Abstract
Oligomeric proanthocyanidins were successfully identified by UHPLC-PDA-HRMSn in a selection of plant-derived materials (jujube fruit, Fuji apple, fruit pericarps of litchi and mangosteen, dark chocolate, and grape seed and cranberry extracts). The identities of 247 proanthocyanidins were theoretically predicted by computing high-accuracy masses based on the degree of polymerization, flavan-3-ol components, and the number of A type linkages and galloyls. MSn fragments allowed characterization on flavan-3-ol based on the monomer, connectivity, and location of A-type bonds. Identification of doubly or triply charged ions of 50 PAs was made on the basis of theoretical calculations. A single catechin standard and molar relative response factors (MRRFs) were used to quantify the well-separated PAs. The ratios of the SIM peak counts were used to quantify each of the unseparated isomers. This is the first report of direct determination of each of the proanthocyanidins in plant-derived foods and proanthocyanidins containing an epifisetinidol unit in grape seeds.
Keywords: oligomeric proanthocyanidins, identification, quantification, plant products, UHPLC-PDA-ESI/HRMSn profiling method
Introduction
Proanthocyanidins (PAs) are various length polymers of flavanols (catechins and their enantiomers) linked through a single C4–C8 or C4–C6 bond (B-type PAs) or with an additional C2–O–C7 or C2–O–C5 bond (A-type PAs) as shown in Figure 1. There are a variety of different classes of PAs, depending on the substitution pattern of the monomeric flavan-3-ols (mainly epicatechins, epigallocatechins, and epiafzelechins to form procyanidins, propelargonidins, and prodelphinidins), acyls (usually galloyl), glycosyls, and other substituents.1−4 The highly polymerized PAs are reported to have molecular weights up to 30000 Da. However, these PAs may not be efficiently extracted from plant materials.1−3
Figure 1.
Structures of flavan-3-ol units and common fragmentation patterns for proanthocyanidins.
PAs are the main polyphenolic components in many different plant-derived foods, such as grains, berries, fruits, nuts, and teas, and are reported to have a variety of health-promoting benefits.1−7 As the degree of polymerization increases, the compounds become less soluble in aqueous solution and less bioavailable in the intestine. Fermentation in the colon, however, leads to absorption of many of the metabolic products. The most absorbed PAs in the intestine have a degree of polymerization (DP) less than or equal to 4 (DP ≤ 4).1−7 Accurate analytical methods for the separation, identification, and quantification of individual oligomeric PAs in foods are necessary to establish the relationship between dietary intake of polyphenols and health outcomes from biological, epidemiological, and clinical studies.
PAs have a high structural diversity with many regioisomeric (order of linkage for the flavan-3-ols) and stereoisomeric (physical structure of individual flavan-3-ols) forms, which makes identification and quantification difficult tasks. In general, analytical methods have focused on each oligomer as a class and have been unable to identify the PAs within each class. Matrix-assisted laser desorption ionization–time-of-flight-mass spectrometry (MALDI-TOF-MS) has been used to detect PA metal adducts and to determine the types and DP values of the compounds.1,8−12 ESI-MSn has also been used to identify PA molecular ions and their fragments.8,13−23 However, neither of these methods can identify the PA isomers.1,7−16
Normal and reverse phase HPLC methods have been used to separate PA oligomers and tandem MS has been used to characterize the PAs for DP ≤ 6 (typically m/z 50–2000).1,12,19−23 Doubly and triply charged negative molecular ions of some higher oligomers (DP > 6) have been detected using negative ionization.1,4,14−19 Reverse phase HPLC-PDS-MS analysis of thiolytically degraded products of PAs has been used to identify the PA terminal (with the C8 connection) and extension units (with the C4 connection) and to determine the mean DP value (mDP).1−4,11−17 Both 1H and 13C NMR analyses have been used to identify PA flavan-3-ols and the cis or trans stereochemistries of PAs.10,11 Until now, however, there has been only limited application of UHPLC-HRMSn to the study of oligomeric PAs.20−23
Total PA concentration has been estimated using colorimetric methods. In addition, total concentrations for each oligomeric class (DP = 2–10) have been estimated using fluorescence detection and relative response factors (based on mass) following separation by normal phase chromatography.2−4,6 HPLC-PDA-MS analysis of PA thiolytic degraded mixtures has also been used for quantification of PAs.1−4,6,10−12 However, direct quantification of the different PAs comprising each oligomeric class is still problematic due to the difficulty of separation and the lack of standards.1−4
As a part of a project to systematically identify and quantify food phenolic compounds, a standardized HPLC-PDA-ESI/MS method was developed for the identification and quantification of food polyphenols, including some PAs.24 Quantification was based on UV absorbance and molar relative response factors (MRRFs).25 This method has been upgraded and now uses ultrahigh-performance liquid chromatography–photodiode array detection–high-resolution mass spectrometry operated in the tandem mode (UHPLC-PDA-ESI/HRMSn).26 In the current study, this method was employed to identify nearly 300 oligomeric PAs in selected plants (fruit pericarps of litchi and mangosteen), extracts (from grape seed and cranberry), and food samples (jujube, Fuji apple, and chocolate) and to quantify PAs in grape seed extract. The main PAs in each of the oligomeric classes were quantified.
Materials and Methods
Chemicals
Formic acid, HPLC grade methanol, and acetonitrile were purchased from VWR International, Inc. (Clarksburg, MD, USA). HPLC grade water was prepared from distilled water using a Milli-Q system (Millipore Laboratory, Bedford, MA, USA).
Standards
(+)-Catechin, (−)-epicatechin, (−)-gallocatechin-3-O-gallate, (−)-epigallocatechin-3-O-gallate, procyanidin B1, procyanidin B2, procyanidin C1, and procyanidin A2 were obtained from Chromadex, Inc. (Irvine, CA, USA). The standards were vacuum-dried using a vacuum drying box (National Appliance Co., Portland, OR, USA) at 110 °C until a constant weight was reached (about 24 h). These dried standards were used to determine the MRRF that were used for calibration.25
Plant Materials and Extraction
Fresh fruits of jujube (Ziziphus jujuba Mill), Fuji apple (Malus domestica Borkh cv. Fuji), litchi (Litchi chinensis Sonn.), and mangosteen (Garcinia mangostana Linn.) were purchased from local food stores. Dark chocolate was purchased from a local Trader Joes store in Maryland, USA. The extracts of grape seed and cranberry were kindly supplied by Triarco Industries, Inc. (Paterson, NJ, USA). The fruit pericarps of litchi and mangosteen and the skins of fresh jujubes and apples were lyophilized, and the dried materials were powdered.24−26
Each of the powdered fruit samples (250 mg) was extracted with 5.000 mL of a methanol/water (60:40, v/v) solvent using sonication for 60 min at room temperature. The slurry mixture was centrifuged at 2500 rpm for 15 min. The supernatant (4.000 mL) was taken from the tube and filtered through a 17 mm (0.45 μm) PVDF syringe filter (VWR Scientific, Seattle, WA, USA) for injections.24−26 A second extraction using acetone/methanol/water (2:2:1, v/v/v, 4.000 mL) was treated in the same way to check the extraction efficiency of the general extraction method. The result showed that >95% of the mass for each main compound was extracted from the plant material by the first extraction.
Powdered chocolate samples (2000 mg) were extracted with 40 mL of the same aqueous methanol and treated as described above, and the supernatant was taken to dryness under vacuum at 40 °C. The approximately 30 mg of the residue was dissolved in water (1 mL) and passed through Sep-PakVac RC (500 mg) C18 cartridge (Waters Corp., Milford, MA, USA). After washing with water (5 mL), the PAs were eluted with methanol (5 mL) and again taken to dryness under vacuum. The residue was dissolved in 1.000 mL of the methanol/water solvent and filtered for injection.
The grape seed (10.80 mg) and cranberry (10.20 mg) extracts were dissolved in the same aqueous methanol (1.0 mL) and filtered. Triplicate injections (1 μL) of each solution were used to determine the average concentration and the relative standard deviation for each of the PAs in the extract. Dried catechin was used as the external calibration standard; 4 mg was placed in a 10 mL volumetric flask, dissolved in the methanol/water (60:40, v/v) solvent, and brought to volume. This stock solution was diluted 1:4 and 1:16. The stock and each dilution were injected onto the column three times and used to construct a calibration curve.
UHPLC-PDA-ESI/HRMSn Conditions
The UHPLC-HRMS system used consisted of an LTQ Orbitrap XL mass spectrometer with an Accela 1250 binary pump, a PAL HTC Accela TMO autosampler, a PDA detector (ThermoScientific, San Jose, CA, USA), and a G1316A column compartment (Agilent, Palo Alto, CA, USA). The separation was carried out on a U-HPLC column (200 mm × 2.1 mm i.d., 1.9 μm, Hypersil Gold AQ RP-C18) (Thermo- Scientific) with an HPLC/UHPLC precolumn filter (UltraShield Analytical Scientific Instruments, Richmond, CA, USA) at a flow rate of 0.3 mL/min. The mobile phase consisted of a combination of A (0.1% formic acid in water, v/v) and B (0.1% formic acid in acetonitrile, v/v). The linear gradient was from 4 to 20% B (v/v) at 40 min, to 35% B at 60 min, and to 100% B at 61 min and held at 100% B to 65 min. The PDA recorded spectra from 200 to 700 nm and provided real-time monitoring at 280 and 330 nm.26
The HRMS was operated in the negative ionization mode using the following conditions: sheath gas at 70 (arbitrary units), aux and sweep gas at 15 (arbitrary units), spray voltage at 4.8 kV, capillary temperature at 300 °C, capillary voltage at 15 V, and tube lens at 70 V. The mass range was from m/z 50 to 2000 with a resolution of 15000, FTMS AGC target at 2e5, FT-MS/MS AGC target at 1e5, isolation width of 1.5 amu, and maximum ion injection time of 500 ms. The most intense ion was selected for the data-dependent scan to provide MS2 to MS5 product ions with a normalized collision energy at 35%.26 The selective ion monitoring (SIM) mode was used to select the molecular ions of the isomers from each of the PA groups in grape seed extract for their quantification.
Results and Discussion
Exact Masses and Molecular Formula for Proanthocyanidins
Chemically, each flavan-3-ol unit of a PA has two stereogenic (or chiral) centers (Figure 1), which can result in four (or 22) stereoisomers, that is, (2R,3S)-catechin or (+)-C, (2R,3R)-epicatechin or (+)-EC, (2S,3R)-catechin, or (−)-C, and (2S,3S)-epicatechin or (−)-EC. In this paper, EC will be used to represent all four isomers in the text, tables, and figures. Similarly, the abbreviations for epiafzelechin (EA), epigallocatechin (EG), epifisetinidol (EF), and robinetinidol (ER) will be used to represent their isomers in PAs. In this paper, the PAs formed with only EA, EC, or EG units are called propelargonidin, procyanidin, or prodelpeinidin, whereas those formed from two different units are called proanthocyanidins.
The B-type PA dimers have two flavan-3-ol units (i.e., four chiral centers) and an additional asymmetric center at C4. Thus, it is possible for them to be connected in two ways, through a C4–C8 or C4–C6 linkage, providing 64 (i.e., 26) possible isomers.1−4,10−23 The formation of B-type trimers and tetramers leads to an exponential increase in the possible number of isomers, which makes the separation and quantification of such complex PA isomers an enormous challenge.1−4
UHPLC columns (with 1.9 μm or smaller particles) provide much better separation of the PA isomers than HPLC columns.1,20−23 In addition, the molecular ions detected by HRMS provide high-resolution molecular weight (HRMW) and exact molecular formula (MF). The HRMW, MF, and singly and multiply charged ions for different PAs are related to the DP, the flavan-3-ols (EC, EA, EF, EG, and ER) in the oligomer, the number of galloyls, and the number of type A bonds as described in the equations
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
where n = degree of polymerization, a = number of ECs that were replaced by EAs or EFs (as regioisomers), b = number of ECs replaced by EGs, c = number of galloyls, and d = number of A-type bonds; 12.0000, 1.0078, and 15.9949 are the accurate masses of carbon, hydrogen, and oxygen, and 15, 12, and 6, and 7, 4, and 4 are the numbers of carbon, hydrogen, and oxygen atoms for each EC (or its regioisomer ER) and galloyl unit, respectively. The equations can be easily modified to accommodate the PAs that contain acyl, glycosyl, or phloroglucinol adducts.1,19
Low-resolution ions are usually expressed to two decimal places in most cases and can be obtained directly from the high-resolution [M – H]− values. Formulas have been described for computing the PA molecular ion metal adduct values (in low resolution) from MALDI-TOF-MS, but the PA mass values can be obtained only after the mass of the metal has been subtracted.11−13,20 Thus, eqs 1–5, for PA mass, are easier to use.
Table 1 presents the HRMW and deprotonated molecule ([M – H]−) (m/z) for most of the PAs (DP = 2–10) detected in common foods in this laboratory and described in the literature.1,4 For each oligomer, the nongalloylated B-type procyanidins (in bold) have the simplest formulas (a = b = c = d = 0), indicating that the PAs have no EC units replaced by EA, EF, or EG and do not contain any galloyls and A-type bonds. To be as systematic as possible, for each oligomer class, the related propelargonidins and proanthocyanidin-containing EA units are listed above the procyanidins, whereas the related prodelphinidins containing EG units are listed below the procyanidins. Similarly, all of the A-type PAs for each oligomer are listed above the B-type PAs and the galloylated PAs are listed below.
Table 1. Computed High-Resolution Mass, Molecular Weight, Molecular Ions, and Composition of Common Oligomeric PAsa.
| DP | proanthocyanidin | HRMW (Da) |
HR [M – H]− (m/z) |
HR [M – 2H]2– (m/z) |
HR [M – 3H]3– (m/z) |
C | H | O | EA/EF | EG | galloyl | A-bond | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dimers | B-type propelargonidin | 546.1518 | 545.1440 | 30 | 26 | 10 | 2 | 0 | 0 | 0 | |||
| B-type proanthocyanidin | 560.1311 | 559.1233 | 30 | 24 | 11 | 1 | 0 | 0 | 1 | ||||
| B-type proanthocyanidin | 562.1467 | 561.1389 | 30 | 26 | 11 | 1 | 0 | 0 | 0 | ||||
| B-type procyanidin | 578.1416 | 577.1338 | 30 | 26 | 12 | 1 | 1 | 0 | 0 | ||||
| A-type procyanidin | 576.1260 | 575.1182 | 30 | 24 | 12 | 0 | 0 | 0 | 1 | ||||
| B-type procyanidin | 578.1416 | 577.1338 | 30 | 26 | 12 | 0 | 0 | 0 | 0 | ||||
| galloylated procyanidin | 730.1524 | 729.1446 | 37 | 30 | 16 | 0 | 0 | 1 | 0 | ||||
| galloylated procyanidin | 882.1632 | 881.1554 | 44 | 34 | 20 | 0 | 0 | 2 | 0 | ||||
| B-type proanthocyanidin | 592.1209 | 591.1131 | 30 | 24 | 13 | 0 | 1 | 0 | 1 | ||||
| B-type proanthocyanidin | 594.1365 | 593.1287 | 30 | 26 | 13 | 0 | 1 | 0 | 0 | ||||
| B-type prodelphinidin | 610.1314 | 609.1236 | 30 | 26 | 14 | 0 | 2 | 0 | 0 | ||||
| galloylated proanthocyanidin | 746.1473 | 745.1395 | 37 | 30 | 17 | 0 | 1 | 1 | 0 | ||||
| galloylated prodelphinidin | 914.1530 | 913.1452 | 44 | 34 | 22 | 0 | 2 | 2 | 0 | ||||
| trimers | B-type propelargonidin | 818.2199 | 817.2121 | 45 | 38 | 15 | 3 | 0 | 0 | 0 | |||
| A-type proanthocyanidin | 832.1992 | 831.1914 | 45 | 36 | 16 | 2 | 0 | 0 | 1 | ||||
| B-type proanthocyanidin | 834.2148 | 833.2070 | 45 | 38 | 16 | 2 | 0 | 0 | 0 | ||||
| A-type proanthocyanidin | 848.1941 | 847.1863 | 45 | 36 | 17 | 1 | 0 | 0 | 1 | ||||
| B-type proanthocyanidin | 850.2097 | 849.2019 | 45 | 38 | 17 | 1 | 0 | 0 | 0 | ||||
| galloylated proanthocyanidin | 986.2256 | 985.2178 | 52 | 42 | 20 | 2 | 0 | 1 | 0 | ||||
| A-type procyanidin | 862.1734 | 861.1656 | 45 | 34 | 18 | 0 | 0 | 0 | 2 | ||||
| A-type procyanidin | 864.1890 | 863.1812 | 45 | 36 | 18 | 0 | 0 | 0 | 1 | ||||
| B-type procyanidin | 866.2046 | 865.1968 | 45 | 38 | 18 | 0 | 0 | 0 | 0 | ||||
| galloylated procyanidin | 1018.2154 | 1017.2076 | 52 | 42 | 22 | 0 | 0 | 1 | 0 | ||||
| galloylated procyanidin | 1170.2262 | 1169.2184 | 59 | 46 | 26 | 0 | 0 | 2 | 0 | ||||
| B-type proanthocyanidin | 882.1995 | 881.1917 | 45 | 38 | 19 | 0 | 1 | 0 | 0 | ||||
| B-type proanthocyanidin | 898.1944 | 897.1866 | 45 | 38 | 20 | 0 | 2 | 0 | 0 | ||||
| B-type prodelphinidin | 914.1893 | 913.1815 | 45 | 38 | 21 | 0 | 3 | 0 | 0 | ||||
| galloylated prodelphinidin | 1034.2103 | 1033.2025 | 52 | 42 | 23 | 0 | 1 | 1 | 0 | ||||
| tetramers | A-type proanthocyanidin | 1120.2622 | 1119.2544 | 60 | 48 | 22 | 2 | 0 | 0 | 1 | |||
| B-type proanthocyanidin | 1122.2778 | 1121.2700 | 60 | 50 | 22 | 2 | 0 | 0 | 0 | ||||
| A-type proanthocyanidin | 1136.2571 | 1135.2493 | 60 | 48 | 23 | 1 | 0 | 0 | 1 | ||||
| B-type proanthocyanidin | 1138.2727 | 1137.2649 | 60 | 50 | 23 | 1 | 0 | 0 | 0 | ||||
| A-type procyanidin | 1148.2208 | 1147.2130 | 60 | 44 | 24 | 0 | 0 | 0 | 3 | ||||
| A-type procyanidin | 1150.2364 | 1149.2286 | 60 | 46 | 24 | 0 | 0 | 0 | 2 | ||||
| A-type procyanidin | 1152.2520 | 1151.2442 | 60 | 48 | 24 | 0 | 0 | 0 | 1 | ||||
| B-type procyanidin | 1154.2676 | 1153.2598 | 60 | 50 | 24 | 0 | 0 | 0 | 0 | ||||
| galloylated procyanidin | 1306.2784 | 1305.2706 | 67 | 54 | 28 | 0 | 0 | 1 | 0 | ||||
| galloylated procyanidin | 1458.2892 | 1457.2814 | 74 | 58 | 32 | 0 | 0 | 2 | 0 | ||||
| A-type proanthocyanidin | 1168.2469 | 1167.2391 | 60 | 48 | 25 | 0 | 1 | 0 | 1 | ||||
| B-type proanthocyanidin | 1170.2625 | 1169.2547 | 60 | 50 | 25 | 0 | 1 | 0 | 0 | ||||
| pentamers | A-type proanthocyanidin | 1392.3303 | 1391.3225 | 695.1574 | 75 | 60 | 27 | 3 | 0 | 0 | 1 | ||
| B-type proanthocyanidin | 1410.3408 | 1409.3330 | 704.1626 | 75 | 62 | 28 | 2 | 0 | 0 | 0 | |||
| A-type proanthocyanidin | 1424.3201 | 1423.3123 | 711.1523 | 75 | 60 | 29 | l | 0 | 0 | 1 | |||
| B-type proanthocyanidin | 1426.3357 | 1425.3279 | 712.1601 | 75 | 62 | 29 | 1 | 0 | 0 | 0 | |||
| A-type procyanidin | 1436.2838 | 1435.2760 | 717.1341 | 75 | 56 | 30 | 0 | 0 | 0 | 3 | |||
| A-type procyanidin | 1438.2994 | 1437.2916 | 718.1419 | 75 | 58 | 30 | 0 | 0 | 0 | 2 | |||
| A-type procyanidin | 1440.3150 | 1439.3072 | 719.1497 | 75 | 60 | 30 | 0 | 0 | 0 | 1 | |||
| B-type procyanidin | 1442.3306 | 1441.3228 | 720.1575 | 75 | 62 | 30 | 0 | 0 | 0 | 0 | |||
| galloylated procyanidin | 1594.3414 | 1593.3336 | 796.1629 | 82 | 66 | 34 | 0 | 0 | 1 | 0 | |||
| B-type proanthocyanidin | 1458.3255 | 1457.3177 | 728.1550 | 75 | 62 | 31 | 0 | 1 | 0 | 0 | |||
| hexamers | B-type proanthocyanidin | 1682.4089 | 1681.4011 | 840.1967 | 90 | 74 | 33 | 3 | 0 | 0 | 0 | ||
| A-type proanthocyanidin | 1696.3882 | 1695.3804 | 847.1863 | 90 | 72 | 34 | 2 | 0 | 0 | 1 | |||
| B-type proanthocyanidin | 1698.4038 | 1697.3960 | 848.1941 | 90 | 74 | 34 | 2 | 0 | 0 | 0 | |||
| A-type proanthocyanidin | 1710.3675 | 1709.3597 | 854.1760 | 90 | 70 | 35 | 1 | 0 | 0 | 2 | |||
| A-type proanthocyanidin | 1712.3831 | 1711.3753 | 855.1838 | 90 | 72 | 35 | 1 | 0 | 0 | 1 | |||
| B-type proanthocyanidin | 1714.3987 | 1713.3909 | 856.1916 | 570.4584 | 90 | 74 | 35 | 1 | 0 | 0 | 0 | ||
| A-type procyanidin | 1724.3468 | 1723.3390 | 861.1656 | 573.7745 | 90 | 68 | 36 | 0 | 0 | 0 | 3 | ||
| A-type procyanidin | 1726.3624 | 1725.3546 | 862.1734 | 574.4463 | 90 | 70 | 36 | 0 | 0 | 0 | 2 | ||
| A-type procyanidin | 1728.3780 | 1727.3702 | 863.1812 | 575.1182 | 90 | 72 | 36 | 0 | 0 | 0 | 1 | ||
| B-type procyanidin | 1730.3936 | 1729.3858 | 864.1890 | 575.7901 | 90 | 74 | 36 | 0 | 0 | 0 | 0 | ||
| galloylated procyanidin | 1882.4044 | 1881.3966 | 940.1944 | 626.4603 | 97 | 78 | 40 | 0 | 0 | 1 | 0 | ||
| B-type proanthocyanidin | 1746.3885 | 1745.3807 | 872.1865 | 581.1217 | 90 | 74 | 37 | 0 | 1 | 0 | 0 | ||
| galloylated proanthocyanidin | 1898.3993 | 1897.3915 | 948.1919 | 631.7920 | 97 | 78 | 41 | 0 | 1 | 1 | 0 | ||
| heptamers | A-type proanthocyanidin | 1980.4200 | 1979.4122 | 989.2022 | 659.1322 | 105 | 80 | 40 | 2 | 0 | 0 | 3 | |
| A-type proanthocyanidin | 1996.4149 | 1995.4071 | 997.1997 | 664.4638 | 105 | 80 | 41 | 1 | 0 | 0 | 3 | ||
| B-type proanthocyanidin | 2002.4617 | 2001.4539 | 1000.2231 | 666.4794 | 105 | 86 | 41 | 1 | 0 | 0 | 0 | ||
| A-type procyanidin | 2012.4098 | 2011.4020 | 1005.1971 | 669.7955 | 105 | 80 | 42 | 0 | 0 | 0 | 3 | ||
| A-type procyanidin | 2014.4254 | 2013.4176 | 1006.2049 | 670.4673 | 105 | 82 | 42 | 0 | 0 | 0 | 2 | ||
| A-type procyanidin | 2016.4410 | 2015.4332 | 1007.2127 | 671.1392 | 105 | 84 | 42 | 0 | 0 | 0 | 1 | ||
| B-type procyanidin | 2018.4566 | 2017.4488 | 1008.2205 | 671.8111 | 105 | 86 | 42 | 0 | 0 | 0 | 0 | ||
| galloylared procyanidin | 2170.4674 | 2169.4596 | 1084.2259 | 722.4813 | 112 | 90 | 46 | 0 | 0 | 1 | 0 | ||
| A-type proanthocyanidin | 2032.4359 | 2031.4281 | 1015.2102 | 676.4708 | 105 | 84 | 43 | 0 | 1 | 0 | 1 | ||
| B-type proanthocyanidin | 2034.4515 | 2033.4437 | 1016.2180 | 677.1427 | 105 | 86 | 43 | 0 | 1 | 0 | 0 | ||
| octamers | B-type proanthocyanidin | 2274.5298 | 2273.5220 | 1136.2571 | 757.1688 | 120 | 98 | 46 | 2 | 0 | 0 | 0 | |
| B-type proanthocyanidin | 2290.5247 | 2289.5169 | 1144.2546 | 762.5004 | 120 | 98 | 47 | 1 | 0 | 0 | 0 | ||
| A-type procynidin | 2302.4884 | 2301.4806 | 1150.2364 | 766.4883 | 120 | 94 | 48 | 0 | 0 | 0 | 2 | ||
| A-type procynidin | 2304.5040 | 2303.4962 | 1151.2442 | 767.1602 | 120 | 96 | 48 | 0 | 0 | 0 | 1 | ||
| B-type procynidin | 2306.5196 | 2305.5118 | 1152.2520 | 767.8321 | 120 | 98 | 48 | 0 | 0 | 0 | 0 | ||
| galloylated procyanidin | 2454.4992 | 2453.4914 | 1226.2418 | 817.1586 | 127 | 98 | 52 | 0 | 0 | 1 | 2 | ||
| B-type proanthocyanidin | 2322.5145 | 2321.5067 | 1160.2495 | 773.1637 | 120 | 98 | 49 | 0 | 1 | 0 | 0 | ||
| nonamers | B-type proanthocyanidin | 2562.5928 | 2561.5850 | 1280.2886 | 853.1898 | 135 | 110 | 52 | 2 | 0 | 0 | 0 | |
| B-type proanthocyanidin | 2578.5877 | 2577.5799 | 1288.2861 | 858.5214 | 135 | 110 | 53 | 1 | 0 | 0 | 0 | ||
| A-type procyanidin | 2592.5670 | 2591.5592 | 1295.2757 | 863.1812 | 135 | 108 | 54 | 0 | 0 | 0 | 1 | ||
| B-type procyanidin | 2594.5826 | 2593.5748 | 1296.2835 | 863.8531 | 135 | 110 | 54 | 0 | 0 | 0 | 0 | ||
| galloylated procyanidin | 2742.5622 | 2741.5544 | 1370.2733 | 913.1796 | 142 | 110 | 58 | 0 | 0 | 1 | 2 | ||
| B-type proanthocyanidin | 2610.5775 | 2609.5697 | 1304.2810 | 869.1847 | 135 | 110 | 55 | 0 | 1 | 0 | 0 | ||
| decamers | B-type proanthocyanidin | 2850.6558 | 2849.6480 | 1424.3201 | 949.2108 | 150 | 122 | 58 | 2 | 0 | 0 | 0 | |
| B-type proanthocyanidin | 2866.6507 | 2865.6429 | 1432.3176 | 954.5424 | 150 | 122 | 59 | 1 | 0 | 0 | 0 | ||
| A-type procyanidin | 2878.6144 | 2877.6066 | 1438.2994 | 958.5303 | 150 | 118 | 60 | 0 | 0 | 0 | 2 | ||
| B-type procyanidin | 2882.6456 | 2881.6378 | 1440.3150 | 959.8741 | 150 | 122 | 60 | 0 | 0 | 0 | 0 | ||
| galloylated procyanidin | 3030.6252 | 3029.6174 | 1514.3048 | 1009.2006 | 157 | 122 | 64 | 0 | 0 | 1 | 2 | ||
| B-type proanthocyanidin | 2898.6405 | 2897.6327 | 1448.3125 | 965.2057 | 150 | 122 | 61 | 0 | 1 | 0 | 0 | ||
| galloylated proanthocyanidin | 3050.6513 | 3049.6435 | 1524.3179 | 1015.8760 | 157 | 126 | 65 | 0 | 1 | 1 | 0 | ||
Composition is used for the numbers of the atoms of carbon, hydrogen, and oxygen of the molecular formula and the numbers of the flavan-3-ol units, A-type bonds, and galloyls. Abbreviations: DP, degree of polymerization; G, galloyl; EC, EA, EG, epicatechin, epiafzelechin, and epigallocatechin, respectively; C, H, O, carbon, hydrogen, and oxygen.
The data in Table 1, calculated from eqs 1–5, were found to agree well with experimentally determined [M – H]− values with an error of <3 ppm in most cases. Consequently, Table 1 can be used to provide the PA structure based on experimental high-resolution [M – H]− values. For example, trace ions recorded in grape seed extract were easily identified as galloylated procyanidin tetramers (1305.2698, error < 3 ppm), pentamers (1441.3250), hexamers (1729.3898), and their gallates (1593.3354 and 1881.4033). Thus, a detailed analysis of plant PAs can be achieved easily without using purified PA or PA-enriched samples.
The data in Table 1 permit a detailed PA oligomeric profile of a sample to be obtained from a single chromatogram using HRMS. Although identification of specific PAs based on the recorded HR [M – H]− values is putative, they are all correctly identified as PAs. It should be noted that nominal MS (typically masses to two decimal places) cannot positively identify them as PAs. The data in Table 1 also provide the opportunity to fully identify interesting or minor PAs (i.e., to specify the flavan-3-ol units and their connectivity) by selecting specific ions for fragmentation as described below.
Identification of Proanthocyanidins in Foods
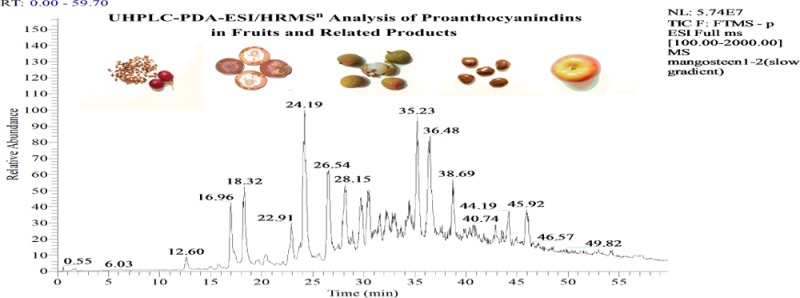
The UHPLC-PDA-ESI-HRMSn profiling method provides retention time, UV, [M – H]−, and MS2–5 product ions for the PAs. Consideration of the product ions, especially MS2 ions, permits easy putative identification of PAs. Table 2 lists 247 proanthocyanidins in 90 isomeric groups from 7 food materials, their plant sources, single-parent ions, formulas, and diagnostic and main MS2 productions. The number of the isomers identified in each sample is in parentheses following the plant name. Approximately 130 of the PAs were detected in the grape seed and mangosteen extracts, and the rest were detected in the other five samples (Fuji apples, cranberry extract, dark chocolate, jujube, and litchi). Many of the PAs were detected in these plants for the first time.
Table 2. Proanthocyanidins Found in Seven Samples.
| DP | proanthocyanidin | plant sourcea | HR [M – H]− (m/z) | mol formula | major MS2 ions (m/z)(%) |
|---|---|---|---|---|---|
| monomer | epiazfelechin | L, C | 273.0761 | C15H13O5 | 167(100) |
| catechin | ALL | 289.0710 | C15H13O6 | 245(100), 205(35), 179(12) | |
| epicatechin | ALL | 289.0714 | C15H13O6 | 245(100), 205(33), 179(11) | |
| epigallocatechin | standard | 305.0665 | C13H13O7 | 305(100), 221(19), 219(29), 179(20) | |
| epicatechin-3-gallate | G | 441.0827 | C22H17O10 | 331(16), 289(100), 271(9), 169(20) | |
| catechin-3-gallate | G, M | 441.0827 | C22H17O10 | 331(19), 289(100), 271(10), 193(6), 169(21) | |
| gallocatechin-3-gallate | standard | 457.0775 | C22H17O11 | 331(67), 305(36), 287(10), 193(10), 169(100) | |
| epigallocatechin-3-gallate | M | 457.0779 | C22H17O10 | 331(53), 305(38), 287(9), 269(7), 193(9), 169(100) | |
| dimer | EA→EC(1) | M(2) | 561.1393 | C30H25O11 | 543(34), 435(50), 425(19), 407(19), 289(100), 271(13), 245(7) |
| EC→EA(1) | M(1) | 561.1380 | C30H25O11 | 543(9)435(100), 409(64), 391(7), 299(44), 287(50), 273(57), 161(8) | |
| EF→EC(1) | G(2) | 561.1382 | C30H25O11 | 451(40), 435(89), 423(100), 409(49), 325(17), 289(13), 271(26) | |
| EF→EC(2) | G(3) | 561.1383 | C30H25O11 | 451(100), 435(78), 423(91), 409(56), 397(17), 299(25), 289(15), 271(48) | |
| EF→EC(3) | G(5) | 561.1389 | C30H25O11 | 451(42), 435(100), 423(100), 409(39), 325(13), 289(15), 271(21) | |
| EC→A→EC(1) | A(2) | 575.1190 | C30H23O12 | 539(23), 449(82), 423(100), 411(13), 407(19), 289(26), 285(18) | |
| EC→A→EC(2) | C(11), D(1), L(8), M(5) | 575.1181 | C30H23O12 | 557(15), 539(30), 453(20), 452(16), 449(100), 447(20), 423(30), 407(20), 289(26), 287(16), 285(27) | |
| EC→A→EC(3) | C(1) | 575.1179 | C30H23O12 | 449(27), 413(13), 395(88), 377(100), 333(21) | |
| EC→A→EC(4) | C(1) | 575.1202 | C30H23O12 | 535(22), 509(47), 391(29), 347(100), 329(84), 285(22) | |
| EC→EC(1) | B(1), B(2), G(10), M(3), A(3) | 577.1345 | C30H25O12 | 559(17), 451(37), 425(100), 407(53), 299(8), 289(26), 287(8) | |
| EC→EC(2) | G(3) | 577.1335 | C30H25O12 | 559(57), 467(20), 451(100), 425(86), 407(59), 289(65) | |
| EC→EC(3) | D(1), M(1) | 577.1340 | C30H25O12 | 559(75), 533(46), 451(29), 439(67), 425(75), 407(20), 393(100), 289(29), 269(35) | |
| EC→EC(4) | D(1), G(1), M(1) | 577.1335 | C30H25O12 | 559(100), 533(31), 451(21), 439(34), 425(32), 407(18), 393(35) | |
| EC→EG(1) | G(1) | 593.1279 | C30H25O13 | 575(13), 525(6), 467(24), 441(100), 427(6), 423(12), 305(16) | |
| (EC→EC)g(1) | G(5) | 729.1434 | C37H29O16 | 603(14), 577(100), 559(46), 451(13), 425(20), 407(50) | |
| (EC→EC)g(2) | G(2) | 729.1435 | C37H29O16 | 711(23), 603(45), 577(99), 559(88), 451(46), 441(42), 407(100), 289(19) | |
| (EC→EC)g(3) | G(1) | 729.1437 | C37H29O16 | 711(35), 619(29), 603(100), 577(80), 559(51), 451(31), 441(29), 433(18), 407(28), 289(15), 245(17) | |
| (EC→EC)2g | G(2) | 881.1541 | C44H33O20 | 729(100), 711(26), 559(20), 407(23) | |
| trimer | EA→EA→EC(1) | M(1) | 833.2083 | C45H37O16 | 816(23), 707(81), 561(91), 543(100), 435(23), 289(35) |
| EA→A→EC→EC(1) | M(1), L(1) | 847.1853 | C45H37O17 | 711(30), 693(12), 557(34), 435(37), 411(100), 289(13) | |
| EA→EC→EC(1) | M(5) | 849.2026 | C45H37O17 | 723(31), 697(31), 577(100), 571(15), 559(51), 451(17), 425(28), 407(23),289(9), 287(15) | |
| EA→EC→EC(2) | G (4) | 849.2014 | C45H37O17 | 831(94), 723(69), 697(26), 679(79), 561(100) | |
| EA→EC→EC(3) | M (2) | 849.2017 | C45H37O17 | 831(45), 723(68), 697(16), 679(70), 561(38), 559(100), 433(36), 407(50), 289(19) | |
| EA→EC→EC(4) | M (2) | 849.2204 | C45H37O17 | 723(100), 697(37), 679(49), 577(51), 571(39), 561(39), 451(43), 425(24), 407(37), 289(32) | |
| EA→EC→EC(5) | G(2) | 849.2010 | C45H37O17 | 831(16), 723(30), 697(100), 679(71), 561(14), 545(12) | |
| EA→EC—EC(6) | G(2) | 849.2018 | C45H37O17 | 831(100), 723(61), 679(35), 561(41) | |
| EF→EC→EC(7) | G(2) | 849.2010 | C45H37O17 | 739(21), 697(100), 679(59), 559(67), 545(26), 527(16), 451(12), 407(11), 397(17), 289(13) | |
| EC→EC→A→EC(1) | L(3), M(6), C(1) | 863.1814 | C45H35O18 | 737(72), 711(62), 693(42), 591(69), 575(100), 573(58), 449(34), 439(32), 289(89), 287(67) | |
| EC→EC→A→EC(2) | M(2) | 863.1823 | C45H35O18 | 845(13), 737(19), 711(100), 693(41), 575(94), 573(15), 451(23), 411(17) | |
| EC→A→EC→EC(1) | C(2) | 863.1804 | C45H35O18 | 737(8), 711(100), 693(8), 575(9), 573(41), 559(7), 531(10), 451(47), 411(43), 299(6), 289(19), 285(7) | |
| EC-(4β-8)-EC-(4β-8)-EC(2) | C(1), A(4) | 865.1971 | C45H37O18 | 847(18), 749(48), 695(100), 577(68), 575(31), 425(27), 407(30) | |
| EC-(4β-8)-EC-(4β-8)-EC(2) | M(12), G(10), J(7), D(3), L(2) | 865.1971 | C45H37O18 | 847(18), 749(48), 695(100), 577(68), 575(31), 425(27), 407(30) | |
| EC→EC→EC(3) | M(4), G(1), A(2), D(2) | 865.1961 | C45H37O18 | 847(40), 779(51), 739(56), 713(57), 695(68), 577(89), 575(100), 449(22), 407(35), 289(27), 287(24) | |
| EC→EC→EC(4) | M(1) | 865.1939 | C45H37O18 | 801(41), 789(49), 779(100), 720(70), 695(51), 577(74), 575(55) | |
| EC→EC→EC(5) | J(2), D(2) | 865.1955 | C45H37O18 | 847(38), 739(100), 713(58), 695(87), 577(64), 575(35), 451(37), 449(26), 407(30), 287(29) | |
| (EC→EC→EC)g(1) | G(4) | 1017.2069 | C52H41O22 | 999(31), 891(47), 865(40), 847(57), 739(19), 729(100), 727(23), 695(28), 677(32), 575(20) | |
| (EC→EC→EC)g(2) | G(1) | 1017.2054 | C52H41O22 | 999(100), 891(48), 865(50), 847(62), 729(40), 695(39), 677(25) | |
| (EC→EC→EC)g(3) | G(2) | 1017.2054 | C52H41O22 | 999(19), 891(54), 865(33), 847(100), 729(83) | |
| (EC→EC→EC)g(4) | G(3) | 1017.2056 | C52H41O22 | 999(20), 891(24), 865(100), 847(53), 727(24), 695(24) | |
| (EC→EC→EC)→2g(1) | G(1) | 1169.2184 | C59H45O26 | not recorded | |
| tetramer | EC→EA→A→EC →EC(1) | L(1) | 1135.2472 | C60H47O23 | 983(36), 965(22), 847(100), 845(30), 829(11), 693(26), 557(22), 411(15) |
| EA→EC→EC→EC(1) | M(3) | 1137.2649 | C60H49O23 | 1119(42), 1011(59), 865(100), 847(51), 739(26), 577(46), 559(33), 407(26) | |
| EA→EC→EC→EC(2) | M(3) | 1137.2666 | C60H49O23 | 1119(42), 1011(67), 985(35), 967(85), 849(87), 847(100), 723(36), 575(32), 561(32) | |
| HA→EC→HC→EC(3) | M(1) | 1137.2651 | C60H49O23 | 1119(32), 1011(56), 985(41), 967(62), 849(74), 847(88), 577(100), 559(58), 407(50) | |
| EC→A→EC→EC→A→EC(1) | L(2), C(1) | 1149.2268 | C60H45O24 | 997(58), 997(19), 979(34), 845(43), 737(22), 575(100), 573(85), 411(85) | |
| EC→A→EC→EC→A→EC(2) | L(1) | 1149.2285 | C60H45O24 | 1131(20), 997(75), 979(35), 845(24), 737(17), 575(80), 573(75), 411(100) | |
| EC→EC→A→EC→EC(1) | L(3) | 1151.2423 | C60H47O24 | 1133(14), 1025(41), 999(45), 981(87), 863(100), 861(45), 711(32), 573(41), 411(34) | |
| EC→A→EC→EC→EC(2) | L(2), C(1) | 1151.2419 | C60H47O24 | 1005(32), 999(48),981(48), 861(100), 739(68), 573(58), 611(61), 407(35) | |
| EC→A→EC→EC→EC(3) | L(1) | 1151.2419 | C60H47O24 | 1133(32), 999(81), 981(70), 863(43), 861(84), 739(100), 699(38), 577(72), 573(49), 411(39), 407(43) | |
| EC→A→EC→EC→EC(3) | L(1) | 1151.2421 | C60H47O24 | 1133(45), 999(100), 981(48), 863(35), 861(90), 739(83), 587(39), 577(45), 573(59), 411(87), 407(38) | |
| EC→EC→EC→A→EC(4) | L(1) | 1151.2416 | C60H49O24 | 999(78), 981(100), 863(86), 861(76), 739(70), 709(35), 577(38), 573(57), 531(38), 451(30),411(38) | |
| EC→EC→EC→.A→EC(5) | L(1) | 1151.2419 | C60H47O24 | 1133(33), 1067(12), 1025(49), 999(21), 981(100), 863(57), 739(12), 737(15), 711(30), 575(48) | |
| EC→EC→EC→EC(1) | J(12), G(3), M(3), A(1), D(2) | 1153.2571 | C60H49O24 | 1135(54), 1027(74), 1002(42), 983(100), 907(21), 865(63), 863(62), 739(35), 695(32), 577(40), 407(21) | |
| EC→EC→EC→EC(2) | D(2), J(2), M(2), G(2) | 1153.2565 | C60H49O24 | 1135(55), 1027(52), 1027(23), 1001(59), 983(96), 865(100), 863(48), 695(21), 577(46), 575(54) | |
| EC→EC→EC→EC(3) | J(2) | 1153.2582 | C60H49O24 | 1135(53), 1027(41), 1001(100), 984(71), 865(79), 863(94), 847(26), 739(50), 701(35), 577(44), 575(65) | |
| EC→EC→EC→EC(4) | J(2) | 1153.2577 | C60H49O24 | 1135(55), 1027(64), 1001(50), 983(77), 907(41), 865(50), 863(100), 701(32), 577(55), 575(73), 407(27) | |
| EC→EC→EC→EC(5) | J(1) | 1153.2577 | C60H49O24 | 1135(89), 1027(78), 1001(56), 983(44), 907(44), 865(44), 863(44), 739(67), 701(67), 577(33), 575(100) | |
| EC→EC→EC→EC(6) | J(2), A(1), D(1) | 1153.2590 | C60H49O24 | 1135(48),1027(100), 1001(30), 983(83), 965(16), 908(36), 865(52), 739(55), 695(26), 575(31) | |
| EC→EC→EC→EC(7) | A(1) | 1153.2590 | C60H49O24 | 1135(100), 1028(72), 983(50), 865(50), 739(39), 737(33) | |
| pentamer | EC→EC→EC→EC→A→EC(1) | L(2) | 1439.3058 | C75H59O30 | 1421(50), 1313(50), 1295(50), 1149(90), 1007(50), 863(100), 861(50) |
| EC→EC→EC→EC→EC(2) | L(1) | 1439.3058 | C75H59O30 | 1421(67), 1287(100), 1151(67), 1113(33), 863(100), 753(100), 711(67), 637(33), 411(33) | |
| EC→EC→EC→EC→EC(3) | L(1) | 1439.3058 | C75H59O30 | 1379(100), 1353(75), 1313(75), 1269(50), 1131(50), 1111(50), 863(75), 857(50), 751 (25) | |
| EC→EC→EC→EC→EC(4) | L(1) | 1439.3058 | C75H59O30 | 1421(100), 1089(33), 1013(67), 997(33), 863(67), 711(33), 589(33), 587(67), 575(33), 531(67) | |
| EC→EC→EC→EC→A→EC(5) | L(1) | 1439.3058 | C75H59O30 | 1421(33), 1395(33), 1285(33), 981(100), 863(50), 665 (50), 445(33) | |
| EC→EC→EC→EC→A→EC(6) | L(2) | 1439.3058 | C75H59O30 | 1314(17), 1269(100), 1149(17), 1117(33), 863(67), 817(17), 737(17), 709(17), 575(17), 453(33) | |
| EC→EC→EC→EC→EC(7) | L(1) | 1439.3058 | C75H59O30 | 1441(14), 1421(43), 1269(100), 1151(14), 1107(14), 957(14), 955(29), 863(43), 829(14), 573(14), 531(14) | |
| EC→EC→A→EC→EC→EC(S) | L(1) | 1439.3058 | C75H59O30 | 1371(50), 1269(50), 1143(50), 987(50), 861(50), 711(100), 671(50), 585(50), 575(50), 573(90), 411(50) | |
| EC→EC→EC→EC→EC(1) | M(1) | 1441.3229 | C75H61O30 | 1421(100), 1315(64), 1271(67), 1153(74), 1153(33), 1151(48), 1027(38), 865(86), 863(43),739(36), 575(36) | |
Abbreviations: A, apple; C, cranberry extract; D, dark chocolate; G, grape seed extract; J, jujube; L, litchi; M, mangosteen; AL, all tested plants (the number of similar peaks in the sample is listed in parentheses); B1, B2, A2, and C1, procyanidin B-type dimers B1, B2, A-type dimers A2, and trimer C1; DP, degree of polymerization; g, galloyl; EC, EA, EG, and EF, epicatechin, epiafzelechin, epigallocatechin, and epifisetinidol, respectively. The signal unit1→unit2 or unit1→A→unit2 expresses the units bonded by B-type (4,8- or 4,6-) bond or A-type (plus additional C2–O–C7– or C2–O–C5−) bond, respectively.
It was noted that catechin and epicatechin showed the same product ions and very similar ratios at MS2 [245 (100%), 205 (35%), and 179 (11–12%)], MS3 [227 (28–30%), 203 (100), and 187 (20–25%)], and even at MS4 [185, (20–37%), 175 (100%), 161 (28–42%)]. Similarly, dimeric procyanidins B1 (EC-4β-8-C) and B2 (EC-4β-8-EC) have very similar MS2 [451 (27–37%), 425 (100%), 407 (41–47%), 289 (17–26%), and 287 (7–8%)], MS3 [407 (100%) and 273 (6–8%)], MS4 [285 (100%), 283 (36–43%), 389 (29–36%), 297 (27–37%), and 255 (17–27%)] and MS5 [257 (100%) and 213 (4–7%)] fragments.
These data indicate that the slight differences in the relative ratio among the fragments might be caused by the stereochemistry of the monomers. However, there are insufficient data to predict the effect of the linkages and the positions of the PA flavan-3-ol units on product ion formation and relative abundance. At present, LC-MSn methods are not able to discriminate between the regioisomeric forms of the PAs or the related stereoisomeric forms.
As shown in Figure 1, the most important MS2 fragments of B- and A-type PA dimers are formed by quinone methide (QM) fission, that is, breaking of the interflavan bond between the monomers to form [MT – H]− and [ME – 3H]− ions for B-type PAs and [ME – 5H] – ions for A-type PAs, where E = extension unit and T = terminal unit. Other typical PA fragments were formed by retro-Diels–Alder (RDA) fission (loss of the whole B-ring with C2–C3 part of the C-ring, i.e., loss of 152, 136, and 168 Da for EC, EA, and EG, respectively) and by heterocyclic ring fission (HRF) of the extension unit (loss of the A-ring, i.e., loss of 126 Da for EC, EA, or EG and loss of 110 Da for EF or ER). Product ions formed by the loss of water (−18 Da), O (−16 Da), CO (−28 Da), HC≡CH (−26 Da), HC≡COH (−42 Da), and HC≡CH—CO (−54 Da) were also observed.4,13,14,19−23,27 For PAs with DP ≥ 3, further fragmentation can occur from repeated QM breaks of interflavan bonds connecting the flavan-3-ols of the extension units. These product ions were also frequently used to identify the PAs. Information obtained from the analysis of thiolytic degradation products of the PAs from similar plants has proven useful for the identification of PAs.1,4,13,14,19−23,27−30
In this study, 247 PAs were identified in 7 tested materials (Table 2) using only the most intense ions among the coeluted ions (each peak) as the target ions. However, the identified PAs can be enhanced by selecting more target ions, such as the second and third most intense ions of each peak. The PAs are denoted as combinations of EC, EA, EG, and EF, and A-type bonds are designated by placing an “-A-” between the flavan-3-ols. Although there are numerous isomers in each oligomeric class, only one isomer was selected to represent all of the remaining isomers (each having the same MS2 base and main fragments). There was no correlation between the PAs found in the different samples. Positive identification was achieved for only some of the PAs in Table 2 based on direct comparison to reference PAs (procyanidins B1, B2, C1, A2) or PAs positively identified in other studies.1,4
Procyanidins with DP = 2–5 have been previously reported in common foods.4,19−23,27 Consequently, special attention was paid to PAs containing different flavan-3-ol units or A-type bonds because these features lead to more regioisomers. For example, 13 PA dimers ([M – H]− at 561.1388) contained two different flavan-3-ols. One of the three detected in mangosteen had MS2 fragments at m/z 435 (−126 Da, HRF from EA or EC), 409 (−152 Da, RDA from EA or EC), 287 ([ME – 3H] –), and 273 ([MT – H]−), suggesting it to be EC-EA, a PA dimer containing an EA unit as the terminal unit. The other two in mangosteen had MS2 fragments at m/z 289 ([MT – H]−, 100%), 435 (−126 Da), 425 (−136 Da), 407 [−(136 + 18) Da), and 271 ([ME – 3H]−), corresponding to EA-EC, the isomers containing EA as extension unit (Figure 1).
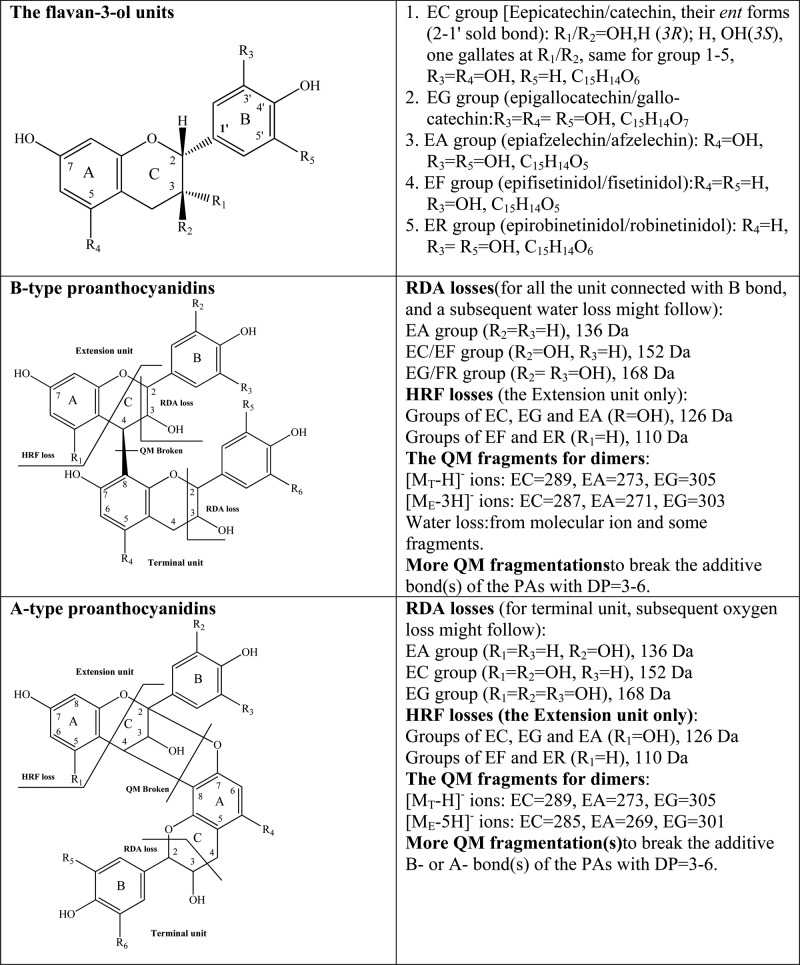
Another 10 interesting dimers were detected in grape seed extract. Three had MS2 fragments at m/z 451 (HRF loss of 110 Da, C6H6O2 for the deoxy-A-ring of EF or ER, 100%), 423 [−(110 +28), 91–98%], 409 (−152 Da, RDA loss, 29–60%), 289 ([MT – H] –, 12–18%), and 271([ME – 3H] –, 20–48%) (Figure 2). Five had the same MS2 fragments but with different intensities; one at m/z 423 (100%), 451 (around 50%), 409 (28–64%), 289, and 271. The remaining two had MS2 fragments at m/z 435 (100) and 451 (40–50%). These fragments suggested that all might be proanthocyanidin dimeric isomers (EF-EC). To date, the PAs containing an EF unit have only been reported to exist in plant woods, such as quebracho (Schinopsis balansae var. chaqueno) wood, but rarely in common foods, such as grapes.1,28−31
Figure 2.
MS2 spectrum of the EF–EC dimer with retention time at 26.98 min.
A PA dimer ([M – H]− at 593.1279 Da) detected in grape seed extract had mass fragments at m/z 441 (−152, RDA) and 305 ([MT – H] –), suggesting it was an EC-EG isomer. A PA trimer containing two EAs ([M – H]− at m/z 833.2083) was found in mangosteen with MS2 fragments at m/z 543 (100%, [ME – 3H]−), 707, and 289 ([MT – H]−) and MS3 fragments at m/z 271 (100%, the second [ME – 3H] –), 417, and 407, indicating that two EA units were formed the extension units and that the EC was the terminal unit. Two PA trimers found in mangostenn and litchi with [M – H]− of 847.1860 Da had one A-type bond, MS2 fragments at m/z 557, 411, and 289, and a MS3 base fragment at m/z 285. This suggested that EA was the extension unit with the A-type bond connected to one of the two ECs and that the remaining EC was the terminal unit.
Nineteen PA trimers (9 in mangosteen and 10 in grape seed extract) had [M – H]− fragments at 849.2030 Da indicating EA or its constitutional isomer EF. Of these, 5 had MS2 fragments at 577 (−272 Da, 100%) and 559 (−290 Da around 40–50%), 1 had MS2 fragments at m/z 561 (−288 Da), and the others had MS2 fragments at 559 (−290 Da), 723 (−126 Da), 697 (−152 Da), and 831 (−18 Da). All of these PAs might be EA-EC-EC isomers. The two detected in grape seed extract (expressed as EF-EC-EF in the last line for this oligomeric class) had a MS2 fragment at m/z 739 (−110 Da, ∼20%) and might have EF instead of EA as the extension unit.
Fourteen procyanidin trimers ([M – H]− at 863.1800 Da) contained one A-type bond. Ten of them (6 from mangosteen, 3 from litchi, and 1 from cranberry) had MS2 fragments at m/z 575 (−288 Da, 21–100%), 711 (42–100%), and 289 (20–89%), indicating they were EC-EC-A-EC isomers. Others (two from litchi and two from cranberry) had MS2 fragments at m/z 573 (−290 Da, 35–62%), 411 (43–100%), and 711 (91–100%) to suggest they were EC-A-EC-EC isomers.
One A-type PA tetramer in litchi ([M – H]− at 1135.2472) contained one EA and one A-type bond and had MS2 fragments at m/z 847 (−288 Da, 100%), 983 (−152 Da, 36%), 845 (−290 Da, −30%), 693 [−(152 + 290) Da, 26%], and 557 [−(288 + 290) Da, 22%]. This suggested it might be an EC-EA-A-EC-EC or EC-EC-A-EA-EC isomer.
Seven PA tetramers in mangosteen ([M – H]− at 1137.2450) contained one EA. Three had MS2 fragments at m/z 865 (−272 Da, 100%), 847 (−290 Da, 30%), and 577 [−(288 + 272) Da, 46%]. Another three had MS2 fragments at m/z 847 (−290 Da, 100%) and 575 [−(288 + 274 or 290 + 272) Da]. The remaining tetramer had MS2 fragments at m/z 1011, 985, 967, 849 (−288 Da), 847, and 577 [−(290 + 274) Da]. These fragments indicated EA was a part of the extension unit with two ECs and might be the final extension unit.
Four procyanidin tetramers (three from Litchi and one from cranberry) with [M – H]− at 1149.2280 had two A-type bonds and MS2 fragment at m/z 575 (80–100%) {−(288 + 286) Da for [MT – H]−} and 573 (75–85%) {−(2 +286 × 2) Da for [ME – 3H]−}, indicating that the A-type bonds were between the first and second and between the third and fourth flavan-3-ols. Ten procyanidin tetramers (9 from Litchi and 1 from cranberry) with [M – H]− at 1151.2415 had one A-type bond. Three (group 1) had MS2 fragments at m/z 863 (100%) (−288 Da for [ME – 3H]−) and 573 (41%) indicating the A-type bond was between the second and third flavan-3-ols. Five (groups 2–4) had MS2 fragments at m/z 861 (84–100%) (−290 Da for [MT – H]−) and 573 (49–59%) {−(2 +286 × 2) Da for [ME – 3H]−} indicating an A-type bond between the first and second flavan-3-ols. Two (groups 4 and 5) had MS2 fragments at m/z 863 (57–86%), 575 (48%) {−(288 + 286) Da for [MT – H]− }, or 577 (38%) and 573 (57%) indicating an A-type bond between the third and fourth flavan-3-ols.14,19 Similarly, the PA pentamers in eight groups (1–8) have one A-type bond, and the PAs of the first six groups (1–6) showed main fragment at m/z 863 (50–100%), indicating the A-type bond between the fourth and fifth flavan-3-ols. The PS of the last group (8) showed the main fragment at m/z 861 (50%), indicating the A-type bond between second and third flavan-3-ols, whereas the remaining one in group 7 showed fragments at m/z 863 and 573 to suggest that this compound might have its A-type bond between the third and fourth flavan-3-ols.14
Ten galloylated dimers and 11 trimers were detected in grape seed extract. The existence of a galloyl connected to a PA with DP ≥ 2 provides the possibility of forming regioisomers; for example, EC-ECg and ECg-EC and EC-EC-ECg, EC-ECg-EC, and ECg- EC-EC. Unfortunately, the ECg position cannot be deduced from the mass fragments because gallate was very easy to lose. Thus, they were expressed as (EC-EC)g or (EC-EC-EC)g, respectively.
Jujube fruit was analyzed because PAs (DP = 2, 3, 5, and 7) consisting of both EA and EG have been isolated from jujube leaves and bark.32 These PAs have the same molecular weight and formula as those of their related procyanidins, but can be easily distinguished from the procyanidins by the noticeable difference in their fragments. For example, the dimers of EA and EG will have QM (271 and 305 Da for EA-EG or 303 and 273 Da for EG-EA) and RDA fragments formed by the loss of 136 Da from EA and 168 Da from EG, whereas the related procyanidin dimers should have QM (289 and 287 Da) and RDA fragments formed by the loss of 152 Da. A careful check confirmed that all 30 of the detected PAs in jujube consisted of EC units only.
Identification of Highly Polymerized PAs Based on the Doubly and Triply Charged Molecular Ions
Negative ionization of many highly polymerized PAs (DP ≥ 5) produces multiply (mainly doubly and triply) charged molecular ions. To date, several dozen multiply charged molecular ions have been reported and used to identify PAs with DP = 7–25.1,4,13−19 With nominal resolution MS, these ions were recognized as doubly or triply charged molecules on the basis of the distance between the 12C and 13C isotope ions. As the charge increases from 1 to 2 to 3, the distance between the isotopes will decrease from 1 to 0.5 to 0.33 amu.17,18 It was noted that the ion masses for PA isotopes were always slightly different.1,4,13−20 This was attributed to the differences in the relative abundances of the 12C and 13C isotopes.
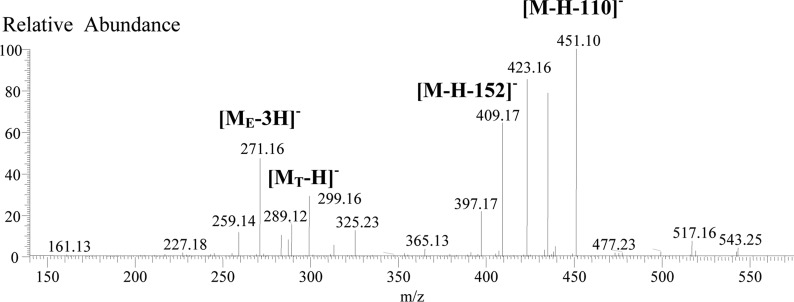
Table 1 contains the accurate [M – 2H] 2– and [M – 3H]3– values for PAs with DP = 5–10, which matched the [M – 2H]2– or [M – 3H]3– of around 50 proanthocyanidins detected in mangosteen and litchi extracts (Table 3). The 12C and 13C isotope ions of each proanthocyanidin were easily found by examining the distance between the two isotopic ion peaks. For example, in mangosteen the main [M – 2H]2– ions were found at m/z 720.1566, 856.1928, 864.1863, 1000.2235, 1008.7217, and 1152.7537 (Figure 3; Tables 1 and 3). The first four values were taken from the 12C isotope ion and perfectly matched (error < 3 ppm) the listed [M – 2H]2– data in Table 1 for the B-type procyanidin pentamer and hexamer, the B-type propelargonidin hexamer containing one EA unit, and the B-type propelargonidin heptamer containing two EAs. The values of the 13C isotope were m/z 0.500 more than that from 12C isotope (Table 3). However, the last two values, m/z 1008.7217 and 1152.7537, were taken from the 13C isotope ions of B-type procyanidin heptamer and octamer, respectively, so these masses were larger than the listed [M – 2H]2– values for the 12C isotope ion by 0.50 amu.
Table 3. Doubly and Triply Charged Proanthocyanidins Found in Mangosteen and Litchi.
| proanthocyanidin | HRMS (Da) | HR [M – 2H]2– (m/z) | 12C isotope (m/z) | 13C isotope (m/z) | plant sourcea (no. of PAs) |
|---|---|---|---|---|---|
| A-type procyanidin pentamers with two A-bonds | 1438.2994 | 718.1419 | 718.1417 | 718.6428 | L(2) |
| A-type procyanidin pentamers with one A-bond | 1440.3150 | 719.1497 | 719.1494 | 719.6509 | M(1), L(1) |
| B-type procyanidin pentamers | 1442.3306 | 720.1575 | 720.1566 | 720.6591 | M(5) |
| B-type proanthocyanidin hexamers with two EA units | 1698.4038 | 848.1941 | 848.1951 | 848.6995 | M(2) |
| B-type proanthocyanidin hexamers with one EA unit | 1714.3987 | 856.1916 | 856.1921 | 856.6930 | M(6) |
| A-type procyanidin hexamers with two A-bond | 1726.3624 | 862.1734 | 862.1740 | 862.6748 | L(2) |
| B-type procyanidin hexamers | 1730.3936 | 864.1890 | 864.1893 | 864.6890 | M(3) |
| B-type proanthocyanidin heptamers with one EA units | 2002.4617 | 1000.2231 | 1000.2235 | 1000.7247 | M(1) |
| A-type procyanidin heptamers with two A-bond | 2014.4254 | 1006.2049 | 1006.0000 | 1006.7057 | L(1) |
| A-type procyanidin heptamers with one A-bond | 2016.4410 | 1007.2127 | 1007.2120 | 1007.7230 | M(1), L(1) |
| B-type procyanidin heptamers | 2018.4566 | 1008.2205 | 1008.2223 | 1008.7228 | M(6) |
| B-type proanthocyanidin octamers with two EA units | 2274.5298 | 1136.2571 | 1136.2540 | 1136.7643 | M(2) |
| B-type procynidin octamers | 2306.5196 | 1152.2520 | 1152.2527 | 1152.7537 | M(7) |
| B-type proanthocyanidin nonamers with one EA unit | 2578.5877 | 1288.2861 | 1288.2840 | 1288.7855 | M(1) |
| B-type procyanidin nonamers | 2594.5826 | 1296.2835 | 1296.2828 | 1296.7859 | M(1) |
| B-type proanthocyanidin decamers with two EA units | 2850.6558 | 1424.3201 | 1424.3212 | 1424.8262 | M(1) |
| B-type proanthocyanidin decaamers | 2866.6507 | 1432.3176 | 1432.3185 | 1432.8220 | M(1) |
| A-type procyanidin decamers with two A-bond | 2878.6144 | 1438.2994 | 1438.2985 | 1438.7967 | M(1) |
| B-type procyanidin decamers | 2882.6456 | 1440.3150 | 1440.3169 | 1440.8147 | M(1) |
| HR [M – 3H]3–m/z | 12C isotope m/z | 13C isotope m/z | |||
|---|---|---|---|---|---|
| B-type procyanidn decamers | 959.8741 | 959.8727 | 960.2103 | M(4) |
Abbreviations: L, litchi; M, mangosteen (number of similar peaks in the sample is listed in parentheses). The value was taken from one of the PA and close to those of the remaining ones.
Figure 3.
Accurate 12C and 13C isotope ion peaks for [M – 2H]2– of m/z 720 and 1152 and for [M – 3H]3– of m/z 960.
Checking the distance between isotopes led to the detection of several minor PA ions in the TIC chromatogram of mangosteen extract. For example, the ions at m/z 1296.2828, 1007.2120, 1136.2540, and 1288.2840 were close matches to the listed values for doubly charged B-type procyanidin octamers, A-type procyanidin heptamers with one A bond, B-type propelargonidin octamers with two EA, and B-type propelargonidin nonamers with one EA (Tables 1 and 3), respectively.
Similarly, checking for 12C and 13C isotopes with a 0.33 amu distance led to the discovery of several [M – 3H]3– ions. However, only one of them (in mangosteen) was for a PA with a DP ≤ 10. As shown in Table 3 and Figure 3, the HRMS values for this PA for the 12C and 13C isotope ions were 959.8727 and 960.2103 (Figure 3), respectively. To date, only five multiply changed ions have been reported in the pericarps of mangosteen.12 This is the first report to use the high-resolution isotope ion values for accurate identification of multiply charged PAs based on the use of 12C and 13C isotope ions.
Quantification of Proanthocyanidin Oligomers
The extraction efficiency of the standardized method for PAs in plant materials was determined by a follow-up extraction using acetone/methanol/water (2:2:1, v/v/v), a solvent frequently used for PA extraction in other studies.2−4,21,22 No additional material was found in the follow-up extractions as determined by the lack of detectable peaks. This indicated that the general extraction method was suitable for the quantification of PAs in jujube, Fuji apple, litchi, and mangosteen.
The UV absorbance of phenolic compounds is widely used for the quantification of PAs.2−4,7,19,20,22 The MRRF of flavan-3-ol (catechin and epicatechin) monomers, dimeric procyanidin B1, B2, and A2, and trimeric procyanidin C1 at 274–280 nm were found to be proportional to the DP number in our previous study.25 This established that, in molar units, the response of the monomers was additive. The MRRF values for catechin, gallocatechin, and gallic acid were determined to be 1.00, 0.31, and 2.8.25 Thus, the MRRF for EC-EC is 2.0, that for EC-EC-A-EC is 3.0, and that for EC-EC-ECg is 4.8. There were no commercial standards for afzelechin or fisetinodol, so an MRRF value of 1.00 was assigned to each. The additivity of the molar absorption coefficient makes it possible to quantify most of the PAs using (+)-catechin as a standard with the MRRF values listed above.
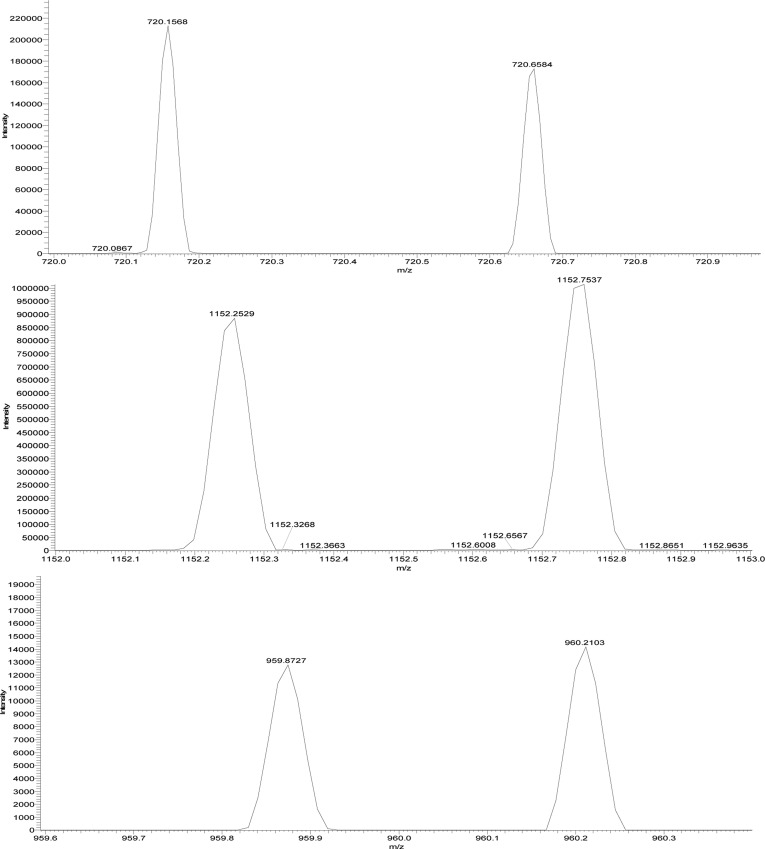
Unfortunately, even with UHPLC, only a few PAs were well separated and could be quantified on the basis of their UV peak area. Most PAs, when viewed with UV or TIC, had peaks that overlapped (coeluted) with other PAs. Selected ion monitoring (SIM) and multiple reaction monitoring (MRM) are the only methods that allow deconvolution of the overlapping peaks, that is, isolation of the ions of interest.33,34 Consequently, concentrations had to be computed on the basis of ion counts obtained from SIM or MRM as reported in previous studies19,22 The few well-separated absorbance peaks were used to equate the peak area in absorbance to integrated counts of specific ions. In other words, MRRF values based on absorbance were converted to MRRF values based on integrated ion counts. This approach allowed catechin and the MRRF values reported above to be used for computing PA concentrations.
Use of MRRFs based on ion counts assumes constant ionization efficiency for all PAs. Unlike absorbance, the ion count of a PA isomer can be expected to be dependent on its structural ionization sensitivity and the mobile phase. The SIM peak intensity might change with the solvent ratio at different retention times, the isomer concentration, and the presence of coeluting PAs (Figure 4). Tests performed with flavan-3-ol monomers and procyanidins B1 and B2 showed the variation in ionization efficiency to be <±10%. Further testing with PAs with DP = 3–5, A-type bond, or galloyls is needed but must wait on the availability of suitable standards.
Figure 4.
PDA (at 278 nm) and SIM chromatograms of grape seed extract.
The PA concentrations in dry weight percent (%) and milligrams per 100 g of dry plant material were calculated using the formulas
| 6 |
| 7 |
where Ax, MWx, Wx, and Vx and As, MWs, Ws, and Vs are the peak area, molecular weight, sample weight, and volume of the extract for the sample and standard, respectively.
As shown in Table 4 for grape seed extract, at least one PA in each of the oligomers was found to have a well-separated peak (no coeluting compounds) that could be used to equate absorbance with ion counts from SIM. The concentrations of monomers, dimers, trimers, and tetramers as percent dry weight were 16.63 ± 0.67, 17.44 ± 0.70, 14.24 ± 0.57, and 0.47 ± 0.20%, respectively. The concentration for PAs with DP > 4 was negligible. The total concentration of PAs was 48.79 ± 1.95%.
Table 4. Retention Time, Molecular Weight, MRRF Value, and Concentration for the Main PAs in Grape Seed Extract.
| compound (or code) (min) | tR (UV) (min) | tR(SIM) (min) | MWx | MRRF | content (%, w/w on dry basis), av ± SD |
|---|---|---|---|---|---|
| catechin | 8.51 | 290 | 1.0 | 6.55 ± 0.26 | |
| epicatechin | 13.30 | 290 | 1.0 | 7.58 ± 0.30 | |
| epicatechin-gallate | 23.34 | 442 | 3.8 | 2.34 ± 0.09 | |
| catechin-gallate | 25.25 | 442 | 3.8 | 0.16 ± 0.01 | |
| monomer concentration | 16.63 ± 0.67 | ||||
| proanthocyanins | |||||
| EF-EC-5 | 30.09 | 30.18 | 562 | 2.0 | 0.07 ± 0 |
| EF-EC-1 | 24.15 | 562 | 2.0 | 0.05 ± 0 | |
| EF-EC-2 | 25.05 | 562 | 2.0 | 0.08 ± 0 | |
| EF-EC-3 | 2619 | 562 | 2.0 | 0.08 ± 0 | |
| EF-EC-4 | 26.98 | 562 | 2.0 | 0.11 ± 0 | |
| EF-EC-6 | 31.34 | 562 | 2.0 | 0.03 ± 0 | |
| EF-EC-7 | 32.00 | 562 | 2.0 | 0.05 ± 0 | |
| EF-EC-8 | 33.18 | 562 | 2.0 | 0.11 ± 0 | |
| EF-EC-9 | 33.46 | 562 | 2 0 | 0 05 ± 0 | |
| EF-EC-10 | 34.78 | 562 | 2.0 | 0.03 ± 0 | |
| EC-EC-1 | 7.76 | 7.83 | 578 | 2.0 | 2.53 ± 0.10 |
| EC-EC-2 | 8 19 | 578 | 2.0 | 1.88 ± 0.08 | |
| EC-EC-3 | 9.86 | 578 | 2.0 | 0.22 ± 0.01 | |
| EC-EC-4 | 10.81 | 578 | 2.0 | 0.87 ± 0.03 | |
| EC-EC-5 | 11.46 | 578 | 2 0 | 0.19 ± 0.01 | |
| EC-EC-6 | 12.10 | 578 | 2.0 | 2.41 ± 0.10 | |
| EC-EC-7 | 16.59 | 578 | 2.0 | 1.60 ± 0.06 | |
| EC-EC-S | 18.91 | 578 | 2.0 | 1.07 ± 0.04 | |
| EC-EC-9 | 26.64 | 578 | 2.0 | 1.18 ± 0.05 | |
| EC-EG | 6.41 | 594 | 2.0 | 0.01 ± 0 | |
| (EC-EC)g-4 | 18.83 | 18.88 | 730 | 4.8 | 1.61 ± 0.06 |
| (EC-EC)g-1 | 15.43 | 730 | 4.8 | 0.53 ± 0.02 | |
| (EC-EQg-2 | 16.59 | 730 | 4.8 | 1.13 ± 0 | |
| (EC-EC)g-3 | 17.40 | 730 | 4.8 | 0.54 ± 0.02 | |
| (EC-EC)g-5 | 27.04 | 730 | 4.8 | 0.23 ± 0.01 | |
| (EC-EC)g-6 | 34.23 | 730 | 4.8 | 0.21 ± 0.01 | |
| (EC-EC)2g-1 | 24.79 | 24.86 | 882 | 7.6 | 0.57 ± 0 |
| dimer concentration | 17.44 ± 0.70 | ||||
| EA-EC-EC-7 | 30.72 | 30.76 | 850 | 3.0 | 0.04 ± 0 |
| EF-EC-EC-1 | 18.55 | 850 | 3.0 | 0.03 ± 07 | |
| EA-EC-EC-2 | 19.67 | 850 | 3.0 | 0.04 ± 0 | |
| EF-EC-EC-3 | 22.80 | 850 | 3.0 | 0.03 ± 0 | |
| EA-EC-EC-4 | 23.67 | 850 | 3.0 | 0.04 ± 0 | |
| EA-EC-EC-5 | 28.04 | 850 | 3.0 | 0.03 ± 0 | |
| EA-EC-EC-6 | 30.03 | 850 | 3.0 | 0.02 ± 0 | |
| EC-EC-EC-4 | 12.04 | 12.28 | 866 | 3.0 | 2.25 ± 0.09 |
| EC-EC-EC-1 | 3.65 | 866 | 3 0 | 1.95 ± 0.08 | |
| EC-EC-EC-2 | 10.04 | 866 | 3.0 | 1.92 ± 0.08 | |
| EC-EC-EC-3 | 11.21 | 866 | 3.0 | 0.78 ± 0.03 | |
| EC-EC-EC-5 | 16.83 | 866 | 3.0 | 1.35 ± 0.05 | |
| EC-EC-EC-6 | 17.65 | 866 | 3.0 | 2.34 ± 0.09 | |
| EC-EC-EC-7 | 20.76 | 866 | 3.0 | 0.89 ± 0.04 | |
| EC-EC-EC-8 | 26.08 | 866 | 3.0 | 1.73 ± 0.07 | |
| (EC-EC-EC)g-2 | 14.14 | 14.21 | 1018 | 5.8 | 0.14 ± 0.01 |
| (EC-EC-EC)g-1 | 9.62 | 1018 | 5.8 | 0 06 ± 0 | |
| (EC-EC-EC)g-3 | 13.43 | 1018 | 5.8 | 0.06 ± 0 | |
| (EC-EC-EC)g-4 | 20.29 | 1018 | 5.8 | 0.16 ± 0.01 | |
| (EC-EC-EC)g-5 | 25.38 | 1018 | 5 8 | 0.07 ± 0 | |
| (EC-EC-EC)g-6 | 26.68 | 1018 | 5.8 | 0.12 ± 0 | |
| (EC-EC-EC)g-7 | 27.46 | 1018 | 5.8 | 0.08 ± 0 | |
| (EC-EC-EC)g-8 | 32.21 | 1018 | 5 8 | 0.10 ± 0 | |
| (EC-EC-EC)2g-1 | 28.23 | 1170 | 8.6 | trace | |
| trimer concentration | 14.24 ± 0.57 | ||||
| EC-EC-EC-EC-5 | 19.34 | 19.40 | 1154 | 4.0 | 0.23 ± 0.01 |
| EC-EC-EC-EC-1 | 9.62 | 1154 | 4.0 | 0.06 ± 0 | |
| EC-EC-EC-EC-2 | 10.63 | 1154 | 4.0 | 0.09 ± 0 | |
| EC-EC-EC -EC-3 | 14.81 | 1154 | 4.0 | 0.06 ± 0 | |
| EC-EC-EC-EC-4 | 17.31 | 1154 | 4.0 | 0.03 ± 0 | |
| EC-EC-EC-EC-6 | 24.64 | 1154 | 4.0 | 0 04 ± 0 | |
| EC-EC-EC-EC-7 | 25.88 | 1154 | 4.0 | 0.03 ± 0 | |
| tetramer concentration | 0.47 ± 0.20 | ||||
| total catechin and PA concentration | 48.79 ± 1.95 | ||||
Highly accurate masses can be computed for PAs on the basis of the degree of polymerization, the specific flavan-3-ol components, the number of A-type bonds, and the number of galloyls. PAs can be identified by comparing experimentally obtained high-accuracy masses to the computed masses. Identifications can be further confirmed by the analysis of fragments from tandem MS. Conversion of MRRF values from UV absorbance to ion counts with SIM was used for the quantification of individual PAs. Thus, this standardized UHPLC-PDA-ESI/HRMSn profiling method was able to offer identification and quantification of oligomeric PAs in plant-derived foods.
This research is supported by the Agricultural Research Service of the U.S. Department of Agriculture and an Interagency Agreement with the Office of Dietary Supplements of the National Institutes of Health.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Hümmer W.; Schreier P. Analysis of proanthocyanidins. Mol. Nutr. Food Res. 2008, 52, 1381–1398. [DOI] [PubMed] [Google Scholar]
- Hellström J. K.; Mattila P. H. HPLC determination of extractable and unextractable proanthocyanidins in plant materials. J. Agric. Food Chem. 2008, 56, 7617–7624. [DOI] [PubMed] [Google Scholar]
- Hellström J. K.; Törrönen A. R.; Mattila P. H. Proanthocyanidins in common food products of plant origin. J. Agric. Food Chem. 2009, 57, 7899–7906. [DOI] [PubMed] [Google Scholar]
- Gu L.; Kelm M. A.; Hammerstone J. F.; Beecher G.; Holden J.; Haytowitz D.; Gebhardt S.; Prior R. L. Screening of foods containing proanthocyanidins and their structural characterization using LC-MS/MS and thiolytic degradation. J. Agric. Food Chem. 2003, 51, 7513–7521. [DOI] [PubMed] [Google Scholar]
- Aron P. M.; Kennedy J. A. Flavan-3-ols: nature, occurrence and biological activity. Mol. Nutr. Food Res. 2008, 52, 79–104. [DOI] [PubMed] [Google Scholar]
- Prior R. L.; Gu L. Occurrence and biological significance of proanthocyanidins in the American diet. Phytochemistry 2005, 66, 2264–2280. [DOI] [PubMed] [Google Scholar]
- Monagas M.; Quintanilla-López J. E.; Gómez-Cordovés C.; Bartolomé B.; Lebrón-Aguilar R. MALDI-TOF MS analysis of plant proanthocyanidins. J. Pharm. Biomed. Anal. 2010, 51, 358–372. [DOI] [PubMed] [Google Scholar]
- Es-Safi N. E.; Guyot S.; Ducrot P. H. NMR, ESI/MS, and MALDI-TOF/MS analysis of pear juice polymeric proanthocyanidins with potent free radical scavenging activity. J. Agric. Food Chem. 2006, 54, 6969–6977. [DOI] [PubMed] [Google Scholar]
- Zhou H.-C.; Lin Y.-M.; Li Y.-Y.; Li M.; Wei S.-D.; Chai W.-M.; Tam F.-Y. Antioxidant properties of polymeric proanthocyanidins from fruit stones and pericarps of Litchi chinensis Sonn. Food Res. Int. 2011, 44, 613–620. [Google Scholar]
- Chai W.-M.; Shi Y.; Feng H.-L.; Qiu L.; Zhou H.-C.; Deng Z.-W.; Yan C.-L.; Chen Q.-S. NMR, HPLC-ESI-MS, and MALDI-TOF MS analysis of condensed tannins from Delonix regia (Bojer ex Hook.) Raf. and their bioactivities. J. Agric. Food Chem. 2012, 60, 5013–5022. [DOI] [PubMed] [Google Scholar]
- Fu C.; Loo A. E.; Chia F. P.; Huang D. Oligomeric proanthocyanidins from mangosteen pericarps. J. Agric. Food Chem. 2007, 55, 7689–7694. [DOI] [PubMed] [Google Scholar]
- Zhou H.-C.; Lin Y.-M.; Wei S.-D.; Tam F.-Y. Structural diversity and antioxidant activity of condensed tannins fractionated from mangosteen pericarp. Food Chem. 2011, 129, 1710–1720. [Google Scholar]
- Mouls L.; Mazauric J.-P.; Sommerer N.; Fulcrand H.; Mazerolles G. Comprehensive study of condensed tannins by ESI mass spectrometry: average degree of polymerisation and polymer distribution determination from mass spectra. Anal. Bioanal. Chem. 2011, 400, 613–623. [DOI] [PubMed] [Google Scholar]
- Gu L.; Kelm M. A.; Hammerstone J. F.; Zhang Z.; Beecher G.; Holden J.; Haytowitz D.; Prior R. L. Liquid chromatographic/electrospray ionization mass spectrometric studies of proanthocyanidins in foods. J. Mass Spectrom. 2003, 38, 1272–1280. [DOI] [PubMed] [Google Scholar]
- Hayasaka Y.; Waters E. J.; Cheynier V.; Herderich M. J.; Vidal S. Characterization of proanthocyanidins in grape seeds using electrospray mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 9–16. [DOI] [PubMed] [Google Scholar]
- Roux E. L.; Doco T.; Sarni-Manchado P.; Lozano Y.; Cheynier V. A-type proanthocyanidins from pericarp of Litchi chinensis. Phytochemistry 1998, 48, 1251–1258. [Google Scholar]
- Wollgast J.; Pallaroni L.; Agazzi M.-E.; Anklam E. Analysis of procyanidins in chocolate by reversed-phase highperformance liquid chromatography with electrospray ionization mass spectrometric and tandem mass spectrometric detection. J. Chromatogr. A 2001, 926, 211–220. [DOI] [PubMed] [Google Scholar]
- Núñez V.; Gómez-Cordovés C.; Bartolomé B.; Hong Y.-J.; Mitchell A. E. Non-galloylated and galloylated proanthocyanidin oligomers in grape seeds from Vitus vinifera L. cv. Graciano, Tempranillo and Cabernet Sauvignon. J. Sci. Food Agric. 2006, 86, 915–921. [Google Scholar]
- Sarnosk P. J.; Johnson J. V.; Reed K. V.; Tanko J. M.; O’Keefe S. F. Separation and characterisation of proanthocyanidins in Virginia type peanut skins by LC–MS. Food Chem. 2012, 131, 927–939. [Google Scholar]
- Appeldoorn M. M.; Vincken J. P.; Sanders M.; Hollman P. C. H.; Gruppen H. Combined normal-phase and reversed-phase liquid chromatography/ESI-MS as a tool to determine the molecular diversity of A-type procyanidins in peanut skins. J. Agric. Food Chem. 2009, 57, 6007–6013. [DOI] [PubMed] [Google Scholar]
- Cooper K. A.; Campos-Giménez E.; Jiménez Alvarez D.; Nagy K.; Donovan J. L.; Williamson G. Rapid reversed phase ultra-performance liquid chromatography analysis of the major cocoa polyphenols and inter-relationships of their concentrations in chocolate. J. Agric. Food Chem. 2007, 55, 2841–2847. [DOI] [PubMed] [Google Scholar]
- Li S.; Xiao J.; Chen L.; Hu C.; Chen P.; Xie B.; Sun Z. Identification of A-series oligomeric procyanidins from pericarp of Litchi chinensis by FT-ICR-MS and LC-MS. Food Chem. 2012, 135, 31–38. [Google Scholar]
- Hokkanen J.; Mattila S.; Jaakola L.; Pirttilä A. M.; Tolonen A. Identification of phenolic compounds from lingonberry (Vaccinium vitis-idaea L.), bilberry (Vaccinium myrtillus L.) and hybrid bilberry (Vaccinium × intermedium Ruthe L.) leaves. J. Agric. Food Chem. 2009, 57, 9437–9447. [DOI] [PubMed] [Google Scholar]
- Lin L.-Z.; Harnly J. M. A screening method for the identification of glycosylated flavonoids and other phenolic compounds using a standard analytical approach for all plant materials. J. Agric. Food Chem. 2007, 55, 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L.-Z.; Harnly J. M. Quantitation of flavanols, proanthocyanidins, isoflavones, flavananes, dihydroxychalcones, stilbens and benzoic acid derivatives using UV absorbance after identification by LC-MS. J. Agric. Food Chem. 2012, 60, 5832–5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L.-Z.; Sun J.; Chen P.; Harnly J. M. U-HPLC-PDA-EIS/HRMS/MSn analysis of anthocyanins, flavonol glycosides and hydroxycinnamic acid derivatives in red mustard green (Brassica juncea Coss variety). J. Agric. Food Chem. 2011, 59, 12059–12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.-J.; Deinzer M. L. The mass spectral analysis of isolated hops A-type proanthocyanidins by electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2008, 43, 1353–1363. [DOI] [PubMed] [Google Scholar]
- Monagas M.; Gómez-Cordovés C.; Bartolomé B.; Laureano O.; Ricardo da Silva J. M. Monomeric, oligomeric, and polymeric flavan-ol composition of wines and grapes from Vitis vinifera L. cv. Graciano, Tempranillo, and Cabernet Sauvignon. J. Agric. Food Chem. 2003, 51, 6475–6481. [DOI] [PubMed] [Google Scholar]
- Escribano-Bailón M. T.; Guitiérrez-Fernández Y.; Rivas-Gonzalo J. C.; Santos-Buelga C. Characterization of procyanidins of Vitisvinifera variety Tinta del Pais grape seeds. J. Agric. Food Chem. 1992, 40, 1794–1799. [Google Scholar]
- Ricardo da Silva J. M.; Rigaud J.; Cheynier V.; Cheminat A.; Moutounet M. Procyanidin dimers and trimers from grape seeds. Phytochemistry 1991, 30, 1259–1264. [Google Scholar]
- Ricardo da Silva J. M.; Rosec J.-Ph.; Bourzeix M.; Mourgues J.; Moutounet M. Dimer and trimer procyanidins in Carignan and Mourvèdre grapes and red wines. Vitis 1992, 31, 55–63. [Google Scholar]
- Malik A.; Kuliev Z. A.; Akhmedov Y. A.; Vdovin A. D.; Abdullaev N. D. Proanthocyanidins of Ziziphus jujuba. Chem. Nat. Compd. 1997, 33, 165–173. [Google Scholar]
- Prasain J. K.; Wang C. C.; Barnes S. Mass spectrometric methods for the determination of flavonoids in biological samples. Free Radical Biol. Med. 2004, 37, 1324–1350. [DOI] [PubMed] [Google Scholar]
- Redeuil K.; Bertholet R.; Kussmann M.; Steiling H.; Rezzi S.; Nagy K. Quantification of flavan-3-ols and phenolic acids in milk-based food products by reversed-phase liquid chromatography–tandem mass spectrometry. J. Chromatogr. A 2009, 1216, 8362–8370. [DOI] [PubMed] [Google Scholar]