In budding yeast, telomerase consists of the catalytic Est2 protein and two regulatory subunits (Est1 and Est3) in association with the TLC1 RNA, with each of the four subunits essential for in vivo telomerase function. Tucey and Lundblad show that a hierarchy of assembly and disassembly results in limiting amounts of the quaternary complex late in the cell cycle, following completion of DNA replication. Telomerase disassembles due to dissociation of the catalytic subunit from the complex in every cell cycle.
Keywords: telomerase, telomeres, Est1, Est2, Est3, TLC1
Abstract
The enzyme telomerase, which elongates chromosome termini, is a critical factor in determining long-term cellular proliferation and tissue renewal. Hence, even small differences in telomerase levels can have substantial consequences for human health. In budding yeast, telomerase consists of the catalytic Est2 protein and two regulatory subunits (Est1 and Est3) in association with the TLC1 RNA, with each of the four subunits essential for in vivo telomerase function. We show here that a hierarchy of assembly and disassembly results in limiting amounts of the quaternary complex late in the cell cycle, following completion of DNA replication. The assembly pathway, which is driven by interaction of the Est3 telomerase subunit with a previously formed Est1–TLC1–Est2 preassembly complex, is highly regulated, involving Est3-binding sites on both Est2 and Est1 as well as an interface on Est3 itself that functions as a toggle switch. Telomerase subsequently disassembles by a mechanistically distinct pathway due to dissociation of the catalytic subunit from the complex in every cell cycle. The balance between the assembly and disassembly pathways, which dictate the levels of the active holoenzyme in the cell, reveals a novel mechanism by which telomerase (and hence telomere homeostasis) is regulated.
In most species, the primary pathway for telomere maintenance relies on the enzyme telomerase, which replenishes chromosome termini to ensure continuous cellular proliferation (Lundblad and Szostak 1989; Bodnar et al. 1998). Even subtle differences in telomerase function as either increases or decreases can have a significant impact on human health. Reduced telomerase activity confers a broad spectrum of disease syndromes, referred to as “syndromes of telomere shortening” (Armanios 2009; Armanios and Blackburn 2012). For example, a multigenerational germline mutation in hTERT (encoding the telomerase reverse transcriptase component) that attenuates one aspect of telomerase enzymatic activity confers a progressive decline in lung function late in life (Alder et al. 2011). Conversely, telomerase activity is up-regulated in many human cancers; this increased expression is often due to mutations in the promoter of hTERT, which can occur in multiple types of both familial and recurrent sporadic cancers (Heidenreich et al. 2014).
Telomerase performs its role at telomeres as a ribonucleoprotein particle (RNP) composed of a highly conserved catalytic core as well as regulatory proteins (Collins 2011). Catalysis relies on the TERT protein subunit bound to an integral RNA, with the latter providing a template that dictates the sequence of the telomeric repeat added onto the 3′ end of chromosome ends. Additional telomerase-associated proteins contribute activities that are essential for holoenzyme function in vivo. This includes factors required for telomerase biogenesis, which are often shared with other RNPs; for example, budding and fission yeast share RNA biogenesis factors with the splicing machinery (Seto et al. 1999; Tang et al. 2012), whereas vertebrate telomerase relies on the ribosome biogenesis factor dyskerin (Mitchell et al. 1999). The ATPases pontin and reptin also contribute as preassembly factors to promote cell cycle-specific assembly of human telomerase but are not associated with the mature telomerase complex (Venteicher et al. 2008). In contrast, TCAB1, which controls transit of telomerase through Cajal bodies, is a stable subunit of the active telomerase holoenzyme (Venteicher et al. 2009), as is dyskerin (Cohen et al. 2007).
Budding yeast telomerase similarly relies on two regulatory subunits: Est1 and Est3 (Lundblad and Szostak 1989; Morris and Lundblad 1997; Hughes et al. 2000; Livengood et al. 2002). Although these two proteins are dispensable for enzyme catalysis (Cohn and Blackburn 1995; Lingner et al. 1997), est1-∆ or est3-∆ mutant strains exhibit a telomere replication defect that is indistinguishable from that of strains bearing mutations in telomerase RNA or the reverse transcriptase (called TLC1 and EST2, respectively) (Singer and Gottschling 1994; Lendvay et al. 1996). In vitro, Est2 and TLC1 are sufficient to reconstitute enzyme activity (Zappulla et al. 2005). However, Est2 and TLC1 do not exist as a discrete subcomplex in vivo; instead, the Est2–TLC1 catalytic core associates with the Est1 regulatory subunit throughout most of the cell cycle (Tucey and Lundblad 2013). This pattern of association for Est1 argues against a model proposing that the catalytic core of telomerase is constitutively bound to telomeres in an inactive state and only becomes “activated” in late S phase due to Est1 association (Taggart et al. 2002). Instead, we proposed that Est1 performs its regulatory function as a telomerase recruitment factor through an interaction with the telomere-bound Cdc13 (Nugent et al. 1996; Evans and Lundblad 1999). These two proteins interact through surfaces defined by the location of reciprocally cosuppressing mutations in EST1 and CDC13 (Pennock et al. 2001); notably, a biochemical association between Est1 and Cdc13 can be observed even in the G1 phase of the cell cycle, when telomerase does not elongate telomeres (Tucey and Lundblad 2013). These studies as well as others (Bianchi et al. 2004; Gallardo et al. 2011) are consistent with a simple model in which telomere elongation is initiated by binding of Cdc13 to telomeres late in the cell cycle, followed by recruitment of a previously assembled telomerase holoenzyme.
Potential insights into the function for the Est3 regulatory subunit have come from a recent study that has provided the first structure of a yeast telomerase protein (Rao et al. 2014). This was coupled with an unbiased mutagenesis of every solvent-accessible residue on the Est3 protein surface, which showed that residues required for telomere replication cluster to a single face. This cluster can be further delineated into two functionally distinct patches: A narrow extended interface mediates association of Est3 with telomerase, with an adjacent surface used for a second function distinct from telomerase binding. The landscape of the Est3 protein surface also reveals features that are shared with the human TPP1 protein; in particular, Est3 and TPP1 use similar surfaces (the “TEL” patch) to associate with the telomerase RNP (Nandakumar et al. 2012; Zhong et al. 2012; Rao et al. 2014). Since TPP1 performs its role as a subunit of the shelterin complex, whereas Est3 is a subunit of telomerase, these two proteins are not obvious orthologs. Nevertheless, their shared structural features suggest that in addition to the TEL patch there may be further telomere-related functions common to both proteins.
In this study, we show that two discrete pathways promote assembly and disassembly of the yeast telomerase RNP. The assembly pathway is driven by binding of the Est3 protein to an Est1–TLC1–Est2 preassembly complex late in the cell cycle to form the holoenzyme. This process is highly regulated, as formation of the quaternary complex relies on sites on both Est1 and Est2 that bind Est3 as well as a toggle switch on the surface of Est3. Telomerase is subsequently subjected to a cell cycle-regulated disassembly process due to dissociation of Est2 from the enzyme to yield an inactive complex composed of Est1, Est3, and TLC1. These mechanistically distinct pathways therefore provide two distinct means by which telomerase biogenesis is regulated: The assembly pathway ensures that the holoenzyme is not formed until late in the cell cycle, and the subsequent disassembly pathway ensures that active enzyme levels remain low. This dynamic balance between the telomerase quaternary complex and its subcomplexes reveals a new regulatory contribution to telomere homeostasis in budding yeast; a similar process of assembly and disassembly may ensure that active telomerase is maintained at low levels in other systems as well.
Results
Assembly of the telomerase quaternary complex occurs late in the cell cycle
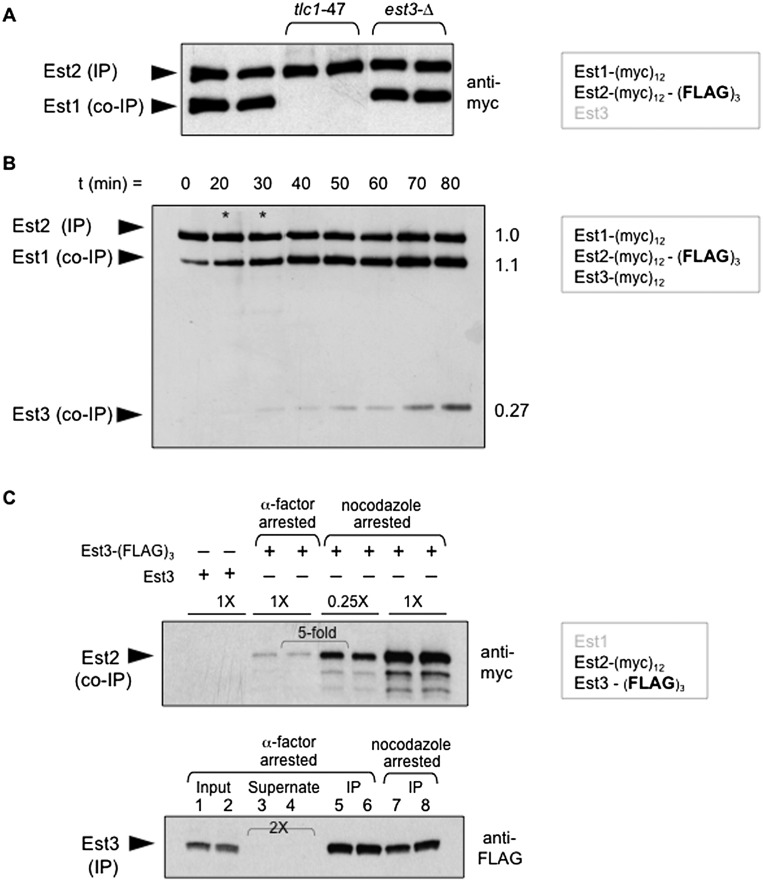
Previous studies have shown that a complex containing the Est1 and Est2 proteins bound to the TLC1 RNA in a 1:1 ratio can be recovered following anti-Est2 immunoprecipitation (Fig. 1A; Lubin et al. 2012). This experiment was performed with a strain in which Est1 and Est2 contained identical (myc)12 epitopes, with an additional (Flag)3 epitope on Est2. Anti-Flag immunoprecipitation targets the limiting protein subunit of the complex (Est2), which is quantitatively depleted from extracts (Supplemental Fig. S1A; Tucey and Lundblad 2013). The presence of (myc)12-tagged Est1 and Est2 proteins in anti-Est2 immunoprecipitates are detected simultaneously on a single anti-myc Western, thereby providing a highly accurate assessment of the relative ratio of these two proteins. This provides a much more sensitive assay for defects in the ability to form the Est1–TLC1–Est2 subcomplex than the conventional approach, which measures the relative levels of the Est1 or Est2 proteins and the TLC1 RNA in separate immunoprecipitates. Using this assay, the Est1:Est2 ratio was unperturbed in an est3-∆ strain (Fig. 1A), indicating that formation of the Est1–TLC1–Est2 intermediate is independent of the Est3 telomerase subunit.
Figure 1.
Est3 associates with the Est1–TLC1–Est2 subcomplex late in the cell cycle. (A) The levels of Est1 and Est2 proteins in anti-Est2 immunoprecipitations (IPs), as assessed by an anti-myc Western of anti-Flag immunoprecipitations prepared from strains bearing identical (myc)12 epitopes present on the C and N termini of Est1 and Est2, respectively, and with the indicated mutations. (B) The pattern of association of the three Est proteins through the cell cycle following anti-Flag immunoprecipitation of Est2 using a strain with all three proteins bearing the same (myc)12 epitope plus an additional (Flag)3 epitope on Est2; epitope-tagged constructs were integrated in place of the endogenous genes. Extracts were isolated from cultures taken at the indicated time points following release from a G1 arrest and analyzed by FACS (Supplemental Fig. S1C) at each time point. Subsequent anti-Flag immunoprecipitations were monitored by anti-myc Westerns; in the two lanes with asterisks, a different exposure was inserted to compensate for the slightly lower amounts of sample loaded in these two lanes. The ratios of Est1 and Est3 proteins in anti-Est2 immunoprecipitations at the 80-min time point (relative to the Est2 protein) are shown at the right; quantitation for all time points is presented in Supplemental Figure S1C. The strain used in this experiment contains a frameshift correction of the Est3 ORF with a corresponding increase in Est3 protein levels (Morris and Lundblad 1997); however, Supplemental Figure S1B demonstrates that the extent of interaction of Est3 with the Est1–TLC1–Est2 subcomplex is comparable in strains with or without the frameshift-corrected allele. (C, top) Anti-myc Western of immunoprecipitation samples monitoring levels of (myc)12-Est2 following anti-Flag immunoprecipitation of Est3-(Flag)3 from two independent cultures arrested with α factor or nocodazole; “0.25X” indicates the amount of immunoprecipitation loaded relative to the amount loaded in “1X” lanes. The mock anti-Flag immunoprecipitation from a strain lacking a (Flag)3 epitope on Est3 (first two lanes) demonstrates that the low levels of the (myc)12-Est2 protein in the α-factor-arrested cultures are biologically relevant and not due to nonspecific binding. (Bottom) The anti-Flag Western shows comparable amounts of Est3 in immunoprecipitations from α-factor-arrested (lanes 5,6) or nocodazole-arrested (lanes 7,8) cells and also shows that the Est3-(Flag)3 protein is depleted from extracts; “2X” indicates that twice as much of the supernate following anti-Flag immunoprecipitation was loaded in lanes 3 and 4 relative to the volume loaded in lanes 1 and 2.
To monitor association of Est3 with the Est1–TLC1–Est2 subcomplex, a variant of the above strain was constructed in which all three Est protein subunits bore (myc)12 epitopes, with the (Flag)3 epitope on Est2. When anti-Est2 immunoprecipitations were examined through the cell cycle, the Est1–TLC1–Est2 intermediate could be detected throughout most of the cell cycle in immunoprecipitations (Fig. 1B), with slightly substoichiometric association of Est1 in G1 phase, as previously observed (Tucey and Lundblad 2013). In contrast, binding of Est3 to the Est1–TLC1–Est2 subcomplex was nearly undetectable in G1 and remained at very low levels through S phase, with appreciable levels of association only as cells progressed into the G2/M phase of the cell cycle (Fig. 1B). Nevertheless, even late in the cell cycle, only a portion (∼25%) of the Est1–TLC1–Est2 precursor complex was bound by Est3 to form the quaternary telomerase holoenzyme (Figs. 1B; Supplemental Fig. S1B,C). This restricted pattern of association as well as the cell cycle dependency could not be attributed to insufficient levels of the Est3 protein, as this subunit was in excess throughout the cell cycle relative to Est1 and Est2 (Supplemental Fig. S1B; Tuzon et al. 2011).
One potential caveat to the above observations was that although a strain with both Est1 and Est2 bearing (myc)12 epitopes has wild-type telomere length (Tucey and Lundblad 2013), the additional (myc)12 epitope on Est3 resulted in substantially impaired function (Supplemental Fig. S1C; Tuzon et al. 2011). To address this concern, the above experiment was repeated in an alternative strain bearing a (Flag)3 epitope on Est3, which has only a minor effect on telomere length (Supplemental Fig. S1C; Rao et al. 2014), with Est2 tagged with (myc)12. Extracts were prepared from cells arrested in G1 or G2/M with α-factor or nocodazole, respectively, and anti-myc Westerns monitored the level of the Est2 protein following anti-Flag immunoprecipitation of Est3. The results once again showed that Est3 exhibited highly regulated association with telomerase, as Est2 protein levels in anti-Est3 immunoprecipitations were ∼20-fold higher in cultures arrested in G2/M versus G1 (Fig. 1C); the association between Est1 and Est3 showed a similar cell cycle dependency (Supplemental Fig. S1D). This pattern of association was not due to variations in immunoprecipitation efficiency at different points in the cell cycle, since the Est3 protein was depleted from extracts in these anti-Flag immunoprecipitations (Fig. 1C); however, the less abundant Est2 protein was not codepleted (Supplemental Fig. S1E), consistent with the substoichiometric association of Est3 with the Est1–TLC1–Est2 subcomplex observed in Figure 1B. Thus, limited Est3 association was observed regardless of whether the epitope used for immunoprecipitation was present on Est3 or a subunit of the Est1–TLC1–Est2 subcomplex. This demonstrates that the Est1–TLC1–Est2 preassembly complex is the primary telomerase species through most of the cell cycle, with the active holoenzyme formed only after genomic DNA replication is essentially complete.
An interaction between Est3 and Est2 requires binding of Est2 to TLC1
Association of Est3 to form the holoenyzme must depend on an interaction between Est3 and one (or more) of the subunits of the Est1–TLC1–Est2 preassembly complex. Genetic studies have suggested that the Est3 subunit associates with telomerase through the Ten domain of Est2 (Friedman et al. 2003), potentially through a direct interaction (Talley et al. 2011; Yen et al. 2011), although these in vitro protein-binding studies are challenged by the propensity of the Est3 protein to form both soluble and insoluble aggregates (Rao et al. 2014). We initially tested this proposed association using Saccharomyces castellii Est2 and Est3 proteins expressed in Saccharomyces cerevisiae in order to minimize the possibility that an interaction between these two S. castellii proteins would be bridged by additional telomerase components. In extracts prepared from a S. cerevisiae strain bearing the appropriate plasmid constructs, the full-length S. castellii Est3 protein and the 186-amino-acid S. castellii Est2 Ten domain with C-terminal (Flag)2 and (myc)11 epitopes, respectively, could be readily detected, indicating that both were stably expressed. However, following immunoprecipitation of the S. castellii Est3 protein, no association with the S. castellii Est2 Ten domain was observed when compared with a mock immunoprecipitation with an untagged version of Est3 (Supplemental Fig. S2A).
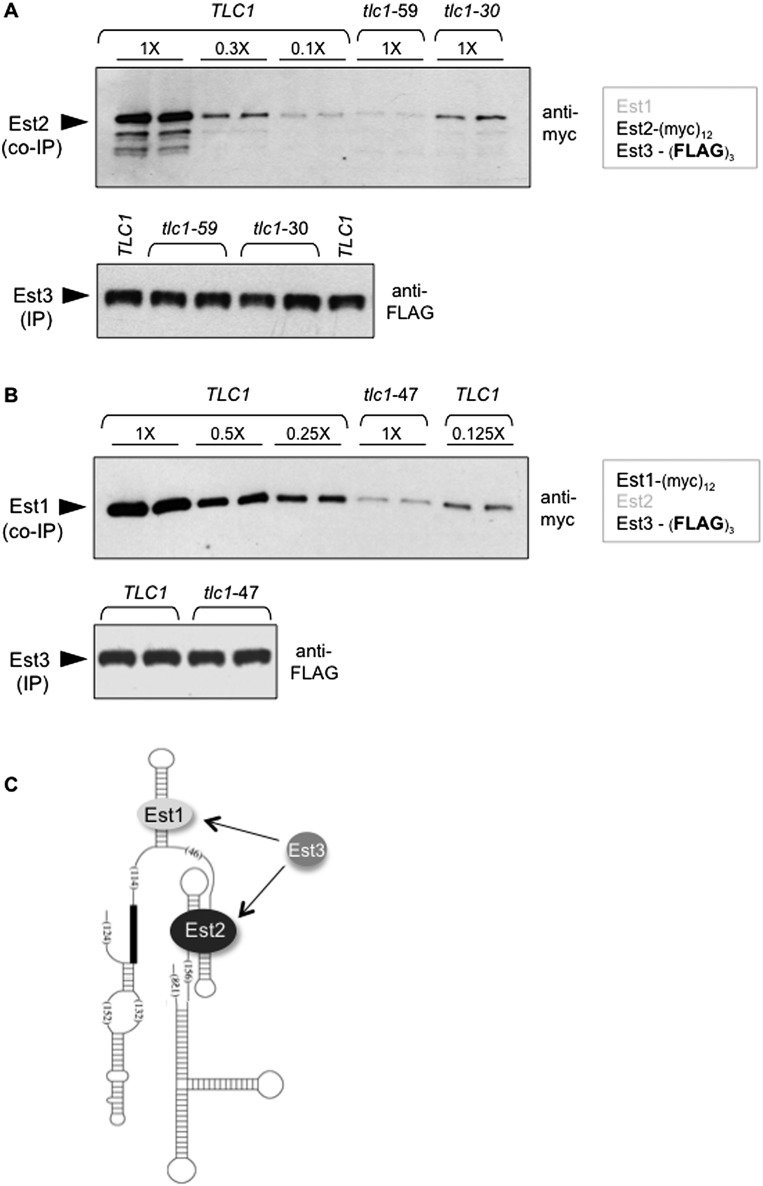
The inability to detect binding of S. castellii Est3 to the 21.5-kDa Est2 Ten domain might be due to improper folding of the 186-amino-acid Ten domain or other potential artifacts stemming from cross-species protein expression. Therefore, as an alternative means of examining this presumed direct interaction, we asked whether an association between the full-length S. cerevisiae Est2 and Est3 proteins could be observed in a strain in which Est2 could not bind the RNA. Two previously characterized tlc1− mutations in a stem–loop of the RNA required for Est2–TLC1 binding were used: a 61-nucleotide (nt) deletion that removes the entire stem–loop structure (tlc1-59) and a 4-nt mutation that prevents base-pairing of an Est2-binding stem (tlc1-30); Supplemental Figure S2B shows the position of these mutations on the TLC1 RNA. These two mutations were previously shown to substantially impair Est2–TLC1 binding (with tlc1-59 exerting a more severe defect than tlc1-30) without affecting the Est1–TLC1 interaction (Chappell and Lundblad 2004). Both tlc1− mutations were integrated into a strain bearing (myc)12 and (Flag)3 epitopes on Est2 and Est3, respectively, and the Est2–Est3 interaction was assessed following anti-Est3 immunoprecipitation with anti-Flag antibodies. Unexpectedly, when Est2 was no longer capable of binding the telomerase RNA, the levels of Est2 in anti-Est3 immunoprecipitations were substantially reduced. A titration series comparing immunoprecipitations from the wild-type TLC1 strain versus the two mutant tlc1− strains demonstrated that the Est2–Est3 interaction was reduced by ∼15-fold in the tlc1-59 strain and approximately threefold in the less impaired tlc1-30 strain (Fig. 2A). This argues that Est3 and Est2 cannot form a stable interaction if Est2 is not bound to the RNA. This result invokes two possibilities: Either prior binding of Est2 to the RNA results in a conformation change that presents a surface that can be recognized by Est3 or binding of Est3 to an additional subunit of telomerase is necessary for assembly of the quaternary complex.
Figure 2.
Association of Est3 with telomerase requires prior binding of both Est1 and Est2 to the telomerase RNA. (A,B) Anti-myc Westerns (top) monitoring levels of (myc)12-Est2 (A) or Est1-(myc)12 (B) following anti-Flag immunoprecipitation (IP) of Est3-(Flag)3 (bottom) in TLC1 versus the indicated tlc1 mutant strains; tlc1 mutations (shown in Supplemental Fig. S2B) were integrated into the genome in place of the wild-type TLC1 gene. Serial dilutions of the anti-Flag immunoprecipitations from the wild-type strains demonstrate the detection range for these Westerns (see also Tucey and Lundblad 2013); Supplemental Figure S2D shows inputs for these strains, and Supplemental Figure S2E shows that the Est3 protein was depleted from extracts for both of these anti-Flag immunoprecipitations. (C) Schematic of the TLC1 RNA secondary structure showing Est1 and Est2 bound to independent sites on the RNA, with Est3 association dependent on both Est1–Est3 and Est2–Est3 protein interactions.
Binding of Est3 to telomerase also depends on an Est1–TLC1 interaction
If Est3 associates with telomerase through a subunit other than or in addition to Est2, the obvious alternative is Est1. Therefore, an experiment analogous to that shown in Figure 2A was performed by assessing whether Est1 and Est3 could interact in a strain in which Est1 was unable to bind the RNA. The rationale for this experiment was the same as described above, which was to determine whether an interaction between Est1 and Est3 could be observed even when Est1 was not physically associated with the telomerase complex. To test this, an Est1-(myc)12 Est3-(Flag)3 strain was constructed that contained a mutation in TLC1 bearing a deletion of the previously well-characterized 5-nt bulge in the Est1-binding arm of TLC1 (Seto et al. 2002). As previously shown, this tlc1− mutation abolishes the ability of Est1 to associate with TLC1 but has no impact on the Est2–TLC1 interaction (Fig. 1A; Seto et al. 2002; Chappell and Lundblad 2004). In this mutant strain, the ability of Est1 and Est3 to coimmunoprecipitate was reduced by ∼10-fold to 15-fold (Fig. 2B), demonstrating that Est1 and Est3 could not interact if the Est1 telomerase subunit was not RNA-bound. Furthermore, Est3 was unable to associate with an Est1–TLC1 complex if Est2 was not bound to the RNA (Supplemental Fig. S2C); reciprocally, Est3 also was unable to interact with a TLC1–Est2 subcomplex lacking Est1 (data not shown). Collectively, these observations establish that stable Est3 association requires prior binding of both Est1 and Est2 to the RNA.
Identification of Est1–Est3 and Est2–Est3 interfaces required for telomerase assembly
The above results suggest two possible models. The first is that a physical interaction between the TLC1 RNA and the Est1 and Est2 proteins induces an RNA structure that becomes competent to bind Est3. Alternatively, association of Est3 to form a stable quaternary complex depends on protein interactions with both Est1 and Est2, as depicted in Figure 2C. Below, we describe single missense mutations in both EST1 and EST2 that reduce the association of Est3 with the Est1–TLC1–Est2 intermediate, consistent with the premise that there are Est3-binding interfaces on two different telomerase protein subunits.
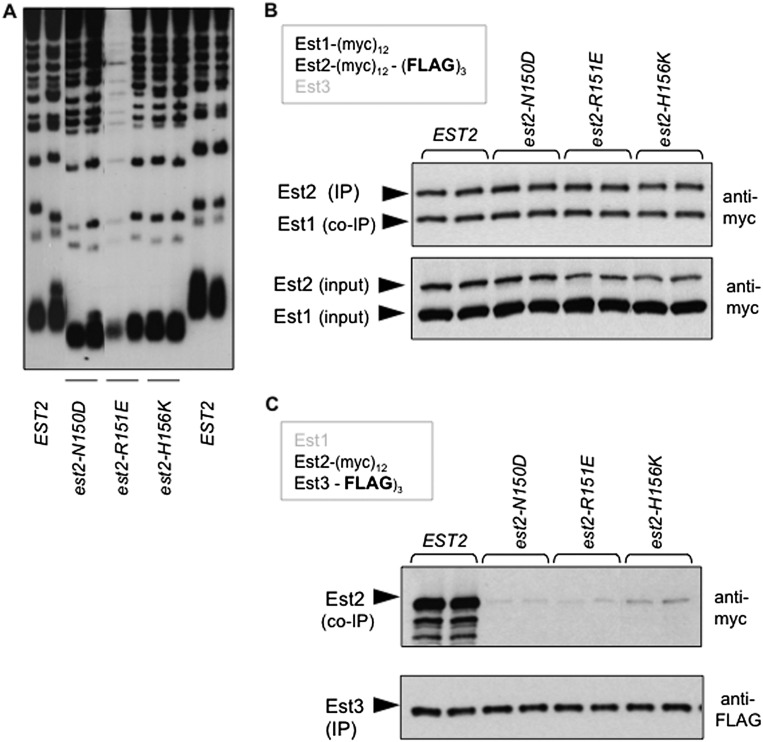
We first identified a cluster of residues in the Ten domain of Est2 that mediate association of Est3 with the telomerase complex as part of an in-progress mutagenesis study of EST2. This group of amino acids resides along an α helix that is immediately adjacent to the highly conserved “GQ” motif, based on the structure of the Ten domain of the Tetrahymena Tert protein (Supplemental Fig. S3A,B; Jacobs et al. 2006). Mutations in comparable residues (Asn150–His156) in the S. cerevisiae EST2 gene resulted in short telomeres (Fig. 3A). These same mutations also disrupted telomere length when overexpressed in a wild-type strain of yeast (Supplemental Fig. S3C), to a degree that correlated well with the loss-of-function phenotypes shown in Figure 3A; we previously argued that such a correlation provides a rigorous means of distinguishing properly folded mutant proteins from unstable proteins (Lubin et al. 2013). Consistent with this premise, mutant protein levels in extracts were unchanged when compared with the wild-type Est2 protein (Fig. 3B), which contrasts with reduced protein stability resulting from mutations in the invariant Gln146 and Gly149 residues that define this motif (Friedman et al. 2003; Lue and Li 2007). These three Est2 mutant proteins were also indistinguishable from the wild-type protein in their ability to form an Est1–TLC1–Est2 complex, as the Est1:Est2 ratio following anti-Est2 immunoprecipitation was not affected by any of these three mutations (Fig. 3B). In contrast, this cluster of est2ten mutations substantially impaired the ability of Est3 to associate with the telomerase complex, which was assayed using a strain bearing (myc)12 and (Flag)3 epitopes on Est2 and Est3, respectively, with each est2ten mutation integrated into the genome. Anti-Est3 immunoprecipitations revealed that the ability of the Est3 protein to bind to the telomerase complex was reduced by as much as 10-fold in est2− strains bearing these three Ten domain mutations (Fig. 3C; Supplemental Fig. S3D). Comparable results for this panel of mutations were obtained in an independent coimmunoprecipitation (co-IP) experiment that used an (HA)3 epitope on Est3 instead of (Flag)3 (data not shown). Based on these observations, we propose that this cluster of amino acids forms an interface that mediates a direct interaction between Est2 and Est3. Notably, this Est2–Est3 interface overlaps with a site on Est2 that helps bind the telomeric primer (Jacobs et al. 2006); the implications of a dual role for this Est2 surface are considered further in the Discussion.
Figure 3.
Identification of an Est3-binding interface on the Ten domain of Est2. (A) Telomere length from two single colonies of the indicated strains, bearing mutations in the Ten domain of EST2, was assessed following ∼75 generations of growth; mutations were integrated into the genome in place of the wild-type EST2 gene in an otherwise wild-type strain. (B) Anti-myc Westerns monitoring the association of Est1 with Est2 following anti-Est2 immunoprecipitations (IPs) (top) and the levels of Est1 and Est2 proteins in extracts (bottom) in wild-type and est2− mutant strains bearing identical (myc)12 epitopes on Est1 and Est2 and an additional (Flag)3 epitope on Est2. (C) Anti-myc Westerns (top) show levels of wild-type and mutant Est2 proteins following anti-Est3 immunoprecipitation with anti-Flag antibodies in a strain with a (Flag)3 epitope on Est3 and a (myc)12 epitope on Est2; equivalent amounts of the Est3-(Flag)3 protein were present in each immunoprecipitation sample, as shown by the anti-Flag Western (bottom).
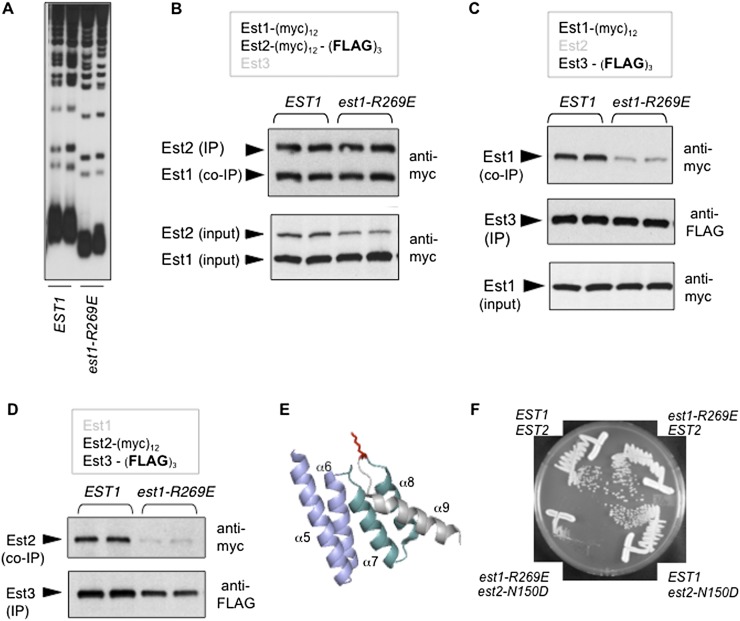
In parallel, as part of a genetic analysis of EST1 (J Lubin, TM Tucey, and V Lundblad, in prep.), we identified an est1− mutation in the N-terminal domain of Est1 (est1-R269E) that exhibited the expected properties for a defect in the predicted Est1–Est3 interaction. The est1-R269E mutation conferred short telomeres in a loss-of-function assay (Fig. 4A) and was also capable of disrupting telomere maintenance when overexpressed in an otherwise wild-type strain (Supplemental Fig. S4A). The Est1-R269E mutant protein was present at wild-type levels in extracts (Fig. 4B) and formed an Est1–TLC1–Est2 complex in a manner comparable with the behavior of the wild-type Est1 protein, as assessed by immunoprecipitation (Fig. 4B); thus, both protein stability and the Est1–TLC1 interaction were unperturbed by this Arg → Glu mutation. However, the ability of Est3 to associate with a telomerase complex containing the Est1-R269E protein was disrupted, as revealed by a comparison of wild-type Est1 versus mutant Est1-R269E protein levels in anti-Est3 immunoprecipitations (Fig. 4C). Similar results were obtained when the ability of Est2 and Est3 to coimmunoprecipitate in an est1-R269E strain was monitored (Fig. 4D); this argued that this Est3–Est1 interface functioned as a part of the Est1–TLC1–Est2 subcomplex (which is also consistent with the results in Fig. 2B). The residue implicated in this analysis (Arg269) is highly conserved (as either arginine or lysine) (Supplemental Fig. S4B). The comparable residue is located on the surface of the human SMG7 protein (Fig. 4E; Fukuhara et al. 2005), which has also been referred to as EST1C (Reichenbach et al. 2003; Snow et al. 2003); given the high degree of sequence conservation between the human SMG7/EST1C and yeast Est1 proteins, this predicts that Arg269 is similarly located on the Est1 protein surface. These findings indicate that we identified an additional interface corresponding to a conserved surface on the Est1 protein, which promotes an interaction with the Est3 telomerase subunit.
Figure 4.
Identification of an Est3-binding interface in the N-terminal domain of Est1. (A) Telomere length assessed as in Figure 3A, comparing EST1 and est1-R269E strains, with the est1-R269E mutation integrated into the genome. (B) Anti-myc Westerns monitoring the association of Est1 with Est2 following anti-Est2 immunoprecipitations (IPs) (top) and the levels of Est1 and Est2 proteins in extracts (bottom) from EST1 and est1-R269E strains, with the same arrangement of epitope tags on Est1 and Est2 as described in Figure 3B. (C) Anti-Flag immunoprecipitations monitoring the association between Est1 and Est3 in EST1 and est1-R269E strains, with Est1 and Est3 tagged with (myc)12 and (Flag)3, respectively. (D) Experiment performed as described in Figure 3C, showing that the est1-R269E mutation also impairs the ability of Est2 to coimmunoprecipitate with Est3. (E) The structure of the human SMG7 protein, amino acids 143–236 (Fukuhara et al. 2005), which corresponds to the second TPR-like domain (composed of helices α5–α6 and α7–α8) and the adjacent helix α9 (indicated in gray); Lys216 (in red), protruding from the hinge between helices α8 and α9, is highly conserved as either lysine or arginine, as shown by the alignment in Supplemental Figure S4B. (F) Single colony streak-outs of isogenic strains with the indicated genotypes, generated following dissection of a diploid strain bearing the integrated est1-R269E and est2-N150D mutations. The results shown here are representative of the behavior of four tetratype tetrads: All four double-mutant est1-R269E est2-N150D strains exhibited a senescence phenotype, whereas none of the eight single-mutant strains showed a growth defect.
Est3-binding sites on both Est1 and Est2 might suggest that two molecules of Est3 associate with the Est1–TLC1–Est2 preassembly complex. However, in a strain expressing Est3-(myc)6 and Est3-(myc)6-(HA)3, the Est3-(myc)6 variant was not detectable in an anti-HA immunoprecipitation, arguing against telomerase complexes with a stoichiometry of 2 for the Est3 subunit (Supplemental Fig. S4C). Alternatively, either Est3-binding site might be sufficient for holoenzyme formation, which was tested by combining defects in these two Est3-binding sites. This resulted in an additive effect, as a strain bearing both the est1-R269E mutation and an est2ten mutation exhibited a more severe phenotype. In single-mutant strains bearing mutations in either of these two binding sites, the telomere length decline (Figs. 3A, 4A) was not severe enough to confer a senescence phenotype (Fig. 4F). In contrast, the double-mutant est1-R269E est2-N150D strain exhibited telomere shortening and an accompanying growth defect that was comparable with that of an est3-∆-null strain (Fig. 4F; data not shown). This was accompanied by an additive defect in the ability of Est3 to bind to a telomerase complex bearing est1-R269E and est2-N150D mutations (Supplemental Fig. S4D). We conclude from these results that binding surfaces on both Est1 and Est2 are required for full association of Est3 with the Est1–TLC1–Est2 preassembly complex.
A toggle switch on the surface of Est3 also regulates telomerase assembly
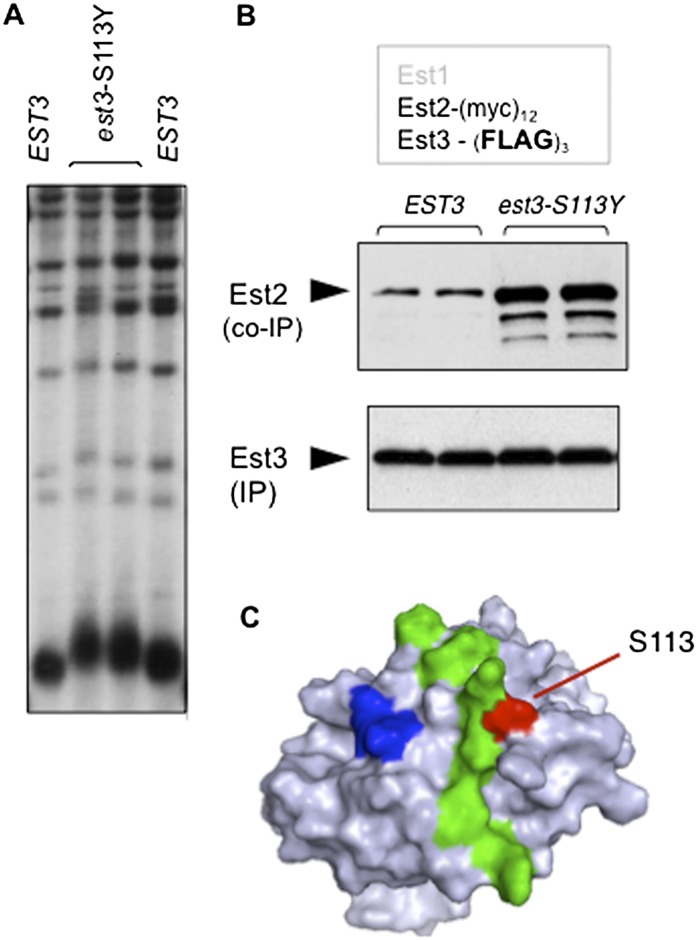
The above analysis provided information about binding surfaces on Est1 and Est2 that recruit Est3 to the telomerase complex. However, left unaddressed is why Est3 shows such limited association with telomerase (Fig. 1B,C) despite the fact that Est3 is in excess relative to the other subunits (Supplemental Fig. S1B). This implies that Est3 binding is actively inhibited. If so, this predicts that relief of a negative regulatory step would increase the interaction of Est3 with telomerase, with a corresponding impact on telomere length. As the result of our ongoing genetic analysis of EST3, we fortuitously identified an est3− mutation at amino acid S113 that fulfilled both of these predictions: The est3-S113Y mutant strain exhibited elongated telomeres (Fig. 5A), which was accompanied by an increased association of the mutant Est3-S113Y protein with telomerase (Fig. 5B). This residue therefore defines an inhibitory site on the surface of Est3 that we call TEL-I (for TEL inhibitory) that regulates the interaction between Est3 and telomerase. Notably, this negative regulatory site is immediately adjacent to an interface on the experimentally determined surface of Est3 that promotes binding (Fig. 5C), as Est3 proteins bearing mutations in residues in this adjoining surface (the TEL patch) show reduced association with telomerase and a corresponding decrease in telomere length (Rao et al. 2014). Thus, these two surfaces appear to act as a toggle switch by which the two sides of the switch either inhibit or promote Est3 binding. Both Est1–Est3 binding and Est2–Est3 binding are regulated by this toggle switch, as evidenced by the partial alleviation of the telomere length defect displayed by est2ten or est1-R269E mutant strains that also contain the est3-S113Y toggle switch mutation (Supplemental Fig. S5A). The role of the TEL-I site in mediating binding of Est3 to telomerase is not restricted to late in the cell cycle, as increased association of Est3-S113Y was also observed in cells arrested in the G1 phase (Supplemental Fig. S5B).
Figure 5.
Adjacent positive and negative regulatory sites on the surface of the Est3 protein. (A) Telomere length, assessed after 75 generations of growth, following introduction of single-copy EST3 or est3-S113Y plasmids into an est3-∆ strain. (B) Anti-Flag immunoprecipitations in a (myc)12-Est2 strain with single-copy plasmids expressing either wild-type Est3-(Flag)3 or mutant Est3-S113Y-(Flag)3; a similar enhancement was observed when association of the Est3-S113Y protein was monitored in an Est1–Est3 co-IP (data not shown). (C) Surface representation of the Est3 protein (Rao et al. 2014) depicting adjacent positive and negative regulatory sites (TEL and TEL-I), indicated in green and red, respectively, that either promote or inhibit association of Est3 with telomerase; a third cluster of residues (in blue) promotes a separate function for Est3, which we previously showed is unrelated to telomerase assembly (Rao et al. 2014).
Assembly of the quaternary complex is followed by disassembly due to dissociation of Est2
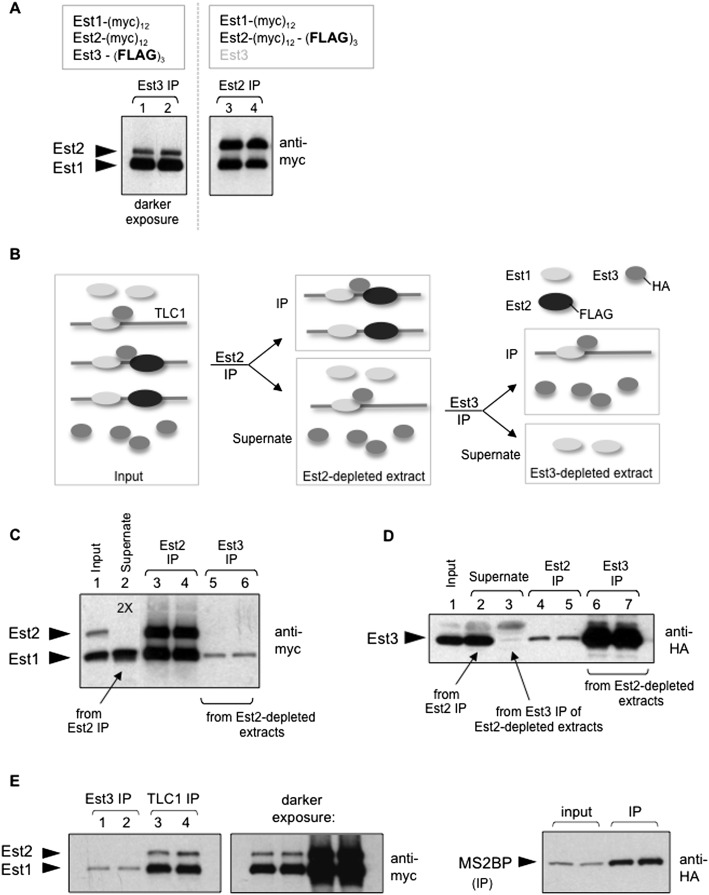
The anti-Est2 immunoprecipitations shown in Figure 1 demonstrated that Est3 exhibited limited association with the Est1–TLC1–Est2 subcomplex. However, in the anti-Est3 immunoprecipitations in Figures 2 and 3 that examined the ability of either Est1 or Est2 to interact with Est3, we consistently observed an increased association of Est1 with Est3 relative to the Est2–Est3 interaction (Supplemental Fig. S6A). To assess this more carefully, anti-Est3 immunoprecipitations were prepared from a single strain in which both Est1 and Est2 were tagged with the same (myc)12 epitope, which revealed an unexpected Est1:Est2 ratio when Est3 was immunoprecipitated. In anti-Est2 immunoprecipitations, the Est1:Est2 ratio was 1:1, but in anti-Est3 immunoprecipitations, the Est1:Est2 ratio was increased to ∼4:1 (Fig. 6A; Supplemental Fig. S6B), indicating that the Est3 telomerase subunit was associating with substantially more Est1 than Est2. This difference could not be easily attributed to a technical artifact (such as a difference in immunoprecipitation efficiency), as the immunoprecipitated protein was depleted from extracts in both immunoprecipitations. These results suggested that the anti-Est3 immunoprecipitations were sampling two distinct Est3-associated complexes: the quaternary telomerase complex and an additional more abundant subcomplex containing Est1 but lacking Est2. We show below that this second subcomplex is derived from the quaternary complex through disassociation of the Est2 subunit to form a catalytically inactive disassembly complex.
Figure 6.
The telomerase quaternary complex disassembles through Est2 dissociation. (A) Comparing anti-Est3 (left panel) or anti-Est2 (right panel) immunoprecipitations (IPs), where Est1 and Est2 proteins were tagged with (myc)12 in both experiments, with a (Flag)3 epitope on either Est3 (lanes 1,2) or Est2 (lanes 3,4). All four samples were run on the same 6% SDS-PAGE gel, but two different exposures are shown to compensate for the reduced association of Est3 with the telomerase complex (a single exposure is shown in Supplemental Fig. S6B); the difference in mobility of the Est2 protein in the left and right panels is due to the absence/presence, respectively, of the (Flag)3 epitope. (B) Schematic depiction of complexes (the telomerase quaternary complex and the Est3–Est1–TLC1 and Est–TLC1–Est2 subcomplexes) and free subunits (Est1 and Est3) in extracts (left panel) and how these various factors behave in consecutive Est2 and Est3 immunoprecipitations, as shown in C and D. For simplicity, the relative amounts of the three complexes in extracts and immunoprecipitations are not indicated here, but see the Discussion and Figure 7C. (C) Results from the experiment depicted in B, showing levels of the (myc)12-tagged Est1 and Est2 proteins in inputs, supernates, and anti-Est2 versus anti-Est3 immunoprecipitations, as indicated. The increased amount of supernate loaded in lane 2 (indicated as “2X”) relative to lane 1 emphasizes that Est2 has been substantially depleted by the anti-Flag immunoprecipitation. In contrast, the Est1 telomerase subunit is not fully depleted by the successive anti-Est2 and anti-Est3 immunoprecipitations (shown in lane 3), consistent with the results shown in Figure 1C and Supplemental Figure S1E. (D) Levels of the Est3-(HA)3 protein in inputs, supernates, and immunoprecipitations corresponding to the experiment shown in C, which shows that the Est3 protein is efficiently immunoprecipitated (lanes 6,7), with a corresponding depletion from the relevant extract (lane 3). The low levels of the Est3 protein detected in the anti-Est2 immunoprecipitations (shown in lanes 4,5) also provide an independent confirmation of the results shown in Figure 1, B and C. (E) Comparison of the relative levels of Est1 and Est2 in anti-Est3 (lanes 1,2) and anti-TLC1 (lanes 3,4) immunoprecipitations from a TLC1-(MS2)10 strain, which contains Est3-(Flag)3 and a plasmid expressing the HA-tagged MS2-binding protein (MS2BP-HA) as well as (myc)12-tagged Est1 and Est2 proteins. Comparable volumes of anti-Flag (anti-Est3) and anti-HA (anti-TLC1) immunoprecipitations prepared from the same extracts were loaded; the differences in immunoprecipitation efficiencies are consistent with the reduced association of Est3 with the Est1–TLC1–Est2 complex, as shown in Figure 1 and Supplemental Figure S7.
As a direct test of the proposal that there were two Est3-containing complexes, we used immunodepletion to fractionate extracts into (1) an Est2-containing fraction and (2) a fraction lacking Est2 in order to ask whether a telomerase subcomplex was present in the latter. To do so, we constructed a strain that would allow successive immunoprecipitations of Est2 and Est3 due to (Flag)3 or (HA)3 epitopes, respectively, on these two proteins. A schematic illustration of the experiment performed with this strain is shown in Figure 6B. Extracts prepared from this strain were initially subjected to anti-Flag immunoprecipitations, thereby depleting Est2-containing complexes from the extract (Fig. 6C, lane 2). Since both Est1 and Est3 are excess relative to the Est2 catalytic subunit (Supplemental Fig. S1B), the levels of these two telomerase subunits were only partially depleted by the anti-Est2 immunoprecipitation (lane 2 in Fig. 6C,D, respectively). This allowed us to assess whether a separate complex containing Est1 and Est3 could be detected in the absence of Est2 by subjecting the Est2-depleted extract to a second immunoprecipitation using anti-HA antibodies targeting Est3. This anti-Est3 immunoprecipitation quantitatively removed the Est3 protein (Fig. 6D, lane 3), and, as expected, the catalytic Est2 subunit was not detectable in these immunoprecipitations (Fig. 6C, lanes 5,6). However, Est1 was readily detected in the anti-HA immunoprecipitation from this Est2-depleted extract (Fig. 6C, lanes 5,6), thereby providing direct support for a subcomplex containing Est1 and Est3 but not Est2.
As an additional step in this analysis, we asked whether the telomerase RNA was a component of this subcomplex. To do so, we used a previously described variant of the yeast TLC1 telomerase RNA containing 10 tandem repeats of the MS2 stem–loop [TLC1-(MS2)10] (Gallardo et al. 2011). These MS2 repeats, which have only a minimal effect on TLC1 function (Supplemental Fig. S6C), can be bound by the MS2-binding protein; in a strain expressing an HA-tagged version of the MS2-binding protein, this allows immunoprecipitation of the TLC1 RNA. In anti-HA immunoprecipitations, both Est1 and Est2 could be detected as expected, since both proteins bind the TLC1 telomerase RNA (Livengood et al. 2002; Seto et al. 2002; Chappell and Lundblad 2004). However, the Est1:Est2 ratio in these anti-TLC1 immunoprecipitations was substantially different from the 1:1 ratio previously observed in anti-Est2 immunoprecipitations (cf. Figs. 6E and 1A). Instead, the Est1:Est2 ratio in anti-TLC1 immunoprecipitations was strikingly similar to the ratio observed in anti-Est3 immunoprecipitations (Fig. 6E). The simple interpretation of this experiment is that both Est2-containing and non-Est2-containing complexes were detected by the anti-TLC1 immunoprecipitations, which is consistent with the presence of a more abundant second complex containing Est1 and TLC1, but lacking Est2, in the anti-TLC1 immunoprecipitations. As a further confirmation of this result, we performed an additional experiment analogous to that shown in Figure 6B where we queried Est2-depleted extracts for the presence of a second TLC1-containing extract using the MS2-tagged TLC1 strain. The results, shown in Supplemental Figure S6D, provide additional support for an inactive Est3–Est1–TLC1 subcomplex that can be recovered from extracts that have been depleted for Est2. As an aside, this experiment also demonstrates that Est2 is the limiting telomerase subunit rather than TLC1, as previously assumed. Thus, fractionation of extracts by immunodepletion has identified two discrete Est3-containing telomerase complexes: the telomerase holoenzyme present in the Est2-containing fraction and a second complex containing Est1, Est3, and TLC1 in the Est2-depleted fraction.
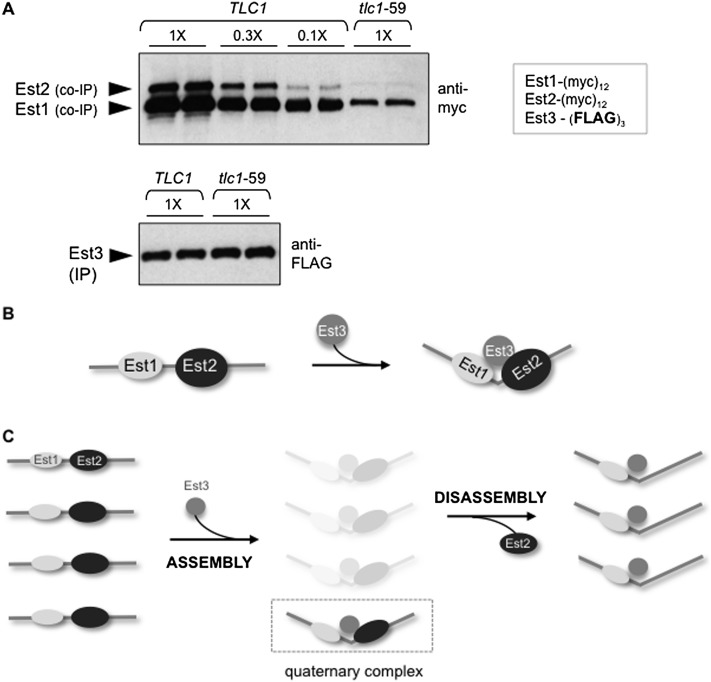
Finally, we asked whether the appearance of this inactive subcomplex depended on prior formation of the quaternary holoenzyme complex. To do so, we examined the relative ratio of the two complexes in anti-Est3 immunoprecipitations in a strain bearing a tlc1-59 mutation. As described in Figure 2A, this mutation blocks the Est2–TLC1 interaction and subsequent formation of the quaternary complex. If the Est3–Est1–TLC1 subcomplex is a product of the quaternary complex (rather than a precursor), both Est1 and Est2 protein levels should be substantially reduced in anti-Est3 immunoprecipitations and to the same degree (see Supplemental Fig. S7 for an illustration of this prediction). Consistent with this expectation, protein levels for Est1 and Est2 were reduced by equal amounts (∼20-fold) in anti-Est3 immunoprecipitations from the tlc1-59 strain as compared with a TLC1 strain (Fig. 7A), demonstrating that formation of both the quaternary complex (containing Est2) and the Est3–Est1–TLC1 disassembly (lacking Est2) were inhibited if Est2 could not bind TLC1. In a similar manner, when formation of the quaternary complex was blocked in est2ten strains that were defective for the Est2–Est3 interaction, levels of both the Est2-containing and the non-Est2-containing complexes were also reduced by comparable amounts in anti-Est3 immunoprecipitations (data not shown). Thus, two Est2-dependent interactions (Est2–TLC1 and Est2–Est3) that were critical for holoenzyme assembly were also required for the formation of a complex that does not contain Est2. This provides additional support for the conclusion that the appearance of this catalytically inactive Est3–Est1–TLC1 subcomplex relies on prior formation of the telomerase holoenzyme complex. Furthermore, the appearance of this disassembly complex was cell cycle-regulated in a manner that parallels the appearance of the quaternary complex, with the levels of both complexes increased by ∼20-fold in G2/M phase relative to G1 phase (Supplemental Fig. S6E,F). These results invoke the possibility that disassembly of the active complex to form this catalytically inactive subcomplex is a key step in telomerase regulation.
Figure 7.
The telomerase assembly and disassembly pathways. (A) Anti-myc and anti-Flag Westerns monitoring the relative levels of (myc)12-tagged Est1 and Est2 proteins following Flag immunoprecipitation (IP) of Est3-(Flag)3 in TLC1 or tlc1-59 strains. Serial dilutions of anti-Flag immunoprecipitations from the wild-type strain were used to demonstrate that the levels of Est1 and Est2 were reduced to the same extent (∼20-fold) in the tlc1-59 mutant strain; see Supplemental Figure S7 for a schematic depiction of the predicted outcomes for this experiment, depending on whether the holoenzyme is a precursor of the Est3–Est1–TLC1 disassembly complex. (B) A model proposing that the Est3-binding surfaces on Est1 and Est2 form a single extended interface; we further speculate that binding of Est3 to form the quaternary holoenzyme complex may be accompanied by a conformational change. (C) A proposed model for the telomerase assembly and disassembly pathways. Assembly of the quaternary complex is driven by binding of Est3 to the previously formed Est1–TLC1–Est2 complex followed by disassembly due to dissociation of the catalytic Est2 subunit. Supplemental Figure S8 illustrates how this model is supported by the immunoprecipitation data reported in this study; we assume that our inability to detect the proposed transient high levels of the quaternary complex is presumably because immunoprecipitation assays primarily provide information on steady-state levels, with a more limited capacity to detect complexes that are present only transiently.
Discussion
In this study, we used a biochemical strategy that uses a high-resolution assessment of the protein levels of telomerase subunits in immunoprecipitates, which has revealed that two subcomplexes exhibit a dynamic balance with the telomerase quaternary complex through the cell cycle. Early in the cell cycle, formation of an Est1–TLC1–Est2 preassembly complex drives the subsequent binding of Est3 to form the active holoenzyme, containing the four components that are required for in vivo telomerase function. Once formed, this quaternary complex disassembles through loss of Est2 to give rise to a catalytically inactive subcomplex composed of Est1, Est3, and the telomerase RNA. By monitoring the relative steady-state levels of the quaternary complex and the two subcomplexes, two notable observations have emerged. The first is that the telomerase holoenzyme is a transient minority species in terms of both abundance and its restricted appearance during a limited period late in the cell cycle. This implies that there is a significant regulatory investment to ensure low levels of active telomerase. The second unexpected conclusion is that assembly and disassembly proceed through different pathways, as assembly relies on Est3 association, whereas disassembly proceeds through disassociation of the catalytic Est2 subunit. Below, we expand on these two observations and discuss several implications of these findings.
Analysis of the assembly pathway indicates that Est3 sits at a regulatory node, as the process by which Est3 joins the Est1–TLC1–Est2 subcomplex is surprisingly elaborate, involving sites on both Est1 and Est2 as well as a regulatory interface on Est3 that functions as an apparent toggle switch. Two alternative models could explain the separate Est3-binding surfaces on Est1 and Est2. One possibility is that there are successive binding events by which Est3 first binds to one subunit followed by transfer of Est3 onto the other subunit. Several observations, however, suggest an alternative model in which the sites on Est1 and Est2 form a single interface that Est3 cobinds, as schematically presented in Figure 7B. First, this proposed single interface is consistent with our findings that both Est1 and Est2 must be physically bound to the telomerase RNA in order for Est3 to associate with the telomerase complex. Furthermore, the telomerase-binding surface of Est3 is itself composed of a single extended interface (Fig. 5C; Rao et al. 2014) rather than two discrete sites that would separately promote Est1–Est3 or Est2–Est3 interactions. A speculative extension of this model is that Est3 binding is accompanied by a conformational change in the structure of the telomerase holoenzyme, as depicted in Figure 7B.
These observations also raise the question of why there is such an investment in regulating telomerase assembly. Since the Est1–TLC1–Est2 subcomplex is present throughout much of the cell cycle and the Est3 protein subunit is in excess, why not have the quaternary complex available throughout the cell cycle? The answer may come from studies that have shown that telomerase can act at nontelomeric sites, as revealed by a GCR (gross chromosomal rearrangement) assay developed by Chen and Kolodner (1999). Roughly 50% of GCR events are due to de novo addition of telomeres at double-strand breaks (DSBs), with such breaks arising most likely as the result of replication errors (Myung et al. 2001). Thus, elongation of DSBs by telomerase could block other repair mechanisms that would be more likely to preserve genome integrity. One means by which these telomerase-mediated events are kept in check is through phosphorylation of Cdc13 by Mec1, which inhibits accumulation of Cdc13 at DSBs (Zhang and Durocher 2010) and subsequent telomerase recruitment to these ectopic sites (Bianchi et al. 2004). We propose that late entry of Est3 into the telomerase complex well after DNA replication has been completed provides a second regulatory barrier to ensure that telomerase is unable to heal DSBs that occur during earlier stages of the cell cycle, when genomic DNA replication is in progress.
This work has also led to the discovery of a pathway by which telomerase disassembles, thereby resulting in an inactive complex due to dissociation of Est2. The identification of this disassembly complex suggests a potential paradox, as Figure 2 demonstrates that an inability of Est2 to bind the telomerase RNA blocks the formation of a complex composed of Est1, Est3, and TLC1. Although this subcomplex cannot form during assembly, it nevertheless is the product of disassembly. This implies that Est1, Est3, and TLC1 must acquire the ability to stably associate during formation of the quaternary complex, which further argues for a conformational change during holoenzyme assembly (as depicted in Fig. 7B). A proposed model for the telomerase assembly and disassembly pathways is presented in Figure 7C. This model incorporates a key observation, which is that the quaternary complex is detected at levels significantly lower than that of both the preassembly and disassembly complexes. This suggests that association of Est3 with the preassembly complex to form the quaternary complex is followed by shuttling of a substantial proportion of the newly formed holoenzyme enzyme into the disassembly pathway, as illustrated in Figure 7C. Direct support for this proposal comes from our analysis of relative amounts of telomerase proteins in the quaternary complex and the two subcomplexes, as revealed by anti-Est2, anti-Est3, and anti-TLC1 immunoprecipitations from extracts as well as anti-Est3 and anti-TLC1 immunoprecipitations from Est2-depleted extracts (see Supplemental Fig. S8 for a summary of these data). The dynamic balance portrayed in Figure 7C may also reflect the proportion of the holoenzyme that is actively engaged at the telomere. Furthermore, the temporally linked appearance of the holoenzyme and disassembly complexes late in the cell cycle (Supplemental Fig. S6) suggest that the appearance of the disassembly complex may be tightly coupled with enzyme synthesis. For example, disassembly may provide a mechanism by which enzyme synthesis is terminated; if so, the overlap between the Est2–Est3-binding site and the previously described “anchor” site on Est2 that binds telomeric DNA (Jacobs et al. 2006) may contribute to this process.
Finally, the observations reported here argue that previous chromatin immunoprecipitation (ChIP) studies that probe the association of individual telomerase subunits with telomeric chromatin may need to be reinterpreted, since these ChIP analyses cannot differentiate between telomeric association of the active holoenzyme and the preassembly or disassembly complexes. In particular, the observation that association of Est1 and Est2 with telomeric chromatin is Est3-independent (Tuzon et al. 2011) may instead be due to a ChIP signal from the more abundant Est1–TLC1–Est2 preassembly complex, which would mask a potential Est3-dependent association of the quaternary complex. This suggests that a role for Est3 in mediating association and/or retention of the active telomerase holoenzyme with chromosome termini (which would further extend the analogy between TPP1 and Est3) (Nandakumar et al. 2012; Zhong et al. 2012) remains an open question. More generally, the inability to distinguish between the quaternary complex and the assembly or disassembly complexes in ChIP experiments complicates the interpretation of how telomeric association of telomerase changes in response to mutations that alter telomere homeostasis; in particular, if such mutations affect the assembly or disassembly pathways, altered ChIP patterns may be a secondary consequence of a change in the level of the quaternary complex relative to its subcomplexes.
Materials and methods
Telomere length was assessed from independent single colonies following propagation for ∼75 generations for each genotype by comparison of isogenic strains (Supplemental Table S1) bearing mutations integrated into the genome (with the exception of Fig. 5A). Mutations were integrated into the genome as URA3 integrants (see Supplemental Table S2 for list of plasmids) followed by screening for 5-FOA-resistant “pop-outs” that retained the mutation; the molecular structure of the resulting “pop-out” mutant strain was assessed by PCR and sequence analysis to confirm that the relevant mutation was integrated without rearrangement of the target gene and by phenotype (telomere length) in every case. The full lists of strains and plasmids are in Supplemental Tables S1 and S2, respectively.
For biochemical experiments, all mutations were integrated into the genome in place of the wild-type gene (with the exception of Fig. 5B) to eliminate possible effects on immunoprecipitation efficiency due to incomplete gene expression by plasmid-borne alleles (as a consequence of variations in plasmid copy number and/or incomplete promoters) (see Lubin et al. 2012 for further discussion). For each immunoprecipitation, two independent cultures were processed in parallel for each genotype from exponentially growing cells unless otherwise noted. Whole-cell extracts were incubated with anti-Flag M2 affinity gel (Sigma) or anti-HA affinity matrix (rat monoclonal antibody clone 3F10; Roche), as described previously (Tucey and Lundblad 2013). Immunoprecipitations containing Myc-, HA-, or Flag-tagged proteins were resolved by 6% (for detection of Est1 or Est2) or 12% (for detection of Est3) SDS-PAGE and probed with anti-myc 2272 (Cell Signaling Technology) at 1:1000 or anti-Flag F7425 (Sigma-Aldrich) at 1:5000 dilution followed by anti-rabbit IgG HRP conjugate (Promega) at 1:10,000 or with HA-11 clone 16B12 (Covance, MMS-101P) at 1:1000 followed by anti-mouse IgG HRP-linked antibody (Cell Signaling Technology) at 1:5000 and subsequent ECL (enhanced chemiluminescence) detection with preflashed film. We showed previously that this protocol can detect even twofold differences with high reproducibility over a 10-fold detection range (Tucey and Lundblad 2013).
Acknowledgments
We are very grateful to Deborah Wuttke and Lou Zumstein for excellent advice and suggestions during the course of these experiments as well as insightful comments on the manuscript. We also thank Johnathan Lubin, who graciously completed several experiments shown in Supplemental Figures S5 and S6 in response to reviewer requests. The pRS413-TLC-10xMS2 and YCp111-HA-MS2-GFP plasmids were generous gifts from the Chartrand and Wellinger laboratories. This work was supported by National Institutes of Health grants T32 GM007240 (to T.M.T.), R37 AG11728 (to V.L.), and P30 CA014195 (to the Salk Institute Cancer Center); the Fritz B. Burns Foundation; and a Rose Hills Foundation Fellowship (to T.M.T.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.246256.114.
References
- Alder JK, Cogan JD, Brown AF, Anderson CJ, Lawson WE, Lansdorp PM, Phillips JA 3rd, Loyd JE, Chen JJ, Armanios M. 2011. Ancestral mutation in telomerase causes defects in repeat addition processivity and manifests as familial pulmonary fibrosis. PLoS Genet 7: e1001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M 2009. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet 10: 45–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Blackburn EH. 2012. The telomere syndromes. Nat Rev Genet 13: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Negrini S, Shore D. 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell 16: 139–146 [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279: 349–352 [DOI] [PubMed] [Google Scholar]
- Chappell AS, Lundblad V. 2004. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol Cell Biol 24: 7720–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 23: 81–85 [DOI] [PubMed] [Google Scholar]
- Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. 2007. Protein composition of catalytically active human telomerase from immortal cells. Science 315: 1850–1853 [DOI] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH. 1995. Telomerase in yeast. Science 269: 396–400 [DOI] [PubMed] [Google Scholar]
- Collins K 2011. Single-stranded DNA repeat synthesis by telomerase. Curr Opin Chem Biol 15: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Friedman KL, Heit JJ, Long DM, Cech TR. 2003. N-terminal domain of yeast telomerase reverse transcriptase: recruitment of Est3p to the telomerase complex. Mol Biol Cell 14: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Gallardo F, Laterreur N, Cusanelli E, Ouenzar F, Querido E, Wellinger RJ, Chartrand P. 2011. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol Cell 44: 819–827 [DOI] [PubMed] [Google Scholar]
- Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. 2014. TERT promoter mutations in cancer development. Curr Opin Genet Dev 24C: 30–37 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. 2000. The Est3 protein is a subunit of yeast telomerase. Curr Biol 10: 809–812 [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. 2006. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol 13: 218–225 [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567 [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR. 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol Cell Biol 22: 2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin J, Tucey TM, Lundblad V. 2012. The interaction between the yeast telomerase RNA and the Est1 protein requires three structural elements. RNA 18: 1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin J, Rao T, Mandell EK, Wuttke DS, Lundblad V. 2013. Dissecting protein function: an efficient protocol for identifying separation-of-function mutations that encode structurally stable proteins. Genetics 193: 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF, Li Z. 2007. Modeling and structure function analysis of the putative anchor site of yeast telomerase. Nucleic Acids Res 35: 5213–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555 [DOI] [PubMed] [Google Scholar]
- Morris DK, Lundblad V. 1997. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr Biol 7: 969–976 [DOI] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent C, Hughes TR, Lue NF, Lundblad V. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Rao T, Lubin J, Armstrong GS, Tucey TM, Lundblad V, Wuttke DS. 2014. Structure of Est3 reveals a bimodal surface with differential roles in telomere replication. Proc Natl Acad Sci 111: 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P, Höss M, Azzalin CM, Nabholz M, Bucher P, Lingner J. 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574 [DOI] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev 16: 2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L. 2003. Functional conservation of the telomerase protein Est1p in humans. Curr Biol 13: 698–704 [DOI] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Talley JM, DeZwaan DC, Maness LD, Freeman BC, Friedman KL. 2011. Stimulation of yeast telomerase activity by the ever shorter telomere 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J Biol Chem 286: 26431–26439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kannan R, Blanchette M, Baumann P. 2012. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484: 260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucey TM, Lundblad V. 2013. A yeast telomerase complex containing the Est1 recruitment protein is assembled early in the cell cycle. Biochem. 52: 1131–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon CT, Wu Y, Chan A, Zakian VA. 2011. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet 7: e1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. 2008. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132: 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WF, Chico L, Lei M, Lue NF. 2011. Telomerase regulatory subunit Est3 in two Candida species physically interacts with the TEN domain of TERT and telomeric DNA. Proc Natl Acad Sci 108: 20370–20375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR. 2005. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol 12: 1072–1077 [DOI] [PubMed] [Google Scholar]
- Zhang W, Durocher D. 2010. De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev 24: 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]