Abstract
This work was conducted to identify mungbean genotypes showing yield stability and resistance to mungbean yellow mosaic virus (MYMV) disease. Sixteen genotypes were evaluated in a randomized complete block design with two replications for two years (2011 and 2012) at three locations (Gazipur, Ishurdi and Madaripur) of the Bangladesh Agricultural Research Institute. An analysis of variance exhibited significant effects of genotype (G), environment (E), and genotype × environment (G×E) on grain yield. Among eight agronomic characters, the principal component 1 (PC1) was always higher than the PC2. Considering G×E interaction, BM6 was the best genotype at all three locations in both years. Based on grain yield and stability performance, BM6 ranked first while the worst performing genotypes were BM1 and G10. Based on discrimination and representation, Gazipur was identified as an ideal environment for these mungbeans. Relationship between soil-plant analysis developments (SPAD) value was positive with yield but negative with MYMV severity. BM6, G1 and G2 were considered as promising sources of resistance for low disease score and stable response across the environments. The environment proved to have an influence on MYMV infection under natural infestation. A positive correlation was observed between disease score and the temperature under natural growing condition.
Keywords: disease score, GGE biplot, genotype × strain interaction, mungbean
Mungbean (Vigna radiata (L.) Wilczek) is an important legume crop, widely grown in South and South-east Asia and is considered the most economically important Vigna crop in Asia. Over 80% of the mungbean is produced in South Asia. Mungbean is grown in crop rotation and relayed cropping with cereals using residual moisture in the soil. Developing mungbean genotypes with improved determinate growth habit and synchronous maturity is essential. Moreover, majority of these cultivars are susceptible to mungbean yellow mosaic virus (MYMV) disease which is a major cause of failure in mungbean cultivation in many growing countries. MYMV, a member of the family Geminiviridae, has long been a great threat to legume crops. Singh (1980) and Marimuthu et al. (1981) reported that yield loss due to MYMV disease in mungbean was about 76 to 100%. Whitefly (Bemisia tabaci) is a responsible vector transmitting MYMV. Incidence of the disease depends on the vector population, which in turn depends on environmental conditions. MYMV disease can be recognized by its typical symptoms. The first symptom appears on young leaves of the plant at vegetative stage in the form of mild yellow spots. Gradually, all of the upper leaves turn to yellow which ultimately reduces plant size, number of flowers and pods. The symptoms become manifest as leaf yellowing affects the content and efficiency of leaf photosynthetic pigments. Pigmentation assessment is an important indicator of breakdown in leaf chlorophyll as a result of environmental stress (Brown et al., 1991). The chlorophyll content meter (or SPAD meter) is a commercially available portable equipment used for measuring greenness based on optical response when a leaf is exposed to light and thus indirectly estimating foliar chlorophyll concentration (Campbell et al., 1990; Kariya et al., 1982; Yadava, 1986).
So far, there is no information on association between SPAD values with disease symptoms and yield. Plant breeders should develop mungbean varieties characterized by MYMV resistance together with other desirable traits. The success of genetic enhancement program hinges on identification of genotypes adapted to the season with disease resistance and stable yield. Stability analysis can be used to identify durable resistance to pathogens (Jenns et al., 1982). Effective breeding for disease resistance relies on a good understanding of host-pathogen relationship. Yan and Falk (2002) reported that genotype and genotype-by-environment (GGE) biplot analysis can be employed to identify the best resistant cultivar and the most virulent pathogen that can be used to differentiate resistance level among cultivars.
GGE biplot is exploited for graphical display of G×E pattern of yield trial data with several advantages. The yield of each cultivar in a tested environment is a result of genotypic main effect (G), environmental main effect (E) and genotype × environment (GE) interaction (Yan and Kang, 2002). GGE biplot methodology allows visual examination of GE interaction pattern of multi-environmental data based on two concepts. First, yield is measured as the combined effect of G, E, and GE. Only G and GE are relevant to and considered in genotype evaluation. Second, GGE biplot technique separates two principal components, PC1 and PC2, which are also referred to as primary and secondary effects, respectively. The principal components are derived from subjecting environment-centered yield data (the yield variation due to GGE) to singular value decomposition. Then the pattern of genotypic response across environments can be graphically determined in a GGE biplot (Yan and Tinker, 2006). For cultivar evaluation, G and GE are important components for explaining a meaningful relationship between genotypes and environments from the GGE Biplot. The horizontal axis (PC1) indicates the main effect of genotype while the vertical axis (PC2) shows the interaction of genotype and environment which is the basic criterion for judging genotypic stability. The lines passing from the origin (0,0) of the coordinate of a location and genotype are referred to as environmental vector and genotype vector, respectively. The average environmental axis (AEA) is the line that passes through the coordinates of all the locations and the biplot origin. The length of the environmental vector from the origin to its coordinate is used to measure the discriminating ability of the location. An ideal genotype should have a large PC1 score (high yielding ability) and a small (absolute) PC2 score (high stability). Similarly, an ideal tested environment should have a large PC1 score (high discrimination effect of the genotypes in term of genotypic main effect) and small (absolute) PC2 score (high representative of the overall environments).
There are only a few reports on GGE biplot analysis on yield performance in mungbean (Asfaw et al., 2012; Ullah et al., 2011, 2012), and none of these considered MYMV disease in the studies. The objectives of this study were (a) to assess the response of mungbean genotypes to MYMV disease, and (b) to determine stability of performance of mungbean genotypes tested across environments.
Materials and Methods
Plant materials
Sixteen mungbean genotypes/varieties, namely G1, G2, G3, G4, G5, G6, G7, G8, G9, G10, G11, G12, G13, G14, BARImung 1 (BM1), and BARImung 6 (BM6) were used in this study (Table 1). BM1 and BM6 are cultivers while G1 and G10 are breeding lines, all developed by Pulses Research Centre, Bangladesh Agricultural Research Institute (BARI), Bangladesh. The rest accessions were developed by AVRDC-The World Vegetable Centre (AVRDC), Taiwan with an objective relevant to MYMV reaction.
Table 1.
Sixteen mungbean genotypes used in this study
| Code | Genotypes | Code | Genotypes |
|---|---|---|---|
| G1 | BMXK205002 | G9 | GK 22 |
| G2 | VC 6148 | G10 | BMXK205006 |
| G3 | VC 6144 | G11 | BD 6897 |
| G4 | VC 6369 | G12 | VC 6153 |
| G5 | VC6173 | G13 | VC 6144 |
| G6 | VC6773 | G14 | BD 6873 |
| G7 | VC6372 | BM1 | BARI Mung1 |
| G8 | SML 134 | BM6 | BARI Mung6 |
Field trails
The mungbean accessions were evaluated at three diverse locations, viz. Gazipur, Ishurdi and Madaripur of BARI during dry seasons (March to May) of 2011 and 2012. These three locations have high MYMV incidence and also represent the main mungbean growing areas of the country. Three major characteristics are presented in Table 2. Each trial was conducted in a randomized complete block design with two replications. In each replication, a mungbean genotype was sown in a size of 4 m long with 4 rows. Row to row distance was 40 cm with 100 cm spacing between the adjacent plots. In each row, spacing between the adjacent plants was 5 cm. After every four rows, two rows of the highly susceptible genotypes BM1 were grown as disease spreader rows. In addition, BM1 was also grown as two border rows around the plot. Prior to soil preparation, fertilizers were applied at 40 kg/ha of urea, 90 kg/ha of triple super phosphate, 60 kg/ha of muriate of potash, and 40 kg/ha of gypsum as per a fertilizer recommendation guide of Bangladesh (Alam et al., 2010). Weeds were controlled manually. Harvesting was done by hand.
Table 2.
Characteristics of the three tested locations
| Location | Soil type | Soil pH | Latitude | Longitude | Altitude above MSL* |
|---|---|---|---|---|---|
| Gazipur | Clay loam | 5.7–6 | 23° 99′N | 90° 42′E | 33 |
| Ishurdi | Silty loam | 7–7.5 | 24° 10′N | 89° 13′E | 49 |
| Madaripur | Clay loam | 6.5–7 | 23° 20′N | 90° 22′E | 13 |
Mean sea level
Trait measurement
Nine characters were recorded, viz. days to flowering, days to maturity, plant height (cm), number of pods per plant, number of seeds per pod, SPAD value, grain (seed) yield (kg), MYMV disease index and MYMV disease score. Since the MYMV causes yellowish and impairs photosynthesis of mungbean, SPAD analysis was used to determine the relative amount of chlorophyll present in mungbean leaves. SPAD was measured by a portable MINOLTA SPAD-502 meter (MINOLTA, Tokyo). Three leaves locating at different positions of each plant from 10 randomly selected plants were used for readings. Then the readings on each leaf were pooled to obtain a SPAD measurement. Grain yield was obtained by converting plot yield to a per hectare basis (kg/ha). For MYMV evaluation, number of MYMV infected plants in each genotypes and disease scoring scale were considered together. Firstly the number of plants infected by MYMV in each genotype was presented in percentage. Then disease scoring was done when more than 80% of the spreader plants (BM1) showed MYMV symptoms. The scoring system was based on 1 to 9 rating scale as suggested by Singh et al., (1992).
Statistical analysis
Statistical analysis was done on grain yield and MYMV scores to identify stable genotypes based on graphical presentation of the G×E interaction. An analysis of variance (ANOVA) was performed across the tested locations and years, treating year and location as random effects, genotype as a fixed effect. Significant test of the fixed treatment effect was done following the method of Satterthwaite (1946) as cited by Petersen (1994). The analysis was carried out using R-program 2.10.0 (R Development Core Team, 2010). The ANOVA provided the effect of genotypes (G), environment (E), and all possible interactions among these factors.
In a GGE biplot, genotype and genotype x environment (G + GE) interaction are usually studied together by dissecting from the observed mean. In this study, the model for constructing a genotype by strain biplot was Ŷij-μ-βj = gi1 e1j + gi2e2j + ɛij. Where Ŷij is the expected value for mungbean entry i grown in environment j; μ is the grand mean for all growing environment; βj is the main effect of environment j; gi1 and e1j are the first principal component (PC1) scores for genotype i and environment j, respectively; gi2 and e2j are the second principal component (PC2) scores for genotype i and environment j, respectively; and ɛij is the residual for each genotype-environment combination not explained by PC1 and PC2. This model was used to explain the relationship between trials, varieties and locations. Mean grain yield of the mungbean varieties tested in the three locations and two years (six environments) were used to generate a two-way table data of genotype and environment for the biplot analysis. The analyses were conducted and the biplot was generated using the GGEbiplot software (Yan, 2001).
Results
SPAD value
High SPAD readings of 47 to 52 were obtained when the disease score is 1, i.e. representing a healthy leaf (data not shown). The readings were 32 to 43 when the disease score was 3 while the readings of 23 to 32 were observed when the disease score was 5. Low SPAD readings of 14–25 were observed when the disease score was 7. SPAD value was found varying among mungbean genotypes and decreasing in proportional to increasing disease score but positive with yield. The cultivars BM 6 had the highest SPAD value and produced the highest yield.
In MYMV infected plants the virus destroys chlorophyll or inhibits its synthesis, thus causing yellowish leaves. SPAD evaluation of infected plants gave significant positive correlation with yield and 100-seed weight, but negative with MYMV disease score (data not shown).
Analysis of variance
Mean yield together with MYMV disease score of the mungbean genotypes over locations for both years are shown in Table 3. Among 16 mungbean genotypes, BM1 showed the highest disease severity, followed by G12, G6 and G10. In contrast, less disease severity was observed in BM6, G1, G2 and G3. An analysis of variance for yield and MYMV disease score of mungbean genotypes is given in Table 4. Locations (L) showed different effect on both yield and MYMV score, while year (Y) affected MYMV incidence. The mungbean accessions showed different reaction in MYMV. These mungbeans showed effect of environment on yield with significance found in G×E interaction. However, their MYMV score was highly influenced by almost all sources of environmental effects. Relatively higher yield was obtained in 2011 compared with 2012. The most favorable location among the tested sites was Gazipur. BM-6 gave the highest yield across the environments at 1374 kg/ha. For MYMV reaction, disease incidence was higher in 2012 than in 2011. Among the three locations, less disease incidence was observed at Ishurdi in both years. Highest MYMV disease incidence was found at Gazipur and Madaripur, also in both years. Correlation coefficients among environmental factors of the tested sites and disease incidence are presented in Table 5. A strong relationship was observed between disease score and maximum temperature, but not other site characters. High positive correlation was observed between disease score and maximum temperature.
Table 3.
Mean grain yield and MYMV disease score of mungbean genotypes grown at three locations in Bangladesh during 2011 and 2012
| Genotypes | Grain yield (kg/ha) | MYMV disease score | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| Gazipur | Ishurdi | Madaripur | Overall mean | Gazipur | Ishurdi | Madaripur | Overall mean | |||||||
|
|
|
|
|
|
|
|||||||||
| 2011 | 2012 | 2011 | 2012 | 2011 | 2012 | 2011 | 2012 | 2011 | 2012 | 2011 | 2012 | |||
| G1 | 1383 | 1329 | 1167 | 1085 | 1120 | 1155 | 1207 | 1 | 3 | 1 | 1 | 3 | 3 | 2.00 |
| G2 | 1182 | 1168 | 1114 | 1169 | 1149 | 1125 | 1151 | 3 | 3 | 1 | 1 | 3 | 3 | 2.34 |
| G3 | 990 | 945 | 1215 | 1159 | 856 | 873 | 1006 | 3 | 5 | 1 | 1 | 3 | 3 | 2.67 |
| G4 | 665 | 677 | 656 | 645 | 923 | 872 | 739 | 3 | 5 | 3 | 3 | 3 | 5 | 3.66 |
| G5 | 855 | 803 | 908 | 858 | 941 | 924 | 881 | 3 | 3 | 1 | 1 | 3 | 3 | 2.34 |
| G6 | 420 | 305 | 531 | 578 | 690 | 701 | 537 | 9 | 9 | 3 | 5 | 7 | 7 | 6.66 |
| G7 | 535 | 325 | 625 | 774 | 812 | 837 | 651 | 7 | 9 | 3 | 5 | 5 | 7 | 6.00 |
| G8 | 412 | 455 | 497 | 512 | 772 | 737 | 564 | 7 | 7 | 3 | 5 | 5 | 7 | 5.66 |
| G9 | 486 | 397 | 537 | 521 | 800 | 702 | 574 | 3 | 7 | 3 | 1 | 7 | 5 | 4.34 |
| G10 | 275 | 331 | 380 | 418 | 670 | 607 | 447 | 9 | 7 | 5 | 5 | 5 | 7 | 6.33 |
| G11 | 944 | 614 | 593 | 577 | 839 | 620 | 698 | 3 | 5 | 3 | 5 | 7 | 5 | 4.33 |
| G12 | 319 | 624 | 841 | 888 | 399 | 360 | 572 | 7 | 9 | 3 | 3 | 7 | 9 | 7.00 |
| G13 | 686 | 355 | 493 | 497 | 597 | 595 | 537 | 7 | 7 | 3 | 3 | 3 | 5 | 4.67 |
| G14 | 378 | 380 | 596 | 599 | 694 | 659 | 551 | 7 | 9 | 3 | 3 | 5 | 7 | 5.66 |
| BM1 | 423 | 409 | 531 | 543 | 628 | 528 | 510 | 9 | 9 | 5 | 7 | 7 | 9 | 7.66 |
| BM6 | 1419 | 1357 | 1343 | 1354 | 1415 | 1357 | 1374 | 1 | 3 | 1 | 1 | 3 | 1 | 1.67 |
|
| ||||||||||||||
| Mean | 711 | 654 | 752 | 761 | 831 | 790 | 750 | 5.25 | 6.12 | 2.62 | 3.12 | 4.87 | 5.25 | 4.56 |
Table 4.
Mixed model analysis of variance for yield data (kg ha−1) and MYMV disease score of mungbean trials conducted at three locations in Bangladesh during 2011–2012
| Sources | df | Mean square | |
|---|---|---|---|
|
| |||
| Yield (kg/ha) | MYMV score | ||
| Locations | 2 | 352905.38** | 138.44** |
| Years | 1 | 0.0 | 97.75** |
| Location × Year | 2 | 2418 ns | 5.88** |
| Rep/Location×Year | 6 | 58836 | 0.50 |
| Genotypes | 15 | 1064425** | 5.83** |
| Genotype × Year | 15 | 10782ns | 3.51** |
| Genotype × Location | 30 | 79324** | 3.42** |
| Genotype × Year × Location | 30 | 8965ns | 1.04ns |
| Pooled error | 90 | 11217 | 1.19 |
Table 5.
Correlations among environmental factors and disease score of mungbean genotypes evaluated over 3 locations during 2011–2012
| Disease score | Elevation | Humidity (%) | Rainfall | MaxTm1 | |
|---|---|---|---|---|---|
| Elevation | −0.832 | ||||
| Humidity (%) | 0.940 | −0.971 | |||
| Rainfall | 0.942 | −0.970 | 0.999** | ||
| MaxTm | 0.979* | 0.859 | 0.957 | −0.957 | |
| MinTm2 | −0.866 | 0.444 | −0.645 | −0.647 | −0.839 |
Maximum temperature
Minimum temperature
Significant at the 0.05 and 0.01 level of probability, respectively
GGE Biplot analysis
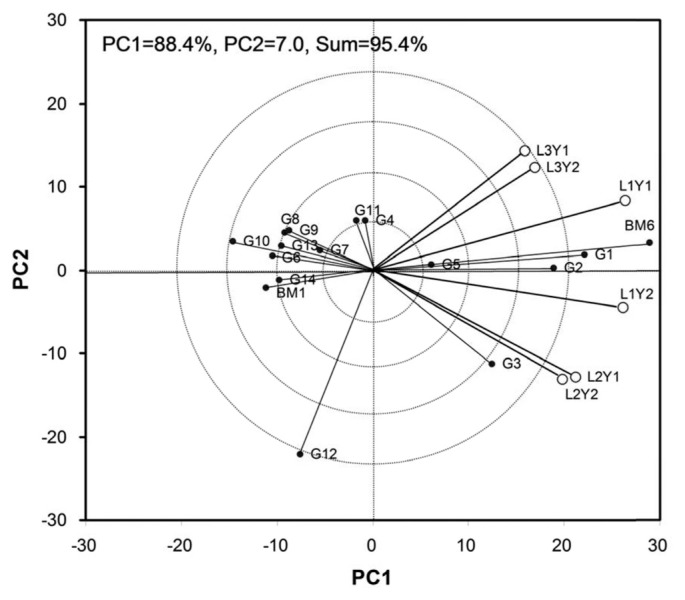
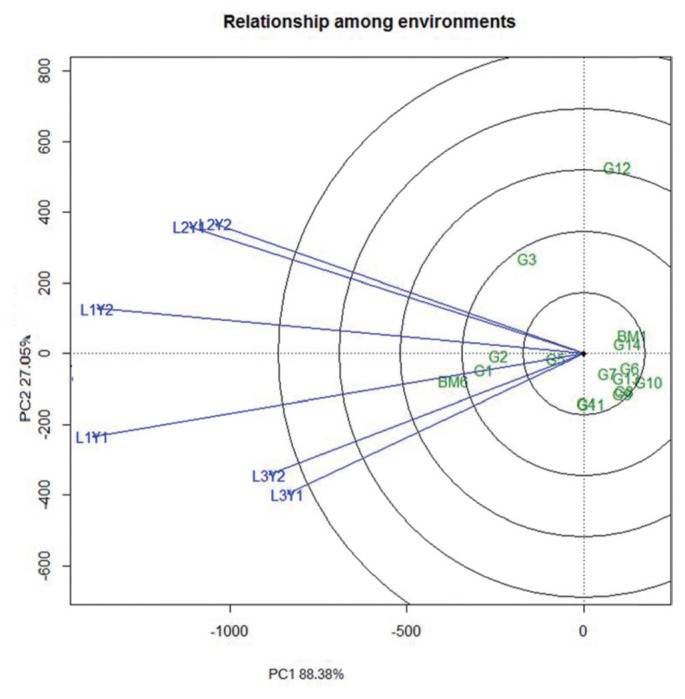
The principal component 1 (PC1) and PC2 obtained from all the eight observed characters accounted for 88.10% and 5.80% for days to flowering, 74.90% and 17.60 for days to maturity, 76% and 16.80 for plant height, 66.90 and 17.20 for pods/plant, 75.80% and 10.10 for seeds/pod, 78% and 13.20 for 100 seed weight, 71.40% and 13.50% for MYMV score, and 88.40% and 7% for yield of the total variation, respectively. Seed yield showed as high as 88.4% variation that can be explained by PC1. The performance of the mungbean genotypes is presented in the biplot based on grain yield data (Fig. 1). All six environments distribute in two sectors. The first sector contains L1Y1 (Gazipur-2011), L3Y2 (Madaripur-2012) and L3Y1 (Madaripur-2011), while the second sector comprises L1Y2 (Gazipur-2012), L2Y1 and L2Y2 (Ishurdi-2011 and -2012) environments. Among the tested locations, L1Y1 and L1Y2 (Gazipur 2011 and 2012) had larger environmental vectors as compared to others. Considering the GGE biplot of yield performance, BM6 had the largest genotypic vector, yet it outclassed all other genotypes at all locations and appeared to be widely adaptive. G1, G2, G3 and G5 also performed relatively well as compared to the rests. These mungbean genotypes are among the most favorable at those locations. Accordingly, location 1 (Gazipur) in 2011 and 2012 provided more information regarding the genotype difference. The most representative location for mungbean grain yield was Gazipur (Fig. 2). All the environments form small angles among each others, implying that they are positively correlated and thus genotypic performances in these environments are similar.
Fig. 1.
GGE biplot of yield (kg/ha) showing the vector of 6 locations and 16 mungbean genotypes. The average environmental axis (AEA) is the straight line that passed through the origin (0,0) and coordinate of the location mean.
Fig. 2.
GGE biplot of relationship among environments based on yield data.
Genotype × Strain interaction
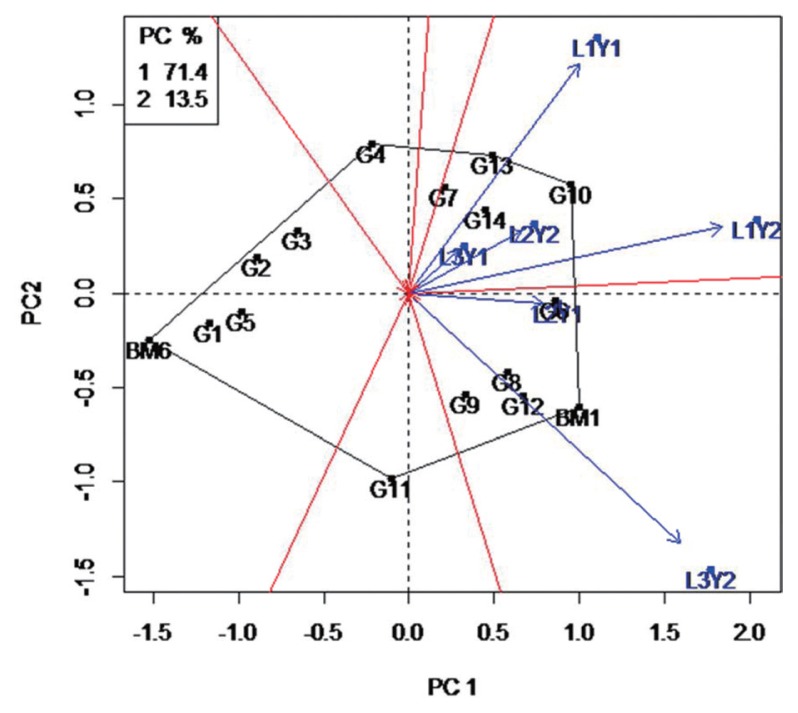
The 16 mungbean genotypes evaluated in different locations and years presented a wide range of response to MYMV. Some of them with low severity values (Table 3). The ANOVA shows significant effect in genotypes, environments and the genotype × environment interaction for MYMV disease severity (Table 4). To evaluate the mungbean genotypes, a biplot based on location specific strain data was constructed (Fig. 3). In this experiment, the first two PCs of the GGE model explained 84.9% of variation in G+GE. The polygon shows that the vertex genotype BM6 contributing the most to the interaction, i.e. showing highest or lowest infection by the disease (Fig. 3). Environments L3Y2 and L1Y1 are highly discriminating in comparison with the other years and locations. These two environments differed greatly from the other four environments, which were more similar to each other. The biplot analysis also shows that disease severity divided genotypes into two or three groups. The first group is represented by YMV strains and the genotypes that showed the strongest interaction with strains. This includes genotypes with the highest disease severity (i.e. BM1, G12, G8, G9 and G6). The second group consists of genotypes in the 3rd quadrant (i.e. G13, G7, G14 and G10) which were susceptible to the strain/disease. The genotypes located nearer to the X-axis show a more stable resistance. The majority of the genotypes are in the 1st and 4th quadrants showing weak interaction with the disease. The genotypes BM6, G1, G3 and G5 can be used as sources for future breeding for MYMV resistance.
Fig. 3.
GGE biplot based on MYMV disease score of 16 mungbean genotypes.
Discussion
Understanding G×E interaction in plant species has implications to increase economic yield. It is necessary for a plant breeder to explore this variation among genotypes and choose materials with desirable traits. The mungbean genotypes in this study expressed significant difference for yield and yield component traits under different environments. The plant materials comprised diverse genotypes of improved varieties, exotic materials and land races. Inclusion of land races is imperative in this type of investigation since they are adaptive to local environments and carrying valuable genetic resources (Anandan et al., 2011). Multi-environment testing revealed not only significant effects of the genotypes but also effects of the environment and genotype × environment interaction for the traits under study. Effect of the environment in this study ascribed largely to the locations rather than the years. When a genotype is tested in multi-location yield trials, a crossover genotype by environment interaction often occurs (Ceccarelli et al., 1995). Such an interaction results in changing of relative ranking of genotypes from one environment to another. Thomas et al. (2004) reported the presence of significant G×E interaction rendering overall means, causing less reliable of data and often confounding attempts to identify high yielding genotypes for specific site as well as for wide adaptability. A sufficiently large variation was observed in principal component analysis of agronomic characters implying the application of GGE biplot analysis in multi-environment data.
Reduction in chlorophyll content has been reported in many host plant infected with different viruses (Dantre et al., 1996; Mali et al., 2000). Sheffield, (1993) found that when aucuba mosaic virus infected a fully developed tomato leaf, it destroyed the chlorophyll and caused chlorosis. Measurements with the SPAD meter produce relative values that are proportional to the amount of chlorophyll present in the leaf (Ling et al., 2011). This relationship has many applications because leaf chlorophyll concentration changes in response to external factors such as light and biotic and abiotic stresses. Quantifying chlorophyll concentration can provide important information about plant growth and physiological plasticity in response to changing environments (Larcher, 1995; Richardson et al., 2002). The amount of solar radiation absorbed by a leaf is largely a function of the foliar concentration of photosynthetic pigments. Low chlorophyll concentration directly limits photosynthetic potential and hence reduces primary products of photosynthesis. In the present study, it was observed that when the plant had high MYMV disease score, the SPAD meter reading significantly decreased together with yield. Low chlorophyll concentration limits photosynthesis, dry matter production, plant growth and yield. Results of this study indicate that SPAD meter potentially offers a useful non-destructive measurement of chlorophyll content to aid evaluation of plant health. High correlations were obtained among SPAD readings with yield and disease score. The difference in yield among the genotypes was likely due to development of reproductive organs being controlled by photoassimilate production and partitioned by the source tissues. This whole process is most critical at this stage (Taiz and Zeiger, 2002; Wahid and Rasul, 2004). Thus high MYMV disease severity should have a pronounced effect on pod development and yield.
GGE biplot allowed discerning the average performance and stability of the accessions. Through a multi-environment evaluation for the resistance in field trials where the disease was present, the accessions showing resistance to MYMV are identified in independent multi-environments. This allowed a fine-tune selection for particular locations in such distant places of Bangladesh with the advantage of testing only promising accessions. The GGE analysis helped identifying a group of accessions with low severity and good stability in their response to MYMV disease. This stability is remarkable since the three evaluated environments differed greatly among them. High yielding genotypes were less responsive to the disease and exhibited low interaction effect. Similarly, poor yielding genotypes were highly responsive to the disease and exhibited high disease severity. Less responsive genotypes BM6, G1 and G2 produced significantly higher grain yield, while the high responsive genotypes BM1, G14, G12, G10 and G6 exhibited lower yield. Among all the location-year tested environments, Ishurdi showed high interaction with years in genotypes and disease reaction. The other two sites displayed lower interaction effect with the above average main effect. Thus locations, Ishurdi (2011 and 2012) were classified as low responsive environments and not suitable for evaluation of MYMV reaction in mungbean. Weather condition and rainfall of the corresponding sites can be explained by similar patterns in both years. The other two locations, Gazipur and Madaripur caused high interaction between genotypes and disease. Ishurdi showed unique soil texture, humidity, and amount of rainfall from the other two environments (Table 2; Fig. S1). The high interaction effect of MYMV disease at Gazipur and Madaripur might probably be due to higher temperature, humidity and rainfall than Ishurdi. MYMV disease is usually transmitted by insect vector white fly. Islam et al. (2008) reported that temperature and humidity accounted for 64% of the factors influencing white fly population. Rahman et al. (2006) reported that whitefly population built up more in the field with higher temperature. Yet, high rainfall relates with high humidity and thus white fly population. Locations with high elevation showed negative correlation with disease score, especially those with low humidity due to distance from the sea and low rainfall. Correlating the external environmental factors with disease score clearly indicated that the higher temperature during evaluation might explain the higher severity of MYMV disease in that environment and humidity is a major factor affecting development of the disease through increasing of white fly population. This underlines the importance of environmental factors influencing the performance of crops against diseases.
Acknowledgements
This research was financially supported by the World Bank. A part of this research was also funded by the Center of Advanced Studies for Agriculture and Food, Institute for Advanced Studies, Kasetsart University under the National Research University Program of Office of the Higher Education Commission, Ministry of Education, Thailand. We acknowledge assistance and support from staffs of Pulses Research Centre, Bangladesh Agricultural Research Institute, Bangladesh.
References
- Alam AKMM, Podder R, Haque AHMM, Hossain AH. Handbook of Pulses. Bangladesh Agricultural Research Institute, Suborno Press; Gazipur, Bangladesh: 2010. Publication No. 28. [Google Scholar]
- Anandan A, Rajiv G, Eswaran R, Prakash M. Genotypic variation and relationships between quality traits and trace elements in traditional and improved rice (Oryza sativa L.) genotypes. J Food Sci. 2011;76:122–130. doi: 10.1111/j.1750-3841.2011.02135.x. [DOI] [PubMed] [Google Scholar]
- Asfaw A, Gurum F, Alemayehu F, Rezene Y. Analysis of multi-environment grain yield trials in mung bean Vigna radiata (L.) Wilczek based on GGE biplot in southern Ethiopia. J Agr Sci Tech. 2012;14:389–398. [Google Scholar]
- Brown SB, Houghton JD, Hendry GAF. Chlorophyll breakdown. In: Scheer H, editor. Chlorophylls. CRC Press; Boca Raton, Florida: 1991. pp. 465–489. [Google Scholar]
- Campbell RJ, Mobley KN, Marini RP, Pfeiffer DG. Growing conditions alter the relationship between SPAD-502 values and apple leaf chlorophyll. Hort Sci. 1990;25:330–331. [Google Scholar]
- Ceccarelli S, Grando S, Booth RH. In: Eyzaguirre P, Iwanaga M, editors. International breeding programs and resource-poor farmers; Proceedings of a workshop on participatory plant breeding IPGRI; Rome, Italy. 1995. pp. 99–116. [Google Scholar]
- Dantre RK, Keshwa RL, Khare MN. Biochemical changes induced by yellow mosaic virus in resistant and susceptible cultivars of soyabean (Glycine max L.) Merrill. Indian J Virol. 1996;12:47–49. [Google Scholar]
- Islam MS, Latif MA, Ali M, Hossain MS. Population dynamics of white fly on some recommended mungbean varieties in Bangladesh and its impact on incidence of mungbean yellow mosaic virus disease and yield. Int J Agri Sci Tech. 2008;2:41–47. [Google Scholar]
- Jenns AE, Leonard KJ, Moll RH. Stability analysis for estimating relative durability of quantitative resistance. Theor Appl Genet. 1982;63:183–192. doi: 10.1007/BF00303707. [DOI] [PubMed] [Google Scholar]
- Kariya K, Matsuzaki A, Machida H. Distribution of chlorophyll content in leaf blade of rice plant. Jpn J Crop Sci. 1982;51:134–135. [Google Scholar]
- Larcher W. Ecophysiology and stress physiology of functional groups. 3rd ed. Springer; New York: 1995. [Google Scholar]
- Ling Q, Huang H, Jarvis P. Use of SPAD-502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana. Photosynth Res. 2011;107:209–214. doi: 10.1007/s11120-010-9606-0. [DOI] [PubMed] [Google Scholar]
- Mali PC, Burman U, Satish L. Effect of planting dates and development of yellow mosaic virus on biochemical constituents of moth bean genotypes. Indian Phytopath. 2000;53:379–383. [Google Scholar]
- Marimuthu T, Subramaniam CL, Mohan R. Assesment of yield loss due to yellow mosaic infection in mungbean. Pulse Crops Newsletter. 1981;1:104. [Google Scholar]
- Petersen RG. Agricultural field experiments: design and analysis. Marcel Dekker Inc; 270 Madison Avenue, New York: 1994. [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- Rahman AHM, Akanda AM, Ashraful AKM. Relationship of whitefly population build up with the spread of TYLCV on eight tomato varieties. J Agric Rural Dev. 2006;4:67–74. [Google Scholar]
- Richardson AD, Shane P, Duigan G, Berlyn P. An evaluation of non-invasive methods to estimate foliar chlorophyll content. New Phytol. 2002;153:185–194. [Google Scholar]
- Satterthwaite FE. An approximate distribution of estimates of variance components. Biom Bull. 1946;2:210–214. [PubMed] [Google Scholar]
- Sheffield FNL. The development of assimilatory tissue of solanaceous host infected with aucuba mosaic to tomato. Ann Appl Biol. 1993;20:57–60. [Google Scholar]
- Singh JP. Effect of virus diseases on growth components and yield of mungbean and urd bean. Indian Phytopath. 1980;33:405–408. [Google Scholar]
- Singh G, Sharma YR, Kaur L. Methods of rating yellow mosaic virus of mungbean and urdbean. Plant Dis Res. 1992;7:1–6. [Google Scholar]
- Taiz L, Zeiger E. Plant Physiology. 3rd ed. Sinauer Associates Inc Publishers; Massachusetts: 2002. [Google Scholar]
- Thomas MJR, Fukai S, Peoples MB. The effect of timing and severity of water deficit on growth, development, yield accumulation and nitrogen fixation of mungbean. Field Crops Res. 2004;86:67–80. [Google Scholar]
- Ullah H, Khalil IF, Khalil IA, Khattak GSS. Performance of mungbean genotypes evaluated in multi-environmental trails using the GGE biplot method. Atlas J Biotech. 2011;1:1–8. [Google Scholar]
- Ullah H, Khalil IF, Durrishahwar I, Khalil IA, Fayaz M, Yan J, Ali F. Selecting high yield and stable mungbean [Vigna radiata (L.) Wilczek) genotypes using GGE biplot techniques. Can J Plant Sci. 2012;92:951–960. [Google Scholar]
- Wahid A, Rasul E. Photosynthesis in leaf, stem, flower and fruit. In: Pessarakli M, editor. Handbook of Photosynthesis. CRC Press; Florida: 2004. pp. 479–497. [Google Scholar]
- Yadava UL. A rapid and non destructive method to determine chlorophyll on intact leaves. Hort Sci. 1986;21:1449–1450. [Google Scholar]
- Yan W. GGEbiplot–A windows application for graphical analysis of multienvironment-trial data and other types of two-way data. Agron J. 2001;93:1111–1118. [Google Scholar]
- Yan W, Kang MS. GGE biplot analysis: A graphical tool for breeders, geneticists, and agronomists. CRC Press; Boca Raton, Florida: 2002. [Google Scholar]
- Yan W, Falk DE. Biplot analysis of host-by-pathogen data. Plant Dis. 2002;86:1396–1401. doi: 10.1094/PDIS.2002.86.12.1396. [DOI] [PubMed] [Google Scholar]
- Yan W, Tinker NA. Biplot analysis of multi-environment trial data: Principles and applications. Can J Plant Sci. 2006;86:623–645. [Google Scholar]