Figure 1.
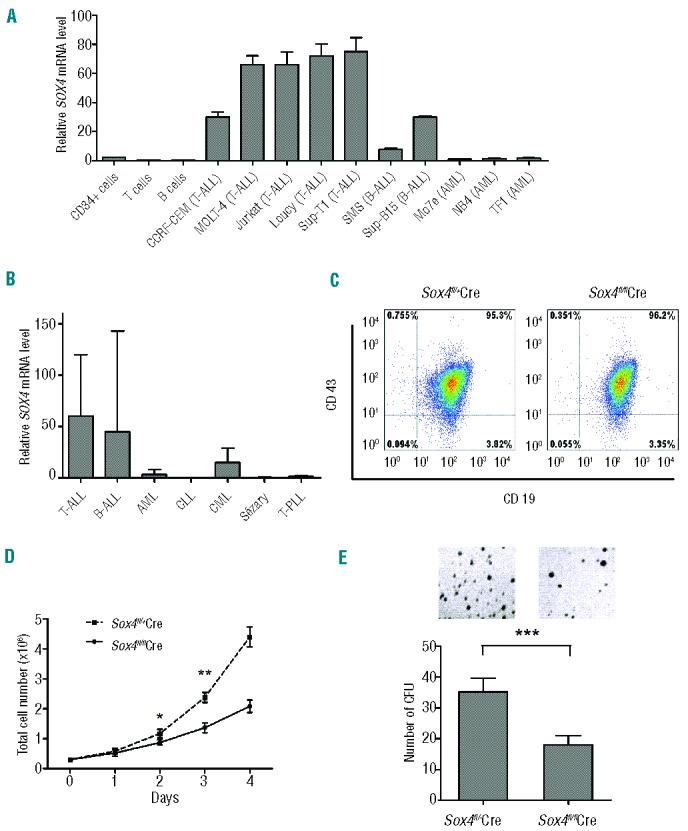
Role of Sox4 in ALL. (A) SOX4 mRNA expression in normal bone marrow CD34+ cells, normal peripheral blood T and B cells, and human ALL and AML cell lines as determined by real-time RT-PCR. The expression level in pooled peripheral blood mononuclear cells was set as 1. Expression of 18S rRNA was used for normalization. (B) SOX4 mRNA expression in leukemic cells from patients with T-cell ALL (n=21), B-cell ALL (n=34), AML (n=22), CLL (n=6), CML (n=7), Sézary disease (n=5), and T-cell prolymphocytic leukemia (T-PLL, n=5). The expression level in pooled peripheral blood mononuclear cells was set as 1. Expression of 18S rRNA was used for normalization. (C) Flow cytometry analysis of p190 BCR-ABL (mCherry+) transformed Sox4fl/flSE-Cre or Sox4fl/+SE-Cre cells for CD43 and CD19 expression. Purified B220+ B cells from bone marrow of Sox4fl/flRosa26-eYFP (experimental) and Sox4fl/+Rosa26-eYFP (control) mice were cultured with OP-9 primary bone marrow stromal cells in the presence of interleukin-7, which yielded pro-B cells of >95% purity.29 Subsequently, the pro-B cells were transformed with p190 BCR-ABL (mCherry+) and transduced with self-excising Cre (SE-Cre) retrovirus.22 Cell surface expression of CD43 and CD19 was examined by flow cytometry. (D) Effect of Sox4fl deletion on the proliferation of ALL cells. p190 BCR-ABL (mCherry+) transformed Sox4fl/flSE-Cre or Sox4fl/+SE-Cre cells sorted out by flow cytometry were cultured, and the cells were enumerated daily for up to 5 consecutive days. Data are representative of three independent experiments. (E) Effect of Sox4 depletion on the colony-forming ability of BCR-ABL-transformed pro-B cells. Ten thousand experimental and control cells, as described in (C), were seeded in semi-solid medium in 3.5-cm dishes and colonies were counted after 14 days. The mean colony numbers were 18 ± 2.6 and 35 ± 3.9 for experimental and control cells, respectively. Data are representative of three independent experiments. Values are means ± SD (n≥3). *P<0.05, **P<0.01, ***P<0.001
