Abstract
Host-parasite interactions can drive rapid, reciprocal genetic changes (coevolution), provided both hosts and parasites have high heritabilities for resistance/infectivity. Similarly, the host’s mating system should also affect the rate of coevolutionary change in host-parasite interactions. Using experimental coevolution, we determined the effect of obligate outcrossing verses partial self-fertilization (mixed mating) on the rate of evolutionary change in a nematode host (Caenorhabditis elegans) and its bacterial parasite (Serratia marcescens). Bacterial populations were derived from a common ancestor. We measured the effects of host mating system on host adaptation to the parasite. We then determined the extent of parasite adaptation to their local host populations. Obligately outcrossing hosts exhibited more rapid adaptation to parasites than did mixed mating hosts. In addition, most of the parasites became adapted to infecting their “local” hosts; but parasites from obligately outcrossing hosts showed a greater level of local adaptation. These results suggest that host populations evolved along separate trajectories, and that outcrossing host populations diverged further than partially selfing populations. Finally, parasites “tracking” outcrossing host populations diverged further than parasites tracking the partially selfing host populations. These results show that the evolutionary trajectories of both hosts and parasites can be shaped by the host’s mating system.
Keywords: inbreeding, outcrossing, self-fertilization, experimental evolution
Coevolutionary interactions are thought to account for a significant amount of the evolutionary change that occurs in nature (Thompson 1982; Thompson 1994; Thompson 2005). Antagonistic interactions between hosts and parasites, in particular, are thought to generate strong selection capable of driving rapid change. The rate of change will depend on the additive genetic variance for both resistance in hosts and infectivity in parasites (Fisher 1930), which may be maintained by frequency-dependent selection in both species (Haldane 1948; Hamilton 1980). The rate of change should also depend on the breeding system for the host and/or parasite (Wright 1921). Additionally, rapid reciprocal evolution could intensify the coevolutionary arms race by increasing the strength of selection imposed upon the antagonist population.
Parasites are generally expected evolve in response to changes in their sympatric host populations, leading to greater infectivity on sympatric hosts than allopatric hosts (Ebert 1994; Gandon et al. 1998; Gandon and Michalakis 2002; Kawecki and Ebert 2004; Lively 1989; Price 1980). Such patterns of specialization, called local adaptation, are indicative of divergent selection and reciprocal evolution between sympatric hosts and parasites (Kawecki and Ebert 2004). Parasite local adaptation has also been interpreted to indicate that the parasites are “ahead” in the coevolutionary race (Roth et al. 2012; Schulte et al. 2011). The term “ahead” is generally used to mean that the parasite population’s relative fitness is greater than that of the hosts’ because the host population has not yet evolved a counter adaptation. However, theoretical studies have shown that the sign for ΔW̄ (change in mean fitness) oscillates over time from positive to negative, even in locally adapted parasite populations (Lively 1999). Hence, local adaptation by parasites, as inferred from reciprocal cross-infection experiments, may not be a good indicator of which of the antagonists is ahead. Nonetheless, reciprocal cross-infection experiments can be very informative on the nature of coevolution especially when combined with time-shift experiments (Koskella, this volume), or when hosts and parasites were experimentally derived from the same stock populations.
In the present study, we utilized the latter method. In a previous study, we experimentally passaged populations of the bacterial parasite, Serratia marcescens, derived from a single ancestral colony, with replicate host populations of a nematode host, Caenorhabditis elegans (Morran et al. 2011). The host populations were either partially outcrossing (mixed mating) or obligately outcrossing. We found that the mixed mating populations evolved greater outcrossing rates in response to coevolving parasites (Morran et al. 2013). In addition, after thirty host generations, both the mixed mating and obligately outcrossing hosts had evolved greater levels of parasite avoidance and/or resistance, and they exhibited lower mortality rates relative to their ancestors. The parasite populations, on the other hand, evolved greater levels of infectivity relative to their ancestral strain (Morran et al. 2011). Thus both hosts and parasites exhibited evolutionary change over the course of the experiment. However, the degree of reciprocal evolutionary change exhibited by both antagonists was not clear.
In the present study, we evaluated the effects of the host mating system on the evolutionary trajectories of both the hosts and parasites. We predicted that host populations with greater rates of outcrossing would evolve more rapidly in response to the parasite, and that this would feed back to generate faster rates of reciprocal evolution in the parasite. We assayed the effect of the host mating system on the change in host fitness over the course of the Morran et al. (2011) experiment. We then conducted reciprocal cross infection experiments to determine whether the parasites became adapted to their individual, “local” replicates of the host population, and whether the degree of adaptation depended on host mating system.
Material and Methods
Host and parasite populations
In our previous experiment (Morran et al. 2011), five replicate populations of obligately outcrossing C. elegans were passaged with populations of Serratia marcescens Sm2170 for 30 generations, thus allowing for coevolution. Five control populations of obligately outcrossing C. elegans were similarly treated, but in the absence of live parasites. Similarly, five replicate populations of partially outcrossing (mixed mating) C. elegans were allowed to coevolve with Serratia marcescens Sm2170 for 30 generations, and five control populations of the host were allowed to evolve in the absence of live parasites. All of the C. elegans populations were derived from a highly inbred PX382 (a CB4856 derivative) strain; but they were independently mutagenized with ethyl-methanesulfonate to infuse genetic variation prior to selection (Morran et al. 2011). The obligately outcrossing C. elegans populations were generated by backcrossing the fog-2 mutant allele into the PX382 background (Morran et al. 2009) prior to mutagenesis. The host and parasite populations were frozen before selection and after generation 30 of coevolution.
Multiple samples from each host and parasite population (approximately 500 nematodes per sample and several thousand bacterial cells per sample) were frozen after 30 generations of passages (Morran et al. 2011). A sample of each population was later thawed for analyses, mortality rates were approximately 50% in the nematode samples. After thawing, host populations were maintained for two generations under standard laboratory conditions to recover from the thawing process (i.e., 10cm Petri dishes filled with NGM Lite seeded with 30μL of OP50 stored at 20°C) (Nematode Growth Medium-Lite, US Biological, Swampscott, MA, USA). Parasite populations were transferred from frozen stock to LB (Luria Broth) and grown overnight at 28°C; they were then used to seed 10cm Petri dishes filled with NGM-Lite and grown at 28°C overnight.
Host adaptation
We measured rates of adaptation in the host populations using competitive fitness assays to compare the relative levels of evolutionary change associated with host mating system. Host fitness was evaluated by putting each coevolved host population in competition with a tester strain of C. elegans. These assays were conducted in the presence of the experimental host’s sympatric parasite population (the S. marcescens population that each nematode population coevolved with during experimental coevolution) to determine each population’s relative reproductive output, a product of survival and reproduction, in the context of the experiment. Host competitive fitness assays were conducted as described in (Morran et al. 2009), using the highly inbred GFP-marked C. elegans strain JK2735 serving as the tester strain. Here, the corresponding experimental, control, and ancestral hosts were exposed to their sympatric parasite population that coevolved with the experimental host population for thirty generations. The ancestral hosts are the progenitor population that was frozen prior to experimental coevolution for each experimental and control host population.
Host and parasite populations that were frozen after thirty generations of coevolution were thawed as described above. At least three replicate competitive fitness assays were conducted for each replicate population (ancestral, control, and coevolved populations). Competitive fitness assays were carried out by mixing 100 hosts from the experimental populations with 100 hosts from the GFP-marked tester strain. These hosts were then exposed to the parasite under conditions that mimicked the selective environment of experimental evolution. After four days of exposure (the same cycle as the experimental regime), we calculated the frequency of offspring that carried the GFP-marker. Given that the marker was introduced at 50%, deviations from 50% were indicative of greater relative fitness in either the experimental hosts (a decrease in frequency) or the tester strain (an increase in frequency). The tester strain carries a dominant marker, therefore hybrid offspring between an experimental host population and the tester strain would express GFP and be counted among the tester strain offspring. In this instance, the measures of host competitive fitness are potentially underestimates of their fitness, with the obligately outcrossing populations more susceptible to underestimate due to the greater frequency of males and more potential outcrossing with the tester strain. Overall, this assay tests the fitness of the experimental populations by requiring that they both survive and reproduce in the environment in which they evolved. The same tester strain is used for each competition assay; therefore each experimental population was measured against the same standard. We present the change in fitness exhibited by the experimental populations relative to the ancestral fitness in Figure (1) as a means of reporting evolutionary change. We also report the experimental to GFP ratios of the populations that were directly obtained from the fitness assays (Figure S1).
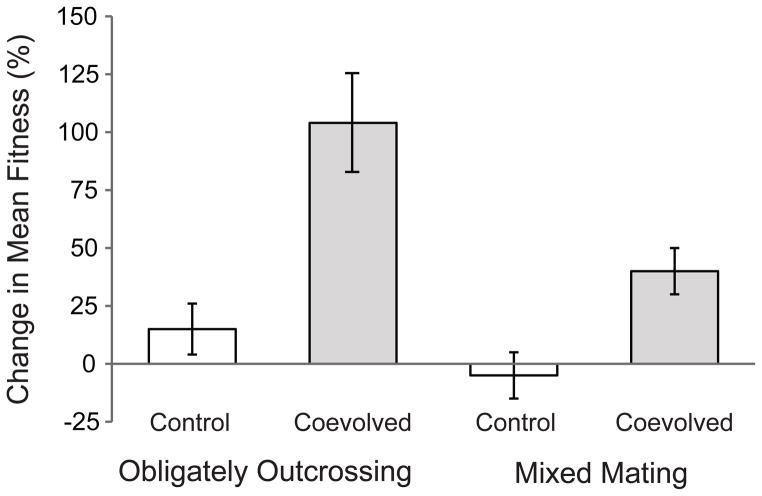
Figure 1.
Host adaptation. Obligately outcrossing hosts adapted more rapidly than mixed mating hosts (F1,8 = 10.35, p = 0.012). The relative fitness of control and coevolved host populations was determined through competitive fitness assays with a GFP-marked tester strain under exposure to the parasite population that coevolved with the host population. The change in fitness over the course of the experiment was calculated relative to the ancestral host population. Error bars indicate ± 1 S.E.M.
We performed an ANOVA in JMP 10 testing the effects of treatment (coevolved or control), mating system (mixed mating or obligately outcrossing), replicate population, and the interaction between treatment and mating system on the percent change in mean fitness relative to the ancestor. Replicate population was nested within mating system and treated as a random effect.
Local adaptation
Host mortality assays were conducted to measure parasite infectivity. Parasite infectivity, a common proxy for parasite fitness (see Kaltz and Shykoff (1998)), was measured as the 24-hour host mortality rate, which was a key component of parasite fitness in our previous experimental coevolution regime (Morran et al. (2011). Within the context of the experiment, the parasite had to kill a host in 24 hours to be passed to the next round of selection. Therefore the parasite’s ability to infect and kill the sympatric host in the allotted time was paramount to parasite fitness.
Host mortality assays were conducted as described in (Morran et al. 2011). However, here host mortality assays were conducted on host-parasite pairs that had coevolved together (sympatric) in addition to those that did not (allopatric). Host-parasite pairings were only done within each mating system group (obligately outcrossing or mixed mating), and each of the five host populations was paired with each of the five parasite populations. For each host population, ~200 L4 C. elegans individuals were suspended in M9 buffer and transferred to a 10 cm Petri dish (NGM-Lite agar), which had been seeded with a lawn of S. marcescens (approximately 6 cm in diameter). The estimated number of nematodes that were transferred to each plate was determined by plating aliquots of nematodes suspended in M9 buffer. The mean of three separate aliquots was calculated for each host populations and used as an estimate of the total number of hosts transferred. We exposed each host population to each parasite population for a total of 24 hours, and then counted the total number of dead nematodes. Twenty-four hour mortality rates were calculated by dividing the number of dead nematodes by the total number of transferred nematodes. Each host-by-parasite combination was replicated four times, with the exception of mixed mating host population # 3 infected by its sympatric parasite # 3, which had only three replicates.
We first tested whether there was a significant interaction effect between our parasite and host populations (Tables 1 and 2). We identified patterns of local adaptation in our parasite populations by conducting the sympatric-versus-allopatic test for local adaptation (Blanquart et al. 2013) on both the mixed mating and obligately outcrossing host populations (Table 3). This is a linear contrast test within an ANOVA that compares the values for all sympatric pairings to those of all allopatric pairings with no regard to the focal species. Host mortality rate values from both data sets were log transformed to meet ANOVA assumptions of normality. Then an ANOVA was performed for each group in JMP 10.0 (SAS Institute) testing the main effects of host population, parasite population, and interaction between the two main effects (Tables 1 and 2), then the subsequent linear contrast test were performed (Tables 3 and 4).
Table 1.
ANOVA table for mixed mating hosts. Main effects and interactions of host mortality rate data from the mixed mating hosts and their corresponding parasite populations.
| Source | Sum of Squares | df | Mean Square | F | P |
|---|---|---|---|---|---|
| Model | 2.29 | 24 | 0.096 | 10.95 | < 0.0001 |
| Error | 0.646 | 74 | 0.009 | ||
| Total | 2.94 | 98 | |||
| Host population | 1.52 | 4 | 0.38 | 43.58 | < 0.0001 |
| Pathogen population | 0.217 | 4 | 0.051 | 6.21 | = 0.0002 |
| Host pop. X Pathogen pop. | 0.675 | 16 | 0.042 | 4.83 | < 0.0001 |
Table 2.
ANOVA table for obligately outcrossing hosts. Main effects and interactions of host mortality rate data from the obligately outcrossing hosts and their corresponding parasite populations.
| Source | Sum of Squares | df | Mean Square | F | P |
|---|---|---|---|---|---|
| Model | 0.489 | 24 | 0.02 | 17.51 | < 0.0001 |
| Error | 0.087 | 75 | 0.001 | ||
| Total | .577 | 99 | |||
| Host population | 0.172 | 4 | 0.043 | 36.86 | < 0.0001 |
| Pathogen population | 0.021 | 4 | 0.005 | 4.49 | = 0.003 |
| Host pop. X Pathogen pop. | 0.297 | 16 | 0.019 | 15.92 | < 0.0001 |
Table 3.
Sympatric vs. Allopatric test of local adaptation. Linear contrast tests of all populations within each mating system group. The sympatric mortality rates were contrasted against the allopatric mortality rates to assess local adaptation.
| Contrast Test | F | DF (N,D) | P | Local Adaptation | ||
|---|---|---|---|---|---|---|
| Host Mating System | Sympatric | Allopatric | ||||
| Mixed | All possible sympatric combinations | All possible allopatric combinations | 41.41 | 1,74 | < 0.001 | Yes |
| Ob. Out | All possible sympatric combinations | All possible allopatric combinations | 182.81 | 1,75 | < 0.001 | Yes |
Table 4.
Fine scale sympatric versus allopatric test for local adaptation. Linear contrast tests for each population within each mating system group. The sympatric host-parasite mortality rates were contrasted against the host mortality rates of both the sympatric host infected by each allopatric parasite and the allopatric hosts infected by the sympatric parasite.
| Contrast Test | F | DF (N,D) | P | Local Adaptation | ||
|---|---|---|---|---|---|---|
| Host Mating System | Sympatric | Allopatric | ||||
| Mixed | Host 1 vs Path 1 | Host 1 vs Paths 2–5 & Path 1 vs Hosts 2–5 | 17.66 | 1,74 | < 0.001 | Yes |
| Mixed | Host 2 vs Path 2 | Host 2 vs Paths 1,3–5 & Path 2 vs Hosts 1,3–5 | 21.01 | 1,74 | < 0.001 | Yes |
| Mixed | Host 3 vs Path 3 | Host 3 vs Paths 1,2,4,5 & Path 3 vs Hosts 1,2,4,5 | 58.78 | 1,74 | < 0.001 | Yes |
| Mixed | Host 4 vs Path 4 | Host 4 vs Paths 1–3,5 & Path 4 vs Hosts 1–3,5 | 6.80 | 1,75 | = 0.011 | No |
| Mixed | Host 5 vs Path 5 | Host 5 vs Paths 1–4 & Path 5 vs Hosts 1–4 | 3.02 | 1,74 | = 0.086 | No |
| Ob. Out | Host 1 vs Path 1 | Host 1 vs Paths 2–5 & Path 1 vs Hosts 2–5 | 126.6 | 1,75 | < 0.001 | Yes |
| Ob. Out | Host 2 vs Path 2 | Host 2 vs Paths 1,3–5 & Path 2 vs Hosts 1,3–5 | 15.85 | 1,75 | < 0.001 | Yes |
| Ob. Out | Host 3 vs Path 3 | Host 3 vs Paths 1,2,4,5 & Path 3 vs Hosts 1,2,4,5 | 99.33 | 1,75 | < 0.001 | Yes |
| Ob. Out | Host 4 vs Path 4 | Host 4 vs Paths 1–3,5 & Path 4 vs Hosts 1–3,5 | 18.37 | 1,75 | < 0.001 | Yes |
| Ob. Out | Host 5 vs Path 5 | Host 5 vs Paths 1–4 & Path 5 vs Hosts 1–4 | 6.25 | 1,75 | = 0.015 | Yes |
We tested for local adaptation on a finer scale by then extending the sympatric vs. allopatric test to level of specific host-parasite population pairs. We performed a least squared mean linear contrast test for each host-parasite sympatric pairing against all possible allopatric pairings for the given host and parasite population (Table 4). Specifically, the transformed host mortality rates exhibited by the sympatric pair were contrasted with the transformed mortality rates induced by the sympatric parasite on the allopatric hosts in addition to the transformed mortality rates induced by allopatric parasites on the sympatric host. Therefore, within the contrast matrix (with host populations as rows and parasite populations as columns) these contrast tests compared the mean of the pairing on the diagonal to the global mean of all the values in the corresponding row and column (Table 4). Given this similarity to the sympatric versus allopatric test, we refer to the current test as the “fine scale sympatric vs. allopatric” test (Table 4). This test is also a linear contrast test with an ANOVA, just like the sympatric-versus-allopatric test. However, the fine scale approach involves performing the analysis for each sympatric species pairing, rather than just analyzing all pairings at one time. Further, we also conducted home versus away and local versus foreign tests (Kawecki and Ebert 2004) for local adaptation on our transformed data (app. A).
We then evaluated the degree of local adaptation under both host-mating systems by comparing the relative difference between sympatric and allopatric mortality rates. The relative sympatric infectivity of parasites against their sympatric hosts versus allopatric hosts was determined by calculating (x – y)/z for each sympatric host-parasite pair, where x is the sympatric pair mean mortality rate, y is the allopatric (as defined above in terms of the contrast tests) mean, and z is the mean of all mortality rates (both sympatric and allopatric) for a sympatric host-parasite pair. Therefore, the relative sympatric infectivity of a parasite population is the proportional change in the mean sympatric host mortality rate relative to the allopatric mortality rates, standardized by the overall mean mortality rate for a given pair of populations. A sum-rank Wilcoxon test was performed in JMP 10.0 to test for difference in the relative infectivity between the mixed mating and obligately outcrossing populations.
Results
Host adaptation
We evaluated the impact of the host mating system on the evolutionary trajectories of the host populations. We assessed fitness by putting each experimental host population, in addition to their respective controls and ancestral populations, in competition against a GFP-marked tester C. elegans strain. These populations were assayed for competitive ability when exposed to the experimental population’s sympatric parasite population to determine relative fitness after thirty host generations of coevolution. The rate of adaptation for each experimental and control host population was then measured by calculating the change in mean fitness over time relative to the fitness of their ancestral population.
Overall, the experimental host populations exhibited a significantly greater rate of adaptation than the control hosts (Fig. 1; F1,12 = 23.28, p < 0.0001), indicating that both the mixed mating and obligately outcrossing experimental host populations adapted to the parasite populations with which they coevolved. Further, the obligately outcrossing experimental host populations became adapted to their coevolving parasites at a greater rate than the mixed mating hosts (Fig. 1; F1,8 = 10.35, p = 0.012). Although the competitive fitness of the obligately outcrossing and mixed mating hosts was not significantly different in direct comparison (Fig. S1; F1,16 = 1.91, p = 0.186), the obligately outcrossing populations exhibited a greater magnitude of change relative to their ancestors and thus a greater rate of adaptation (Fig. 1). Therefore, the host mating system influenced the evolutionary trajectories of the host populations, with obligate outcrossing facilitating greater rates of host adaptation.
Local adaptation
We then tested for parasite local adaptation within each host-mating type (mixed mating or obligately outcrossing) to determine if host mating system had an effect on the evolutionary trajectories of the parasite populations. Within a host mating group, we exposed each of the five parasite populations to each of the five mixed mating host populations. These reciprocal exposures generated one sympatric pairing (host-parasite populations that coevolved together) and eight allopatric pairings (host-parasite populations that coevolved with a different antagonistic population) per host-parasite sympatric combination. The eight allopatric pairings were comprised of the focal parasite against the four allopatric host populations and the focal host against the four allopatric parasite populations. We then measured the twenty-four hour host mortality rates exhibited by each pair to identify local adaptation, as indicated by greater sympatric host mortality rates. Host killing at twenty-four hours of exposure was the parasite trait under selection during experimental evolution and a proxy for parasite infectivity (Morran et al. 2011).
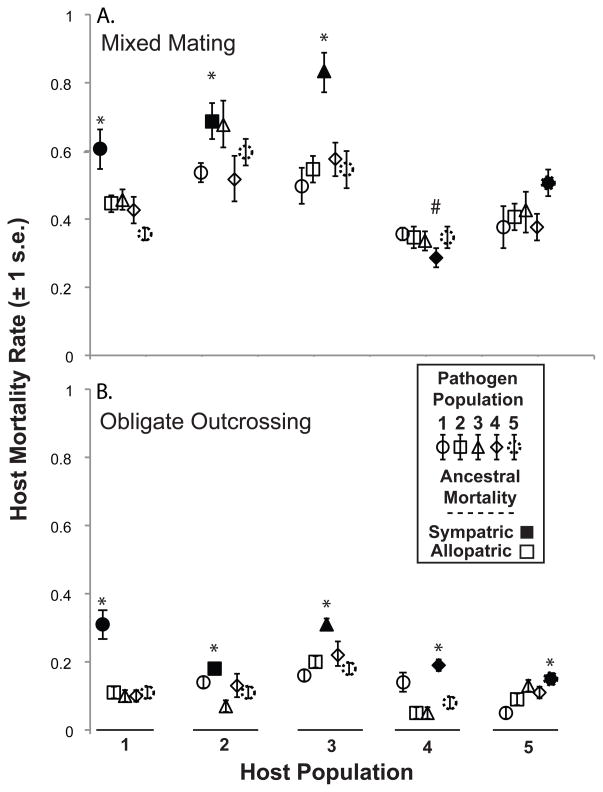
We detected significant levels of local adaptation in both the mixed mating (Tables 1 and 3) and obligately outcrossing populations (Tables 2 and 3) using the sympatric-vs-allopatric test (Blanquart et al. 2013). Then, we extended the sympatric vs. allopatric test to the level of population in order to assess local adaptation at a finer scale. We defined local adaptation as the case for which the focal parasite population induced significantly greater host mortality against its sympatric host population than its allopatric hosts combined with the mortality rates for all allopatric parasites exposed to the focal host. Using this method, we detected local adaptation in three-of-five parasite populations that coevolved with mixed mating hosts (Fig. 2A and Table 4). In the parasite populations that did not exhibit local adaptation, one population was roughly equally infective relative to the allopatric populations in comparison to their sympatric hosts, while the other population induced significantly lower mortality rates relative to allopatric populations (Fig. 2A and Table 4). In the obligately outcrossing populations, we found that all five parasite populations exhibited significant levels of local adaptation (Fig. 2B and Table 4).
Figure 2.
Parasite local adaptation. (A) Three of the five parasite populations that coevolved with mixed mating hosts exhibited local adaptation, while one of the populations was locally maladapted (Tables 1 and A1). (B) Each of the five parasite populations that coevolved with obligately outcrossing hosts exhibited local adaptation (Tables 1 and A2). * indicates local adaptation and # indicates significantly greater mortality in allopatric pairs.
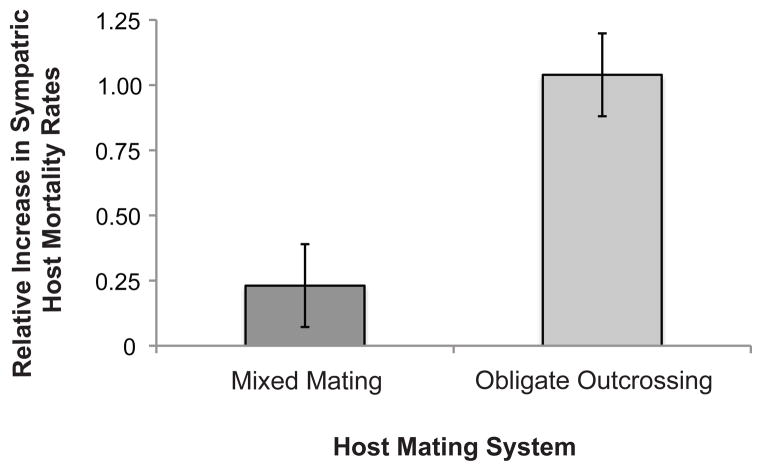
Next, we compare the degree of parasite local adaptation in the mixed mating and obligately outcrossing groups. The parasites that coevolved with obligately outcrossing hosts exhibited a greater degree of local adaptation than those that coevolved with mixed mating hosts. Specifically, parasites from sympatric host pairings in the obligately outcrossing group exhibited proportionally greater increases in host mortality, measured as the difference between the sympatric and allopatric mean mortality rates relative to the mean mortality rate for a given host population, than the mixed mating group (Fig. 3; χ21 = 5.77, p = 0.0163). Therefore, the parasites that coevolved with the obligately outcrossing hosts exhibited a greater degree of divergence in their evolutionary trajectories from their origin of a common clonal ancestor.
Figure 3.
Relative infectivity. Parasites that coevolved with obligately outcrossing hosts induced proportionally greater mortality rates against sympatric hosts than parasites that coevolved with mixed mating hosts (χ21 = 5.77, p = 0.0163). The relative increase in mortality rate was determined by standardizing the difference between the sympatric and mean allopatric mortality rate by the overall mean for each host-parasite pair. Error bars indicate ± 1 S.E.M.
Discussion
Our results illustrate the rapid rate of coevolutionary change that can be driven by host-parasite interactions. Despite evolving from a single ancestral colony, eight-of-ten parasite populations exhibited local adaptation as a result of reciprocal coevolution with their respective host population over a span of just thirty host generations (Fig. 2). These parasites rapidly evolved host specialization, indicating that natural selection generated by coevolution was likely quite strong, and that the replicate parasite populations traveled different evolutionary paths from a common ancestral starting point. The different evolutionary trajectories of the parasite populations are presumably due to different mutations that arose in each population throughout the experiment and subsequent reciprocal evolution in response to different host populations that each followed their own unique evolutionary path to resistance or parasite avoidance.
The coevolutionary dynamics of the host-parasite interactions were greatly affected by the host’s mating system, which also exerted influence over the evolutionary trajectories of both the host and parasite populations. Specifically, the obligately outcrossing hosts exhibited more rapid adaptation than mixed mating hosts (Fig. 1), and coevolution with obligately outcrossing hosts resulted in a greater degree of divergent evolution between parasite populations (Fig. 3). Thus, it seems that obligate outcrossing indirectly stimulated a greater degree of divergent evolution between the parasite populations by facilitating more rapid evolution on the part of the hosts. Greater rates of adaptation in obligately outcrossing hosts likely increased the strength of selection against their coevolving parasites. Consequently, the parasites coevolving with obligately outcrossing host populations exhibited greater rates of divergent evolution and more rapid host specialization than parasites that coevolved with mixed mating hosts (Fig. 2 and Fig. 3). We suggest that, compared to mixed mating, obligate outcrossing facilitated more rapid host evolution, leading to greater divergence among replicate populations. Parasite tracking of these host populations then led to greater divergence among the replicate parasite populations, producing a stronger signal of local adaptiation.
Mating systems are particularly influential traits from an evolutionary perspective. They have the potential to shape the distribution of genetic variation within populations and therefore alter evolutionary trajectories in response to natural selection (Bell 1982; Charlesworth and Charlesworth 1987; Fisher 1930; Lande and Schemske 1985; Stebbins 1957). Obligate outcrossing has previously been shown to facilitate more rapid adaptation to a fixed parasite population and to reduce rates of host infection more rapidly than mixed mating in this C. elegans-S. marcescens host-parasite system (Morran et al. 2009). Here, we demonstrated that obligate outcrossing also facilitated more rapid adaptation to coevolving parasites than mixed mating (Fig. 1).
Given that our host populations were subjected to mutagenesis prior to experimental evolution (Morran et al. 2011), it is possible that a portion of this adaptive response is due to the purging of deleterious mutations, rather than selection for specific beneficial alleles (Glemin 2003; Roze and Rousset 2004; Whitlock 2002). However, the lack of significant change in the fitness of control hosts over the course of the experiment (Fig. 1) indicates that a large portion of any such purging had to be parasite-mediated. If the parasites facilitated purging, then the hosts adapted to coevolving parasites and the change in host fitness is still indicative of the rate of evolutionary change in the hosts. Nonetheless, we believe that the adaptation we observed (Fig. 1) was largely due to the assembly of novel beneficial genotypes for two reasons. First, the mixed mating populations should be capable of purging at a much greater rate than the obligately outcrossing populations (Charlesworth and Charlesworth 1987; Charlesworth et al. 1993; Lande and Schemske 1985). The results (Fig. 1) are not consistent with this expectation because the obligately outcrossing populations exhibited a greater increase in fitness. Second, the mixed mating populations showed elevated levels of avoidance/resistance against S. marcescens relative to a non-mutated strain with the same genetic background (CB4856) (L. Morran, pers. observation, 2013). Overall, these results further demonstrate the advantage of outcrossing relative to self-fertilization under host-parasite coevolution. These results may also have broad implications for explaining patterns of host and parasite local adaptation.
Many factors, both biotic and abiotic, are known to influence parasite local adaptation. However, patterns of local adaptation, or the lack thereof, have proven difficult to predict in many cases. Spatial structure (Forde et al. 2004; King et al. 2009; Koskella et al. 2011; Laine 2005; Lively 1999; Lively and Jokela 1996; Thrall et al. 2012; Vogwill et al. 2008; Vogwill et al. 2010), temporal dynamics (Berenos et al. 2012; Forde et al. 2004; Lively 1999; Thrall et al. 2012), host-parasite infection dynamics (Berenos et al. 2012; Koskella et al. 2012; Koskella et al. 2011; Lively 1999; Lively and Dybdahl 2000; Schulte et al. 2011; Thrall et al. 2012), temperature (Laine 2008), nutrient levels (Lopez Pascua et al. 2012), and even the lab environment (Laine 2007) can play a role in determining the extent of parasite local adaptation. Each of these factors plays a role in either determining the consistency and strength of divergent selection and/or the ability of host or parasite populations to respond to divergent selection. Our study indicates that the host mating system, and consequently the rate of host adaptation, is a key factor in establishing parasite local adaptation. Specifically, we find that the host mating system acts as a biotic factor that both determines the strength of selection on the parasite and also influences the ability of the host to respond to reciprocal selection. Although the host’s mating system itself may not always be a predictor of local adaptation, the rate of adaptation in the host population could be quite influential.
Some degree of coevolutionary change is certainly necessary before patterns of local adaptation can emerge. But, highly asymmetrical rates of evolution between hosts and parasites are not conducive for local adaptation (Morgan and Buckling 2006). When hosts are under weaker selection relative to their coevolving parasites, the rate of adaptation in the host population might often be the limiting factor for reciprocal coevolution and the establishment of local adaptation. In that case, any factors that might increase the rate of reciprocal evolution on the part of the host could enhance the degree of local adaptation exhibited by parasites by simply driving divergent evolution between allopatric parasite populations. As sympatric hosts and parasites travel farther along their unique evolutionary trajectories they are likely develop greater levels of specificity towards their sympatric antagonists, potentially at the cost of the ability to infect generally. Parasites are capable of enhancing the rate of divergent evolution between allopatric host populations (Buckling and Rainey 2002), which should ultimately serve to reinforce parasite local adaptation no matter their standing in the coevolutionary arms race.
Parasite local adaptation has been interpreted to mean that the parasites are ahead of their hosts in their coevolutionary arms race (Roth et al. 2012; Schulte et al. 2011). However, we found that the parasites that coevolved with the obligately outcrossing hosts induced lower rates of mortality in their contemporary hosts after thirty generations of coevolution than the ancestral parasites did against their ancestral hosts (Morran et al. 2013; Morran et al. 2011). This means that the hosts evolved greater resistance and/or avoidance faster than the parasites evolved greater infectivity and virulence. We interpret this to mean the hosts were ahead of their coevolving parasites after thirty generations of experimental coevolution. Despite the host’s apparent advantage, the parasite populations consistently exhibited local adaptation (Fig. 2B) and a high degree of specificity (Fig. 3). Conversely the mixed mating hosts exhibited approximately equal mean susceptibility to their coevolving parasites after thirty generations of coevolution relative to their ancestral starting points (Morran et al. 2013; Morran et al. 2011), and exhibited a mix of local adaptation and no local specificity or susceptibility (Fig. 2A), while exhibiting a lesser overall degree of specificity relative to the obligately outcrossing hosts (Fig. 3). Similar results were observed in an experimental study in which the bacterial hosts adapted more rapidly than their phage parasites, yet they also observed parasite local adaptation (Gomez and Buckling 2011). Lopez Pascua et al. (2012) also observed this lack of congruence between local adaptation and changes in fitness over time. Therefore, parasites can exhibit local adaptation even though their hosts are “ahead,” meaning hosts are evolving resistance at a greater rate than the parasites are evolving infectivity, in the coevolutionary arms race. Thus, caution should be exercised when interpreting local adaptation as an indicator of a host or parasite population’s standing in a coevolutionary arms race.
In conclusion, our results demonstrate the ability of host-parasite coevolution to generate rapid evolutionary change. Additionally, we found that a specific host trait, mating system, influenced the evolutionary trajectories of both the hosts and parasites, altering the rate of reciprocal adaptation in the host-parasite interaction. And, although patterns of local adaptation are often difficult to predict, the rate of adaptation in the host population may be a key factor in determining the rate and degree of reciprocal evolution in host-parasite interactions.
Supplementary Material
Acknowledgments
We thank J. Bronstein for helpful comments on the manuscript and assistance with this symposium issue. We thank H. Hundley for the use of her laboratory space and equipment. We also thank M. Greischar, the members of the Lively, Hall, and Wade labs for beneficial discussion pertaining to this work, and several anonymous reviewers for helpful comments. Funding was provided by the NSF (DEB-0640639 to CML) and the NIH (1F32GM096482-01 to LTM). The nematode strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). LTM and CML conceived and designed the experiments. LTM, RCP, IAG, and MBA performed the experiments. LTM and CML analyzed the data. LTM and CML wrote the paper.
Contributor Information
Levi T. Morran, Email: levi.morran@emory.edu.
Raymond C. Parrish, II, Email: raycparr@umail.iu.edu.
Ian A. Gelarden, Email: iagelard@imail.iu.edu.
Michael B. Allen, Email: allenmb@imail.iu.edu.
Curtis M. Lively, Email: clively@indiana.edu.
Literature Cited
- Bell G. The Masterpiece of Nature: The Evolution and Genetics of Sexuality. Berkley, CA: University of California Press; 1982. [Google Scholar]
- Berenos C, Schmid-Hempel P, Wegner KM. Complex adaptive responses during antagonistic coevolution between Tribolium castaneum and its natural parasite Nosema whitei revealed by multiple fitness components. BMC Evol Biol. 2012;12:11. doi: 10.1186/1471-2148-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A, Rainey PB. The role of parasites in sympatric and allopatric host diversification. Nature. 2002;420:496–499. doi: 10.1038/nature01164. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Syst. 1987;18:237–268. [Google Scholar]
- Charlesworth D, Morgan MT, Charlesworth B. Mutation accumulation in finite, outbreeding, and inbreeding populations. Genetics Research. 1993;61:39–56. [Google Scholar]
- Ebert D. Virulence and local adaptation of a horizontally transmitted parasite. Science. 1994;265:1084–1086. doi: 10.1126/science.265.5175.1084. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Oxford: Clarendon Press; 1930. [Google Scholar]
- Forde SE, Thompson JN, Bohannan BJ. Adaptation varies through space and time in a coevolving host-parasitoid interaction. Nature. 2004;431:841–844. doi: 10.1038/nature02906. [DOI] [PubMed] [Google Scholar]
- Gandon S, Ebert D, Olivieri I, Michalakis Y. Differential adaptation in spatially hetergeneous environments and host-parasite coevolution. In: Mopper S, Strauss S, editors. Genetic Structure and Local Adaptation in Natural Insect Populations. London: Chapman & Hall; 1998. [Google Scholar]
- Gandon S, Michalakis Y. Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, mutation, population size, and generation time. J Evol Biol. 2002;15:451–462. [Google Scholar]
- Glemin S. How are deleterious mutations purged? Drift versus nonrandom mating. Evolution. 2003;57:2678–2687. doi: 10.1111/j.0014-3820.2003.tb01512.x. [DOI] [PubMed] [Google Scholar]
- Gomez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. The theory of a cline. J Genet. 1948;48:277–284. doi: 10.1007/BF02986626. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. Sex Versus Non-Sex Versus Parasite. Oikos. 1980;35:282–290. [Google Scholar]
- Kaltz O, Shykoff JA. Local adaptation in host-parasite systems. Heredity (Edinb) 1998;81:361–370. [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- King KC, Delph LF, Jokela J, Lively CM. The Geographic Mosaic of Sex and the Red Queen. Curr Biol. 2009;19:1438–1441. doi: 10.1016/j.cub.2009.06.062. [DOI] [PubMed] [Google Scholar]
- Koskella B, Lin DM, Buckling A, Thompson JN. The costs of evolving resistance in heterogeneous parasite environments. Proc R Soc B. 2012;279:1896–1903. doi: 10.1098/rspb.2011.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B, Thompson JN, Presont GM, Buckling A. Local biotic environment shapes the spatial scale of bacteriophage adaptation to bacteria. Am Nat. 2011;177:440–451. doi: 10.1086/658991. [DOI] [PubMed] [Google Scholar]
- Laine AL. Spatial scale of local adaptation in a plant-pathogen metapopulation. J Evol Biol. 2005;18:930–938. doi: 10.1111/j.1420-9101.2005.00933.x. [DOI] [PubMed] [Google Scholar]
- Laine AL. Detecting local adaptation in a natural plant-pathogen metapopulation: a laboratory vs. field transplant approach. J Evol Biol. 2007;20:1665–1673. doi: 10.1111/j.1420-9101.2007.01359.x. [DOI] [PubMed] [Google Scholar]
- Laine AL. Temperature-mediated patterns of local adaptation in a natural plant-pathogen metapopulation. Ecol Lett. 2008;11:327–337. doi: 10.1111/j.1461-0248.2007.01146.x. [DOI] [PubMed] [Google Scholar]
- Lande R, Schemske DW. The evolution of self-fertilization and inbreeding depression in plants. 1. genetic models. Evolution. 1985;39:24–40. doi: 10.1111/j.1558-5646.1985.tb04077.x. [DOI] [PubMed] [Google Scholar]
- Lively CM. Adaptation by a parasitic trematode to local populations of its snail host. Evolution. 1989;43:1663–1671. doi: 10.1111/j.1558-5646.1989.tb02616.x. [DOI] [PubMed] [Google Scholar]
- Lively CM. Migration, virulence, and the geographic mosaic of adaptation by parasites. Am Nat. 1999;153:S34–S47. doi: 10.1086/303210. [DOI] [PubMed] [Google Scholar]
- Lively CM, Dybdahl MF. Parasite adaptation to locally common host genotypes. Nature. 2000;405:679–681. doi: 10.1038/35015069. [DOI] [PubMed] [Google Scholar]
- Lively CM, Jokela J. Clinal variation for local adaptation in a host-parasite interaction. Proc R Soc Lond B. 1996;263:891–897. [Google Scholar]
- Lopez Pascua L, Gandon S, Buckling A. Abiotic heterogeneity drives parasite local adaptation in coevolving bacteria and phages. J Evol Biol. 2012;25:187–195. doi: 10.1111/j.1420-9101.2011.02416.x. [DOI] [PubMed] [Google Scholar]
- Morgan AD, Buckling A. Relative number of generations of hosts and parasites does not influence parasite local adaptation in coevolving populations of bacteria and phages. Journal of Evolutionary Biology. 2006;19:1956–1963. doi: 10.1111/j.1420-9101.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- Morran LT, Parmenter MD, Phillips PC. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature. 2009;462:350–352. doi: 10.1038/nature08496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Parrish RC, 2nd, Gelarden IA, Lively CM. Temporal dynamics of outcrossing and host mortality rates in host-pathogen experimental coevolution. Evolution. 2013;67:1860–1868. doi: 10.1111/evo.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Schmidt OG, Gelarden IA, Parrish RC, 2nd, Lively CM. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science. 2011;333:216–218. doi: 10.1126/science.1206360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PW. Evolutionary Biology of Parasites. Princeton: Princeton University Press; 1980. [Google Scholar]
- Roth O, Keller I, Landis SH, Salzburger W, Reusch TB. Hosts are ahead in a marine host-parasite coevolutionary arms race: innate immune system adaptation in pipefish Syngnathus typhle against Vibrio phylotypes. Evolution. 2012;66:2528–2539. doi: 10.1111/j.1558-5646.2012.01614.x. [DOI] [PubMed] [Google Scholar]
- Roze D, Rousset FO. Joint effects of self-fertilization and population structure on mutation load, inbreeding depression and heterosis. Genetics. 2004;167:1001–1015. doi: 10.1534/genetics.103.025148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. Host-parasite local adaptation after experimental coevolution of Caenorhabditis elegans and its microparasite Bacillus thuringiensis. Proc Biol Sci. 2011;278:2832–2839. doi: 10.1098/rspb.2011.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. Self-fertilization and population variation in higher plants. American Naturalist. 1957;91:337–354. [Google Scholar]
- Thompson JN. Interaction and coevolution. New York: John Wiley and Sons; 1982. [Google Scholar]
- Thompson JN. The coevolutionary process. Chicago: University of Chicago Press; 1994. [Google Scholar]
- Thompson JN. The geographic mosaic of coevolution. Chicago: University of Chicago Press; 2005. [Google Scholar]
- Thrall PH, Laine AL, Ravensdale M, Nemri A, Dodds PN, Barrett LG, Burdon JJ. Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation. Ecol Lett. 2012;15:425–435. doi: 10.1111/j.1461-0248.2012.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogwill T, Fenton A, Brockhurst MA. The impact of parasite dispersal on antagonistic host-parasite coevolution. Journal of Evolutionary Biology. 2008;21:1252–1258. doi: 10.1111/j.1420-9101.2008.01574.x. [DOI] [PubMed] [Google Scholar]
- Vogwill T, Fenton A, Brockhurst MA. How Does Spatial Dispersal Network Affect the Evolution of Parasite Local Adaptation? Evolution. 2010;64:1795–1801. doi: 10.1111/j.1558-5646.2009.00937.x. [DOI] [PubMed] [Google Scholar]
- Whitlock MC. Selection, load and inbreeding depression in a large metapopullation. Genetics. 2002;160:1191–1202. doi: 10.1093/genetics/160.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Systems of mating. IV. The effects of selection. Genetics. 1921;6:162–166. doi: 10.1093/genetics/6.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.