik+Bildverarbeitung GmbH) following a dispersion compensation unit (DCU). A pair of achromat doublets (AC300-080-B and AC254-500-B, Thorlabs Inc.) are used to conjugate the DMD or SDM to a liquid-crystal phase-only spatial light modulator (SLM, PLUTO-NIR, Holoeye Photonics AG), which is further conjugated to a pair of galvanometers (X galvo and Y galvo, 3 mm beam aperture, 6215H, Cambridge Technology Inc.) and the back pupil plane of a 40× or a 16× NA 0.8 water-dipping objective (CFI Apo 40XW NIR, CFI LWD 16XW, Nikon Inc.) using three pairs of achromat doublets (AC254-150-B and AC254-060-B, AC508-080-B and AC508-080-B, AC508-75-B and 014-1310, Thorlabs Inc. and OptoSigma). A field stop (FS) is located at the intermediate image plane between the DMD/SDM and the SLM to block unwanted diffraction orders. The microscope objective is mounted on a single-axis piezo stage (P-725.4CD, Physik Instrumente L.P.) for taking axial images. In the detection path, fluorescence signal reflected from a dichroic long-pass beamsplitter (FF665-Di02-25x36, Semrock Inc.) immediately above the objective gets collected by a photomultiplier tube (PMT, H7422-40, Hamamatsu) after passing through an emission filter (FF01-680/SP, Semrock). Ray-tracing software Zemax (Zemax 12 EE, Radiant Zemax Corp.) is used to optimize the design of optical path. With the DMD, two SLM patterns are used for AO correction (i.e., two pupil-segmentation schemes) (Supplementary Fig. 1d), with the active area of the SLM inscribed to the back pupil. With SDM, the hexagon formed by 37 mirror segments is slightly larger than the back pupil.
Operation of the digital micromirror device (DMD) and the segmented deformable mirror (SDM)
DMD is a bistable spatial light modulator capable of intensity modulation, consisting of 1024×768 movable micromirrors, where each mirror can be individually deflected at +/−12° about a hinged diagonal axis. We rotate the DMD by 45° around its surface normal, so that the diagonal axes are parallel to the optical table, simplifying the direction and polarization control for the deflected light. The DMD is then divided into segments through precision mapping to the SLM segments. The DMD is capable of a maximum binary switching rate of 22,727 Hz, but to generate a variety of modulation frequencies for different pupil segments, we utilize pulse width modulation to have grayscale control for each DMD segments. Throughout this paper, we operate the DMD at a bit-depth of 5, which provide 32 gray levels and a switching rate of 2,016 Hz. We choose the signal integration time at each gray level to be the reverse of the switching rate (0.5 ms), so that the resulting fluorescent signal reflects the gray level value.
Optically, the DMD acts as a blazed grating, thus the coherent light from the Ti:Sapphire laser bouncing off the DMD travels along discrete directions dictated by diffraction. We adjust the incident angle of the laser beam to maximize the energy in the brightest diffraction order, and achieve 30% diffraction efficiency at 900 nm. A further complication arises because femtosecond pulses are not monochromatic, thus are spatially dispersed by the DMD in the incidence plane at ~0.026°/nm around 900 nm. Without compensation, the angular spread of the different frequency components leads to a focus elongated in the direction of dispersion (Supplementary Fig. 1c). For dispersion compensation, we build a dispersion compensation unit (DCU) based on a single-prism approach16, 17 (Supplementary Fig. 1b). An equilateral dispersive prism (EDP, PS855, Thorlabs Inc.) with an incident angle of 49.3° is used for dispersion compensation. To counter the one-dimensional demagnification the beam experienced while traveling through the EDP, an anamorphic prism pair (APP, NT47-244, Edmund Optics Inc.) is used to expand the beam ~5× in the same dimension. The lateral image profiles of 2 μm beads become much more circular after dispersion compensation (Supplementary Fig. 1c).
The SDM is made of piston-tip-tilt mirror segments and operated in two modes: by modulating the piston value of each mirror segment, we modulate the phase of the light ray at the corresponding pupil segment; by tilting each mirror segment so that the reflected ray gets blocked by the field stop, we modulate the light intensity at its pupil segment. Both types of modulation affect the interference between the light ray and the reference focus. Without the need for dispersion compensation, the SDM provides better power throughput. It also has the potential to replace the SLM for aberration correction, thus could simplify the setup substantially. However, once the magnification between the SDM and the SLM is determined, the number of pupil segment is fixed at 37 in our experiment, while DMD, with vastly more independent actuators, allows us to vary the number of pupil segments.
Details on the multiplexed aberration measurement method
For half of all pupil segments, we apply a phase ramp Rikjk to pupil segment k via the SLM ((1) in Supplementary Fig. 2). For each pupil segment, the phase ramp Rikjk is randomly chosen from a set of total NR phase ramps, which together lead to a 2D grid displacement of the deflected ray in the focal plane around its original location (Rij=0). This way, we prevent the modulated light rays from landing at the same location in the focal plane and forming a focus of similar intensity to the reference focus, which would interfere with the reference focus at near-zero displacements and lead to spurious signal near the center of the final maps. Using DMD or SDM, we modulate the illumination intensity or phase of rays impinging on these pupil segments at distinct frequency ωk’s for time duration T, while monitoring the signal change. The amount of interference that each pupil segment has with the reference focus is found by reading out the magnitude Mikjk at its distinct modulation frequency ωk from the Fourier transform of the signal trace (inspired by the multidither coherent optical adaptive technique5, although in our implementation we read out the magnitudes rather than the phases).
Repeating (1) NR times, the whole set of phase ramps is applied each segment and the resulting Fourier magnitudes Mij’s are measured. “Signal vs. displacement” maps are then formed for these modulated pupil segments, by plotting the magnitude Mij’s according to the displacements of the rays in the focal plane ((2) in Supplementary Fig. 2). The wavefront tilt for each particular pupil segment can then be measured from the shift of the peak from the center, just like in a Shack-Hartmann wavefront sensor18. Whereas a peak modulation magnitude always exists for phase modulation, for intensity modulation, if there is a ~90° phase shift between the ray and the reference focus, a clear maximum would be absent from the resulting map; a 90° phase offset is then added to the corresponding local wavefront and the measurement repeated.
Repeating (2) one more time for the other half of the pupil segments, we obtain the local wavefront gradients for all pupil segments ((3) in Supplementary Fig. 2), with an overall signal integration time of 2×NR×T. Followed by either phase reconstruction algorithms or phase measurement methods4, we then obtain the corrective wavefront. Depending on the sample, NR varies between 100 and 256 (10×10 to 16×16 2D grid), and T varies between 45 and 360 ms (and can be smaller if higher signal level is used). Additional overheads due to the hardware (e.g., SLM update time) and software (e.g., time to calculate DMD and SLM patterns) usually increase the overall correction time by 50%. For large initial aberrations, additional rounds of correction can improve the correction by having an increasingly aberration-free reference focus. For all biological samples shown in the paper, two or three rounds are used to obtain the final corrective wavefront.
Typical operational parameters
As an example, we describe here the detailed imaging parameters for the data shown in Figure 1. The same integration time per phase ramp (T = 180 ms) is used in all measurements. 15×15 phase ramps (NR = 225) that displaced rays by ±15 μm in X and Y directions in the focal plane are applied. With 6×6 pupil segments, (NP = 36), the serial method (i.e., the DC method with overall integration time of NP×NR×T) has an overall integration time of 1458 seconds or 24 minutes (and an overall correction time of 3568 seconds), while the multiplexed method have an overall integration time of 81 seconds (and an overall correction time of 127 seconds).
System aberration is corrected before all measurements and the resulting diffraction-limited microscope is used to take all data labeled with “No AO”. Experimentally, we park the laser focus at one sample location, and use the fluorescence signal from this location for AO correction. (More generally, we can scan the laser focus across a user-defined area or volume, and use the integrated signal for AO correction. This will lead to an AO correction that is applicable to a large area/volume of the sample, but would slow down the correction speed because of longer scanning time.) Because full-pupil illumination is used, no extra excitation power increase is needed during AO correction. Depending on the samples, the excitation power post objective varies from 0.2 mW (fluorescent beads), 9.1 mW (layer 2/3 in mouse cortex), to 75 mW (layer 5 in mouse cortex). For all in vivo samples, care is taken to make sure that no observable photobleaching or photodamage occurs. For mouse, images are often collected from the same mouse over multiple days at similar excitation power, and no obvious photo-induced morphological changes are observed. Supplementary Table 1 lists the excitation power and time used for different samples.
Comparison with the single-segment illumination method
The “signal vs. displacement” maps presented in Figure 1 only reflect the focal intensity variation relative to the ray displacement in focal plane, and do not reflect or rely on specific fluorescent structures of the sample. Therefore, we can now achieve AO correction from fluorescent samples of arbitrary structural complexity, including densely labeled fluorescent samples, for which the single-segment illumination method3 fails, because the reduction of excitation NA during single-segment illumination allows the originally out-of-focus fluorescent structures to appear in the image and makes it difficult to determine wavefront gradients2. For the same densely labeled mouse brain in vivo, the single-segment illumination method fails to improve signal quality, while the multiplexed method, which operates at the full NA of the objective, is able to improve image quality notably (Supplementary Fig. 5).
An extreme example of a densely labeled fluorescent sample is a fluorescent solution (“fluorescent sea”). Without any structural feature, fluorescent sea sample cannot provide the image shift information that the single-segment illumination method needs to work. In contrast, the multiplexed aberration measurement method can be used to recover diffraction-limited resolution using signal in a fluorescein solution (Supplementary Fig. 6). Here an aberration was introduced to the system by overlaying a phase pattern on the SLM, which reduced the signal from a fluorescent sea by 1.7×. Parking the laser at a fixed spot inside the fluorescent sea and after five iterations with parallel intensity modulation, we fully recover the signal from the fluorescent sea (Supplementary Fig. 6b). Since signal from a smaller object is much more sensitive to aberration than a fluorescent sea7, we test how well this AO correction works by imaging a 2-μm-diameter fluorescent bead without and with AO correction obtained from the fluorescent sea, and compare the resulting images with that from the bead under ideal, aberration-free imaging condition (Supplementary Fig. 6c,e). The multiplexed method improves signal from this bead by 7.5×, almost fully recovers the imaging performance under ideal condition.
Finally, deep inside scattering samples where excitation power becomes limited, the multiplexed method would perform better even for sparsely labeled samples, because it does not require more power than what is necessary to visualize fluorescent structures, whereas the single-segment illumination method needs higher excitation power during aberration measurement to compensate for the reduction of NA.
Multiplexed aberration measurement improves imaging quality in C. elegans and zebrafish in vivo
The ultimate test of any AO method is in live biological samples, where microscopic and sometimes macroscopic motions abound and photodamage as well as photobleaching limit the amount of light exposure. In addition to the mouse brain, we also tested our method in C. elegans and zebrafish, two of the most popular model organisms for bioimaging. Two examples of AO correction on GFP-labeled neurons in live C. elegans are shown In both cases, we imaged neurons through the aberrating cylindrical bodies of the worms. In one example (Supplementary Fig. 3b,c), AO correction after two iterations improves both the signal (1.8×) and resolution of two neurons in the deeper body wall. In the other (Supplementary Fig. 3d–h), we corrected the aberration through the more densely labeled worm head, by parking the focus at a bottom neuron (crosses in Supplementary Fig. 3d). After one iteration of AO correction, the improvement in signal (2×) and resolution is most prominent for the neuron on which the AO correction is done, because the large surface curvature of the worm leads to a highly localized aberration (Supplementary Fig. 3f,g,h).
As an example of a very densely labeled sample, we applied the multiplexed aberration measurement method to in vivo imaging of 4-day-old zebrafish embryos, where all cell membranes are labeled with EGFP. The axial views through the body of one embryo show notable signal and resolution deterioration for myotomes below the notochord (Supplementary Fig. 4a,b). After three rounds of AO correction at a myotome 170 μm inside the embryo (laser focus parked on crosses in Supplementary Fig. 4), the signal increases up to 2.3× in both lateral and axial imaging planes. More importantly than mere signal increase, the resolution improvement leaves various structural features much better resolved (Supplementary Fig. 4c,e). For example, the membrane folds (yellow arrowheads) can be easily observed after AO correction, the aberration-induced ghosts (red arrowheads) are eradicated, the individual membranes are much better separated (blue arrowhead, middle panels in Supplementary Fig. 4c,e), and the periodic structures of the myotome are much better visualized (last panels in Supplementary Fig. 4c,e).
Functional imaging in the mouse brain in vivo
We transfected neurons in the mouse primary visual cortex with GCaMP6s and recorded the fluorescence signal variation when mice were shown visual stimulations composed of grating patterns moving in 8 different directions. For all functional imaging data, each direction lasts for 10 image frames separated by 10 frames with no visual stimulation present (i.e., 160 frames for one round of calcium imaging). 5 trials of measurement (i.e., 800 frames total) were repeated for each imaging condition (“AO” or “No AO”) for all functional imaging sessions. The trial-averaged results are shown. All movies have their brightest pixels saturated to improve the visibility of structures with weaker signals.
Digital Image Processing
All images are presented with no digital manipulation except those listed below: Due to brain motion at depth during functional imaging, TurboReg plugin in ImageJ was used for image registration. Images are presented using either the gray scale or the green-hot lookup table (to make the weaker feature more visible) in ImageJ. For images where the signal was too weak before AO correction, a linear scaling factor was applied to all pixels to improve visibility with the scaling factor listed on the image (e.g., Fig. 3a, right panel). No deconvolution or nonlinear scaling such as Gamma correction was used on the data.
Sample preparation
Beads
Carboxylate-modified fluorescent microspheres of 1-μm and 2-μm diameters (F-8821 and F-8826, Invitrogen) were immobilized on poly-L-lysine-coated microscope slides (12-550-12, Fisher Scientific). Under our two-photon fluorescence microscope, they generate red fluorescence when excited at 900 nm.
C. elegans
C. elegans (Caenorhabditis elegans) larvae of strain ST2, with green fluorescence protein (GFP) expressed in nearly all neurons, were used for all experiments. Before imaging, the worms were transferred to a 2% agar pad made with M9 buffer. 5 μL of 100 mM sodium azide was then added to anesthetize the worms, before a No. 1.5 coverslip was gently laid over the worms. The worms stayed immobilized for 2–3 hours during imaging.
Zebrafish embryos
Zebrafish (Danio rerio) embryos of Tg(β-actin:HRAS-EGFP) line were grown at 28°C in E3 zebrafish embryo medium. Between 10–16 hours postfertilization (hpf), the embryos were transferred into 1× phenylthiourea (PTU) solution in E3 medium to inhibit melanin formation and render the embryos optically transparent19. Before imaging, the chorions, if present, were manually removed using forceps under a stereomicroscope. The embryos were then anesthetized in E3 medium containing 1× tricaine, and immobilized on a microscope slide by embedding them in 1% low-melting-point agarose with 1× PTU and 1× Tricaine. During imaging, E3 medium containing 1× PTU and 1× Tricaine was used as immersion medium.
Mouse
Procedures for viral infection and craniotomy were described previously7. Briefly, wild-type mice older than P60 of both sexes were anesthetized with isoflurane (2% vol isoflurane/vol O2). Following a craniotomy, 20 nL of suspended AAV2/1.CB7.CI.EGFP.rBG virus (Penn Vector Core) or AAV1-syn-GCaMP6s-WPRE-SV40 virus was slowly injected into the neocortex at 300 μm and 500 μm below the pia of anesthetized adult mice. A cranial window made of one or two No. 1.5 coverslips was then gently placed over the craniotomy and sealed onto the skull with dental cement. Three-to-twelve weeks after viral injection, mice were anesthetized and mounted under the AO two-photon microscope for imaging.
Supplementary Material
Acknowledgments
We thank our colleagues at Janelia Farm Research Campus, Howard Hughes Medical Institute: E. Betzig for helpful discussions; H. Dana, B. MacLennan, G. Ranganathan, K. Smith for help with mice samples; P. Keller and M. Ahrens for providing zebrafish samples. We thank C. Fang-Yen at University of Pennsylvania for advice on C. elegans samples. This work was supported by Howard Hughes Medical Institute.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
N. J. conceived of the project and designed the experiments; C. W., R. L., and N. J. designed and built the optical setup, D. E. M. and N. J. developed the instrument control program; C. W. and R. L. debugged the program; C. W. (intensity modulation with DMD and SDM, morphological and functional imaging), R. L. (phase and intensity modulation with SDM, spine morphological imaging), and N. J. performed the experiments and analyzed the data; W. S., Z. T., A. K., R. L. prepared the mice for in vivo imaging; T.-W. C. and D. S. K. provided GCaMP6s reagents; and N. J. wrote the paper with input from all co-authors.
References
- 1.Kubby J, editor. Adaptive Optics for Biological Imaging. CRC Press; 2013. [Google Scholar]
- 2.Milkie DE, Betzig E, Ji N. Opt Lett. 2011;36:4206–4208. doi: 10.1364/OL.36.004206. [DOI] [PubMed] [Google Scholar]
- 3.Ji N, Milkie DE, Betzig E. Nat Methods. 2010;7:141–147. doi: 10.1038/nmeth.1411. [DOI] [PubMed] [Google Scholar]
- 4.Liu R, Milkie DE, Kerlin A, MacLennan B, Ji N. Opt Express. 2014;22:1619–1628. doi: 10.1364/OE.22.001619. [DOI] [PubMed] [Google Scholar]
- 5.Bridges WB, et al. Appl Opt. 1974;13:291–300. doi: 10.1364/AO.13.000291. [DOI] [PubMed] [Google Scholar]
- 6.Chen TW, et al. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji N, Sato TR, Betzig E. Proc Natl Acad Sci U S A. 2012;109:22–27. doi: 10.1073/pnas.1109202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Göbel W, Helmchen F. Physiology. 2007;22:358–365. doi: 10.1152/physiol.00032.2007. [DOI] [PubMed] [Google Scholar]
- 9.Theer P, Hasan MT, Denk W. Opt Lett. 2003;28:1022–1024. doi: 10.1364/ol.28.001022. [DOI] [PubMed] [Google Scholar]
- 10.Leray A, Lillis K, Mertz J. Biophys J. 2008;94:1449–1458. doi: 10.1529/biophysj.107.111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vellekoop IM, Mosk AP. Opt Lett. 2007;32:2309–2311. doi: 10.1364/ol.32.002309. [DOI] [PubMed] [Google Scholar]
- 12.Yaqoob Z, Psaltis D, Feld MS, Yang C. Nat Photon. 2008;2:110–115. doi: 10.1038/nphoton.2007.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang J, Germain RN, Cui M. Proc Natl Acad Sci U S A. 2012;109:8434–8439. doi: 10.1073/pnas.1119590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui M, McDowell EJ, Yang C. Opt Express. 2010;18:25–30. doi: 10.1364/OE.18.000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, et al. Nat Methods. 2014;11:625–628. doi: 10.1038/nmeth.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng S, et al. Opt Lett. 2006;31:1091–1093. doi: 10.1364/ol.31.001091. [DOI] [PubMed] [Google Scholar]
- 17.Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F. Nat Methods. 2010;7:399–405. doi: 10.1038/nmeth.1453. [DOI] [PubMed] [Google Scholar]
- 18.Hardy JW. Adaptive Optics for Astronomical Telescopes. Oxford University Press; 1998. [Google Scholar]
- 19.Godinho L. Cold Spring Harbor Protocols. 2011;2011 doi: 10.1101/pdb.prot5644. pdb.prot5647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.