Abstract
The Krüppel-like transcription factors KLF1 and KLF2 are essential for embryonic erythropoiesis. They can partially compensate for each other during mouse development, and coordinately regulate numerous erythroid genes, including the β-like globins. Simultaneous ablation of KLF1 and KLF2 results in earlier embryonic lethality and severe anemia. In this study, we determine that this anemia is caused by a paucity of blood cells, and exacerbated by diminished β-like globin gene expression. The anemia phenotype is dose-dependent, and, interestingly, can be ameliorated by a single copy of the KLF2, but not the KLF1 gene. The roles of KLF1 and KLF2 in maintaining normal peripheral blood cell numbers and globin mRNA amounts are erythroid cell-specific. Mechanistic studies led to the discovery that KLF2 has an essential function in erythroid precursor maintenance. KLF1 can partially compensate for KLF2 in this role, but is uniquely crucial for erythroid precursor proliferation through its regulation of G1- to S-phase cell cycle transition. A more drastic impairment of primitive erythroid colony formation from embryonic progenitor cells occurs with simultaneous loss of KLF1 and KLF2 than with loss of a single factor. KLF1 and KLF2 coordinately regulate several proliferation-associated genes, including Foxm1. Differential expression of FoxM1, in particular, correlates with the observed KLF1 and KLF2 gene dosage effects on anemia. Furthermore, KLF1 binds to the FoxM1 gene promoter in blood cells. Thus KLF1 and KLF2 coordinately regulate embryonic erythroid precursor maturation through the regulation of multiple homeostasis-associated genes, and KLF2 has a novel and essential role in this process.
Introduction
Erythropoiesis in the mouse and other mammals proceeds in two distinct waves during development.1 First, primitive erythroid cells appear in the yolk sac, an extra-embryonic membrane, in regions known as blood islands.2,3 Large, nucleated primitive erythroid cells begin to circulate at about mouse embryonic Day 8.25 (E8.25).4,5 Mouse primitive erythroid progenitor cells form colonies (EryP-CFC) in semi-solid culture media4 which can first be detected at E7. Primitive erythroid cells mature in circulation in a synchronous manner and eventually undergo enucleation.6 The second wave, or definitive erythropoiesis, begins in the fetal liver and results in mature, enucleated definitive erythroid cells by around E12.1
The erythroid developmental program is regulated by a network of key transcription factors,7 including Krüppel-like factor 1 (KLF1 or EKLF, Erythroid Krüppel-like factor). Members of the KLF family have three Cys2-His2 zinc fingers near their C-terminus that recognize a core 5′-CACCC-3′ DNA sequence.8 KLF1 is erythroid-specific and required for the normal expression of the embryonic and adult β-like globin genes.9–11 KLF1−/− mice develop β-thalassemia and die by E16.9,10 KLF1 regulates the expression of numerous other erythroid genes, including those encoding cytoskeletal and membrane proteins, and heme synthesis enzymes.2,13 In addition, KLF1 regulates cell cycle control, proliferation and apoptosis.14–16 Pathogenic mutations in KLF1 can cause congenital dyserythropoietic anemia in humans.17,18
To date, 17 mammalian KLFs have been identified, and they have diverse roles in development.8 The zinc finger domain of KLF2 is 89% similar to KLF1. KLF2 is necessary for primitive erythropoiesis, and positively regulates embryonic globin gene expression, as shown in KLF2 knockout and erythroid-conditional knockout mouse embryos.11,19,20 Global gene expression assays indicate that many genes that are important in the processes of development, differentiation and cell migration are dysregulated in KLF2−/− compared to wild-type primitive erythroid cells.21 Ablation of KLF2 causes lethality between E11.5 and E14.5,22,23 which can be due to intra-embryonic hemorrhaging24 or heart failure.23,25
KLF1 and KLF2 share functional roles in globin gene regulation. KLF1 and KLF2 can partially compensate for each other, as is evident from the reduced embryonic β-like globin gene expression of KLF1/KLF2 double (KLF1−/−KLF2−/−) compared to single knockout embryos.11 There is also more aberrant morphology, consistent with increased apoptosis, in double compared to single knockout primitive red blood cells.
In addition, E10.5 KLF1−/− KLF2−/− embryos are strikingly pale and appear anemic, whereas E10.5 KLF1−/− and E10.5 KLF2−/− mice are grossly normal. KLF1−/− KLF2−/− mice die by E11.5, which is earlier than KLF1−/− or KLF2−/− embryos. Gene expression profiling of E9.5 erythroid cells was used to identify genes that are synergistically regulated by KLF1 and KLF2.12 These genes are principally involved in cellular pathways that control homeostasis, hemopoiesis, apoptosis and proliferation. Erythroid conditional knockout and other functional studies identified Myc as an important downstream target of KLF1 and KLF2. Additional direct or indirect targets include other genes governing proliferation and/or apoptosis, such as Cd24a antigen,26 forkhead box M1 (FoxM1),27 sphingosine kinase 1 (Sphk1)28 and parathyroid hormone 1 receptor (Pthr).29
In this work, we further assess how KLF1 and KLF2 coordinately control primitive erythropoiesis. E10.5 KLF1/KLF2 double erythroid-conditional knockout embryos are anemic like KLF1−/− KLF2−/− mice. A surprising new discovery was made in that restoring one copy of the KLF2 (KLF1−/− KLF2+/−) but not the KLF1 gene (KLF1+/−KLF2−/−) ameliorates this phenotype. Anemia in E10.5 KLF1+/−KLF2−/− and KLF1−/− KLF2−/− mice correlates with a drastic reduction in the number of peripheral blood cells. EryP-CFC assays identified a novel role for KLF2 in the maintenance of primitive erythroid progenitor potential. KLF1 can partially compensate for KLF2 in this function, but its pivotal role is in the regulation of erythroid precursor cell proliferation. KLF1−/− KLF2−/− embryos have more severely impaired colony forming ability than single knockouts. The data indicate that KLF1 and KLF2 coordinately control the maturation of primitive erythroid precursors through the regulation of multiple cell cycle- and proliferation-associated genes, including FoxM1.
Methods
Generation of mice
The KLF1 and KLF2 knockout (KO) mouse models have been described previously.9,22 The KLF1 and KLF2 loci are linked on mouse chromosome 8. A model with the KLF1 and KLF2 null alleles on the same chromosome, KLF1+/−KLF2+/−(R), was developed.11 Mice with a floxed KLF2 allele were obtained from Dr. Jerry Lingrel.30 Mice carrying the KLF1 null allele and the KLF2 floxed allele on the same chromosome were created by screening for recombinants as previously described.11 The Tie2-Cre mouse model has been described previously.31 All experiments involving mice were approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee (VCU IACUC) under protocol number AM10347. The mice used for this study are of mixed genetic background.
Tissue collection and processing
Peripheral blood cells were collected by severing the vitelline and umbilical vessels of E9.5 or E10.5 mouse embryos.20 Whole-mount embryos were photographed with an Olympus SZ2-ILSTmicroscope (Olympus America, Center Valley, PA, USA), using an Olympus Q-Color 3 camera and QCapture 2.81.0 software (Quantitative Imaging, Surrey, BC, USA). Embryonic yolk sacs were embedded and sectioned as previously described.11 Images of yolk sac sections were captured with an Olympus BX41 compound microscope and Olympus DP71 digital camera. cDNA synthesis and quantitative reverse-transcriptase PCR (qRT-PCR) were performed as previously described.11,12,19,20 Primer sequences for qRT-PCR are listed in Online Supplementary Table S1.
Erythroid progenitor assays
To study primitive erythroid progenitors, E8.25–E8.5 (6–12 somite pairs) implants including embryo and yolk sac were dissected in PB2,32 and dissociated into single cells using 0.01% trypsin (Worthington Biochemical Corporation, NJ, USA) and mechanical trituration. Half of the cells from each implant were plated in 1% methylcellulose supplemented with 10% plasma derived serum (Animal Technologies, TX, USA), 5% protein-free hybridoma medium (Gibco/BRL), IL-3 (20 ng/mL), IL-6 (20 ng/mL), stem cell factor (60 ng/mL) (IL-3, IL-6 and SCF-Peprotech, NJ, USA), erythropoietin (2 U/mL), MTG and L-glutamine. The remaining cells were used for genotyping. Erythroid colonies were stained using benzidine and counted either on Day 2 or Day 6 of culture. Colony images were captured using an Olympus IX70 microscope and Q-Color 3 camera (Olympus America). Two-dimensional colony areas were calculated using Image J software.
Cell cycle analysis
Cell cycle profiles were assessed using the APC BrdU Flow kit (BD Biosciences). The protocol was adapted from Malik et al.33 Briefly, E9.5 erythroblasts were cultured in erythroid maturation medium containing BrdU for 90 min, fixed, and stained with APC-conjugated anti-BrdU antibody and 7-amino-actinomycin D (7-AAD). The BD FACSCanto™ II Analyzer was used to measure total DNA content and BrdU incorporation using 7-AAD and APC fluorescence intensity, respectively. Gating was applied to include only single cells and exclude clumped cells.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed as previously described.20 Primer sequences for qPCR are listed in Online Supplementary Table S2.
Statistical analysis
The Student’s t-test was used for statistical analyses. P<0.05 was considered significant.
Results
KLF1/KLF2 double conditional KO embryos recapitulate the KLF1−/− KLF2−/− anemia phenotype
KLF1 is erythroid-specific, but KLF2 has a more widespread tissue distribution. In order to determine whether the anemia observed in KLF1−/− KLF2−/− embryos results from erythroid cell-specific ablation of KLF2, the ideal mouse model would be an erythroid conditional knockout of KLF2 on a KLF1 null background. Unfortunately, Cre expression under the control of the erythroid-specific erythropoietin receptor (EpoR) promoter in ErGFP-Cre mice20,34 is dependent on KLF1 (Online Supplementary Figure S1). Consequently, this model was untenable. β-Cre,35 with the β-globin promoter, was also unworkable because it is linked to the KLF1 gene. Tie2-Cre mice express Cre in erythroid cells by E7.5, using the Tie2 promoter.36 Therefore, this model was used, with the caveat that KLF2 is ablated in both erythroid and endothelial cells.
KLF1+/−KLF2F/+,Tie2-Cre and KLF1+/−KLF2F/F mice were mated to obtain E10.5 embryos of the following genotypes: KLF1−/− KLF2F/F,Tie2-Cre (KLF1/KLF2 double conditional KO or dcKO), KLF1−/− KLF2F/F (KLF1 KO) and KLF1+/+KLF2F/+ (wild-type control). E10.5 KLF1/KLF2 dcKO embryonic blood cells have substantially reduced KLF2 mRNA compared to wild-type (Online Supplementary Figure S2). At E10.5, wild-type (Figure 1A), KLF1 KO (Figure 1B) and KLF2 KO11 embryos are surrounded by yolk sacs with distinctly red blood vessels. In comparison, E10.5 KLF1/KLF2 dcKO mice (Figure 1C) are pale and appear anemic, similar to KLF1−/− KLF2−/− embryos.11 As a control to rule out Cre toxicity, it was determined that E10.5 whole-mount Tie2-Cre embryos do not show an anemia phenotype (data not shown).
Figure 1.
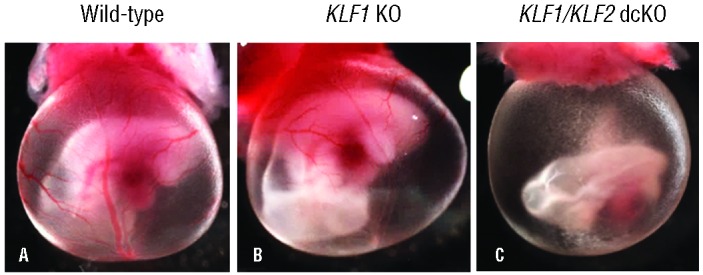
KLF1/KLF2 double conditional KO embryos recapitulate the KLF1−/− KLF2−/− anemia phenotype. E10.5 whole-mount embryos surrounded by yolk sacs. (A) Wild-type embryos (KLF1+/+KLF2F/+, n=3) have distinct red blood vessels. (B) KLF1 KO (KLF1−/− KLF2F/F, n=4) embryos are similar in appearance to wild-type embryos. (C) KLF1/KLF2 double conditional KO embryos (KLF1−/− KLF2F/F,Tie2-Cre, n=4) appear anemic in contrast to A and B. Photographs were taken at 15Χ magnification.
To determine whether the pale appearance of KLF1/KLF2 dcKO embryos is due to a defect in the development of blood vessels, yolk sac tissue sections were studied. Blood islands consisting of blood cells (Ery) surrounded by endothelial cells (En) are situated between the epithelium (Ep) and mesothelium (Me), as shown for wild-type in Figure 2A and D. The yolk sac vasculature of KLF2 KO,19 KLF1 KO (Figure 2B and E) and KLF1/KLF2 dcKO embryos (Figure 2C and F) appears normal, with well-defined blood vessels lined by endothelial cells. This suggests that aberrant erythroid rather than vascular development causes the anemia phenotype. Although it is difficult to accurately assess the relative number of blood cells in yolk sac sections, KLF1/KLF2 dcKO vessels appear to contain fewer cells than the other genotypes.
Figure 2.
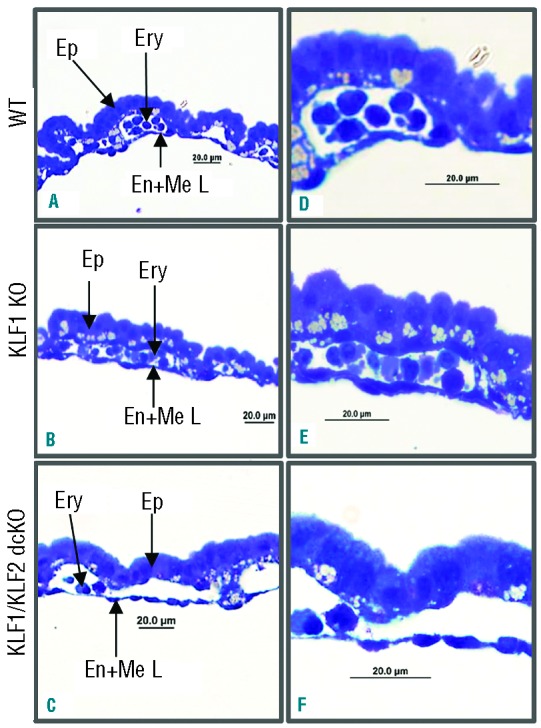
KLF1/KLF2 double conditional KO embryos have normal yolk sac blood vessels. Blood islands from wild-type (A and D), KLF1 KO (B and E) and KLF1/KLF2 double conditional KO (C and F) yolk sacs. n=3 for each genotype. A, B and C are 400Χ; and D, E and F are 1000Χ magnification. The abbreviations used are columnar epithelium (Ep), blood cells (Ery) and endothelial/mesothelial layer (En + Me L).
The globin mRNA amounts in E10.5 wild-type, KLF1−/− and KLF1/KLF2 dcKO peripheral blood cells were determined. E10.5 KLF2 erythroid conditional KO (KLF2F/F,ErGFP-Cre) blood cells have approximately 20% less embryonic globin mRNA than wild-type.20,34 KLF1−/− embryos have an approximately 35% reduction in βh1-and Ey-globin mRNA (Online Supplementary Figure S3A and B), while βh1- and Ey-globin mRNA in KLF1/KLF2 dcKO embryos is reduced to approximately 50% of wild-type (Online Supplementary Figure S3A and B). Surprisingly, KLF1/KLF2 dcKO embryos do not display drastic decreases in globin mRNA correlating with the observed anemia phenotype.
E10.5 KLF1+/−KLF2−/− and KLF1−/−KLF2−/− embryos have fewer peripheral blood cells than KLF1−/− KLF2+/−
Rare recombinant embryos of the genotypes KLF1−/− KLF2F/+,Tie2-Cre and KLF1+/−KLF2F/F,Tie2-Cre were obtained. E10.5 KLF1−/− KLF2F/+,Tie2-Cre embryos, with one KLF2 allele, have yolk sacs with visibly red blood vessels like wild-type (Online Supplementary Figure S4A), but a KLF1+/−KLF2F/F,Tie2-Cre embryo, with one KLF1 allele, appears anemic like KLF1/KLF2 dcKO (Online Supplementary Figure S4B). To extend this surprising finding to a system that is more easily manipulated, several E10.5 KLF1−/− KLF2+/− and KLF1+/−KLF2−/− embryos were obtained. KLF1−/− KLF2+/− whole mount embryos appear similar to wild-type, with distinct red blood vessels (Figure 3A), but KLF1+/−KLF2−/− embryos appear anemic (Figure 3B). These findings confirm that one functional KLF2, but not KLF1, allele can ameliorate the anemia phenotype.
Figure 3.
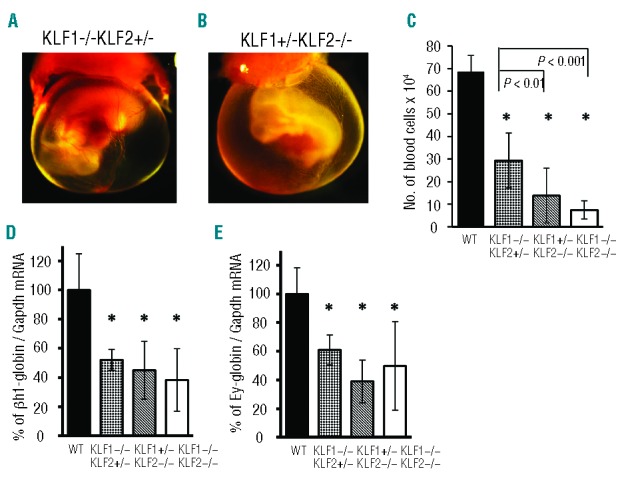
E10.5 KLF1+/−KLF2−/− and KLF1−/− KLF2−/− embryos have fewer peripheral blood cells than KLF1−/− KLF2+/− embryos. (A and B) E10.5 whole-mount embryos surrounded by yolk sacs. Embryos with one normal KLF2 gene, (A) KLF1−/− KLF2+/− (n=3) appear grossly normal; whereas (B) KLF1+/−KLF2−/− (n=3) embryos, with one functional KLF1 gene, appear anemic. Photographs were taken at 15Χ magnification. (C) KLF1+/−KLF2−/− and KLF1−/− KLF2−/− embryos have fewer peripheral blood cells than KLF1−/− KLF2+/−, correlating with the anemia phenotype. *P<0.005 compared to wild-type. Other significant P-values are as shown by brackets. The amount of mouse embryonic (D) βh1-globin and (E) Ey-globin mRNA is reduced in KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/− blood cells compared to wild-type (WT) (* = P<0.05). Gapdh mRNA was used as an internal standard for qRT-PCR. The globin-to-Gapdh mRNA ratio for WT was taken as 100%. n=4–10 for each genotype. Error bars indicate standard deviation.
E10.5 KLF1−/− embryos have approximately 2-fold fewer peripheral blood cells than wild-type.12 In order to understand the contributions of KLF1 and KLF2 to the starkly different erythropoietic phenotypes of E10.5 KLF1+/−KLF2−/− and KLF1−/− KLF2+/− embryos, circulating blood cell numbers were determined. KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/− embryos have fewer peripheral blood cells than wild-type (Figure 3C). Of particular interest was the observation that KLF1+/−KLF2−/− embryos have significantly fewer peripheral blood cells than KLF1−/− KLF2+/− embryos (Figure 3C). The number of blood cells in E10.5 KLF1+/−KLF2−/− and KLF1−/− KLF2−/− embryos, for which whole-mounts appear anemic, is similar and is several-fold less than wild-type.
The amount of embryonic βh1- and Ey-globin mRNA in blood cells from these embryos was determined using qRT-PCR. KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/− blood cells have approximately 50% of the βh1-globin (Figure 3D) and Ey-globin mRNA (Figure 3E) compared to wild-type. The KLF1−/− KLF2−/− amount correlates with the findings in KLF1/KLF2 dcKO embryos (Online Supplementary Figure S3). There are no significant differences in the amounts of βh1- and Ey-globin mRNA between KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/− embryos. Therefore, disparities in globin gene expression do not explain the major difference in the anemia phenotypes of KLF1+/−KLF2−/− and KLF1−/− KLF2−/− compared to KLF1−/− KLF2+/− whole-mount embryos. The measurements of the amount of globin mRNA per cell and the number of blood cells per embryo were multiplied to calculate the total embryonic β-like globin mRNA per embryo (Online Supplementary Table S3). KLF1−/− KLF2+/− embryos have approximately 4-fold less, whereas KLF1+/−KLF2−/− and KLF1−/− KLF2−/− embryos have 10- to 15-fold less total globin mRNA per embryo than wild-type. Based on these findings, a combined reduction in globin per cell and in number of blood cells contributes to the anemia phenotype of E10.5 KLF1+/−KLF2−/− and KLF1−/− KLF2−/− embryos.
E10.5 KLF1−/− KLF2+/− embryos may escape anemia because there is a compensatory increase in KLF2 mRNA in the absence of KLF1, ameliorating the phenotype. To test this hypothesis, the amount of KLF2 mRNA in blood cells of E10.5 wild-type and KLF1−/− embryos was quantified by qRT-PCR. An approximate 2.5-fold increase in KLF2 mRNA amount was observed in KLF1−/− blood cells compared to wild-type (Online Supplementary Figure S5), supporting this postulate.
KLF1−/− and KLF1−/− KLF2−/− erythroblasts display impaired G1- to S-phase cell cycle progression
Primitive and definitive KLF1−/− blood cells show cell cycle defects.14,15 Furthermore, KLF1 directly regulates E2f2 and p18, genes that control cell cycle progression.14,15,37 It was, therefore, of interest to determine if KLF1 and KLF2 have overlapping functions in cell cycle regulation, and whether decreased proliferation is responsible for the reduced blood cell number in KLF1−/− KLF2−/− embryos. Therefore, the cell cycle profiles of E9.5 erythroblasts from KLF1−/− KLF2−/−, KLF1−/−, KLF2−/− and wild-type embryos were compared. After 90 min in cell culture medium containing BrdU, an average of 18.8% of wild-type erythroblasts are in G0/G1, 79.7% are in S-phase and 1.5% are in G2/M (Figure 4A). Similar results have been reported by Malik et al. for E10.5 erythroblasts.33 The cell cycle distribution of KLF2−/− erythroblasts is similar to wild-type, suggesting that KLF2 ablation does not affect cell cycle progression. KLF1−/− and KLF1−/− KLF2−/− embryos have an approximately 20% reduction in cells in S-phase compared to wild-type, and a concomitant 2-fold increase in cells in G0/G1 phase, suggesting a block in S-phase entry. There were no significant differences between the cell cycle profiles of KLF1−/− and KLF1−/− KLF2−/− E9.5 erythroblasts, suggesting that KLF1 has a unique role in regulating cell cycle progression that is not shared by KLF2. Simultaneous ablation of KLF2 does not exacerbate the cell cycle disruption in KLF1−/− red blood cells. This work with E9.5 KLF1−/− erythroblasts extends and confirms other reports of cell cycle defects in KLF1−/− blood cells.14,15
Figure 4.
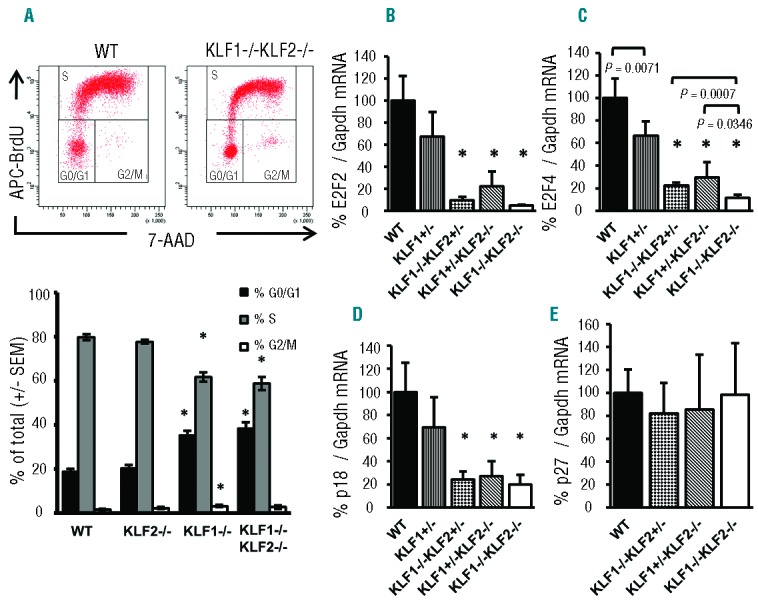
KLF1−/− and KLF1−/− KLF2−/− blood cells show aberrations in G1- to S-phase progression during cell division. (A) Representative flow cytometry plots measuring DNA content (7-AAD) and BrdU incorporation (APC-BrdU) are shown for WT and KLF1−/− KLF2−/− embryos. The graph below depicts analyses of the percentage of cells in G0/G1, S and G2/M phases of the cell cycle in wild-type (WT), KLF2−/−, KLF1−/− and KLF1−/− KLF2−/− E9.5 erythroblasts. n = 3–9 for each genotype. Error bars indicate standard error. *P-value <0.01 compared to wild-type. Expression of genes (B) E2f2 (C) E2f4 (D) p18 and (E) p27 was compared in wild-type (WT), KLF1+/−, KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/− blood cells by qRT-PCR. Gapdh was used as an internal standard for qRT-PCR. The gene-to-Gapdh mRNA ratio for WT was taken as 100%. n = 4–10 for each genotype. *P-value <0.01 compared to wild-type and KLF1+/−. Other significant P-values are shown using brackets. Error bars indicate standard deviation.
The relative expression of certain genes important for G1- to S-phase progression, including E2f2, E2f4, p18 and p27, was assessed in wild-type, KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/− blood cells, to assess whether the observed gross phenotypic differences are associated with differential gene regulation. Recent studies have indicated that KLF1 haploinsufficiency results in dysregulation of a subset of its target genes and hence KLF1+/− blood cells were included as an additional control.38,39 The amount of E2f2 (Figure 4B), E2f4 (Figure 4C) and p18 (Figure 4D) mRNA is significantly reduced in KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/− blood cells compared to both wild-type and KLF1+/− blood cells. KLF1 and KLF2 both contribute to the expression of these three cell cycle-related genes. The expression of p27 mRNA in the mutant genotypes is not different than wild-type (Figure 4E). None of these genes show significantly different expression in KLF1+/−KLF2−/− and KLF1−/− KLF2+/− blood cells, so there is no correlation with the gross phenotypic differences between these embryos.
KLF1−/− KLF2−/− embryos have defects in primitive erythroid precursor maturation
The paucity of peripheral blood cells in KLF1−/− KLF2−/− embryos may result from a decrease in the number or a defect in the differentiation of erythroid progenitors. To determine whether KLF1 and KLF2 control the primitive erythroid progenitor (EryP-CFC) compartment, we performed colony-forming assays on E8.25–E8.5 wild-type, KLF1−/−, KLF2−/− and KLF1−/− KLF2−/− embryos. At this developmental stage, EryP-CFC colony numbers reflect the number of primitive erythroid progenitors present in the yolk sac. On Day 4 of culture, visibly red primitive erythroid colonies are observed in wild-type (n=10) but not KLF1−/− KLF2−/− (n=4) cultures, indicating a defect in the erythroid compartment in the absence of KLF1 and KLF2 (data not shown).
To further assess this defect, colonies in methylcellulose were stained with benzidine for hemoglobin, at Day 6 of culture. KLF1−/− embryos gave rise to normal numbers of EryP-CFCs (Figure 5A), but the colonies are approximately 10-fold smaller than wild-type (Figure 5B). This finding correlates with the data indicating that KLF1−/− erythroid cells exhibit cell cycle defects (Figure 4A). Interestingly, KLF2−/− embryos produce significantly fewer EryP-CFCs than wild- type (Figure 5A), but colony size is indistinguishable from wild-type (Figure 5B). The number of EryP-CFCs derived from KLF1−/− KLF2−/− embryos is significantly less than wild-type, KLF2−/− and KLF1−/− (Figure 5A). The KLF1−/− KLF2−/− colonies are also smaller than wild-type, although similar in size to KLF1−/−. It was of interest to distinguish whether there are fewer than normal progenitors in KLF1−/− KLF2−/− embryos, or if they are unable to produce colonies that survive to Day 6 of culture. Therefore the method of Malik et al. was used to count nascent colonies on Day 2 of culture.33 There are similar numbers of EryP-CFC in KLF1−/− KLF2−/− and KLF1+/−KLF2+/− on Day 2 (Figure 5C). This suggests that normal numbers of primitive erythroid progenitors are specified in KLF1−/− KLF2−/− embryos, but the simultaneous ablation of KLF1 and KLF2 leads to a stochastic defect in maturation of erythroid precursors, leading to fewer colonies on Day 6. The KLF1 and KLF2 genes interact to synergistically control colony number. Thus, aberrant precursor potential and proliferation capacity likely contribute to the reduced number of blood cells in E10.5 KLF1−/− KLF2−/− embryos.
Figure 5.
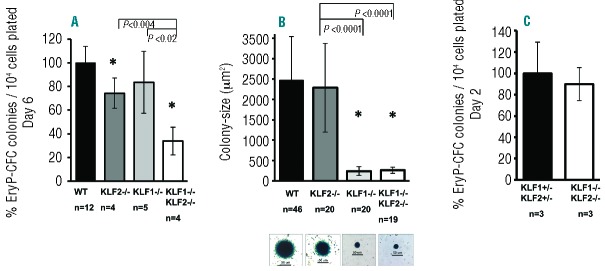
KLF1−/− KLF2−/− embryos have defects in primitive erythroid precursor maturation. E8.25–E8.5 implants were dissociated into single cells using trypsin and plated in methylcellulose supplemented with growth factors. Erythroid colonies were stained with benzidine and counted on Day 2 or Day 6 of culture. (A) Comparison of number of EryP-CFC colonies formed per 104 cells plated from wild-type (WT), KLF2−/−, KLF1−/− and KLF1−/− KLF2−/− implants on day 6 of culture. The average number of colonies formed per 104 cells plated from wild type implants was set to 100% for each litter. ‘n’ indicates number of embryos. (B) The average colony size was determined by measuring the two-dimensional area of colonies from captured images. Average colony size of KLF2−/−, KLF1−/− and KLF1−/− KLF2−/− colonies was compared to wild-type colonies. ‘n’ indicates number of colonies analyzed from 4 to 6 embryos of each genotype. Representative colonies from each genotype are shown below the respective genotype (100Χ magnification, scale bar is 50 μm). (C) Comparison of number of EryP-CFC colonies formed per 104 cells plated from KLF1+/−KLF2+/− and KLF1−/− KLF2−/− implants on day 2 of culture. The average number of colonies formed per 104 cells plated from KLF1+/−KLF2+/− implants was set to 100. n: indicates number of embryos. *P-value <0.01 compared to wild-type. Other significant p-values are shown using brackets. Error bars indicate standard deviation.
KLF1 and KLF2 modulate the expression of genes controling proliferation
Microarray analyses were used previously to demonstrate that the expression of five proliferation-associated genes, Foxm1, Cd24a, Myc, Sphk1 and Pthr, is progressively reduced in wild-type, KLF1−/− and KLF1−/− KLF2−/− E10.5 blood cells.12 Therefore, qRT-PCR was used to assess expression of these genes in wild-type, KLF1−/− KLF2+/−, KLF1+/KLF2−/− and KLF1−/− KLF2−/− E10.5 blood cells. The amounts of all five mRNAs are significantly reduced in KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/− blood cells compared to wild-type and KLF1+/− blood cells, verifying the results of the microarray analyses (Figure 6A–C and Online Supplementary Figure S6A and B). Only Foxm1 mRNA is significantly reduced in KLF1+/−KLF2−/− and KLF1−/− KLF2−/− compared to KLF1−/− KLF2+/− (Figure 6A), specifically correlating with the genotypes that display the anemia phenotype and reduced peripheral blood cell number. However, it seems unlikely that dysregulation of a single downstream target gene is responsible for the anemia phenotype in E10.5 KLF1+/−KLF2−/− and KLF1−/− KLF2−/− embryos. Rather it is probably due to the cumulative effects of reduced expression of multiple proliferation-associated genes, including FoxM1.
Figure 6.
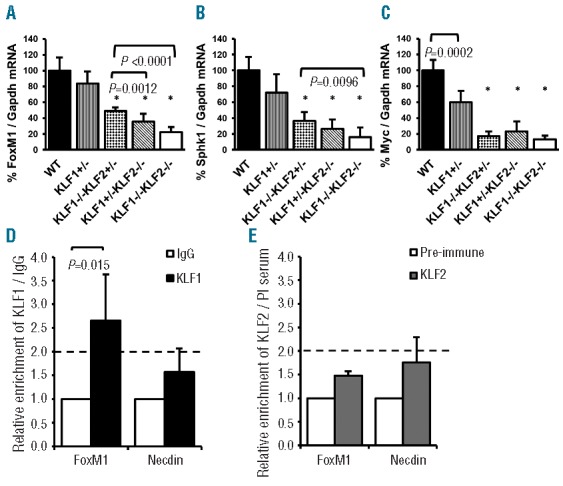
KLF1 and KLF2 modulate the expression of genes involved in proliferation. qRT-PCR was used to determine the amount of (A) Foxm1, (B) Sphk1 and (C) cMyc mRNA in wild-type (WT), KLF1+/−, KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/− E10.5 blood cells. Gapdh mRNA was used as an internal standard. Wild-type was taken as 100%. n= 4–10 per genotype. All of the test genotypes (KLF1−/− KLF2+/−, KLF1+/−KLF2−/− and KLF1−/− KLF2−/−) were significantly different from wild-type and KLF1+/− (*P< 0.0001). Other significant p-values are as shown by brackets. Error bars indicate standard deviation. ChIP assays were performed on E10.5 wild-type erythroid cells. Polyclonal antibodies specific for (D) KLF1 and (E) KLF2 were used to determine binding/enrichment of the respective transcription factor at the FoxM1 gene promoter. Non-specific IgG or pre-immune serum (PI serum) were used as controls for antibody specificity as indicated. The y-axis represents the relative fold enrichment. The mean IgG or PI serum enrichment was set to 1.0, and the enrichment of KLF1 and KLF2 were scaled appropriately. Based on the negative controls, a greater than 2-fold enrichment was considered a minimal requirement for binding, indicated by the dotted line. The x-axis indicates the gene promoter primers used for qPCR (described in Online Supplementary Table 2). Primers specific for the Necdin gene were used as a negative control. n = 3, error bars indicate standard deviation.
The Foxm1 gene has two consensus CACCC KLF binding sites within 500 bp upstream of the transcriptional start site. To assess whether KLF1 and KLF2 directly bind to the Foxm1 promoter, ChIP assays were performed using wild-type E10.5 blood cells, followed by qPCR to quantitate enrichment of KLF1 and KLF2. Based on the negative controls, a greater than 2-fold enrichment was considered a minimal requirement for binding. KLF1 is approximately 2.5-fold enriched compared to the negative control (Figure 6D), suggesting that it directly binds and regulates the Foxm1 gene. Binding of KLF2 was not detected within this Foxm1 promoter region (Figure 6E); however, it could bind to the gene at another site.
Discussion
The KLF1 and KLF2 genes interact to control globin gene regulation and primitive erythroid precursor maturation. Interactions between related transcription factor genes in erythroid cell development have previously been reported. Gata1 null embryos have normal primitive erythroid cell numbers.40 Gata2 knockout embryos have fewer primitive erythroid cells than wild-type.41 Gata1/Gata2 double knockout embryos show a more severe defect in primitive erythropoiesis than either single knockout, with yolk sac blood islands almost completely devoid of erythroid cells.42 Functional overlap has also been described for KLF3 and KLF8, which act as transcriptional repressors. KLF3 knockout mice have a mild erythroid phenotype.43,44 KLF8 knockout mice have normal erythroid parameters.45 The KLF3/KLF8 double knockout causes embryonic lethality, and greater dysregulation of fetal liver gene expression than either single knockout.45
The reduced number of E10.5 KLF1−/− KLF2−/− peripheral blood cells stems from a defect in erythroid precursor maintenance and maturation. Primitive erythroid progenitor colonies (EryP-CFC) derived from E8.5 KLF1−/− embryos are similar in frequency but significantly smaller in size than wild-type. Studies of definitive fetal liver (adult) erythroid progenitors have produced inconsistent results with respect to the frequency of BFU-E and CFU-E colony forming units. Perkins et al. found a similar number of BFU-E and CFU-E in E15 KLF1−/− and wild-type mice.9 Pilon et al. used sorted E13.5 KLF1−/− fetal liver erythroid progenitors (CD71LO, Ter119NEG), and also found normal numbers of BFU-E.14
However, Pilon et al. observed an increased frequency of CFU-E in KLF1−/− compared to wild-type, which was attributed to a block in erythropoiesis. Interestingly, they observed that KLF1−/− fetal livers have fewer cells than wild-type.14 In addition, KLF1−/− BFU-E and CFU-E colonies contain less hemoglobin and require 24–48 h longer to reach the same size as wild-type, suggesting a defect in proliferation, analogous to our findings for embryonic progenitors.14 Thus KLF1, though present in a lower amount in primitive than in definitive erythroid cells,20,46 plays a similar role in proliferation at both stages.
A novel role for KLF2 in the maintenance of mouse primitive erythroid precursors was identified in this work. This correlates with ENCODE data from the Ross Hardison laboratory, which indicates that KLF2 is robustly expressed in mouse adult megakaryocyte-erythroid progenitors (MEPs) (UCSC Accession: wgEncodeEM003184). Investigations into the role of KLF2 in definitive erythroid progenitors have produced discordant results. Wani et al. reported a severe reduction in the number of CFU-E derived from E11.5 KLF2−/− fetal liver progenitors compared to wild-type.22 However, Kuo et al. found normal numbers of erythroid colonies in KLF2−/− fetal liver cultures.24 These studies could be complicated because the demise of the KLF2−/− embryos is imminent. Our studies reveal that E8.25–E8.5 KLF2−/− embryos have fewer primitive erythroid progenitor colonies than wild-type embryos on Day 6 of culture. KLF1 can partially compensate for the lack of KLF2, because fewer EryP-CFC are obtained from KLF1−/− KLF2−/− than from KLF2−/− embryos. Interestingly, there are normal numbers of erythroid colonies derived from KLF1−/− KLF2−/− embryos on Day 2 of culture. It is possible that a stochastic selection event occurs beyond Day 2, causing only some KLF1−/− KLF2−/− colonies to be maintained. KLF2 ablation leads to increased apoptosis in E10.5 yolk sac erythroid cells,19 and there is some evidence of a modest increase in apoptosis in KLF1−/− definitive erythroid cells.16 Although it is not possible to study apoptosis directly in E8.5 erythroid progenitor cells, the stochastic event in KLF1−/− KLF2−/− cultures might logically involve apoptosis.
KLF1 and KLF2 gene dosage is important for the normal regulation of several proliferation genes. The transcription factor Foxm1 stimulates proliferation by promoting S-phase entry in the cell cycle.47 Foxm1−/− embryos have approximately 3-fold fewer cardiomyocytes, with defects in DNA replication and mitosis,48 but no increase in apoptosis was noted.27,48 Similarly, embryos with KLF1 mutations have fewer blood cells that display aberrant S-phase entry, coincident with reduced Foxm1. Foxm1 mRNA is present in lower amounts in KLF1+/−KLF2−/− than in KLF1−/− KLF2+/− embryonic erythroid cells, suggesting that it is more responsive to KLF2 than KLF1, and correlating with the anemia phenotype in E10.5 KLF1+/−KLF2−/− but not KLF1−/− KLF2+/− whole-mount embryos. Although the difference in Foxm1 expression between KLF1−/− KLF2+/− and KLF1+/−KLF2−/− embryos is modest, we speculate that small changes could have large effects in a milieu where the expression of multiple proliferation genes is dysregulated. In addition, there is a modest increase in KLF2 mRNA in KLF1−/− blood cells, possibly ameliorating the KLF1−/− KLF2+/− phenotype.
In summary, the Krüppel-like transcription factors KLF1 and KLF2 are critical for embryonic erythropoiesis. They are expressed at similar amounts and display functional compensation in primitive erythroid cells. This is evident in the dramatic anemia phenotype observed in KLF1/KLF2 double knockout but not in KLF1 or KLF2 single knockout embryos at E10.5. In this study, we show that E10.5 anemia is primarily due to a reduction in the number of peripheral blood cells. Surprisingly, the anemia and the number of erythroid cells observed is dose-dependent, with more peripheral blood cells in KLF1−/− KLF2+/− than in KLF1+/−KLF2−/− embryos. KLF1 and KLF2 have a greater than additive positive effect on maintaining the colony forming ability of E8.5 erythroid progenitor cells, but KLF2 depletion alone leads to a reduced number of colonies compared to wild-type. These data suggest that certain genes involved in the maturation or proliferation of red blood cells are preferentially responsive to regulation by KLF2, as opposed to KLF1. Further studies of the roles of KLF1 and KLF2 in erythroid precursor maturation may lead to novel therapeutic strategies for the anemias.
Acknowledgments
The KLF2 knockout and conditional knockout mice were a gift from Dr. Jerry Lingrel. Susan Walker and Dr. John Povlishock provided expertise and resources for plastic embedding of tissue. Julie Farnsworth provided exceptional technical guidance for flow cytometry. We thank Dr. James Palis and Anne Koniski for invaluable technical advice on colony forming assays. We appreciate the input and suggestions of past and current members of the Lloyd, Ginder and Landry laboratories.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
Services and products in support of the research project were generated by the VCU Massey Cancer Center Flow Cytometry Shared Resource, supported, in part, with funding from NIH-NCI Cancer Center Support Grant P30 CA016059.This work was supported by two grants from the National Institutes of Health, R01DK074694 and R56DK097907.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.McGrath K, Palis J. Ontogeny of erythropoiesis in the mammalian embryo. Curr Top Dev Biol. 2008;82:1–22 [DOI] [PubMed] [Google Scholar]
- 2.Haar JL, Ackerman GA. A phase and electron microscopic study of vasculogenesis and erythropoiesis in the yolk sac of the mouse. Anat Rec. 1971;170(2):199–223 [DOI] [PubMed] [Google Scholar]
- 3.Ferkowicz MJ, Yoder MC. Blood island formation: Longstanding observations and modern interpretations. Exp Hematol. 2005;33(9):1041–7 [DOI] [PubMed] [Google Scholar]
- 4.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126(22):5073–84 [DOI] [PubMed] [Google Scholar]
- 5.Palis J. Ontogeny of erythropoiesis. Curr Opin Hematol. 2008;15(3):155–61 [DOI] [PubMed] [Google Scholar]
- 6.Kingsley PD, Malik J, Fantauzzo KA, Palis J. Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood. 2004;104(1):19–25 [DOI] [PubMed] [Google Scholar]
- 7.Baron MH, Vacaru A, Nieves J. Erythroid development in the mammalian embryo. Blood Cells Mol Dis. 2013;51(4):213–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010;90(4):1337–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375(6529):318–22 [DOI] [PubMed] [Google Scholar]
- 10.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375(6529):316–8 [DOI] [PubMed] [Google Scholar]
- 11.Basu P, Lung TK, Lemsaddek W, Sargent TG, Williams DC, Jr., Basu M, et al. EKLF and KLF2 have compensatory roles in embryonic beta-globin gene expression and primitive erythropoiesis. Blood. 2007;110(9):3417–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang CJ, Lemsaddek W, Alhashem YN, Bondzi C, Redmond LC, Ah-Son N, et al. Krüppel-like factor 1 (KLF1), KLF2, and myc control a regulatory network essential for embryonic erythropoiesis. Mol Cell Biol. 2012;32(13):2628–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107(8):3359–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilon AM, Arcasoy MO, Dressman HK, Vayda SE, Maksimova YD, Sangerman JI, et al. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol Cell Biol. 2008;28(24):7394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tallack MR, Keys JR, Humbert PO, Perkins AC. EKLF/KLF1 controls cell cycle entry via direct regulation of E2f2. J Biol Chem. 2009;284(31):20966–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tallack MR, Magor GW, Dartigues B, Sun L, Huang S, Fittock JM, et al. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res. 2012;22(12):2385–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnaud L, Saison C, Helias V, Lucien N, Steschenko D, Giarratana MC, et al. A dominant mutation in the gene encoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am J Hum Genet. 2010;87(5):721–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffray JA, Mitchell WB, Gnanapragasam MN, Seshan SV, Guo X, Westhoff CM, et al. Erythroid transcription factor EKLF/KLF1 mutation causing congenital dyserythropoietic anemia type IV in a patient of Taiwanese origin: Review of all reported cases and development of a clinical diagnostic paradigm. Blood Cells Mol Dis. 2013;51(2):71–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu P, Morris PE, Haar JL, Wani MA, Lingrel JB, Gaensler KM, et al. KLF2 is essential for primitive erythropoiesis and regulates the human and murine embryonic beta-like globin genes in vivo. Blood. 2005;106(7):2566–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alhashem YN, Vinjamur DS, Basu M, Klingmüller U, Gaensler KM, Lloyd JA. Transcription factors KLF1 and KLF2 positively regulate embryonic and fetal beta-globin genes through direct promoter binding. J Biol Chem. 2011;286(28):24819–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Redmond LC, Dumur CI, Archer KJ, Grayson DR, Haar JL, Lloyd JA. Krüppel-like factor 2 regulated gene expression in mouse embryonic yolk sac erythroid cells. Blood Cells Mol Dis. 2011;47(1):1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wani MA, Means RT, Jr, Lingrel JB. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 1998;7(4):229–38 [DOI] [PubMed] [Google Scholar]
- 23.Chiplunkar AR, Lung TK, Alhashem Y, Koppenhaver BA, Salloum FN, Kukreja RC, et al. Krüppel-like factor 2 is required for normal mouse cardiac development. PLoS One. 2013;8(2):e54891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11(22):2996–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JS, Yu Q, Shin JT, Sebzda E, Bertozzi C, Chen M, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11(6):845–57 [DOI] [PubMed] [Google Scholar]
- 26.Nielsen PJ, Lorenz B, Muller AM, Wenger RH, Brombacher F, Simon M, et al. Altered erythrocytes and a leaky block in B-cell development in CD24/HSA-deficient mice. Blood. 1997;89(3):1058–67 [PubMed] [Google Scholar]
- 27.Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388(12):1257–74 [DOI] [PubMed] [Google Scholar]
- 28.Le Scolan E, Pchejetski D, Banno Y, Denis N, Mayeux P, Vainchenker W, et al. Overexpression of sphingosine kinase 1 is an oncogenic event in erythroleukemic progression. Blood. 2005;106(5):1808–16 [DOI] [PubMed] [Google Scholar]
- 29.Qian J, Colbert MC, Witte D, Kuan CY, Gruenstein E, Osinska H, et al. Midgestational lethality in mice lacking the parathyroid hormone (PTH)/PTH-related peptide receptor is associated with abrupt cardiomyocyte death. Endocrinology. 2003;144(3):1053–61 [DOI] [PubMed] [Google Scholar]
- 30.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31(1):122–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230(2):230–42 [DOI] [PubMed] [Google Scholar]
- 32.England SJ, McGrath KE, Frame JM, Palis J. Immature erythroblasts with extensive ex vivo self-renewal capacity emerge from the early mammalian fetus. Blood. 2011;117(9):2708–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik J, Kim AR, Tyre KA, Cherukuri AR, Palis J. Erythropoietin critically regulates the terminal maturation of murine and human primitive erythroblasts. Haematologica. 2013;98(11):1778–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinrich AC, Pelanda R, Klingmüller U. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood. 2004;104(3):659–66 [DOI] [PubMed] [Google Scholar]
- 35.Peterson KR, Fedosyuk H, Zelenchuk L, Nakamoto B, Yannaki E, Stamatoyannopoulos G, et al. Transgenic cre expression mice for generation of erythroid-specific gene alterations. Genesis. 2004;39(1):1–9 [DOI] [PubMed] [Google Scholar]
- 36.Tang Y, Harrington A, Yang X, Friesel RE, Liaw L. The contribution of the Tie2+ lineage to primitive and definitive hematopoietic cells. Genesis. 2010;48(9):563–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tallack MR, Keys JR, Perkins AC. Erythroid Krüppel-like factor regulates the G1 cyclin dependent kinase inhibitor p18INK4c. J Mol Biol. 2007;369(2):313–21 [DOI] [PubMed] [Google Scholar]
- 38.Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat Genet. 2010;42(9):801–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helias V, Saison C, Peyrard T, Vera E, Prehu C, Cartron JP, et al. Molecular analysis of the rare in(lu) blood type: Toward decoding the phenotypic outcome of haploinsufficiency for the transcription factor KLF1. Hum Mutat. 2013;34(1):221–8 [DOI] [PubMed] [Google Scholar]
- 40.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93(22):12355–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371(6494):221–6 [DOI] [PubMed] [Google Scholar]
- 42.Fujiwara Y, Chang AN, Williams AM, Orkin SH. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103(2):583–5 [DOI] [PubMed] [Google Scholar]
- 43.Sue N, Jack BH, Eaton SA, Pearson RC, Funnell AP, Turner J, et al. Targeted disruption of the basic Krüppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol Cell Biol. 2008;28(12):3967–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Funnell AP, Norton LJ, Mak KS, Burdach J, Artuz CM, Twine NA, et al. The CACCC-binding protein KLF3/BKLF represses a subset of KLF1/EKLF target genes and is required for proper erythroid maturation in vivo. Mol Cell Biol. 2012;32(16):3281–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Funnell AP, Mak KS, Twine NA, Pelka GJ, Norton LJ, Radziewic T, et al. Generation of mice deficient in both KLF3/BKLF and KLF8 reveals a genetic interaction and a role for these factors in embryonic globin gene silencing. Mol Cell Biol. 2013;33(15):2976–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou D, Pawlik KM, Ren J, Sun CW, Townes TM. Differential binding of erythroid Krüppel-like factor to embryonic/fetal globin gene promoters during development. J Biol Chem. 2006;281(23):16052–7 [DOI] [PubMed] [Google Scholar]
- 47.Wierstra I. The transcription factor FOXM1 (forkhead box M1): Proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res. 2013;118:97–398 [DOI] [PubMed] [Google Scholar]
- 48.Ramakrishna S, Kim IM, Petrovic V, Malin D, Wang IC, Kalin TV, et al. Myocardium defects and ventricular hypoplasia in mice homozygous null for the forkhead box M1 transcription factor. Dev Dyn. 2007;236(4):1000–13 [DOI] [PubMed] [Google Scholar]


