Using CRISPR/Cas9 technology, Lu et al. generated mouse embryonic stem cells (ESCs) that are deficient for all three Tet proteins. Functional characterization of these ESCs revealed a role for Tet proteins in regulating the two-cell embryo (2C)-like state under ESC culture conditions. The knockout ESCs exhibited increased telomere–sister chromatid exchange and elongated telomeres.
Keywords: DNA methylation, demethylation, enhancer, embryonic stem cell, telomere
Abstract
DNA methylation at the C-5 position of cytosine (5mC) is one of the best-studied epigenetic modifications and plays important roles in diverse biological processes. Iterative oxidation of 5mC by the ten-eleven translocation (Tet) family of proteins generates 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). 5fC and 5caC are selectively recognized and excised by thymine DNA glycosylase (TDG), leading to DNA demethylation. Functional characterization of Tet proteins has been complicated by the redundancy between the three family members. Using CRISPR/Cas9 technology, we generated mouse embryonic stem cells (ESCs) deficient for all three Tet proteins (Tet triple knockout [TKO]). Whole-genome bisulfite sequencing (WGBS) analysis revealed that Tet-mediated DNA demethylation mainly occurs at distally located enhancers and fine-tunes the transcription of genes associated with these regions. Functional characterization of Tet TKO ESCs revealed a role for Tet proteins in regulating the two-cell embryo (2C)-like state under ESC culture conditions. In addition, Tet TKO ESCs exhibited increased telomere–sister chromatid exchange and elongated telomeres. Collectively, our study reveals a role for Tet proteins in not only DNA demethylation at enhancers but also regulating the 2C-like state and telomere homeostasis.
DNA methylation at the C-5 position of cytosine (5mC) plays an important role in diverse biological processes (Goll and Bestor 2005; Ooi and Bestor 2008; Wu and Zhang 2010). DNA methylation is established and maintained by a family of DNA methyltransferases (Goll and Bestor 2005). Growing evidence supports a biochemical pathway of active DNA demethylation involving ten-eleven translocation (Tet) family proteins and thymine DNA glycosylase (TDG) (Tahiliani et al. 2009; He et al. 2011; Ito et al. 2011; Pastor et al. 2013; Wu and Zhang 2014). Tet proteins can oxidize 5mC to 5-hydroxymethylcytosine (5hmC) (Tahiliani et al. 2009; Ito et al. 2010), which in turn can be further oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) and then excised by TDG followed by base excision repair to complete the demethylation cycle (He et al. 2011; Ito et al. 2011; Shen et al. 2013).
Mouse embryonic stem cells (mESCs) are derived from preimplantation epiblasts and have the capacity for self-renewal and the ability to differentiate into all embryonic lineages (Rossant 2008; Silva and Smith 2008; Hanna et al. 2010; Boroviak et al. 2014). ESCs are heterogeneous and consist of subpopulations bearing distinct characteristics. A small population (<0.5%) of ESCs, called two-cell embryo (2C)-like cells, has the ability to contribute to both embryonic and extraembryonic tissues (Macfarlan et al. 2012). ESCs enter the transient 2C-like state regularly (Macfarlan et al. 2012). Physiologically, the activation of Zscan4, one of the 2C-specific genes, is important for telomerase-independent telomere elongation and restoration of developmental capacity, two properties required for long-term maintenance of ESC pluripotency (Zalzman et al. 2010; Amano et al. 2013).
Tet1 and Tet2 are highly expressed in mESCs, which is consistent with the relative high level of 5hmC and detectable levels of 5fC and 5caC in these cells (Ito et al. 2010, 2011; Ficz et al. 2011; Koh et al. 2011). Knockdown of Tet1 in mESCs leads to reduced 5hmC levels and substantial transcriptional and promoter DNA methylation changes (Ito et al. 2010; Ficz et al. 2011; Koh et al. 2011; Williams et al. 2011; Wu et al. 2011; Xu et al. 2011). While Tet1-deficient mESCs maintain pluripotency and are largely normal, they showed skewed differentiation in vitro and in the teratoma assay (Dawlaty et al. 2011; Koh et al. 2011). Similarly, both Tet1/2 double-knockout (DKO) and Tet triple-knockout (TKO) mESCs are largely normal under ESC culture conditions, but severe abnormalities in lineage differentiation of Tet DKO and Tet TKO mESCs have been reported (Dawlaty et al. 2013, 2014; Hu et al. 2014). In contrast to the accumulating knowledge regarding the loss-of-function phenotypes of Tet proteins, it remains unclear where and to what extent Tet proteins contribute to DNA demethylation within the genome. Thus, quantitative and high-resolution mapping of DNA methylation changes in the genome of mESCs deficient for all Tet proteins will provide insights into the role of these 5mC oxidases in regulating the methylome of ESCs.
To identify targets of Tet-mediated active DNA demethylation in the genome and their biological significance, we generated Tet TKO mESCs and performed base-resolution DNA methylome and transcriptome analyses. We found that DNA hypermethylation occurred extensively at distal enhancers in Tet TKO mESCs. Parallel transcriptome analysis demonstrated that Tet TKO cells showed activation of 2C-specific genes, suggesting an important role for Tet proteins in fine-tuning ESC states. Up-regulation of these 2C-specific genes in Tet TKO cells was at least partly caused by impairment of Kap1-mediated gene repression. Consistent with the activation of 2C-specific genes, telomere elongation through telomere–sister chromatid exchange (T-SCE) was observed in Tet TKO cells. Therefore, our study uncovers a critical role of Tet proteins in enhancer function and reveals a new function for Tet proteins in repressing the 2C-like state and telomere length regulation.
Results
Tet TKO does not affect mESC maintenance
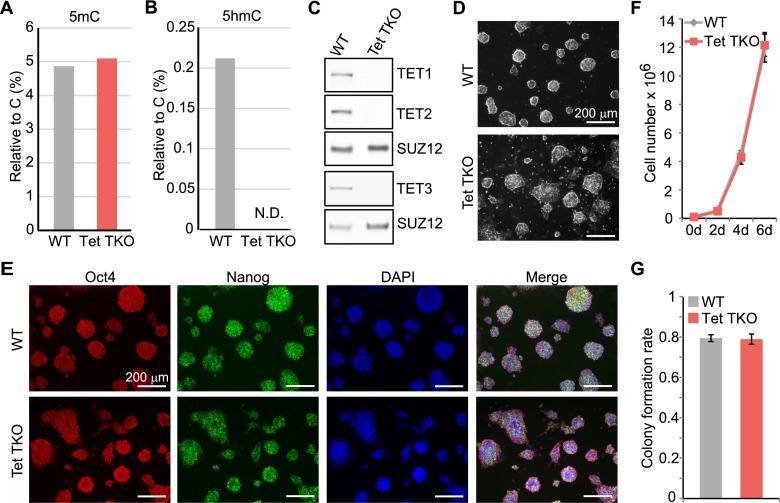
To better understand the role of Tet proteins, we generated Tet1 and Tet2 DKO or Tet1, Tet2, and Tet3 TKO mESCs using CRISPR/Cas9 technology as previously described (Wang et al. 2013). Mutations were first screened using restriction digestion within the CRISPR/Cas9 targeting sites. 5hmC levels of genomic DNA from positive clones were then assayed by slot blot to identify the clones with a significant reduction of 5hmC. Genomic DNA samples from candidate clones were then digested to a single nucleotide, and levels of 5mC and 5hmC were quantified by mass spectrometry. Even though the bulk 5mC remained at a similar level in Tet DKO and Tet TKO cells (Fig. 1A; Supplemental Fig. 1A), ∼2% of 5hmC still remained in Tet DKO cell lines compared with that in wild-type E14 cells. In contrast, Tet TKO cell lines showed no detectable 5hmC (Fig. 1B; Supplemental Fig. 1B). 5hmC levels in Tet DKO and Tet TKO mESCs are consistent with Tet DKO and Tet TKO ESC lines obtained by conventional gene targeting methods (Hu et al. 2014). Despite its low expression level, Tet3 contributes ∼2% of 5hmC in mESCs, suggesting a minor role of Tet3 in these cells. To avoid any possible functional redundancy, we chose to focus our studies on Tet TKO cells devoid of any Tet-mediated oxidation products. All Tet alleles detected in Tet TKO cells harbored frameshift mutations, resulting in premature stop codons (Supplemental Fig. 1C). Western blot analysis confirmed that both Tet1 and Tet2 proteins were not detectable in the TKO ESCs (Fig. 1C). Given that Tet3 is minimally expressed in ESCs, it is undetectable by Western blot, even in wild-type cells (data not shown). We therefore sought to assess Tet3 expression in cells subjected to embryoid body (EB) differentiation and found that Tet3 was detectable upon differentiation in wild-type cells but not in Tet TKO cells (Fig. 1C), thereby confirming successful Tet3 knockout. Characterization of Tet TKO cells indicated that TKO colonies appeared flatter compared with wild-type colonies when cultured on a feeder-free gelatin-coated plate in serum-containing medium with LIF (Fig. 1D). Nevertheless, the Tet TKO cells appeared largely unaltered with the expression of normal levels of pluripotent factors, such as Oct4, Nanog, and Sox2 (Fig. 1E; Supplemental Fig. 1D; Hu et al. 2014), a proliferation rate similar to that of wild-type control cells (Fig. 1F), and a similar colony formation rate from dissociated single cells compared with that of the wild-type control cells (Fig. 1G). Collectively, these data suggest that ESCs deficient for all three Tet proteins exhibit no obvious defects in ESC maintenance, similar to previous findings in Tet TKO cells generated through conventional knockout strategies (Dawlaty et al. 2014; Hu et al. 2014).
Figure 1.
Tet TKO ESCs are depleted of 5hmC but retain ESC characteristics. (A,B) While 5mC level was not globally altered by Tet TKO (A), 5hmC was undetectable (B) in Tet TKO ESCs. Bars are 5mC and 5hmC levels (relative to unmodified C) in control and TKO mESCs as measured by mass spectrometry. (C) Western blot analysis demonstrated depletion of Tet1, Tet2, and Tet3 proteins in Tet TKO cells, with Suz12 serving as a loading control for all blots. ESC nuclear extracts were used for Tet1, Tet2, and Suz12 blots, while nuclear extracts from differentiated cells (EBs) were used for the Tet3 and Suz12 blots. (D) Tet TKO mESCs exhibited relatively flatter morphology when compared with wild-type (WT) mESCs. (E) Pluripotent marker genes were expressed normally in Tet TKO mESCs. Data shown are images of wild-type and Tet TKO mESCs costained with Oct4 and Nanog antibodies. Nuclei were visualized by DAPI staining. (F) Growth curves of wild-type and Tet TKO mESCs. Data shown are mean of three biological replicates, with error bars representing standard deviation. (G) Tet TKO did not affect ESC colony formation capacity. Data shown are mean single-cell colony formation rates of three biological replicates, with error bars representing standard deviation.
Tet-dependent DNA demethylation mainly occurs at distal regulatory regions
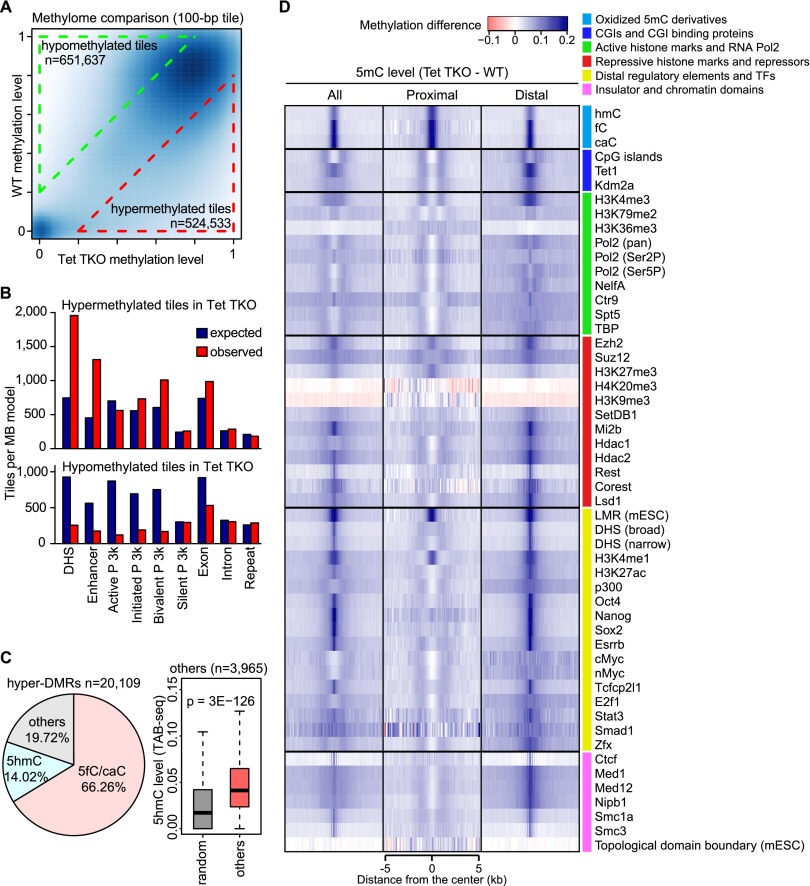
To determine the genomic locations where Tet proteins regulate DNA methylation in mESCs, we performed whole-genome bisulfite sequencing (WGBS) analyses in wild-type and Tet TKO ESCs. Since bisulfite sequencing cannot distinguish 5hmC from 5mC in wild-type ESCs, we excluded the 5hmC signal from wild-type WGBS data by subtracting the 5hmC level obtained by Tet-assisted bisulfite sequencing (TAB-seq) (Yu et al. 2012). The WGBS data from Tet TKO cells represent 5mC exclusively, as there is no oxidized 5mC in Tet TKO cells. Similar to the wild-type methylome, the overall methylation pattern of Tet TKO cells showed a bimodal distribution. Unexpectedly, after binning WGBS signals into 100-base-pair (bp) tiles (fixed window-based), Tet TKO cells showed higher numbers of hypomethylated tiles (651,637) than hypermethylated tiles (524,533), using 20% difference in 5mC level as the cutoff (Fig. 2A). Additionally, the average CpG methylation level was lower in Tet TKO ESCs (64.7%) than in wild-type cells (69.0%), as sites nearly fully methylated in wild-type tend to have lower methylation levels in Tet TKO ESCs (Supplemental Fig. 2). Further analysis of various genomic features revealed that the hypermethylated sites in Tet TKO ESCs were enriched mostly in DNase I-hypersensitive sites (DHSs), enhancer regions, and bivalent promoters (Fig. 2B, top). In contrast, hypomethylated sites were not highly enriched for any genomic features (Fig. 2B, bottom). With Methpipe, a hidden Markov model (HMM; nonfixed window)-based algorithm for detecting differentially methylated regions (DMRs) (Song et al. 2013), we identified 20,109 hypermethylated DMRs (hyper-DMRs) and only 315 hypomethylated DMRs (hypo-DMRs) in Tet TKO ESCs (Supplemental Table 1). Taken together, these data indicate that hypermethylated CpGs tend to cluster together at specific genomic loci with gene regulatory potentials (promoter, enhancers, and gene bodies). In contrast, hypomethylated CpGs are randomly distributed across the genome. We found that 66% of hyper-DMRs overlapped with 5fC/5caC DNA immunoprecipitation (DIP) sequencing (DIP-seq) peaks in TDG knockdown ESCs, and another 14% of hyper-DMRs overlapped with 5hmC DIP-seq peaks when not covered by 5fC/5caC peaks (Fig. 2C, left; Shen et al. 2013). Although the remaining 20% of hyper-DMRs did not overlap with 5hmC, 5fC, or 5caC peaks, there was significant 5hmC enrichment at these regions when looking into the high-confidence base-resolution 5hmC data generated by TAB-seq analysis (Fig. 2C, right; Yu et al. 2012). Therefore, these data strongly suggest that the majority of hyper-DMRs identified are targets of Tet-mediated oxidation-dependent DNA demethylation in mESCs.
Figure 2.
Tet-dependent DNA demethylation mainly occurs at distal regulatory regions involving 5mC oxidation. (A) Global methylation comparison between wild-type (WT) and Tet TKO mESCs revealed both hypermethylated and hypomethylated loci. Data shown are a heat map comparison of 100-bp tile-binned methylome between wild-type and Tet TKO mESCs. Tiles with >20% methylation change were identified as differentially methylated. (B) Distribution of the relative enrichment of hypermethylated tiles and hypomethylated tiles at various genomic features. (DHS) DNase I-hypersensitive sites; (P) promoters, which are divided into four groups based on histone methylation pattern. Details of promoter definition can be found in “Promoter Classification by Chromatin States” in the Materials and Methods. (C) Hyper-DMRs were mainly located at loci with Tet-mediated 5mC oxidization. (Left) The pie chart shows distribution of hyper-DMRs in Tet TKO mESCs. (Right) The box plot shows 5hmC signal from TAB-seq in loci with hyper-DMR but not 5hmC or 5fc/5caC peaks. (D) Data shown are heat maps of average 5mC level differences between Tet TKO and wild-type ESCs (Tet TKO − WT) at centers of annotated genomic features or enriched regions for oxidized 5mC derivatives, transcriptional regulators, histone modifications, pluripotency transcription factors (TFs), and distal regulatory regions (derived from published data sets in mESCs). The difference is shown for all, proximal (overlapping with ±1 kb flanking the TSSs), and distal features. Each row is centered at midpoint of the feature with a 5-kb flanking sequence.
To further characterize the differential methylation in Tet TKO cells, we calculated the average levels of 5mC change within genomic features derived from published genome-wide mapping data sets for 5mC oxidative derivatives, DNA-binding factors, and major histone modifications (Fig. 2D). DNA hypermethylation was detected at the center of 5fC, 5caC, and, to a lesser extent, 5hmC DIP-seq peaks (Fig. 2D, cyan color group; Shen et al. 2013) regardless of proximal or distal localization. Although 5fC and 5caC accumulated to high abundance, 5mC levels remained similar to TDG knockdown cells (Shen et al. 2013). Additionally, hypermethylation levels at the center of 5hmC peaks were lower than those for 5fC and 5caC (Fig. 2D). These data therefore provide the first direct evidence of 5fC and 5caC serving as demethylation intermediates in cells.
To determine whether 5hmCs serves as an intermediate of the DNA demethylation process or as a stable epigenetic mark, we analyzed the relationship between hypermethylated CpGs in Tet TKO cells and the previously reported high-confidence 5hmC (Yu et al. 2012). This analysis revealed no clear correlation between hypermethylation in Tet TKO cells and high-confidence 5hmC in wild-type ESCs. Indeed, a significant portion of hypermethylated cytosines had no or a very low level of 5hmC in wild-type ESCs (Supplemental Fig. 3A). Consistently, only 27% of the hypermethylated cytosines in Tet TKO ESCs had a 5hmC signal in wild-type ESCs when cytosines with high coverage in both our WGBS data and the TAB-seq data were analyzed (at least covered by 10 reads per strand) (Supplemental Fig. 3B). These results suggest that stable 5hmC may serve as an epigenetic mark rather than as an intermediate of active DNA demethylation. Hypermethylation was preferentially detected at binding sites of core transcription factors in ESCs (Oct4, Nanog, Sox2, and Esrrb) (Chen et al. 2008; Marson et al. 2008) and histone marks (H3K4me1 and H3K27ac) associated with enhancers (Fig. 2D, yellow group; Meissner et al. 2008; Creyghton et al. 2010), the mediator complex, and the cohesion complex (Fig. 2D, pink color group; Kagey et al. 2010). Furthermore, hypermethylation was detected at binding sites of complexes involved in decommissioning active enhancers during differentiation, such as the LSD1–NuRD complex (Fig. 2D, red color group; Whyte et al. 2012). These features are all known to be associated with enhancers, indicating that Tet-dependent DNA demethylation mainly occurs at enhancer regions. Features associated with basal transcriptional machineries (Pol2, NelfA, Ctr9, Spt5, and TBP) and actively transcribing gene bodies (H3K36me3 and H3K79me2) were minimally enriched for hypermethylation in Tet TKO ESCs (Fig. 2D, green color group; Kagey et al. 2010; Rahl et al. 2010). Minimal hypomethylation was seen at regions with heterochromatin-associated histone marks (H3K9me3 and H4K20me3) and was distributed evenly across the regions without visible peaks at the center of the marks (Fig. 2D, red color group). For most features analyzed, distally located elements were preferentially hypermethylated at the center in Tet TKO ESCs (Fig. 2D). Most of the proximally located elements (within 1 kb from transcriptional start sites [TSSs]) possessed small amounts of hypermethylation flanking the center of the features (Fig. 2D). Besides 5mC oxidative derivatives, hypermethylation at proximally located elements was also seen in Polycomb-binding sites (EZH2 and H3K27me3) (Fig. 2D, red color group; Mikkelsen et al. 2007; Ku et al. 2008), indicating that bivalent promoters are the major type of promoters regulated by Tet-mediated DNA demethylation. In summary, these results demonstrate that Tet-mediated DNA demethylation mainly occurs at distal regulatory elements.
Enhancers and promoters are the main targets of Tet-mediated DNA demethylation
The above data indicate that DNA hypermethylation is enriched at enhancers and promoters. Indeed, within the 20,109 hyper-DMRs that we called in Tet TKO ESCs, 45.3% overlapped with enhancers (Kieffer-Kwon et al. 2013), 17.1% overlapped with promoters, and 27.7% overlapped with other DHSs (Fig. 3A). The enhancer regions used here were defined by DHSs with cohesion, mediator, or p300 binding outside of promoter regions (1 kb away from TSSs) (Kieffer-Kwon et al. 2013). We chose this enhancer data set because cohesion and mediator-demarcated enhancers tethered with core promoters and because the enhancer–promoter interaction data were available for this data set (Kagey et al. 2010; Kieffer-Kwon et al. 2013). With regard to chromatin modifications, H3K4me1 and H3K27ac are two typical markers of enhancers (Heintzman et al. 2007; Creyghton et al. 2010; Rada-Iglesias et al. 2011). Indeed, the 9785 hypermethylated enhancers were highly enriched for H3K4me1 and H3K27ac (Fig. 3B), consistent with the notion that they are typical marks of enhancers. More strikingly, the mean DNA methylation level of all enhancers (n = 62,766) increased ∼15% at the center of the enhancer in the absence of Tet proteins (Fig. 3C), highlighting the important role of Tet proteins in regulating enhancer DNA methylation levels. As H3K27ac level is indicative of enhancer activity (Creyghton et al. 2010), we ranked and grouped enhancers by H3K27ac level in wild-type ESCs and compared the levels of DNA methylation increase in Tet TKO ESCs between each group. We found an inverse correlation between DNA methylation and H3K27ac levels as well as all 10 groups of enhancers exhibiting a significant increase in DNA methylation in Tet TKO cells (Supplemental Fig. 4). Our analysis indicates that proximal features associated with promoters show relatively lower hypermethylation at the shores of the center and that Polycomb-binding sites show hypermethylation at the center of the elements (Fig. 2D). Indeed, bivalent promoters showed the largest average methylation increase among promoters (Supplemental Fig. 5). For example, we detected a significant increase in DNA methylation at both the promoter and one nearby enhancer of T (also named Brachyury) in Tet TKO ESCs (Fig. 3D). The methylation change was validated by targeted bisulfite sequencing of an independent batch of samples with at least 1750× coverage (Fig. 3D). The average methylation level at active and initiated promoters also increased to a lesser extent, whereas silent promoters were heavily methylated even in wild-type cells, with levels comparable with those of repeat sequences (Supplemental Fig. 5). These results suggest a general enrichment of Tet-mediated DNA demethylation at regulatory sequences, especially at enhancers and promoter regions. Taken together, our data reveal the location of genome-wide Tet-dependent DNA demethylation in mESCs, which mainly include enhancers, promoters, and other distal regulatory elements.
Figure 3.
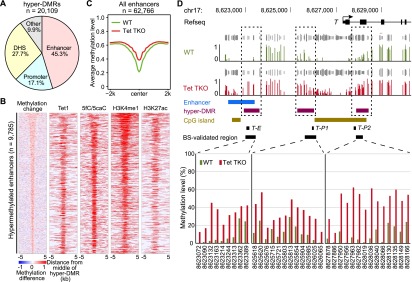
Enhancers are the main targets of Tet-mediated DNA demethylation. (A) Pie chart distribution of hyper-DMRs in Tet TKO mESCs. Hyper-DMRs were mainly localized to enhancers, promoters, and other DHSs (defined by at least 1 bp overlap). For regions belonging to more than one category, the classification follows the priority of enhancer, promoter, and DHSs. (B) Heat maps of methylation difference; peak regions of Tet1 binding, 5fC/5caC in TDG knockdown cells, H3K4me1, and H3K27ac at hypermethylated enhancers in Tet TKO cells. Heat maps are ranked by the size of hyper-DMRs that overlap with enhancers (top, largest; bottom, smallest). Each row represents one enhancer that overlaps with a hyper-DMR in Tet TKO cells and is centered at the midpoint of the enhancer with a 5-kb flanking sequence. In the methylation change column, the colors in the legend indicate the value of the methylation difference. In the other columns, the colored regions indicate peaks of the selected features. (C) Plot of the average methylation level at all annotated enhancers showing general hypermethylation at enhancers in Tet TKO ESCs. (D) Representative genome browser view of hypermethylation of the T locus. Both the enhancer and the promoter of T were hypermethylated in Tet TKO mESCs. Track information is shown on the left side of each track. Gray tracks above DNA methylation tracks are sequencing coverage data (CpGs covered for at least 5× in both control and TKO samples). T-E, T-P1, and T-P2 indicate regions analyzed in Supplemental Figure 7. Targeted bisulfite sequencing validation is shown at the bottom. The X-axis numbers indicate position of CpG sites in mm9 genome assembly. The CpGs were covered at least 1750×.
Dual function of Tet-mediated DNA demethylation in transcriptional regulation
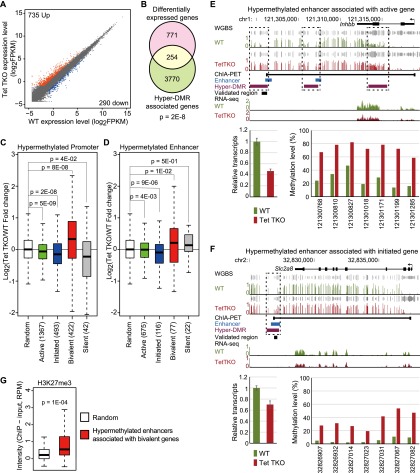
To examine the transcriptional effect of Tet TKO, we performed RNA sequencing (RNA-seq) in wild-type and Tet TKO ESCs and identified 735 up-regulated and 290 down-regulated genes, respectively (fold change > 2, FPKM [fragments per kilobase of exon per million mapped fragments] > 0.5 in at least one sample) (Fig. 4A; Supplemental Table 2). In order to analyze the effect of DNA hypermethylation on transcriptional change, we analyzed hypermethylated genes exhibiting hypermethylation in either promoters or associated enhancers. Previous chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) analysis has experimentally identified promoter–enhancer interactions representing 3764 genes, among which 998 showed hypermethylation in at least one enhancer in Tet TKO cells (Kieffer-Kwon et al. 2013). When compared with DNA methylation change, we found that hypermethylated genes (3296 promoter hypermethylated genes and 998 enhancer hypermethylated genes with known enhancer–promoter interaction, of which 270 genes were shared) overlapped significantly with differentially expressed genes (Fig. 4B; Supplemental Table 3), indicating that change in DNA methylation is likely one of the primary causes of the expression change in Tet TKO ESCs.
Figure 4.
Dual function of Tet-mediated DNA demethylation in transcriptional regulation. (A) Scatter plot of RNA-seq data illustrates transcriptional change in Tet TKO cells. Genes up-regulated are shown in red dots, while down-regulated genes are shown in blue dots. Genes showing an expression difference larger than twofold were considered differentially expressed. (B) Venn diagram depicting the overlap between differentially expressed genes and differentially methylated genes. Statistical significance was calculated by hypergeometric test. (C) Dual role of promoter methylation in transcriptional regulation. Expression of genes (FPKM > 0.5 in at least one sample) with hypermethylated promoters was compared with that of randomly selected genes. For each category of genes, the P-value of the two-tailed t-test was calculated. (D) Dual role of enhancer methylation in transcriptional regulation. Expression of genes (FPKM > 0.5 in at least one sample) with known promoter–enhancer interaction was compared with that of randomly selected genes. For each category of genes, the P-value of the two-tailed t-test was calculated. (E,F) Genome browser view of enhancer hypermethylation and gene expression decrease at Inhbb (active gene) (E) and Slc2a8 (initiated gene) (F) loci. Track information is shown on the left side of each track. Gray tracks above DNA methylation tracks are sequencing coverage information. RT-qPCR (left) and targeted bisulfite sequencing validation (right) are shown at the bottom of each panel. Actb (β-actin) was used as internal control for expression analysis. Error bars represent standard error of the mean. The X-axis numbers indicate position of CpG sites in mm9 genome assembly. The CpGs were covered at least 2400×. (G) Bivalent gene-associated enhancers were modified by H3K27me3. The H3K27me3 level in wild-type (WT) ESCs at hypermethylated enhancers that interact with bivalent genes is shown in the box plot. Randomly sampled regions were used as a control. Statistical significance was calculated by Student’s t-test. (RPM) Reads per million mapped reads.
To further examine this relationship, we analyzed promoter methylation and enhancer methylation separately. Promoters can be classified into four groups based on their associated histone modification (Whyte et al. 2012). The four groups include (1) active promoters that contain H3K4me3 and downstream H3K79me2/3, (2) initiated promoters that contain H3K4me3 only, (3) bivalent promoters that contain H3K4me3 and H3K27me3, and (4) silent promoters that do not have H3K4me3. Analysis of the relationship between transcription and promoter hypermethylation for each gene group revealed that transcription from active promoters and initiated promoters decreased significantly when they were hypermethylated (Fig. 4C; Supplemental Fig. 6). This is consistent with the notion that promoter methylation generally correlates with transcriptional repression (Klose and Bird 2006). In contrast, hypermethylation of bivalent promoters associated with significant up-regulation of transcription (Fig. 4C), which is consistent with the previous finding that increased DNA methylation at bivalent genes positively correlates with increased transcription, possibly due to impaired recruitment of Polycomb repression complexes (PRCs) (Wu et al. 2011).
Enhancers are generally hypomethylated when they are bound by cell type-specific transcription factors, suggesting that active demethylation at active enhancers might be important for their activity (Stadler et al. 2011; Kieffer-Kwon et al. 2013; Ziller et al. 2013). However, these studies were carried out by comparing enhancer methylation and enhancer activity in different cell types, making the correlations difficult to interpret. Tet TKO cells are largely normal and exhibit aberrant methylation at a large cohort of enhancers, which provide a unique opportunity for studying the role of enhancer methylation in transcriptional regulation. We found that expression of active genes and initiated genes was significantly decreased when their associated enhancers were hypermethylated (Fig. 4D–F). In contrast, bivalent genes showed increased expression when their enhancers became hypermethylated in the absence of Tet proteins (Fig. 4D). Activation of this group of genes might be caused by either methylation-mediated repelling of PRCs similar to that observed at hypermethylated bivalent promoters or gene bodies (Wu et al. 2010, 2011), or Polycomb binding necessitating nonmethylated CpG (Farcas et al. 2012; He et al. 2013; Wu et al. 2013). Clearly, these enhancers are modified by H3K27me3 in wild-type cells (Fig. 4G). Using T as an example, we found that expression of T was increased in Tet TKO (Supplemental Fig. 7A). ChIP-qPCR (chromatin immunoprecipitation [ChIP] coupled with quantitative PCR [qPCR]) analysis of hypermethylated regions within promoter and enhancer regions showed that binding of both Ring1B and Ezh2, core components of the PRC1 and PRC2 complexes, decreased upon hypermethylation (Supplemental Fig. 7B). The number of silent genes with hypermethylated enhancers and the number of silent genes with hypermethylated promoters were small, so the transcriptional changes associated with these hypermethylation events were not statistically significant (Fig. 4C,D). Taken together, we found that hypermethylation at promoters/enhancers of active/initiated genes is generally associated with gene repression, while the opposite is observed at bivalent gene promoters and associated enhancers, possibly due to the negative effect of 5mC on the binding of Polycomb group proteins.
Increased 2C-like population in Tet TKO ESCs
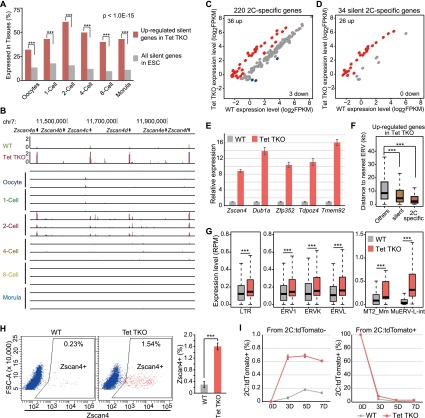
During transcriptome analysis, we noticed that many silent genes up-regulated in Tet TKO cells were highly enriched in genes specifically expressed during preimplantation development (Fig. 5A), including genes known to be specifically expressed in 2Cs (Xue et al. 2013), such as the Zscan4 cluster of genes (Fig. 5B; Falco et al. 2007). In contrast, up-regulated genes in the other three groups did not show such enrichment (Supplemental Fig. 8A). By reanalyzing published transcriptomes of mouse preimplantation embryos (Xue et al. 2013), we identified 220 2C-specific genes (Supplemental Fig. 8B; Supplemental Table 4), of which 34 were classified as silent genes in mESCs. Among the 220 2C-specific genes, 36 were up-regulated in Tet TKO ESCs, while only three showed decreased expression (Fig. 5C; Supplemental Table 4). More strikingly, 26 of the 34 silent 2C-specific genes showed increased expression, while none of these showed decreased expression in Tet TKO cells (Fig. 5D; Supplemental Table 4). The activation of 2C-specific genes was confirmed by RT-qPCR using an independent batch of samples (Fig. 5E).
Figure 5.
2C-specific genes are up-regulated, and the 2C-like population is increased in Tet TKO ESCs. (A) Silent genes up-regulated in Tet TKO ESCs were enriched in oocytes and all stages of preimplantation development. The percentage of up-regulated silent genes in Tet TKO cells and all silent genes expressed at a given developmental stage is shown. Statistical significance was performed by proportion test. (B) Zscan4 cluster is shown as an example of 2C-specific genes up-regulated in Tet TKO ESCs. Bar, 50 kb. (Top) RNA-seq results of wild-type (WT) and Tet TKO ESCs. (Bottom) RNA-seq results of mouse oocytes and embryos at different stages of preimplantation development. (C,D) Scatter plot illustrating the enrichment of increased expression of 2C-specific genes (C) and silent 2C-specific genes (D) in Tet TKO ESCs. Up-regulated genes and down-regulated genes are shown as red and blue dots, respectively. The number of up-regulated genes and down-regulated genes is shown at the top left and bottom right, respectively. (E) Activation of 2C-specific genes validated by RT-qPCR. Actb was used as internal control for expression analysis. Error bars represent standard error of the mean. (F) Up-regulated 2C-specific genes in Tet TKO ESCs were closer to ERVs. The average distance between TSSs and ERVs was plotted for different categories of genes. Statistical significance was calculated by Wilcoxon test; (***) P-value < 0.001. (G) Derepression of ERVs, especially type III Mu-ERVLs in Tet TKO ESCs. The expression of the LTRs of all ERVs, the three subtypes of ERV (ERV1, ERVK and ERVL), and two subregions of MuERV-L (MT2_Mm and MuERV-L-int) in wild-type and Tet TKO ESCs is shown in the box plot. Statistical significance was calculated by Wilcoxon test; (***) P-value < 0.001. (H) Tet TKO resulted in an increase in the Zscan4-positive 2C-like cell population. Zscan4 staining analyzed by FACS is shown in the scatter plot. Approximately 20,000 cells were counted for each experiment. The bar graph on the right shows average data from three experiments. Error bars represent standard deviation. (I) Plot of 2C:tdTomato-positive cells in the culture at the indicated times. 2C:tdTomato-negative cells (left) and 2C:tdTomato-positive cells (right) sorted by FACS were plated at day 0. Error bars represent standard deviation.
Previous studies have established a link between the expression of the 2C-specific genes and the activation of repeat sequences, especially type III endogenous retrovirus (ERV), murine ERV with leucine tRNA primer (MuERV-L, also known as MERVL), and Mt2_mm, the corresponding long terminal repeat (LTR) promoters of MuERV-L (Macfarlan et al. 2011, 2012). In support of this link, we found that up-regulated 2C-specific genes were significantly closer to ERVs than other up-regulated genes (Fig. 5F). This observation prompted us to look into the expression of different repeat sequences. We found that expression of LTR-type repeats showed significant increase (Fig. 5G), while expression of LINE and SINE elements was also increased to a lesser extent in Tet TKO ESCs (Supplemental Fig. 8C). Detailed analysis of the three subtypes of ERVs (ERV1, ERVK, and ERVL) revealed that ERVL exhibited the highest activation in Tet TKO ESCs (Fig. 5G). Both the LTR region (MT2_Mm) and the internal region (MuERV-L-int) of MuERV-L showed significant activation in Tet TKO ESCs (Fig. 5G). Collectively, these results suggest that Tet proteins may play an important role in repressing type III ERV and 2C-specific gene expression in mESCs.
Because Zscan4 and other 2C-specific genes are expressed in a small population of cells in standard ESC culture (Zalzman et al. 2010; Macfarlan et al. 2012; Amano et al. 2013), we asked whether activation of Zscan4 in Tet TKO ESCs correlates with an increase in the Zscan4-positive cell population. To this end, we stained cells with Zscan4 antibody and then counted the number of Zscan4-positive cells by fluorescence-activated cell sorting (FACS), which revealed that Zscan4-positive cells in Tet TKO ESCs increased to around sixfold of that in wild-type ESCs (Fig. 5H; Supplemental Fig. 9). Given that the staining intensity of Zscan4-positive cells in Tet TKO ESCs is similar to that of wild-type ESCs, we conclude that a significantly higher fraction of Tet TKO cells exist in a 2C-like state when compared with the wild-type cells. In addition, we labeled Tet TKO ESCs with a 2C:tdTomato reporter and sorted out the 2C:tdTomato-positive and 2C:tdTomato-negative populations. When these populations were placed in culture, the reporter was quickly activated in 2C:tdTomato-negative cells, while the reporter was quickly shut down in 2C:tdTomato-positive cells, indicating that the 2C-like population maintained the dynamic manner in Tet TKO ESCs (Fig. 5I). Taken together, our data suggest that Tet TKO ESCs maintain a higher population of 2C-like cells compared with that in wild-type ESCs.
Regulation of Kap1 in repressing 2C-specific genes by Tet proteins
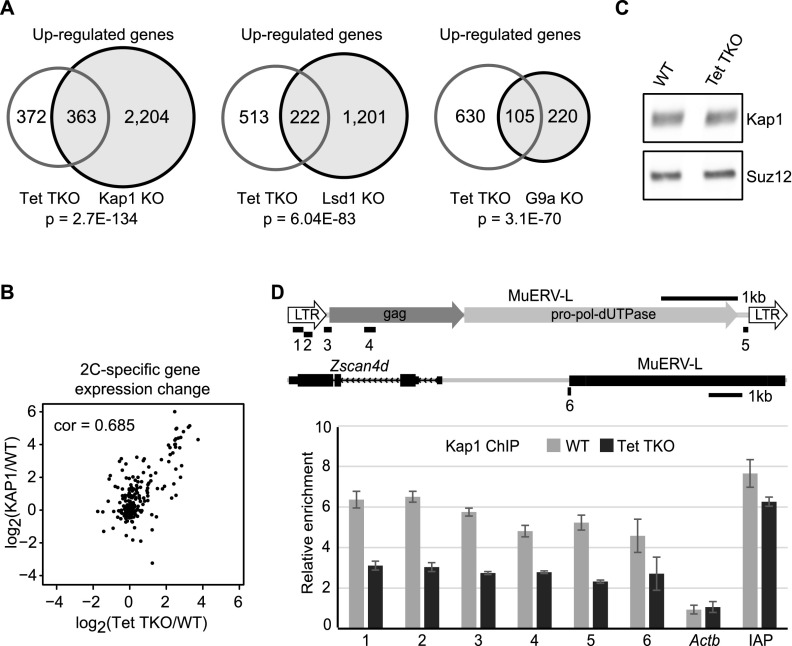
Previous studies have shown that expression of 2C-specific genes is repressed by Kap1, LSD1, and G9a in mESCs (Macfarlan et al. 2011, 2012; Maksakova et al. 2013). To explore whether Tet-mediated 2C gene repression was linked to any of these factors, we compared differentially expressed genes in Tet TKO cells with those in Kap1, LSD1, and G9a mutant cells. This analysis revealed that up-regulated genes in Tet TKO cells significantly overlapped with genes up-regulated in Kap1, LSD1, or G9a mutant cells (Fig. 6A; Supplemental Table 5). Since up-regulated genes in Tet TKO cells showed the most overlap with those up-regulated in KAP knockout cells and since expression of 2C-specific genes in Tet TKO ESCs correlated well with those in KAP1 knockout cells (Pearson correlation coefficient = 0.685) (Fig. 6A,B), we explored the possibility of whether Tet proteins and Kap1 function in the same pathway or cooperate in repressing 2C-specific genes. By reanalyzing published Kap1 ChIP sequencing (ChIP-seq) data (Quenneville et al. 2011; Rowe et al. 2013), we found that Kap1 was significantly enriched at MuERV-L repeats (Supplemental Fig. 10A,B). Since Kap1 mRNA level (FPKM: wild-type, 1013; Tet TKO, 990) and protein level (Fig. 6C) were not significantly altered in Tet TKO cells, we explored whether Tet protein depletion would affect KAP1 binding to chromatin. We performed ChIP-qPCR to analyze Kap1 binding by comparing DNA captured by anti-Kap1 with that captured by IgG control. The result revealed that KAP1 binding at MuERV-L repeats (regions 1–5) was significantly decreased in Tet TKO cells (Fig. 6D). Similarly, ChIP-qPCR analysis of a single-copy region (region 6) upstream of Zscan4d also showed decreased Kap1 binding in Tet TKO cells (Fig. 6D). In contrast, Kap1 binding at IAP regions was minimally affected in Tet TKO ESCs (Fig. 6D). This is consistent with the minimal IAP up-regulation detected in Tet TKO ESCs compared with that in Kap1 knockout cells (Supplemental Fig. 10C,D). Collectively, these data suggest that Tet proteins are likely required for efficient binding of Kap1 to MuERV-L and repressing transcription of MuERV-L as well as the nearby 2C-specific genes.
Figure 6.
Tet proteins repress 2C-specific genes and LTRs by facilitating Kap1 recruitment. (A) Venn diagram showing that genes up-regulated in Tet TKO ESCs significantly overlap with those up-regulated in Kap1, Lsd1, or G9a knockout (KO) ESCs. (B) Scatter plot showing strong expression correlation of the 220 2C-specific genes in Tet TKO and Kap1 knockout ESCs. (cor) Pearson coefficiency. (C) Western blot analysis demonstrated that the Kap1 protein level was not significantly altered in Tet TKO ESCs. Suz12 served as a loading control. (D) ChIP analysis demonstrated reduced KAP1 binding at MuERV-L elements in Tet TKO ESCs. The relative enrichment is fold enrichment of relative DNA captured by Kap1 antibody compared with DNA captured by IgG control. ChIP-qPCR-analyzed genomic regions are indicated in the diagram in the top panel. One-kilobase scale bars are shown for each of the two loci. Amplicons 1–5 are within MuERV-L repeats. Amplicon 6 is a single-copy region that partly overlaps with the MuERV-L repeat upstream of Zscan4d. Amplicon IAP is one region in the LTR of the IAP elements. The promoter region of Actb was used as a negative control. Error bars represent standard error of the mean.
Increased telomere length in Tet TKO ESCs
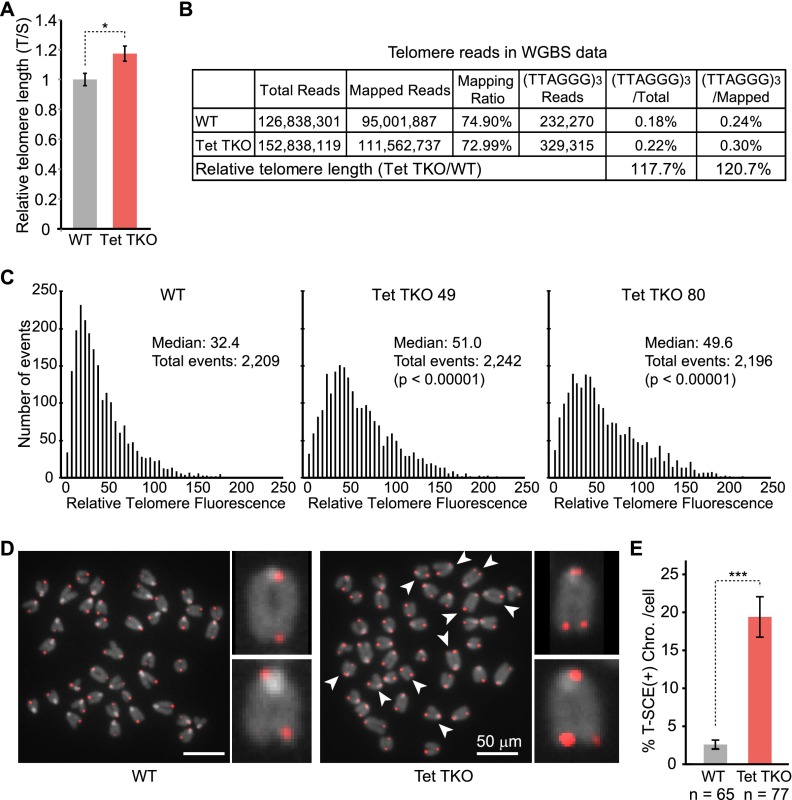
Since the transient Zscan4-positive state is known to promote telomere elongation through telomere recombination (Zalzman et al. 2010), we asked whether telomere length is altered in Tet TKO cells. Using a well-established qPCR method (Callicott and Womack 2006), we found that the average telomere length increased ∼18% in Tet TKO cells (Fig. 7A). Additionally, our WGBS data provided an independent means to quantify the relative telomere lengths. By counting the (TTAGGG)3 reads in the raw sequencing data, we estimated ∼20% more telomere-associated reads in Tet TKO ESCs compared with that in wild-type ESCs (Fig. 7B). To better understand the distribution of telomere length in Tet TKO ESCs and rule out the potential clonal effect of Tet TKO ESCs, we performed telomere quantitative fluorescence in situ hybridization (Q-FISH) in wild-type and two independent Tet TKO cell lines. The result indicated that telomeres in both Tet TKO cell lines exhibited wider length distribution with an overall shift in addition to increased average length, which was similar to that seen in Zscan4-overexpressing cells (Fig. 7C; Supplemental Fig. 11; Zalzman et al. 2010). Since Zscan4 promotes telomere elongation through increased telomere recombination (Zalzman et al. 2010), we asked whether Zscan4 promoted telomere elongation using a recombination-based mechanism in Tet TKO ESCs. To this end, we measured the frequency of telomere recombination using the chromosome orientation FISH (CO-FISH) technique (Williams and Bailey 2009). Consistent with the previous demonstration that the frequency of T-SCE events is significantly increased in Zscan4-overexpressed ESCs (Zalzman et al. 2010), we found that the frequency of T-SCE was significantly increased in Tet TKO ESCs compared with that of wild-type cells (Fig. 7D,E). Collectively, multiple lines of evidence suggest that Tet TKO cells enter the 2C-like state more frequently and acquire 2C-like features such as increased telomere length.
Figure 7.
Tet TKO results in telomere elongation along with high frequency of T-SCE. (A) Increased telomere length in Tet TKO ESCs. Average telomere length was assessed by qPCR with a telomere repeat-specific primer set and normalized to a single-copy control primer set. (B) Increased telomere length in Tet TKO ESCs as measured by WGBS reads. The table summarizes WGBS reads and uniquely mapped reads. (TTAGGG)3-containing reads were counted as telomere reads in the raw data and normalized to either total sequencing reads or uniquely mapped reads. (C) Distribution histogram of relative telomere length analyzed by telomere Q-FISH and TFL-TELO software. (D,E) Increased T-SCE in Tet TKO cells. T-SCE events were measured by CO-FISH. (D) Representative images are shown. (E) Quantification of chromosomes with T-SCE events are shown in the bar graph. Statistical significance was calculated by student's t-test; (*) P-value < 0.05; (***) P-value < 0.001.
Discussion
Tet-mediated successive oxidation of 5mC to 5hmC, 5fC, and 5caC leads to either passive dilution of 5hmC or active removal of 5fC and 5caC by TDG and base excision repair (Pastor et al. 2013; Wu and Zhang 2014). However, where and to what extent Tet proteins mediate DNA demethylation still remains poorly understood. Previous studies using antibody-based methylated DIP (MeDIP) followed by microarray analysis (MeDIP–chip) or targeted bisulfite sequencing of wild-type and Tet1-depleted ESCs revealed DNA hypermethylation at a cohort of Tet1-bound gene promoters (Wu et al. 2011; Xu et al. 2011). Furthermore, MeDIP-coupled sequencing (MeDIP-seq) of several tissues in Tet DKO mice showed an increase of 5mC signal across all chromosomes, suggesting a widespread contribution of Tet proteins in DNA methylation in vivo (Dawlaty et al. 2013). Recently, Dawlaty et al. (2014) reported that the EBs derived from Tet TKO mESCs also showed increased baseline 5mC across all chromosomes through MeDIP-seq analysis but failed to identify more distribution features of the hypermethylated regions. In addition, the absolute level of 5mC change could not be determined by the MeDIP, and targeted bisulfite sequencing only provided limited information of selected promoter regions. Additionally, methylation analyses of highly heterogeneous cell populations, such as tissues or EBs, further complicate meaningful interpretation of the data. Here, we took advantage of Tet TKO mESCs that are completely free of 5mC oxidative derivatives and revealed (1) the exact locations where Tet-mediated active DNA demethylation occurs at base resolution and (2) the physiological function of Tet proteins in ESCs.
Methylation changes in Tet TKO ESCs
Since Tet TKO cells do not contain 5hmC, 5fC, and 5caC, WGBS signals from these cells represent only 5mC. Comparing WGBS data from Tet TKO ESCs with those from wild-type cells with 5hmC signal subtracted, we defined Tet deficiency-induced 5mC change at base resolution. Unexpectedly, we found the overall CpG methylation level in Tet TKO cells (64.7%) to be slightly lower than that in wild-type cells (69%). This is likely due to decreased methylation within highly methylated CpGs (Fig. 2A; Supplemental Fig. 2), such as those located in heterochromatin regions (Fig. 2D, H3K9me3 and H4K20me3). This suggests that DNA methylation is not fully maintained in Tet TKO ESCs. It would be interesting to investigate the relationship between 5mC oxidation and maintenance in the future.
Nevertheless, in Tet TKO cells, we found 60 times more hyper-DMRs across the genome than hypo-DMRs, indicating that hypomethylated CpG sites are randomly distributed, whereas hypermethylated CpG sites tend to cluster at specific genomic loci. Apart from a subset of hypermethylated CpGs that are 5hmC-modified in wild-type ESCs, a significant fraction of hypermethylated CpGs are not associated with stable 5hmC signals detected in wild-type cells. It appears that a cohort of these 5hmC-free hypermethylated CpGs resides within 5fC/5caC peaks, suggesting that active demethylation (conversion of 5mC to C) and stable accumulation of 5hmC modification may represent distinct molecular events. Thus, genome-wide mapping of 5hmC may not be sufficient to identify all cytosines undergoing active DNA demethylation. Mapping 5fC/5caC accumulation in TDG-deficient cells at single-base resolution may contribute to a better understanding of Tet and TDG-dependent active DNA demethylation.
Previous studies have shown that Tet proteins are required for the maintenance of low methylation status at a subset of promoters in ESCs (Wu et al. 2011; Xu et al. 2011; Dawlaty et al. 2014). In this study, we not only provided a complete list of promoters undergoing active DNA demethylation but also found that enhancer regions are the most profound Tet-dependent active demethylation loci. Proximal CpG islands colocalize with the majority of promoters in the mouse genome and are mostly unmethylated (Deaton and Bird 2011). We found that DNA methylation increased significantly at the CGI shores but minimally at the center of CGIs (Fig. 2D), indicating that Tet proteins are not the major players in preventing CGIs from methylation. The main players in preventing CGI methylation remain undefined. Enhancers are generally lower in DNA methylation (Stadler et al. 2011; Kieffer-Kwon et al. 2013; Ziller et al. 2013). Here we provide evidence indicating that Tet-mediated active DNA demethylation is one of the major contributing factors in maintaining this phenomenon. Since enhancers are well known to be cell type-specific, it will be interesting to study the roles of Tet proteins in enhancer reconfiguration during cell fate transition.
DNA methylation at enhancers fine-tune enhancer activity in ESCs
Previous studies have revealed that active enhancers are generally hypomethylated, suggesting that hypomethylation may be required for enhancer activity (Stadler et al. 2011; Kieffer-Kwon et al. 2013; Ziller et al. 2013). However, this hypothesis has not been confirmed, as all of the studies so far have compared enhancer methylation and activity from different cell types and have not ruled out a possible secondary effect resulting from different cell identities. Here, we found widespread hypermethylation at enhancers in Tet TKO cells without disrupting their identity. This provided us with a unique opportunity to examine the effect of DNA methylation on enhancer activity. We found that increased methylation at the enhancer region of actively transcribed genes (Fig. 4D, active/initiated) was negatively correlated with transcription in general. Hypermethylation in enhancer regions resulted in small changes in transcription, likely due to the minor role played by DNA methylation in gene repression of early embryos and ESCs (Fouse et al. 2008; Bogdanovic et al. 2011). In contrast, increased enhancer methylation of bivalent genes (Fig. 4D, bivalent) correlated with transcriptional activation, supporting the mutual exclusion of DNA methylation and Polycomb protein binding (Wu et al. 2010, 2011, 2013; Farcas et al. 2012; He et al. 2013) and the notion that removal of Polycomb repression is required for enhancer activation (Rada-Iglesias et al. 2011). These results suggest that Tet-dependent DNA demethylation at enhancers fine-tunes transcription of genes interacting with these enhancers in ESCs. Further studies will be needed to determine whether Tet-dependent DNA demethylation plays a larger role in regulating enhancer activity in somatic cells where DNA methylation is heavily associated with gene repression.
Role of Tet proteins in repeat silencing and telomere regulation
Temporal acquisition of a 2C-specific condition is critical to restoring the developmental capacity in ESCs (Amano et al. 2013). Previous studies have demonstrated that a 2C-like cell population increases in cells deficient for Kap1, Lsd1, or G9a (Macfarlan et al. 2012). We found that 2C-specific genes are up-regulated in Tet TKO cells. Although Tet1 ChIP-seq did not reveal stable occupancy of Tet1 protein at these regions, the presence of high-confidence 5hmC signals at 2C-specific gene loci indicates that Tet proteins indeed function at these loci. Tet2 might also occupy these loci, as it possesses intrinsic DNA-binding activity (Hu et al. 2013). However, currently, no reliable Tet2 ChIP-seq data are available for analyzing the contribution of Tet2. Derepression of 2C-specific genes in Tet TKO cells correlated well with that in Kap1 knockout cells, although the change was to a lesser extent in Tet TKO ESCs than that in Kap1 knockout cells (Fig. 6B). This was consistent with the observation that binding of Kap1 to MuERV-L was decreased but not completely absent in Tet TKO ESCs (Fig. 6D). The activation of MuERV-L was also to a lesser extent in Tet TKO ESCs when compared with Kap1 knockout ESCs, although it was comparable with G9a knockout ESCs (Fig. 5G; Supplemental Fig. 10C,E). Therefore, it is possible that Tet proteins and/or 5hmC marks may help recruit/stabilize Kap1 binding at these loci. We also noticed that the effect of Tet proteins in Kap1 binding was not global, as Kap1 binding at IAP regions was minimally affected (Fig. 6D). Future studies may provide new insights into the function of Tet proteins in repressing type III ERVs and 2C-specific genes.
Among the 2C-specific genes, Zscan4 is of particular interest. Activation of Zscan4 in ESCs is associated with rapid telomere extension by promoting T-SCE (Zalzman et al. 2010; Dan et al. 2014). Zscan4 is specifically expressed in 2Cs, and telomere elongation takes place most dramatically at the 2C stage through a T-SCE-based mechanism (Liu et al. 2007). However, how Zscan4 is activated at the two-cell stage remains unclear. We found that Tet TKO ESCs exhibit activation of Zscan4, which led to increased T-SCE and elongated telomeres (Fig. 7). Among the three Tet proteins, Tet3 is a maternal factor that is highly expressed in the one-cell embryo and degrades in the 2C, while Tet1 and Tet2 commence expression at the 2C and morula stages, respectively (Tan and Shi 2012). Therefore, 2Cs lack all of the Tet proteins. The various phenotypes that we observed in this study suggest that loss of Tet proteins may be required for the establishment of a 2C-specific state, including ERV activation, Zscan4 expression, and subsequent telomere elongation. It will be interesting to determine whether forced expression of Tet proteins in 2Cs will result in disruption of preimplantation development in the future.
In summary, we generated a base-resolution DNA methylation map in Tet TKO ESCs and identified genome-wide Tet-dependent DNA demethylation events in ESCs. We found that Tet-mediated DNA demethylation takes place mostly at enhancers and distal regulatory sequences. We also provided evidence that enhancer methylation regulates enhancer activity. Furthermore, we demonstrated that Tet TKO ESCs show an increased 2C-like population and elongated telomeres. Taken together, these findings not only substantially broaden our understanding of the role of Tet proteins in DNA demethylation but also reveal new functions for Tet proteins in repressing repeat sequences as well as nearby 2C-specific genes and telomere length regulation.
Materials and methods
Cell culture and antibodies
We used the mESC line E14Tg2a in this study. Cells were cultured on 0.1% gelatin-coated (Millipore) plates in DMEM (Life Technologies) supplemented with 15% fetal bovine serum (Sigma), penicillin/streptomycin, nonessential amino acid, sodium pyruvate, GlutaMax, β-mercaptoethanol (Life Technologies), and 1000 U/mL LIF (ESGRO, Millipore) unless specified. Antibodies used in this study were 5hmC (Active Motif, 39769), Oct4 (Santa Cruz Biotechnology, sc-8628), Nanog (Bethyl Laboratories, IHC-00205), Sox2 (Millipore, Ab5603), Tet1 (Ito et al. 2010), Tet2 (described below), Tet3 (Gu et al. 2011), Zscan4 (Millipore, Ab4340), Suz12 (Cao et al. 2002), Ring1b (Cell Signaling, 5694), Ezh2 (Cell Signaling, 5246), and Kap1 (Cell Signaling, 4123). Tet2 antibody was generated using HIS-tagged recombinant Tet2 2-374 expressed in Escherichia coli and purified using Talon superflow metal affinity resin (Clontech). Rabbit immunization was carried out by Pocono Rabbit Farm and Laboratory, Inc. The antiserum was affinity-purified using immunization antigen. The specificity of the affinity-purified antibody was confirmed using extracts from the control and Tet2 knockdown mESCs.
Generation of Tet TKO cells
Tet TKO cells were generated using the CRISPR/Cas9 method with the previously published guide RNA sequences (Wang et al. 2013). E14 cells cultured in 2i medium (DMEM/F12 supplemented with penicillin/streptomycin, GlutaMax, 1 mM MEK inhibitor PD0325901, 3 mM GSK3 inhibitor CHIR99021, 1000 U/mL LIF) on Matrigel-coated (BD Biosciences) plates were transiently transfected with a mixture of px330 vectors containing guide RNAs for Tet1, Tet2, and Tet3 along with one puromycin resistance gene-containing empty vector using the Fugene 6 transfection reagent (Promega). The cells were subcultured to a low density 24 h later. Two days following transfection, the cells were selected with 2 μg/mL puromycin for 48 h. Viable clones were grown larger and picked up for mutation analysis. Mutations were first screened using PCR followed by restriction analysis as restriction sites were presented at CRISPR/Cas9 cutting sites. 5hmC levels of DNA from mutant clones were then analyzed by slot blot. Clones with a significantly lower level of 5hmC along with wild-type ESCs were kept and adapted to serum-based culture conditions for at least 17 d. DNA samples from these clones were then digested to single nucleotide and analyzed by mass spectrometry to quantify absolute 5hmC levels.
5hmC level analysis by mass spectrometry
Genomic DNA (1 μg) prepared using a DNeasy blood and tissue kit (Qiagen) was hydrolyzed with 90 U of nuclease S1 (Sigma) in digestion buffer (14 mM NaAc, 0.5 mM ZnSO4 at pH 4.6) for 1 h at 37°C followed by treatment with 8 mU of phosphodiesterase I (VPH) and 1.6 U of CIAP (New England Biolabs) for an additional 1 h. The digested samples were then filtered with Nanosep3K (PALL) and subjected to mass spectrometry analysis.
WGBS
DNA (1 μg) with 0.5% nonmethylated λ DNA spike-in was sheared by Covaris M220 instrument to an average size of 350 bp in length. DNA was then end-repaired, dA-tailed, and ligated with methylated adaptors. Bisulfite conversion was carried out using an EpiTect fast bisulfite conversion kit (Qiagen) according to the manufacturer’s instructions. Twenty minutes were used in both conversion steps. Bisulfite-converted DNA was then amplified with five PCR cycles to obtain the final library. The WGBS libraries were subjected to pair-end (2 × 110 bp) sequencing on a HiSeq 2500 (Illumina) with or without a PhiX spike in.
After sequencing, low-quality bases and adapter-containing reads were trimmed from raw data by trim_galore with default parameters, and reads aligned to the PhiX genome were removed. Bismark 0.7.9 was used to align reads to a reference genome (mm9). Analyzing λ DNA reads confirmed that >99.7% Cs were successfully bisulfite-converted. The coverage depth and methylation level of each cytosine were extracted by bismark_methylation_extractor and samtools from SAM files. DMR analysis was performed by methpipe-3.0.1. False discovery rate (FDR) was estimated by randomly shuffling the Tet TKO and wild-type methylome to call DMRs with the same pipeline and parameters. For hyper-DMRs, the FDR was estimated as 0.0499. For hypo-DMRs, the DMR number was too small to be statistically analyzed, and thus hypo-DMRs were not analyzed further. When calculating methylation level for CpG sites, information from both strands was combined, and a coverage of at least five reads was required. For regions, all reads within one specific region were pooled together, and a coverage of at least 10 reads was required. Since bisulfite sequencing cannot distinguish 5mC and 5hmC, 5mC levels in wild-type cells were corrected by subtracting 5hmC levels obtained from TAB-seq (Yu et al. 2012). For both 100-bp tiles and DMRs, regions with a methylation difference >20% were considered as differentially methylated.
The following procedure was used to generate the heat maps in Figure 2D. The average methylation profile of both wild type and TKOs was calculated for both 5 kb upstream of and downstream from the center of each feather in 100-bp bins. Methylation differences were calculated by subtracting average methylation in wild type from those in Tet TKO. For all of the features, peaks overlapping with ±1 kb TSS sequences were considered as proximal elements, with the rest of the peaks considered as distal elements. Regions within ±1.5 kb from the TSS were considered as promoter regions unless specified. The data of different genomic features are described in ”Published Data Sets Used in This Study.” Heat maps were generated by R.
Targeted bisulfite sequencing
Two micrograms of genomic DNA was bisulfite-treated with an EpiTect fast bisulfite conversion kit (Qiagen) according to the manufacturer’s instructions. Twenty minutes were used in both conversion steps. Targeted regions were amplified with the primers listed in Supplemental Table 6 using Kapa Hifi Uracil+ enzyme (Kapa Biosystems). The amplified DNA samples were purified and mixed at equal molar concentrations for each genotype. The mixture of DNA was then fragmented by sonication (Covaris). DNA sequencing libraries were then constructed from the fragmented DNA samples using NEBnext ultra DNA library prep kit for Illumina (New England Biolabs) with different barcodes. Libraries were sequenced on a HiSeq 2500 (Illumina).
RNA-seq
RNA samples were prepared by RNeasy minikit (Qiagen). Depletion of rRNA was carried out using ribo-zero kit (Epicentre). Barcoded RNA-seq libraries were constructed using NEBNext ultra directional RNA library prep kit for Illumina according to the manufacturer’s instructions (New England Biolabs). Single-end 50-bp sequencing was performed on a HiSeq 2500 (Illumina). Sequencing reads were mapped against the reference genome (mm9) with Bowtie/TopHat version 2.0.2, which allowed mapping across splicing junctions by read segmentation. All programs were performed with default settings unless otherwise specified. The unique mapped reads (∼77% of total reads) were subsequently assembled into transcripts guided by reference annotation (University of California at Santa Cruz [UCSC] gene models) with Cufflinks version 2.0.2. The expression level of each gene was quantified with normalized FPKM. Differentially expressed genes were identified by asking for a fold change >2 with FPKM > 0.5 in at least one sample. For repeat sequence annotation, the RepeatMasker track (RMSK) from UCSC Genome Browser was used. For repeat analyses, we counted the unique mapped reads in each repeat element by custom Perl script and then calculated the expression level (reads per million unique mapped reads [RPM]).
Published data sets used in this study
Single-cell RNA-seq data in the mouse early embryos were downloaded from GSE44183 (Xue et al. 2013). We used the following published RNA-seq data sets for Figure 6 and Supplemental Figure 10: G9a knockout ESCs (GSE33923) (Macfarlan et al. 2012), Lsd1 knockout (Macfarlan et al. 2011), and Kap1 knockout ESCs (GSE41903) (Rowe et al. 2013). The lists of up-regulated genes identified here are included in Supplemental Table 5.
We used the following published data sets for Figure 2D: 5fC/5caC (Shen et al. 2013); Tet1 (Wu et al. 2011); Kdm2a (Blackledge et al. 2010); H3K4me3, H3K36me3, H3K27me3, H3K9me3, and H4K20me3 (Mikkelsen et al. 2007); H3K4me1 (Meissner et al. 2008); Pol2 (pan), Pol2 (Ser2), Pol2 (Ser5), NelfA, Ctr9, and Spt5 (Rahl et al. 2010); Ezh2 and Suz12 (Ku et al. 2008); Oct4, Nanog, Sox2, and H3K79me2 (Marson et al. 2008); Med1, Med12, Nipbl, Smc1a, Smc3, and TBP (Kagey et al. 2010); LSD1, Mi2b, Hdac1, Hdac2, Rest, and Corest (Whyte et al. 2012); LMR (Stadler et al. 2011); H3K27ac and p300 (Creyghton et al. 2010); Esrrb, cMyc, nMyc, Tcfcp2l1, E2f1, Stat3, Smad1, and Zfx (Chen et al. 2008); CTCF (Shen et al. 2012); DHSs (ENCODE project at genome.ucsc.edu); and topological domain boundary (Dixon et al. 2012). Two published Kap1 ChIP-seq data sets were used for Supplemental Figure 10 (Quenneville et al. 2011; Rowe et al. 2013).
Early embryonic development stage specificity analysis
To quantify the stage specificity of genes in early embryos, the expression level of samples at a given stage versus the other samples was compared. The stage specificity score of each gene is defined as score = meanA − (meanOther + 2 × sdOther), where meanA is the mean expression of the samples in a certain stage, and meanOther and sdOther are the mean and standard deviation of the expression levels in the other samples, respectively. Therefore, a positive score indicates that the gene is expressed in a certain stage at a considerably higher level than in the other stages. The gene with a score >0 was considered as specifically expressed in certain stage.
Statistical analyses
Statistical analyses were implemented with R (http://www.r-project.org). The χ2 test was used to compare the frequency of an occurrence between two groups by analyzing the two-by-two contingency tables using the chisq.test function in R. Independent two-group Wilcoxon rank sum tests were used to compare distributions using the wilcox.test function in R. A Pearson’s R coefficient was calculated using the cor function with default parameters. Student’s t-test (two-tailed, equal variance) was performed to obtain P-values for qPCR experiments and ChIP-seq enrichment.
Promoter classification by chromatin states
Gene promoters are classified into four groups (active, initiated, bivalent, and silent) by histone modifications as previously described (Whyte et al. 2012). Specifically, (1) active promoters are associated with H3K4me3 (±1.5 kb flanking the TSS) and H3K79me2 (5 kb downstream from the TSS), (2) initiated promoters are associated with H3K4me3 (±1.5 kb flanking the TSS) only, (3) bivalent promoters are associated with H3K4me3 (±1.5 kb flanking the TSS) and H3K27me3 (±1.5 kb flanking the TSS), and (4) silent promoters are not associated with H3K4me3/H3K79me2/H3K27me3.
Immunostaining and FACS analysis
Cells were fixed with 4% paraformaldehyde for 15 min at room temperature. After washing with PBS twice, the cells were permeabilized with PBS containing 0.3% Triton X-100 for 10 min and blocked with PBS with 1% BSA for 30 min. The cells were then incubated with primary antibodies diluted in PBS with 1% BSA for 1 h at room temperature. After washing three times with PBS, cells were incubated with Alexa Fluor 488-, Alexa Fluor 594-, or Alexa Fluor 647-conjugated secondary antibodies (Molecular Probes) for 1 h at room temperature. The cells were washed three times with PBS, stained with DAPI, and visualized under an Axio Observer Z1 microscope (Carl Zeiss). For FACS analysis of Zscan4 staining, cells were dissociated with trypsin to single-cell suspension, washed with PBS, and fixed in 4% paraformaldehyde for 15 min at room temperature. After washing with PBS twice, permeabilization was done by resuspending cells in 90% cold methanol for 30 min. After washing with PBS twice, staining was done with procedures described above. Stained cells were analyzed on a BD FACSaria machine (BD Biosciences).
Telomere length qPCR quantification
Telomere length quantification was carried out as previously described (Callicott and Womack 2006; Liu et al. 2007), with the exception that 1 ng of DNA was used for each amplification, and 4, 2, 1, 0.5, and 0.25 ng of DNA per sample was used for standard curve generation.
Telomere Q-FISH
Telomere Q-FISH was carried out as previously described with minor modifications (Herrera et al. 1999). Briefly, ESCs were cultured with 0.2 μg/mL Colcemid for 1 h and 40 min to enrich cells at metaphases. Harvested cells were treated with 75 mM KCl solution followed by fixation with Carnoy’s fixative solution (methanol:acetic acid, 3:1) and spread onto clean glass slides. Slides were then washed with PBS and fixed with 4% paraformaldehyde/PBS followed by the incubation with 1 mg/mL pepsin (pH 2.0). Slides were then post-fixed with 4% paraformaldehyde/PBS followed by dehydration in ethanol series and air-dried. Telomeres were denatured for 3 min at 80°C and hybridized with telomere-specific peptide nucleic acid (PNA) probe (PNA Bio, F1002) for 4 h at room temperature. Chromosomes were counterstained with DAPI and mounted with Slowfade Gold (Life Technologies). Telomere images were taken by confocal microscope with fixed exposure parameters. For quantitative measurement of telomere length, fluorescence intensity was analyzed using the TFL-TELO software (a gift kindly provided by P. Lansdorp, Terry Fox Laboratory) and calibrated using standard fluorescent beads.
Telomere CO-FISH
CO-FISH was carried out as previously described (Williams and Bailey 2009; Falconer et al. 2010) with minor modifications. In brief, cells were cultured with ESC culture medium containing 10 μM 5′-bromo-2′-deoxyuridine (BrdU) for 12 h. Colcemid (0.2 μg/mL final) was added for the final 1 h and 40 min. ESCs were then harvested and treated with hypotonic solution followed by fixation with Carnoy’s fixative solution. Chromosome spreads were prepared and treated with 100 μg/mL RNase A for 10 min at room temperature. After fixation with 4% paraformaldehyde in PBS for 10 min at room temperature, slides were dehydrated with ethanol and stained with 1.0 μg/mL Hoechst 33258 for 15 min at room temperature, followed by washing in 2× SSC. Slides were then mounted with 2× SSC and exposed to 365 nm UV light (Spectronics) for 35 min. After a brief rinse with 2× SSC, BrdU-substituted DNA was digested by exonuclease III for 15 min at room temperature followed by washing in 2× SSC. DNA was denatured with 70% formamide for 1 min at 70°C followed by dehydration in ethanol. A Cy3-conjugated PNA Tel-C (CCCTAA)3 probe (PNA Bio, F1002) was hybridized in the hybridization buffer containing denatured herring sperm DNA (Sigma) for 3 h at 37°C. Slides were washed twice with formamide wash solution for 15 min at 32°C and twice in Tris-NaCl-Tween (TNT) wash solution for 15 min at room temperature. Chromosomes were counterstained with DAPI (Life Technologies) and imaged under confocal microscopy.
ChIP analysis
Cells were fixed in 2 mM ethylene glycol bis (succinimidylsuccinate; Thermo Scientific) for 30 min followed by 10 min in 1% formaldehyde. The fixation was quenched by adding glycine to 0.25 M for 5 min at room temperature. Chromatin was lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris at pH 8.0) and sonicated to ∼200–400 bp (Branson Sonifier 450). Immunoprecipitation was performed with anti-Kap1 (Cell Signaling, 4123), anti-Ring1b (Cell Signaling, 5694), anti-Ezh2 (Cell Signaling, 5246), or rabbit IgG (Santa Cruz Biotechnology, SC-2027). The bound chromatin was washed, eluted, and reverse cross-linked. DNA was extracted by phenol/chloroform and precipitated. Primers used in qPCR analysis are listed in Supplemental Table 6. IAP is “IAP U3” from published literature (Rowe et al. 2010). Fold enrichment was calculated by comparing DNA captured by anti-Kap1 with that captured by IgG control, which avoided the possible bias caused by copy number difference.
Accession numbers
The WGBS and RNA-seq data sets generated in this study have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE56986.
Acknowledgments
We thank Dr. Hao Wu and Dr. Shogo Matoba for their helpful discussions; Dr. Li Shen, Dr. Wei Jiang, and Dr. Kumiko A. Iwabuchi for technical assistance; and Dr. Ana D’Alessio for generating the Tet2 antibody. We are grateful to Dr. Luis M. Tuesta and Dr. Hao Wu for critical reading of the manuscript. This project is supported by National Institutes of Health grant U01DK089565. S.Y. is supported by a post-doctoral fellowship from the Japan Society for the Promotion of Science (JSPS). Y.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.248005.114.
References
- Amano T, Hirata T, Falco G, Monti M, Sharova LV, Amano M, Sheer S, Hoang HG, Piao Y, Stagg CA, et al. 2013. Zscan4 restores the developmental potency of embryonic stem cells. Nat Commun 4: 1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. 2010. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Long SW, van Heeringen SJ, Brinkman AB, Gomez-Skarmeta JL, Stunnenberg HG, Jones PL, Veenstra GJ. 2011. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Res 21: 1313–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, Nichols J. 2014. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol 16: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott RJ, Womack JE. 2006. Real-time PCR assay for measurement of mouse telomeres. Comp Med 56: 17–22 [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Liu Y, Liu N, Chiourea M, Okuka M, Wu T, Ye X, Mou C, Wang L, Wang L, et al. 2014. Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev Cell 29: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. 2011. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9: 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. 2013. Combined deficiency of tet1 and tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 24: 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell Benjamin E, Cheng Albert W, Faull Kym F, Lyko F, et al. 2014. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 29: 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. 2007. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol 307: 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer E, Chavez E, Henderson A, Lansdorp PM. 2010. Chromosome orientation fluorescence in situ hybridization to study sister chromatid segregation in vivo. Nat Protoc 5: 1362–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. 2012. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife 1: e00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. 2011. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473: 398–402 [DOI] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. 2008. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2: 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477: 606–610 [DOI] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. 2010. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143: 508–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. 2013. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 15: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Herrera E, Samper E, Martin-Caballero J, Flores JM, Lee HW, Blasco MA. 1999. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J 18: 2950–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. 2013. Crystal structure of TET2–DNA complex: insight into TET-mediated 5mC oxidation. Cell 155: 1545–1555 [DOI] [PubMed] [Google Scholar]
- Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR, Wu HP, Gao J, Guo F, Liu W, et al. 2014. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 14: 512–522 [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, et al. 2013. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155: 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. 2011. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8: 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, et al. 2007. Telomere lengthening early in development. Nat Cell Biol 9: 1436–1441 [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. 2011. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev 25: 594–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Thompson PJ, Goyal P, Jones SJ, Singh PB, Karimi MM, Lorincz MC. 2013. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. 2008. The colorful history of active DNA demethylation. Cell 133: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. 2013. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 14: 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J 2008. Stem cells and early lineage development. Cell 132: 527–531 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. 2013. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res 23: 452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K, Zhang Y. 2013. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153: 692–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Smith A. 2008. Capturing pluripotency. Cell 132: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, Smith AD. 2013. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS ONE 8: e81148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Shi YG. 2012. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 139: 1895–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. 2012. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ES, Bailey SM. 2009. Chromosome orientation fluorescence in situ hybridization (CO-FISH). Cold Spring Harb Protoc 2009: pdb prot5269. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. 2010. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. 2014. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156: 45–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. 2011. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K. 2013. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 49: 1134–1146 [DOI] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. 2011. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42: 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. 2013. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. 2012. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149: 1368–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. 2010. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464: 858–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 500: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]