A hallmark of the inflammatory response to pathogen exposure is the production of TNF. Caldwell et al. used a systems biology approach to develop a quantitative understanding of the regulatory modules that control TNF production. They predict and then experimentally confirm that in response to lipopolysaccaride, TNF does not have an autocrine function in amplifying the NFκB response, although it plays a potent paracrine role in neighboring cells. However, in response to CpG DNA, autocrine TNF extends the duration of NFκB activity and shapes CpG-induced gene expression programs.
Keywords: inflammation, innate immune response, pathogen sensors, MAP kinase, NFκB
Abstract
A hallmark of the inflammatory response to pathogen exposure is the production of tumor necrosis factor (TNF) that coordinates innate and adaptive immune responses by functioning in an autocrine or paracrine manner. Numerous molecular mechanisms contributing to TNF production have been identified, but how they function together in macrophages remains unclear. Here, we pursued an iterative systems biology approach to develop a quantitative understanding of the regulatory modules that control TNF mRNA synthesis and processing, mRNA half-life and translation, and protein processing and secretion. By linking the resulting model of TNF production to models of the TLR-, the TNFR-, and the NFκB signaling modules, we were able to study TNF’s functions during the inflammatory response to diverse TLR agonists. Contrary to expectation, we predicted and then experimentally confirmed that in response to lipopolysaccaride, TNF does not have an autocrine function in amplifying the NFκB response, although it plays a potent paracrine role in neighboring cells. However, in response to CpG DNA, autocrine TNF extends the duration of NFκB activity and shapes CpG-induced gene expression programs. Our systems biology approach revealed that network dynamics of MyD88 and TRIF signaling and of cytokine production and response govern the stimulus-specific autocrine and paracrine functions of TNF.
Tumor necrosis factor (TNF) is a key inflammatory cytokine produced by macrophages exposed to pathogens. Toll-like receptors (TLR) recognize a variety of molecular substances derived from pathogens such as bacteria, viruses, and fungi, eliciting signaling events that coordinate inflammatory and innate immune responses (Takeuchi and Akira 2010). TLRs are expressed in many cell types, but perhaps one of the most relevant types for the innate immune response is the classically activated macrophage. A hallmark of activated macrophages is the production of proinflammatory cytokines, including TNF (Mosser and Edwards 2008; Parameswaran and Patial 2010). Proper control of the TLR-responsive signaling pathways is of particular importance, as aberrant signaling and/or TNF production can contribute to disease states such as Crohn’s disease, rheumatoid arthritis, and cancer (MacDonald et al. 1990; Murch et al. 1993; McInnes and Schett 2007; Waters et al. 2013).
In macrophages, TLRs use two adaptors that mediate the signaling events leading to proinflammatory cytokine production: the plasma membrane-proximal MyD88 and the endosomal membrane-proximal TRIF (Kawai et al. 1999; Häcker et al. 2000; Hoebe et al. 2003; Sato et al. 2003; Yamamoto et al. 2003). Whereas TLR9, the receptor for unmethylated CpG DNA, engages MyD88, TLR3, the receptor for dsRNA engages TRIF, and TLR4, the receptor for lipopolysaccharide (LPS) engages both (Takeuchi and Akira 2010). These adaptors mediate the activation of transcription factors such as NFκB and IRF3, both of which have been implicated in the control of TNF production (Drouet et al. 1991; Wesche et al. 1997; Yamamoto et al. 2003; Covert et al. 2005; Lee et al. 2009).
Gene transcription is not the only means of controlling TNF production; control of TNF mRNA half-life and protein translation and secretion has been reported (Han et al. 1991a,b; Black et al. 1997; Andersson and Sundler 2006). In unstimulated macrophages, TNF mRNA has a relatively short half-life, but upon LPS stimulation, TNF mRNA half-life increases as much as sixfold through the down-regulation of mRNA degradation pathways (Carballo et al. 1998; Kontoyiannis et al. 1999; Kotlyarov et al. 1999; Lai et al. 1999; MacKenzie et al. 2002; Stoecklin et al. 2004; Hitti et al. 2006; Ronkina et al. 2007; Hao and Baltimore 2009). At the level of protein translation, TNF production is modulated in response to LPS through the activation of the translation initiation factor eIF4E. The resulting membrane-bound pro-TNF must then be cleaved and secreted by the catalase TACE, the activity of which is up-regulated following LPS stimulation (Black et al. 1997; Soond et al. 2005; Xu and Derynck 2010). A number of studies have sought to characterize the signaling control of one or a subset of these mechanisms (Datta et al. 2004; Hitti et al. 2006; Ronkina et al. 2007; Gais et al. 2010; Wang et al. 2011), but a coherent picture remains elusive in part because such control appears to be cell type-specific. Here we sought to develop a quantitative understanding of each of the TRIF and MyD88-mediated mechanisms of TNF production control in primary macrophages and then evaluate their relative contributions within the network.
Cytokines, such as TNF, have both autocrine and paracrine functions. Autocrine functions amplify or shape the signaling or gene expression response of cells responding to pathogen exposure and are typically studied in monocell culture. TNF has been implicated as an autocrine regulator of TLR-induced inflammatory signaling (Wu et al. 1993; Blasi et al. 1994; Xaus et al. 2000; Coward et al. 2002; Kuno et al. 2005; Lombardo et al. 2007). Paracrine signaling functions are key to the amplification and control of an inflammatory response within a tissue or organ and the activation of a coordinated immune response that involves diverse cell types. It remains remarkably unclear what the determinants are for autocrine or paracrine cytokine functions and whether one or the other may be more likely triggered by different classes of pathogen signals.
The systems biology approach of combining experimental studies with computational models to achieve quantitative and qualitative insights has been particularly fruitful in the study of signal transduction (Ozaki et al. 2005; Basak et al. 2012; Purvis and Lahav 2013), including signaling in response to TNF stimulation (Werner et al. 2008). In order to develop a predictive understanding of TNF production, we considered each regulatory mechanism as a regulatory module (with a defined and experimentally measurable input and output) (Hartwell et al. 1999) and constructed and parameterized simple mathematical models to represent these modules. By further linking the resulting TNF production model with models of the TLR, TNFR, and NFκB signaling modules (Werner et al. 2005, 2008; Z Cheng, B Taylor, D Rios, and A Hoffmann, in prep.), we reveal that TNF’s autocrine and paracrine functions are stimulus- or TLR-specific, determined by the underlying signaling network dynamics of TNF production and NFκB response.
Results
Multitier control of TNF production depends on TRIF and MyD88
To investigate the TRIF- and MyD88-specific control mechanisms of TNF production in TLR signaling, we used the TLR agonist LPS, as it engages both TRIF and MyD88 (Poltorak et al. 1998; Kawai et al. 1999; Alexopoulou et al. 2001; Yamamoto et al. 2003). Both adaptors have been shown to activate multiple kinase signaling pathways (Fig. 1A), including IKK and TBK1, which lead to the activation of the transcription factors NFκB and IRF3, respectively, as well as the MAP kinases p38 and ERK. In the context of TNF production, p38 and ERK have been implicated in activating MK2, the kinase that phosphorylates and inactivates TTP, the ARE-binding destabilizer of mRNAs (Carballo et al. 1998; Lai et al. 1999; Stoecklin et al. 2004; Hitti et al. 2006; Ronkina et al. 2007; Deleault et al. 2008). Furthermore, p38 and ERK have been shown to control TNF translation through the initiation factor eIF4E and TNF secretion through the enzyme TACE (Black et al. 1997; Soond et al. 2005; Andersson and Sundler 2006; Xu and Derynck 2010). However, prior studies of one or a subset of these mechanisms used different cell types and experimental systems. Here we used primary murine macrophages to develop a quantitative understanding of each mechanism and assess its contribution to TNF production in that cell type.
Figure 1.
TNF production is regulated by both MyD88 and TRIF pathways, but only NFκB, not IRF3/7, is required for its transcriptional induction. (A) Diagram illustrating molecular mechanisms potentially regulating the production of TNF; solid lines indicate mechanisms known to play a role in macrophages, and dashed lines indicate mechanism that have been reported in the literature in different cell systems. (B) Schematic of the three regulatory modules that control TNF expression, highlighting the experimentally quantifiable input and output of each module. (C) Secretion of TNF as measured by ELISA of cell media from wild-type, trif −/−, or myd88−/− bone marrow-derived macrophages (BMDMs). Cells were stimulated with 10 ng/mL LPS; n = 3. (D) Levels of TNF mRNA (log2 fold) produced by wild-type, trif−/−, or myd88−/− BMDMs stimulated with 10 ng/mL LPS, measured by RT-qPCR. Wild type, n = 5. trif−/−, n = 3. myd88−/−, n = 3. (E) Levels of nascent TNF RNA (log2 fold) produced by wild-type, trif−/−, or myd88−/− BMDMs stimulated with 10 ng/mL LPS, measured by RT–PCR. Wild type, n = 3. Nascent transcripts measured by RT-qPCR with intron–exon junction-spanning primers. (F) TNF mRNA levels (fold) measured by RT-qPCR in wild-type or rela−/−relb−/−crel−/− fetal liver-derived macrophages (FLDMs) stimulated with 100 ng/mL LPS. (G) Levels of TNF mRNA (log2 fold) produced by wild-type or irf3−/−irf7−/− BMDMs stimulated with 10 ng/mL LPS, measured by RT–PCR (wild type, n = 5; irf3−/−irf7-/-, n = 3). (H) Levels of nascent TNF RNA (log2 fold) produced by wild-type or irf3−/−irf7−/− BMDMs stimulated with 10 ng/mL LPS, measured by RT-qPCR (n = 3). (I) Quantified activation of NFκB measured by EMSA (κB-site-containing HIV probe) in wild-type, trif−/−, or myd88−/− BMDMs stimulated with 10 ng/mL LPS. Quantification of NFκB EMSA bands normalized to peak activity (n = 3). (J) Schematic of regulatory module 1 that determines nascent TNF mRNA; input is quantified NFκB activation data (shown in H), and output is nascent TNF mRNA. (K) Levels of nascent mRNA simulated by the mathematical model in I (solid lines) and determined experimentally in E (data points) for wild-type, trif−/−, or myd88−/− genotypes. For all graphs, error bars indicate one standard deviation. The combined root mean square difference (RMSD) between all model simulations and experimental data points shown are indicated.
To investigate the processes of TNF production controlled by TRIF and MyD88, wild-type, trif−/−, and myd88−/− bone marrow-derived macrophages (BMDMs) were stimulated with LPS, and TNF secretion in the supernatant was measured by ELISA. While both trif−/− and myd88−/− have significant defects (P-values ranging from 7.1 × 10−4 to 3.5 × 10−8 for 1-h to 4-h time points) in TNF secretion (Fig. 1C), trif−/− showed a more severe defect at later time points than myd88−/− (Supplemental Fig. 1A). However, when measuring TNF production at the level of mature mRNA by RT–PCR, myd88−/− showed a severe defect at early time points (P-values < 0.003), whereas trif−/− BMDMs showed a smaller defect (Fig. 1D). These discrepancies were not entirely due to gene transcription: Measuring nascent, intron-containing transcripts, we found substantially reduced levels for the first 25 min in myd88−/− (P-values < 0.05) but slightly increased levels in trif−/− BMDMs (Fig. 1E). These data are consistent with TRIF-dependent control of post-transcriptional events and both MyD88- and TRIF-dependent control of translational and post-translational events.
Transcriptional control of TNF expression is regulated by the MyD88–NFκB axis but not IRF3/7
Whereas MyD88 primarily mediates the activation of NFκB, TRIF is also a potent mediator of interferon regulatory factors IRF3 and IRF7, which were suggested to control TNF mRNA synthesis in response to LPS (Covert et al. 2005). To investigate the genetic requirements for TNF mRNA production, we produced fetal liver-derived macrophages (FLDMs) from wild-type and rela−/−relb−/−crel−/− mouse embryos (which lack all activation domain-containing NFκB family members) and wild-type and irf3−/−irf7−/− BMDMs and stimulated them with LPS. Remarkably, no TNF mRNA was detected in rela−/−relb−/−crel−/− FLDMs (Fig. 1F), but irf3−/−irf7−/− BMDMs showed no defects in either mature or nascent TNF mRNA levels (Fig. 1G,H). These data support a model in which TNF mRNA synthesis in macrophages is controlled by NFκB but not IRF3/7.
As NFκB was confirmed to be the transcription factor responsible for TNF transcription, we sought to quantitate the roles of MyD88 and TRIF in activating NFκB in response to LPS. Electrophoretic mobility shift assay (EMSA) revealed that NFκB activation is decreased in both trif−/− and myd88−/− yet at different times (Fig. 1I). The trif−/− BMDMs have normal early activation (0–30 min) but defects at late times; conversely, myd88−/− BMDMs have decreased early activation (P-value < 0.025 at 15 min; P-value < 0.005 at 30 min), but late activation (45 min to 4 h) is indistinguishable from wild-type. These results informed the formulation of a mathematical model of the TNF mRNA production module (Fig. 1J) in which MyD88- and TRIF-induced NFκB up-regulates nascent TNF mRNA synthesis, which is then processed into mature mRNA (Osman et al. 1999). Following parameterization (Supplemental Material), model simulations recapitulated the experimentally determined nascent TNF mRNA levels in wild-type, trif−/−, and myd88−/− BMDMs with root mean square deviation (RMSD) = 0.16 (Fig. 1K).
Post-transcriptional control of TNF expression is mediated by TRIF
As summarized above, previous reports have shown that TNF mRNA half-life and translation and proprotein processing and secretion can be modulated by TLR signaling. The discrepancies between nascent and mature TNF mRNA and secreted protein levels (Fig. 1C–E) suggested that these mechanisms may substantially contribute to TNF protein production.
To investigate the LPS-responsive half-life control of TNF mRNA, wild-type BMDMs were stimulated with TNF alone, which induces TNF mRNA synthesis to detectable levels but not TNF mRNA stabilization, or with TNF and LPS prior to treatment with actinomycin-D, a drug that intercalates into DNA and arrests transcription. Subsequent decay of TNF mRNA levels was tracked by RT–PCR in 15-min increments to which exponential decay curves were fitted to estimate mRNA half-lives. Stimulation with TNF set a baseline of a constitutive TNF mRNA half-life of ∼10 min (Fig. 2A). When stimulated in conjunction with LPS, the half-life of TNF mRNA increased 3.5-fold to 35 min. Using trif−/− and myd88−/− BMDMs, we determined whether this LPS-induced stabilization of TNF mRNA was TRIF- or MyD88-dependent. Our data showed no decrease in LPS-induced TNF mRNA half-life in myd88−/− but a complete loss of LPS-induced half-life stabilization in trif−/− BMDMs compared with wild-type controls. To determine whether this TRIF-mediated stabilization was p38-dependent, we used the p38 inhibitor SB203580. After actinomycin-D treatment, p38 inhibitor-treated TNF mRNA half-life was determined by RT–PCR to be ∼13 min, near the baseline value with TNF stimulation alone (Fig. 2B). We examined the activity of the MAP kinases p38 and ERK by immunoblotting for phospho-p38 and phospho-ERK following time courses of LPS-stimulated wild-type, trif−/−, and myd88−/− BMDMs (Fig. 2C). Our data revealed that trif−/− BMDMs show decreased p38 activation from 30 to 75 min and decreased ERK activation from 30 to 60 min. Indeed, the MAP kinase target MK2 showed more severe deficiencies in phosphorylation in trif−/− than myd88−/− BMDMs (Fig. 2C), and this phosphorylation was p38- and not ERK-dependent (Fig. 2D). Similarly, phosphorylation of TTP, which leads to its inactivation and prevents TNF mRNA degradation, was decreased in p38 but not ERK inhibitor-treated wild-type BMDMs stimulated with LPS (Fig. 2D).
Figure 2.
TRIF regulates TNF mRNA half-life and translation and protein processing and secretion. (A) TNF mRNA half-life measured by RT-qPCR following actinomycin-D time course in wild-type, trif−/−, or myd88−/− BMDMs prestimulated for 30 min with 10 ng/mL TNF alone (purple) or 10 ng/mL TNF and 10 ng/mL LPS (blue) (wild-type, n = 5; trif−/−, n = 4; myd88−/−, n = 3). (B) TNF mRNA half-life measured as in A in wild-type BMDMs following prestimulation with 10 ng/mL TNF alone (purple); 10 ng/mL TNF and 10 ng/mL LPS (blue); or 10 ng/mL TNF, 10 ng/mL LPS, and 10µM p38 inhibitor for 30 min (white) (TNF, n = 5; LPS, n = 5; p38, n = 1). (C) Immunoblots for phospho-p38, phospho-ERK, phospho-MK2, and actin of whole-cell extracts made from wild-type, trif−/−, or myd88−/− BMDMs stimulated with 10 ng/mL LPS. Blots shown are representative of four experiments. (Below) Quantification of immunoblots normalized to wild-type peak phosphorylation. Error bars indicate one standard deviation; (*) P-value < 0.05; (**) P-value < 0.02 for difference between wild-type and trif−/− time points. (D) Immunoblots for phospho-MK2, phospho-TTP, and actin of whole-cell extracts made from wild-type BMDMs pretreated with DMSO, 10 µM p38 inhibitor, or 10 µM ERK inhibitor for 1 h followed by stimulation with 10 ng/mL LPS. (E) Schematic of regulatory module 2 that controls TNF mRNA half-life via TRIF. (Input) Nascent TNF mRNA experimental data; (output) mature TNF mRNA abundance. (F) Computational simulations of module 2 (solid lines) for TNF mRNA production in the wild-type, trif−/−, or myd88−/− genotype in response to 10 ng/mL LPS with either no stabilization control (top left), stabilization by TRIF and MyD88 (top right), stabilization by MyD88 alone (bottom left), or stabilization by TRIF alone (bottom right). Data points indicate experimental data for TNF mRNA in wild-type, trif−/−, or myd88−/− BMDMs stimulated with 10 ng/mL LPS as reported in Figure 1C. The combined RMSD between model simulations for each half-life control mechanism and experimental data points shown are indicated.
Therefore, our data support a model in which TRIF mediates LPS-responsive TNF mRNA stabilization through p38, allowing us to represent these mechanisms in mathematical form within the TNF mRNA stabilization module (Fig. 2E). This module uses the experimentally determined nascent TNF RNA levels as inputs, predicting mature TNF mRNA levels as the output. The effect of TNF mRNA stabilization can be illustrated by contrasting the output of total TNF mRNA simulations without or with TRIF-mediated stabilization. Considering four potential half-life control mechanisms provided independent evidence that the aforementioned TRIF–p38 axis best recapitulates the experimental data (Fig. 2F, with lowest RMSD). Thus, our model for the TNF mRNA stabilization module quantitatively accounts for LPS-responsive TNF mRNA stabilization via the TRIF signaling axis.
Translational and protein processing control of TNF expression is mediated by TRIF
While post-transcriptional control of TNF mRNA stabilization by TRIF accounts for the discrepancy between nascent TNF RNA and whole-cell TNF mRNA levels in trif−/− BMDMs, these cells show a greater deficiency in secreted TNF than is apparent at the level of mRNA abundance (Fig. 1C,D). In order to characterize the control of TNF translation, LPS-stimulated wild-type, trif−/−, and myd88−/− BMDMs were pretreated with TACE inhibitor TAPI-1 to block processing and secretion. Immunoblots revealed that while wild-type cells produce significant amounts of pro-TNF peaking at 60 min, trif−/− and myd88−/− have substantial defects in pro-TNF expression (Fig. 3A). In the case of myd88−/−, the pro-TNF protein expression defect at 60 min correlates with a defect in transcriptional induction at 30 min (Fig. 1D). However, the defect in trif−/− BMDMs suggests that TRIF regulates TNF translation.
Figure 3.
TRIF regulates TNF translation, protein processing, and secretion. (A) Immunoblot for pro-TNF and actin in wild-type, trif−/−, and myd88−/− BMDMs stimulated with 10 ng/mL LPS. Data are representative of three experiments. (Right) Quantification of pro-TNF bands normalized to peak wild-type protein levels. Error bars indicate one standard deviation; (*) P-value < 0.05; (**) P-value < 0.02 for difference between wild-type and trif−/− time points. (B) Immunoblot for pro-TNF and actin of whole-cell extracts from wild-type BMDMs pretreated with DMSO, 10 µM p38 inhibitor, or 10 µM ERK inhibitor for 1 h followed by stimulation with 10 ng/mL LPS. The blot is representative of two experiments. (C) Immunoblot for phospho-eIF4E, phospho-TACE, and actin of whole-cell extracts from wild-type, trif−/−, and myd88−/− BMDMs stimulated with 10 ng/mL. Phospho-eIF4E, n = 2; p-TACE, n = 3. (Below) Quantification of p-eIF4E and p-TACE immunoblot bands normalized to peak wild-type levels; (*) P-value < 0.05 for a difference between wild-type and trif−/− time points. (D) Immunoblot for phospho-eIF4E, eIF4E, phospho-TACE, TACE, and actin of whole-cell extracts from wild-type BMDMs pretreated with DMSO, 10 µM p38 inhibitor, or 10 µM ERK inhibitor for 1 h followed by stimulation with 10 ng/mL LPS. The blots are representative of two experiments. (E) Schematic of regulatory module 3 describing the promotion of TNF translation and secretion by TRIF. (Input) Experimental data for mature TNF mRNA; (output) secreted TNF. (F) Computational simulations (solid lines) of module 3 for pro-TNF expression without (left) and with (right) the promotion of TNF procession through TRIF-mediated translation regulation. Data points indicate experimental data for pro-TNF expression in wild-type, trif−/−, or myd88−/− BMDMs stimulated with 10 ng/mL LPS as reported in A, respectively. RMSD between model simulations and experimental data points are indicated for each genotype. (G) Computational simulations (solid lines) of module 3 for secreted TNF without (left) and with (right) the promotion of TNF secretion through TRIF-mediated secretion regulation. Data points indicate experimental data for TNF secretion in wild-type, trif−/−, or myd88−/− BMDMs stimulated with 10 ng/mL LPS as reported in Figure 1C, respectively.
Next, we sought to determine the role of MAP kinases in TNF translational control by TRIF. Inhibitor studies indicated a role for p38 and not ERK, as the ERK inhibitor condition showed no decrease in pro-TNF expression (Fig. 3B). Given that eIF4E and TACE have been shown to be necessary for TNF translation (Andersson and Sundler 2006) and secretion (Black et al. 1997), respectively, we next sought to characterize their activation by phosphorylation (Wang et al. 1998; Fan and Derynck 1999; Díaz-Rodríguez et al. 2002; Topisirovic et al. 2004; Xu and Derynck 2010). Immunoblots indicated a modest decrease in eIF4E phosphorylation at 30–45 min in myd88−/−, whereas trif−/− showed a decrease at 60–75 min (Fig. 3C). Similarly, immunoblots for TACE phosphorylation showed substantial signal in wild-type and myd88−/− cells peaking at 75 min but reduced signal in trif−/− cells (Fig. 3C). To determine whether TRIF-controlled phosphorylation of eIF4E and TACE was mediated through p38 or ERK MAP kinase, wild-type BMDMs were pretreated with p38 or ERK inhibitor for 1 h followed by stimulation with LPS (Fig. 3D). This analysis suggested that p38 has an effect on eIF4E phosphorylation but not eIF4E protein levels; in contrast, both p38 and ERK have an effect on TACE phosphorylation, but the ERK inhibitor condition resulted in more severe inhibition. A TRIF–ERK–TACE axis for the control of TNF secretion is further supported by the significant decrease in ERK activity by p-ERK in trif−/− but not myd88−/− cells (Fig. 2C).
These experimental results informed the construction of a mathematical model for the TNF translation and secretion control module. In this module, TRIF leads to the activation of p38 and ERK pathways; p38 controls the activity of eIF4E through phosphorylation, which promotes TNF translation, whereas ERK and, to a lesser extent, p38 control the activity of TACE through phosphorylation, which promotes the cleavage and secretion of TNF (Fig. 3E). Computational simulations confirmed that models involving TRIF regulation of TNF capture the data better (Fig. 3F,G).
A mathematical model of TLR-responsive TNF production
To develop a predictive understanding of TLR-induced TNF production, we connected models of the three previously described regulatory modules and linked them with a model that accounts for TLR-induced NFκB activation via MyD88 and TRIF signaling activities (Fig. 4A; Z Cheng, B Taylor, D Rios, and A Hoffmann, in prep.). Computational simulations of LPS-induced TNF production recapitulates the experimental data in wild-type cells at the level of nascent RNA, mRNA, pro-TNF production, and TNF secretion (Fig. 4B). Expanding the computational simulations to other TLR ligands, we used the model to predict the TNF production dynamics of two other TLR agonists: PolyI:C (TLR3) and CpG DNA (TLR9/MyD88 agonist). Here, the model was able to successfully predict PolyI:C-induced TNF production at the level of mRNA and protein secretion with small RMSDs of 0.08 and 0.07, respectively (Fig. 4C). However, it failed to recapitulate CpG-induced dynamics of TNF mRNA production or protein secretion (RMSDs of 0.63 for mRNA and 0.30 for secretion due to discrepancies at late time points). A key difference between these TLRs is their use of signaling adaptors; whereas TLR3 uses TRIF, TLR9 uses MyD88. We asked whether the failure of the model was due to differential adaptor use by using trif−/− and myd88−/− BMDMs in LPS stimulations. These conditions confirmed that while the model is able to capture TRIF dynamics (in myd88−/−), it is not able to accurately capture MyD88 dynamics (in trif−/−), particularly at the level of nascent and mature mRNA expression (Fig. 4D). Given previous reports of TNF’s potential to signal in an autocrine manner, we posited that perhaps autocrine TNF augments the cellular response to MyD88-dependent stimuli such as CpG.
Figure 4.
A multimodular model of the TNF production network accounts for experimental data for some TLR agonists but not others. (A) Schematic of the computational model combining models of the regulatory modules for TLR activation of adaptors TRIF and MyD88 converging on IKK, activation of NFκB by IKK, and the three modules for TNF production. (B) Model simulations and experimental data for nascent mRNA, mature mRNA, pro-TNF, and secreted TNF protein in wild-type cells in response to 10 ng/mL LPS; solid lines indicate values of model simulations, and points represent experimental data represented in previous figures. (C) Model simulations and experimental data for mature TNF mRNA and secreted TNF for wild-type cells in response to 500 nM CpG and 50 µg/mL PolyI:C. (D) Model simulations and experimental data for same molecular species for trif−/− and myd88−/− cells in response to 10 ng/mL LPS. For experimental data points, n = 3. Error bars are one standard deviation from the mean. RMSDs are indicated for each panel in B–D.
TNF’s autocrine function is stimulus-specific, augmenting NFκB activation in response to CpG
To examine TNF’s potential as an autocrine regulator to enhance NFκB activation, we linked the model of TLR-responsive TNF production with a model of TNFR-mediated NFκB activation (Fig. 5A; Werner et al. 2008). We then simulated the model for LPS, CpG, and PolyI:C stimulation conditions to determine whether autocrine feedback in the model would allow for a better recapitulation of experimental data. Interestingly, we found that autocrine TNF signaling had little effect on levels of the LPS and PolyI:C conditions but dramatically improved the ability of the model to predict CpG-induced TNF mRNA production and protein secretion (Fig. 5B). To determine whether this prolonging of CpG-induced dynamics was due to augmented NFκB activity induced by autocrine TNF, we simulated NFκB activation with and without TNF autocrine signaling by allowing for or blocking TNFR generation in the model. In this scenario, the model predicted that while LPS-induced NFκB would not be affected by the loss of autocrine TNF signaling, CpG-induced NFκB activation depends on autocrine TNF, particularly at late time points (Fig. 5C). To test this prediction experimentally, NFκB activity was measured in the absence of TNF autocrine signaling using tnf−/− BMDMs stimulated with either LPS or CpG. EMSAs for NFκB activation revealed no reduction in tnf−/− BMDMs in NFκB activation in response to LPS but did show substantially reduced NFκB activation in response to CpG during the 4- to 8-h time points (Fig. 5D). We found that this trend was observed in both high- and low-density conditions (although the variability of individual experiments may require more than three replicates to determine statistical significance) (Supplemental Fig. 2). Interestingly, TNFR1 deficiency generally resulted in a partial defect (compared with TNF deficiency) (Supplemental Fig. 2, orange bars), consistent with a role of membrane-bound TNF (which is recognized by TNFR2) in autocrine signaling (Rodríguez et al. 2011). In sum, these experimental results confirm the model’s prediction that TNF’s autocrine functions are stimulus-specific such that cellular responses to CpG are more dependent on TNF autocrine feedback than cellular responses to LPS.
Figure 5.
TLR-responsive TNF production functions in an autocrine manner in response to some TLR agonists but not others. (A) Schematic of the expanded multimodular computational model, incorporating the TNFR-to-NFκB signaling module. (B) Model simulations and experimental data for mature TNF mRNA and secreted TNF protein in wild-type cells stimulated with 10 ng/mL LPS, 500 nM CpG, or 50 µg/mL PolyI:C; solid lines indicate values of model simulations, and points represent experimental data presented in previous figures. RMSDs are shown for each panel. (C) Model simulations for NFκB activity in wild-type or tnf−/− stimulated by 10 ng/mL LPS or 100 nM CpG. Solid lines indicate wild-type simulation, and dashed lines indicate tnf−/−. (D) Experimental validation of model simulations in C. Activation of NFκB measured by EMSA in wild-type and tnf−/− BMDMs stimulated with 10 ng/mL LPS or 100 nM CpG. Graphs are quantification of experimental data normalized to peak wild-type NFκB activation (n = 4). The gel image is representative of four experiments. (**) P-value < 0.02 for a difference between wild-type and tnf−/− time points.
We examined to what degree this conclusion was dependent on the specific parameter values that we obtained by training our models to BMDM experimental data. We considered six key parameters that might be cell type-specific, such as those controlling TLR or TNFR abundance, the strength of TNF mRNA synthesis, half-life control, translation, or secretion. While 10-fold changes of these parameters led to changes in NFκB activation dynamics, we found that CpG-induced NFκB activation was consistently more dependent on TNF feedback than LPS responses (Supplemental Fig. 3).
TLR-specific autocrine and paracrine functions of TNF
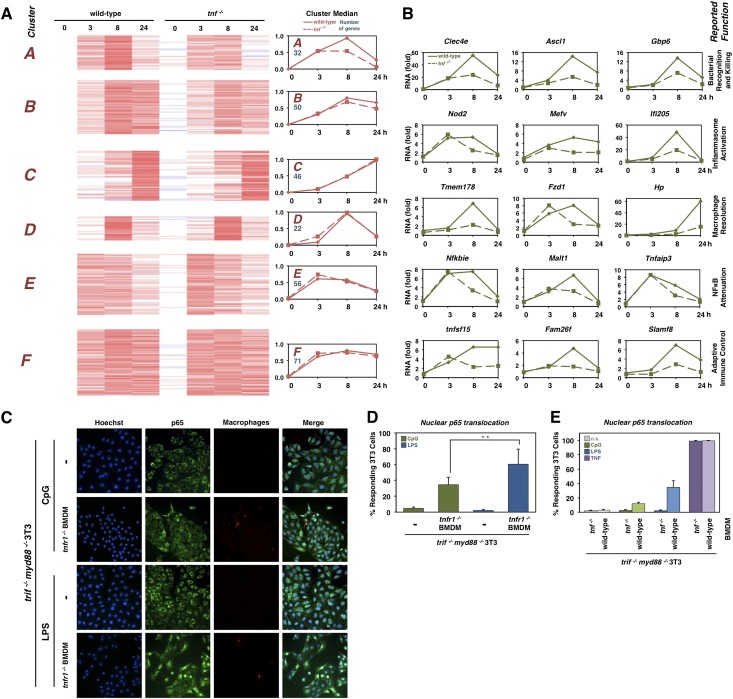
To determine TNF’s autocrine role in augmenting NFκB’s gene expression response in macrophages, we performed transcriptomic analysis using wild-type and tnf−/− BMDMs stimulated with CpG. From the RNA sequencing (RNA-seq) data, we selected 267 genes that were substantially up-regulated by CpG stimulation (Fig. 6A). K-means clustering identified groups of genes that showed either a strong early peak induction (cluster E), a peak at 8 h but persistent induction (cluster B and F), a peak at 8 h followed by a decrease by 24 h (clusters A and D), or a slow induction resulting in a peak at 24 h of stimulation (cluster C). Of these clusters, A and B contained many genes that were found to be significantly decreased in the tnf−/− condition. Interestingly, the gene ontology process term “response to cytokine” was top-ranked in clusters A/B, but “defense response” was top-ranked in clusters C/D/E/F (data not shown). “Immune response” was ranked third in both groups. We examined a few genes specifically whose expression was partially dependent on TNF (Fig. 6B). These included genes involved in bacterial recognition and killing (Clec4e, Ascl1, and Gbp6), inflammasome activation (nod2, Mefv, and Ifi205), macrophage resolution (Tmem178, Fzd1, and Hp), NFκB attenuation (Nfkbie, Mlt1, and Tnfaip3), and adaptive immune control (tnfsf15, Fam26f, and Slamf8).
Figure 6.
TNF’s paracrine- and autocrine-specific functions are TLR-specific. (A) Transcriptomic analysis of wild-type and tnf−/− BMDMs responding to 100 nM CpG. K-means clustering of 267 gene expression profiles. (Right) Cluster median reveals TNF dependency of clusters A and B. (B) Select genes whose expression is TNF-dependent in response to CpG, grouped by their known roles in inflammation and macrophage function. (C) Immunofluorescence images of coculture of tnfr1−/− BMDMs with trif−/−myd88−/− 3T3s. The top row shows p65 staining of trif−/−myd88−/− 3T3s stimulated with 1 µM CpG for 75 min. The second row shows p65 staining of coculture of tnfr−/− BMDMs with trif−/−myd88−/− 3T3s stimulated with 1 µM CpG for 75 min. The third row shows p65 staining of trif−/−myd88−/− 3T3s stimulated with 1 µg/mL LPS for 75 min. The fourth row shows p65 staining of coculture of tnfr−/− BMDMs with trif−/−myd88−/− 3T3s stimulated with 1 µg/mL LPS for 75. Images are representative of three separate experiments. (D) Bar graphs showing the average percentage of 3T3 cells with nuclear p65 in conditions described in C. Three-hundred to 500 cells were counted over three experiments. Error bars indicate standard error of the mean. (**) P-value = 0.02. (E) Bar graphs of the percentage of trif−/−myd88−/− 3T3 cells with nuclear p65 when cocultured with wild-type or tnf−/− BMDMs and stimulated as indicated. More than 1000 cells were counted over three experiments.
Although macrophages responding to LPS secrete substantial amounts of TNF, we found no evidence that TNF functions in an autocrine manner to augment NFκB activation. Tissue-resident macrophages exist in an environment in which they secret cytokines and signal to other cell types in the tissue, such as fibroblasts. To investigate the possibility that LPS-produced TNF may function in a paracrine manner, we cocultured BMDMs with myd88−/−trif−/− fibroblasts that are unable to respond to TLR agonists such as LPS or CpG but may respond to TNF. Indeed, our studies show that NFκB activation (scored by RelA nuclear translocation) in fibroblasts was dependent on the presence of cocultured BMDMs (marked by the dye CellTracker Red CPTMX) (Fig. 6C). When tnfr1 −/− BMDMs were cocultured at a ratio of 2.5% and stimulated with LPS, we found that more than half of the fibroblasts activated NFκB in response to the TNF signal (Fig. 6D). However, in the CpG-stimulated condition, a smaller percentage of fibroblasts showed NFκB activation, corresponding to lower ELISA measurements (Fig. 5B). Furthermore, we found that fibroblast activation was dependent on both BMDM ratio and fibroblast seeding densities, but the trend of higher activation in the LPS stimulation condition was consistent (Supplemental Fig. 4). To demonstrate that the fibroblast activation was due to TNF produced by the BMDMs, we cocultured either wild-type or tnf −/− BMDMs with fibroblasts and found that neither LPS nor CpG stimulation resulted in fibroblast NFκB activation when tnf −/− BMDMs were used (Fig. 6E). These results indicate that substantial TNF production elicited by LPS-stimulated macrophages has primarily paracrine functions during the inflammatory/immune response, whereas the smaller amount of CpG-induced TNF also functions in an autocrine manner to enhance the macrophage activation of NFκB.
Discussion
Macrophages play an early, key role in the innate immune response to a variety of pathogens through the activation of TLRs. In this study, we investigated the mechanisms of production of one potent cytokine secreted by macrophages—TNF—and the autocrine and paracrine roles that it plays during the immune response. Based on previous qualitative studies of TNF mRNA induction and mRNA stabilization, translation, and secretion, we here quantitatively characterized to what degree TRIF and MyD88 control the associated regulatory modules in order to develop a systems model that accounts for the contributions of each module and signaling pathway to TNF production in response to TLR agonists. The resulting mathematical model led to the computational prediction—validated experimentally—that TNF functions are autocrine or paracrine, depending on the specific TLR agonist. Indeed, whereas CpG-induced autocrine TNF signaling led to sustained NFκB activity and enhanced expression of inflammatory genes (Fig. 5), LPS-induced TNF functions in a potent paracrine manner (Fig. 6).
While there have been many reports concerning the mechanisms by which TNF production is modulated in response to TLR agonists, the cell systems used were diverse; as a result, it was unclear whether these mechanisms are cell- or context-dependent. Using a consistent cell system, we demonstrate that in primary, naïve macrophages, MyD88 is primarily responsible for early transcriptional induction of TNF mRNA (Fig. 1E), while the TRIF pathway is essential for the activation of post-transcriptional mechanisms that promote mRNA half-life stabilization (Fig. 2) as well as pro-TNF translation and processing and TNF secretion (Fig. 3). Stabilization of TNF mRNA is controlled through a TRIF–p38–MK2 axis that down-regulates mRNA degradation mechanisms via, for example, TTP. Translation of TNF mRNA to pro-TNF is controlled by the ERK and p38 via eIF4E, and TNF secretion is regulated through protein processing via the enzyme TACE. These three TRIF-controlled post-transcriptional mechanisms are essential for full TNF production and secretion. However, they also affect the timing, as post-transcriptional mechanisms provide for faster responses than those involving de novo synthesis of mRNA. Indeed, ELISAs detect substantial amounts of TNF in response to LPS at 1 h, whereas, in response to CpG, equivalent amounts are detected at 2 h (Fig. 5B).
The kinetics of TLR-induced NFκB activation and TNF production are also essential to understanding the TLR-specific autocrine and paracrine functions of TNF. Whereas MyD88-mediated NFκB activation occurs early with a peak (15–45 min), TRIF-mediated NFκB activation occurs later and is sustained for considerably longer (45 min to 4 h). Thus, in the case of LPS, rapid production of TNF interfaces with sustained NFκB activity, thus diminishing any potential autocrine enhancement of NFκB. However, in the case of CpG, a delayed production of TNF interfaces with a transient NFκB activity profile triggered by the MyD88 pathway, thus allowing for a potent autocrine signaling effect. In reference to NFκB signaling, CpG-induced autocrine TNF serves a role analogous to the TRIF signaling axis following LPS exposure: to augment and prolong NFκB activity. Thus, both TLR agonists may not differ substantially in the degree to which they activate NFκB (although by different means) but differ primarily in whether the TRIF-specific IRF3-induced network is activated.
We might then ask why prolonged NFκB activation has evolved to be dependent on secreted cytokine in response to some TLR agonists but is hardwired in response to others. Autocrine TNF signaling was proposed in the study of intracellular bacteria infections (Kindler et al. 1989), which are sensed by TLRs located in cellular compartments, such as the endosomal TLR9. Indeed, mice with a bioactive transmembrane-bound TNF, which is not secreted and cannot function in a paracrine manner, are still able to survive physiological doses of Listeria monocytogenes (Alexopoulou et al. 2006) but not all intracellular bacteria (McIlwain et al. 2012). We may hypothesize that prolonging NFκB activation, which counteracts programmed cell death, only makes physiological sense if cells infected with intracellular bacteria remain healthy enough to have a fighting chance to suppress the infection and thus healthy enough to produce and secrete TNF. Cells that can no longer muster these regulatory mechanisms will not be able to produce TNF and thus may not extend NFκB activity and consequential survival signals, therefore allowing for the possibility of stemming the infection via programmed cell death. Although speculative, this hypothesis of regulatory “design principles” is based on the network dynamics of TNF production and signaling that were revealed by the present systems-level analysis.
A goal of the systems biology approach is to develop quantitatively predictive models of regulatory networks. However, the vast and interconnected nature of immune signaling networks presents a challenge to developing mechanistic models. Here we used the strategy of “modular” biology (Hartwell et al. 1999; Kitano 2002; Mallavarapu et al. 2009) by focusing our experimental tools on separable regulatory modules and parameterizing corresponding mechanistic yet simple coarse-grained ordinary differential equation (ODE)-based models. By linking these models and then connecting them to previously established models of TLR and TNFR signaling, we developed a TNF signaling network model that represents a research tool for studies of not only the dynamics and underlying mechanisms of TNF production but also the signaling functions of TNF. We focused on specific predictions and insights regarding autocrine and paracrine roles of this cytokine, but numerous other questions may be addressed, such as dose responses, duration, or inflammatory spread within a tissue. The model formulation may be further fine-grained to represent additional mechanistic details, altered to study the role of biological extrinsic or intrinsic noise, or embedded within an agent-based formulation to study cell-to-cell variability and the role of paracrine TNF in tissue behavior. As such, models of increasing scope may bring the wealth of mechanistic knowledge of inflammatory networks to benefit therapeutic development and clinical decision-making.
Materials and methods
Animals, cell culture, and reagents
Wild-type and gene-deficient C57BL/6 mice were housed at University of California at San Diego in accordance with protocols authorized by the Institutional Animal Care and Use Committee. BMDMs were made through the isolation of 6 × 106 BM cells from mouse femurs. FLDMs were isolated from the FLs of embryonic day 13 (E13)–E14 embryos. BM and FL cells were cultured in suspension in L929-conditioned DMEM on 15-cm suspension plates for 7 d and were replated for experiment on day 8 at a density of 7.1 × 10−2 cells per square centimeter. We used recombinant mouse TNFa (Roche), LPS (Sigma, B5:055), CpG DNA (InvivoGen, ODN 1668), and PolyI:C (InvivoGen). Drugs used were actinomycin-D (Sigma), p38 inhibitor SB 203580 (Tocris), and ERK1/2 inhibitor FR180204 (Sigma). Antibodies against RelA/p65 (sc-372), eIF4E (sc-9976), TACE (sc-6416), and actin (sc-1615) were from Santa Cruz Biotechnology. Antibodies against p-p38 Thr180/Tyr182 (#4511), p-ERK1/2 Thr202/Tyr204 (#4370), p-MK2 Thr222 (#3316), p-eIF4E Ser209 (#9741), and TNF (#3042) were from Cell Signaling. Antibodies against p-TACE Thr735 were from Abcam (Ab60996) and Assay Biotech (A0763). Goat anti-rabbit Alexa Fluor 488 antibody was from Life Technologies. Antibody against p-TTP Ser178 was a gift from Dr. Paul Anderson at Brigham and Women’s Hospital.
Tnfr1−/− and tnf−/− BMDMs were stained with CellTracker Red (Invitrogen), mixed with mouse trif−/− myd88−/− 3T3 cells at a 2.5:97.5 ratio, and plated onto glass coverslips (Fisher), or mouse trif−/− myd88−/− cells alone (control) were stained and fixed in 4% PFA (EM Sciences) for 15 min. Slides were blocked with blocking buffer (5% normal goat serum, 0.2% Triton-X100 in PBS) and stained with rabbit sc-372 relA/p65 primary antibody (Santa Cruz Biotechnology) in blocking buffer at a 1:200 dilution overnight at 4°C. Secondary antibody goat anti-rabbit Alexa Fluor 488 (Invitrogen) was incubated at 1:1000 for 1 h at room temperature. Nuclei were counterstained with Hoechst, and images were acquired on an Axio Observer Z1 inverted microscope (Carl Zeiss Microscopy) with a 20×, 0.8 NA air immersion objective to a Coolsnap HQ2 CCD camera (Photometrics) using ZEN imaging software (Carl Zeiss Microscopy). Cell nuclei were counted and scored for nuclear translocation of p65.
RNA analysis
Mature mRNA extracts were prepared using the QIAshredder kit and purified using the RNEasy kit according to the manufacturer’s instructions (Qiagen). Nascent mRNA extracts were prepared from chromatin-associated RNA using TRIzol according to the manufacturer’s instructions (Life Technologies) and purified using the RNEasy kit (Qiagen). Nuclei were purified from BMDMs by lysis of cytoplasmic membrane with lysis buffer (10 mM Tris at pH 7.5, 150 mM NaCl, 0.5 mM EDTA at pH 8.0, 0.15% NP-40, 1 mM DTT, 1 mM PMSF, 10 µg/mL aprotinin, 5 µg/mL leupeptin, 1 mM pepstatin) followed by nuclei pellet isolation by centrifugation in 0.9 M sucrose in lysis buffer. Nuclei pellets were resuspended in glycerol buffer (20 mM Tris at pH 7.9, 75 mM NaCl, 50% glycerol, 0.5 mM EDTA, 1 mM DTT, 1 mM PMSF, 10 µg/mL aprotinin, 5 µg/mL leupeptin, 1 mM pepstatin) and lysed with nuclei lysis buffer (10 mM HEPES at pH 7.6, 75 mM MgCl2, 0.3 mM NaCl, 1 mM urea, 0.2 mM EDTA, 1% NP-40, 1 mM DTT, 1 mM PMSF, 10 µg/mL aprotinin, 5 µg/mL leupeptin, 1 mM pepstatin). Chromatin was isolated following centrifugation. cDNA libraries were created from RNA using iScript cDNA synthesis kit according to the manufacturer’s instructions (Bio-Rad). RT–PCR was performed on a Bio-Rad CFX384 real-time detection system in triplicates for each sample using the following primers: TNF: 5′-CACCACGCTCTTCTGTCTAC-3′ forward and 5′-AGAAGATGATCTGAGTGTGAGG-3′ reverse, and GAPDH: 5′-AACTTTGGCATTGTGGAAGG-3′ forward and 5′-GGATGCAGGGATGATGTTCT-3′ reverse. The intron–exon junction-spanning primers used for nascent analysis were ntTNF: 5′-CCCAGACCCTCACACTCAGTA-3′ forward and 5′-AACTGCCCTTCCTCCATCTT-3′ reverse, and ntActin: 5′-CTGTATTCCCCTCCATCGTG-3′ forward and 5′-GCTTGCCACTCCCAAAGTAA-3′ reverse. Data were analyzed using the Bio-Rad CFX manager software version 1.6.
Transcriptome analysis
Total RNA was isolated from CpG-stimulated (100 nM) wild-type and tnf −/− BMDMs using the QIAshredder kit and purified using the RNEasy kit according to the manufacturer’s instructions (Qiagen). cDNA libraries were prepared for RNA-seq using the TruSeq Stranded mRNA HT kit according to the manufacturer’s instructions (Illumina, reference no. 15032623). Quantitation was performed using the Roche LightCycler 480. Sequencing was performed on Illumina’s HiSeq 2000 according to the manufacturer’s recommendations and prepared for RNA sequencing analysis by the Broad Stem Cell Research Center High-Throughput Sequencing Core at the University of California at Los Angeles. Reads were aligned to the ENSEMBL NCBI m37 mouse genome build, release 66 (Flicek et al. 2012), with the STAR RNA-seq aligner (Dobin et al. 2013). HTSeq count from the HTSeq Python package (Anders et al. 2014) was used to determine raw gene read counts. The total number of reads mapping to features in each sample was used to normalize to counts per million (cpm). Genes not induced or with <10 cpm in all low-dose wild-type conditions were removed from consideration. After a 25 cpm pseudocount was added to all data, the log2 fold change was calculated relative to the wild-type 0-h time point. For K-means clustering, the time-point data of each gene were divided by its maximum expression level.
Protein analysis
Nuclear extracts were prepared using high-salt HEPES extraction buffer. Whole-cell extracts were prepared using RIPA buffer. EMSA and immunoblotting were done as previously described (Hoffmann et al. 2002; Werner et al. 2005; Kearns et al. 2006). EMSA bands were quantified using the GE Life Sciences ImageQuant software. Immunoblot bands were quantified using the Bio-Rad ImageLab 4.01 software.
Mathematical modeling
Simple ODEs were written for each step in the production of TNF (nascent gene transcription, mRNA half-life stabilization, and translation/secretion) based on the modules identified through experimental approaches. Experimentally derived values of inputs and outputs for each mathematical model were used in parameterization. For TRIF-mediated TNF, mRNA half-life begins at 10 min, linearly increases upon stimulation to 35 min following 30 min of stimulation, and then decreases linearly to 10 min of half-life after 1 h of stimulation. The three TNF production models were then connected, where the output of one model serves as the input for the subsequent model. The fitness of each mathematical model to match experimental data was determined and scored by RMSD, which was calculated after normalizing the experimental data and simulation to their own maximum value. This TNF production model was then connected with a model for TLR-induced IKK activation (Z Cheng, B Taylor, D Rios, and A Hoffmann, in prep.) and IKK-induced NFκB activation (Hoffmann et al. 2002; Werner et al. 2005) to produce a model for TLR-induced TNF production in the context of NFκB signaling. Iterative simulation and experimentation led to the inclusion of the TNFR model (Werner et al. 2008) to allow for autocrine TNF signaling. Matlab version R2013a (The MathWorks, Inc.) was used to numerically solve ODEs with the subroutine ode15s. Matlab model codes are available on request.
Acknowledgments
We thank Kim Ngo for assistance in cDNA library preparation for transcriptome analysis, Christine Cheng for FLDM preparation and RNA extraction, and the University of California at Los Angeles Broad Stem Cell Research Center for support. This study was funded by P50 GM085764, R01 GM071573, and the University of California at San Diego Molecular Biophysics Training Program NIH 5 T32 GM008326 (A.B.C.). A.B.C. carried out experimental macrophage work, and Z.C. carried out the mathematical modeling work. J.D.V. carried out microscopy work and image analysis. H.A.B. and A.B.C. performed the transcriptome analysis. A.B.C., Z.C., and A.H. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.244749.114.
References
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413: 732–738 [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Kranidioti K, Xanthoulea S, Denis M, Kotanidou A, Douni E, Blackshear PJ, Kontoyiannis DL, Kollias G. 2006. Transmembrane TNF protects mutant mice against intracellular bacterial infections, chronic inflammation and autoimmunity. Eur J Immunol 36: 2768–2780 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2014. HTSeq—a Python framework to work with high-throughput sequencing data. bioRxiv doi: 10.1101/002824 [DOI] [PMC free article] [PubMed]
- Andersson K, Sundler R. 2006. Posttranscriptional regulation of TNFα expression via eukaryotic initiation factor 4E (eIF4E) phosphorylation in mouse macrophages. Cytokine 33: 52–57 [DOI] [PubMed] [Google Scholar]
- Basak S, Behar M, Hoffmann A. 2012. Lessons from mathematically modeling the NF-κB pathway. Immunol Rev 246: 221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, et al. 1997. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature 385: 729–733 [DOI] [PubMed] [Google Scholar]
- Blasi E, Pitzurra L, Bartoli A, Puliti M, Bistoni F. 1994. Tumor necrosis factor as an autocrine and paracrine signal controlling the macrophage secretory response to Candida albicans. Infect Immun 62: 1199–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Covert MW, Leung TH, Gaston JE, Baltimore D. 2005. Achieving stability of lipopolysaccharide-induced NF-κB activation. Science 309: 1854–1857 [DOI] [PubMed] [Google Scholar]
- Coward WR, Okayama Y, Sagara H, Wilson SJ, Holgate ST, Church MK. 2002. NF-κ B and TNF-α: a positive autocrine loop in human lung mast cells? J Immunol 169: 5287–5293 [DOI] [PubMed] [Google Scholar]
- Datta S, Novotny M, Li X, Tebo J, Hamilton TA. 2004. Toll IL-1 receptors differ in their ability to promote the stabilization of adenosine and uridine-rich elements containing mRNA. J Immunol 173: 2755–2761 [DOI] [PubMed] [Google Scholar]
- Deleault KM, Skinner SJ, Brooks SA. 2008. Tristetraprolin regulates TNF TNF-α mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol Immunol 45: 13–24 [DOI] [PubMed] [Google Scholar]
- Díaz-Rodríguez E, Montero JC, Esparís-Ogando A, Yuste L, Pandiella A. 2002. Extracellular signal-regulated kinase phosphorylates tumor necrosis factor α-converting enzyme at threonine 735: a potential role in regulated shedding. Mol Biol Cell 13: 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet C, Shakhov AN, Jongeneel CV. 1991. Enhancers and transcription factors controlling the inducibility of the tumor necrosis factor-α promoter in primary macrophages. J Immunol 147: 1694–1700 [PubMed] [Google Scholar]
- Fan H, Derynck R. 1999. Ectodomain shedding of TGF-α and other transmembrane proteins is induced by receptor tyrosine kinase activation and MAP kinase signaling cascades. EMBO J 18: 6962–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2012. Ensembl 2012. Nucleic Acids Res 40: D84–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gais P, Tiedje C, Altmayr F, Gaestel M, Weighardt H, Holzmann B. 2010. TRIF signaling stimulates translation of TNF-α mRNA via prolonged activation of MK2. J Immunol 184: 5842–5848 [DOI] [PubMed] [Google Scholar]
- Häcker H, Vabulas RM, Takeuchi O, Hoshino K, Akira S, Wagner H. 2000. Immune cell activation by bacterial CpG-DNA through myeloid differentiation marker 88 and tumor necrosis factor receptor-associated factor (TRAF)6. J Exp Med 192: 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Huez G, Beutler B. 1991a. Interactive effects of the tumor necrosis factor promoter and 3′-untranslated regions. J Immunol 146: 1843–1848 [PubMed] [Google Scholar]
- Han JH, Beutler B, Huez G. 1991b. Complex regulation of tumor necrosis factor mRNA turnover in lipopolysaccharide-activated macrophages. Biochim Biophys Acta 1090: 22–28 [DOI] [PubMed] [Google Scholar]
- Hao S, Baltimore D. 2009. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 10: 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. 1999. From molecular to modular cell biology. Nature 402: C47–C52 [DOI] [PubMed] [Google Scholar]
- Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. 2006. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol Cell Biol 26: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe K, Janssen EM, Kim SO, Alexopoulou L, Flavell RA, Han J, Beutler B. 2003. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat Immunol 4: 1223–1229 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. 2002. The IκB-NF-κB signaling module: temporal control and selective gene activation. Science 298: 1241–1245 [DOI] [PubMed] [Google Scholar]
- Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. 1999. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11: 115–122 [DOI] [PubMed] [Google Scholar]
- Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. 2006. IκBɛ provides negative feedback to control NF-κB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol 173: 659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler V, Sappino AP, Grau GE, Piguet PF, Vassalli P. 1989. The inducing role of tumor necrosis factor in the development of bactericidal granulomas during BCG infection. Cell 56: 731–740 [DOI] [PubMed] [Google Scholar]
- Kitano H 2002. Computational systems biology. Nature 420: 206–210 [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10: 387–398 [DOI] [PubMed] [Google Scholar]
- Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk HD, Gaestel M. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-α biosynthesis. Nat Cell Biol 1: 94–97 [DOI] [PubMed] [Google Scholar]
- Kuno R, Wang J, Kawanokuchi J, Takeuchi H, Mizuno T, Suzumura A. 2005. Autocrine activation of microglia by tumor necrosis factor-α. J Neuroimmunol 162: 89–96 [DOI] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol Cell Biol 19: 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, Denny EM, Sanghvi JC, Gaston JE, Maynard ND, Hughey JJ, Covert MW. 2009. A noisy paracrine signal determines the cellular NF-κB response to lipopolysaccharide. Sci Signal 2: ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo E, Alvarez-Barrientos A, Maroto B, Boscá L, Knaus UG. 2007. TLR4-mediated survival of macrophages is MyD88 dependent and requires TNF-α autocrine signalling. J Immunol 178: 3731–3739 [DOI] [PubMed] [Google Scholar]
- MacDonald TT, Hutchings P, Choy MY, Murch S, Cooke A. 1990. Tumour necrosis factor-α and interferon-γ production measured at the single cell level in normal and inflamed human intestine. Clin Exp Immunol 81: 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie S, Fernàndez-Troy N, Espel E. 2002. Post-transcriptional regulation of TNF-α during in vitro differentiation of human monocytes/macrophages in primary culture. J Leukoc Biol 71: 1026–1032 [PubMed] [Google Scholar]
- Mallavarapu A, Thomson M, Ullian B, Gunawardena J. 2009. Programming with models: modularity and abstraction provide powerful capabilities for systems biology. J R Soc Interface R Soc 6: 257–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, et al. 2012. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science 335: 229–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes IB, Schett G. 2007. Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol 7: 429–442 [DOI] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP. 2008. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8: 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murch SH, Braegger CP, Walker-Smith JA, MacDonald TT. 1993. Location of tumour necrosis factor α by immunohistochemistry in chronic inflammatory bowel disease. Gut 34: 1705–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Jarrous N, Ben-Asouli Y, Kaempfer R. 1999. A cis-acting element in the 3′-untranslated region of human TNF-α mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev 13: 3280–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki Y, Sasagawa S, Kuroda S. 2005. Dynamic characteristics of transient responses. J Biochem 137: 659–663 [DOI] [PubMed] [Google Scholar]
- Parameswaran N, Patial S. 2010. Tumor necrosis factor-α signaling in macrophages. Crit Rev Eukaryot Gene Expr 20: 87–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282: 2085–2088 [DOI] [PubMed] [Google Scholar]
- Purvis JE, Lahav G. 2013. Encoding and decoding cellular information through signaling dynamics. Cell 152: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez M, Cabal-Hierro L, Carcedo MT, Iglesias JM, Artime N, Darnay BG, Lazo PS. 2011. NF-κB signal triggering and termination by tumor necrosis factor receptor 2. J Biol Chem 286: 22814–22824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronkina N, Kotlyarov A, Dittrich-Breiholz O, Kracht M, Hitti E, Milarski K, Askew R, Marusic S, Lin L-L, Gaestel M, et al. 2007. The mitogen-activated protein kinase (MAPK)-activated protein kinases MK2 and MK3 cooperate in stimulation of tumor necrosis factor biosynthesis and stabilization of p38 MAPK. Mol Cell Biol 27: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, Akira S. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κ B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol 171: 4304–4310 [DOI] [PubMed] [Google Scholar]
- Soond SM, Everson B, Riches DWH, Murphy G. 2005. ERK-mediated phosphorylation of Thr735 in TNFα-converting enzyme and its potential role in TACE protein trafficking. J Cell Sci 118: 2371–2380 [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WFC, Blackwell TK, Anderson P. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23: 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140: 805–820 [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Ruiz-Gutierrez M, Borden KLB. 2004. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res 64: 8639–8642 [DOI] [PubMed] [Google Scholar]
- Wang X, Flynn A, Waskiewicz AJ, Webb BL, Vries RG, Baines IA, Cooper JA, Proud CG. 1998. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J Biol Chem 273: 9373–9377 [DOI] [PubMed] [Google Scholar]
- Wang L, Trebicka E, Fu Y, Waggoner L, Akira S, Fitzgerald KA, Kagan JC, Cherayil BJ. 2011. Regulation of lipopolysaccharide-induced translation of tumor necrosis factor-α by the toll-like receptor 4 adaptor protein TRAM. J Innate Immun 3: 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JP, Pober JS, Bradley JR. 2013. Tumour necrosis factor and cancer. J Pathol 230: 241–248 [DOI] [PubMed] [Google Scholar]
- Werner SL, Barken D, Hoffmann A. 2005. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 309: 1857–1861 [DOI] [PubMed] [Google Scholar]
- Werner SL, Kearns JD, Zadorozhnaya V, Lynch C, O’Dea E, Boldin MP, Ma A, Baltimore D, Hoffmann A. 2008. Encoding NF-κB temporal control in response to TNF: distinct roles for the negative regulators IκBα and A20. Genes Dev 22: 2093–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. 1997. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity 7: 837–847 [DOI] [PubMed] [Google Scholar]
- Wu S, Boyer CM, Whitaker RS, Berchuck A, Wiener JR, Weinberg JB, Bast RC Jr. 1993. Tumor necrosis factor α as an autocrine and paracrine growth factor for ovarian cancer: monokine induction of tumor cell proliferation and tumor necrosis factor α expression. Cancer Res 53: 1939–1944 [PubMed] [Google Scholar]
- Xaus J, Comalada M, Valledor AF, Lloberas J, López-Soriano F, Argilés JM, Bogdan C, Celada A. 2000. LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-α. Blood 95: 3823–3831 [PubMed] [Google Scholar]
- Xu P, Derynck R. 2010. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol Cell 37: 551–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, et al. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301: 640–643 [DOI] [PubMed] [Google Scholar]