During erythroid differentiation, co-operative activities of erythroid transcription factors co-ordinate transcript levels of genes required for correct maturation.1 These genes include the eight heme biosynthetic enzymes that are sequentially up-regulated to ensure augmented heme synthesis essential for the formation of hemoglobin.
The transcription factor nuclear factor erythroid 2 (NFE2) plays a pivotal role in erythroid maturation and, together with other erythroid transcription factors such as EKLF and GATA1,1 controls the transcription of erythroid-specific genes, including β-globin.2 Furthermore, NFE2 is necessary for the transcriptional activation of two heme biosynthetic enzymes: porphobilinogen deaminase (PBGD)3 and ferrochelatase (FECH).4
This led us to investigate the role of NFE2 in the regulation of the remaining heme biosynthetic enzymes. In silico analysis revealed potential NFE2 consensus sites within promoter and upstream regions of five enzymes besides PBGD and FECH: aminolevulinic acid dehydratase (ALAD), uroporphyrinogen III synthase (UROS), uroporphyrinogen III decarboxylase (UROD), coproporphyrinogen oxidase (CPOX), and protoporphyrinogen oxidase (PPOX) (Figure 1A). Chromatin immunoprecipitation (ChIP) assays using K562 cells5 confirmed in vivo binding of NFE2 to the UROS erythroid promoter at the -27 bp binding site (Figure 1B, second row). NFE2 also bound the sequence between bp -452 and bp -183 of the UROD promoter, encompassing two predicted consensus sites at bp -391 and bp -311 (Figure 1B, third row). Finally, we observed strong NFE2 binding in the sequence containing the -5161 bp site of the CPOX promoter (Figure 1B, fourth row). The other sites investigated were not bound by NFE2. This suggests that UROS, UROD and CPOX constitute novel direct NFE2 target genes and raises the question as to whether NFE2 activity is required for transcription of these genes.
Figure 1.
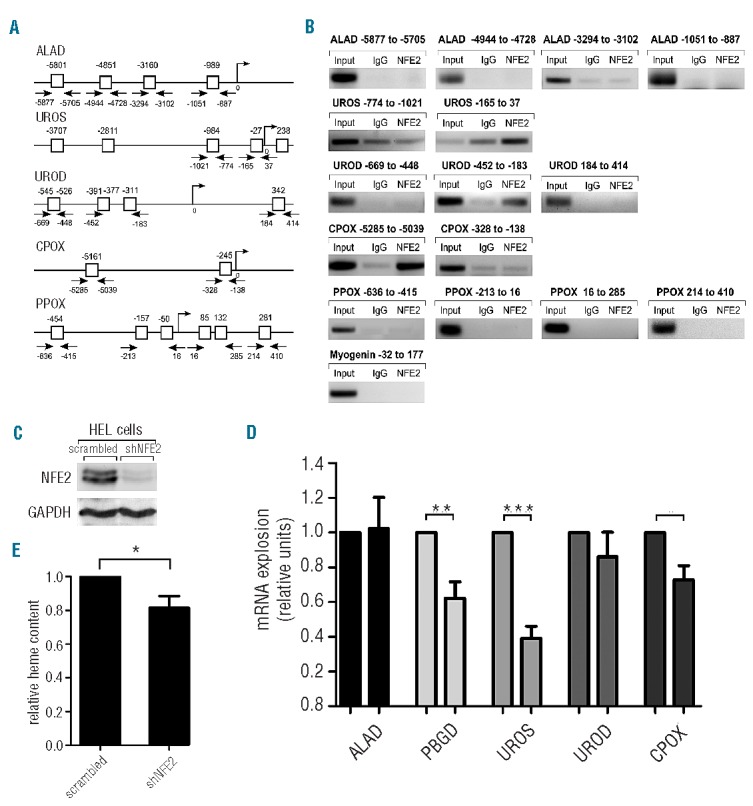
(A) Predicted NFE2 consensus sites in the promoters of heme biosynthetic enzymes. (B) Chromatin immunoprecipitation of K562 cells was performed with either an NFE2 antibody or an unrelated IgG control, as indicated. Precipitated DNA was amplified using primers covering the predicted binding sites shown in A. In the lane marked “Input”, a 1:25 dilution of the input DNA was used. As an additional control, a genomic region of myogenin known not to interact with NFE2 was amplified (bottom panel). (C–E) HEL cells were treated with a lentiviral construct containing either shRNA against NFE2 (shNFE2) or scrambled RNA as negative control. (C) Western blot analysis of NFE2 protein expression. Equal protein loading was assessed by reprobing with an antibody against GAPDH. (D) mRNA expression of ALAD, PBGD, UROS, UROD, CPOX and the housekeeping gene β-2-microglobulin (β-2-M) was quantified by qRT-PCR. Expression levels of the heme biosynthetic enzymes were normalized to β-2-M expression and the expression in cells treated with scrambled RNA set to 1. Values are reported as fold change shRNA versus scrambled RNA-treated cells and represent the means and standard error of the mean of four individual experiments measured in duplicates. *P<0.05; **P<0.01; ***P<0.001 one sample t-test. (E) Heme content of the cells was determined by measuring the fluorescence at an excitation wavelength of 400 nm and an emission wavelength of 662 nm. Values are shown relative to heme content in cells treated with scrambled RNA, which was set to one. The results represent the means and standard error of the mean of two independent transfections with five independent measurements each. *P<0.05 paired t-test.
To investigate this, we depleted human erythroleukemia (HEL) cells of NFE2 by transduction with a lentiviral construct containing either an shRNA against NFE2 (shNFE2)6 or a scrambled RNA as a negative control. Western blot analysis demonstrated a strong reduction in NFE2 protein expression after transduction with shNFE2 (Figure 1C). The expression levels of ALAD, PBGD, UROS, UROD and CPOX were determined by quantitative real-time PCR (Figure 1D). mRNA expression of both UROS and CPOX was decreased by 60% and 25%, respectively, in HEL cells silenced for NFE2 expression. UROD expression showed a slight, but reproducible reduction, which did not reach statistical significance. As expected, ALAD mRNA levels remained unchanged while the mRNA levels of PBGD, which is a known NFE2 target,3 were reduced by approximately 40% in HEL cells following NFE2 knockdown. These data demonstrate that NFE2 activity is required for full expression of UROS and CPOX and contributes to UROD expression, confirming these enzymes as novel NFE2 target genes. Consistent with this decrease in mRNA levels, HEL cells silenced for NFE2 displayed a reduction in overall cellular heme content by 20% as determined by a microfluorometric assay7 (Figure 1E). These data underscore the critical role of NFE2 in controlling heme biosynthesis.
Because UROS expression was most strongly reduced upon NFE2 knockdown, we wished to further delineate the effect of NFE2 on the UROS promoter. We therefore cloned either a region encompassing the proximal 1438 bp of the UROS promoter (UROS -1438), which includes both sites at bp -27 and at bp -984, or a fragment encoding 790 bp (UROS -790), which only includes the site at bp 27, into luciferase reporter gene vectors (Figure 2A). The resulting constructs were transfected into HEK293T cells in the presence or absence of expression vectors encoding NFE2 and MafG, as previously described.5 Co-transfection of both NFE2 and MafG with the UROS 1438 construct resulted in a 2.5-fold increase in UROS-promoter-driven luciferase activity (Figure 2B). Co-transfection of NFE2 and MafG with the UROS -790 construct likewise achieved a 2.2-fold increase, showing that sequences upstream of -bp 790 do not contribute to the transactivation of the UROS promoter by NFE2 (Figure 2C). Mutation of the NFE2 binding site at bp -27 completely abrogates NFE2 transactivation of the UROS promoter (Figure 2D), identifying this site as the NFE2 target sequence. These data demonstrate that NFE2 is able to directly transactivate the UROS erythroid promoter and corroborate the ChIP assays, demonstrating NFE2 binding to this sequence in vivo (Figure 1B).
Figure 2.
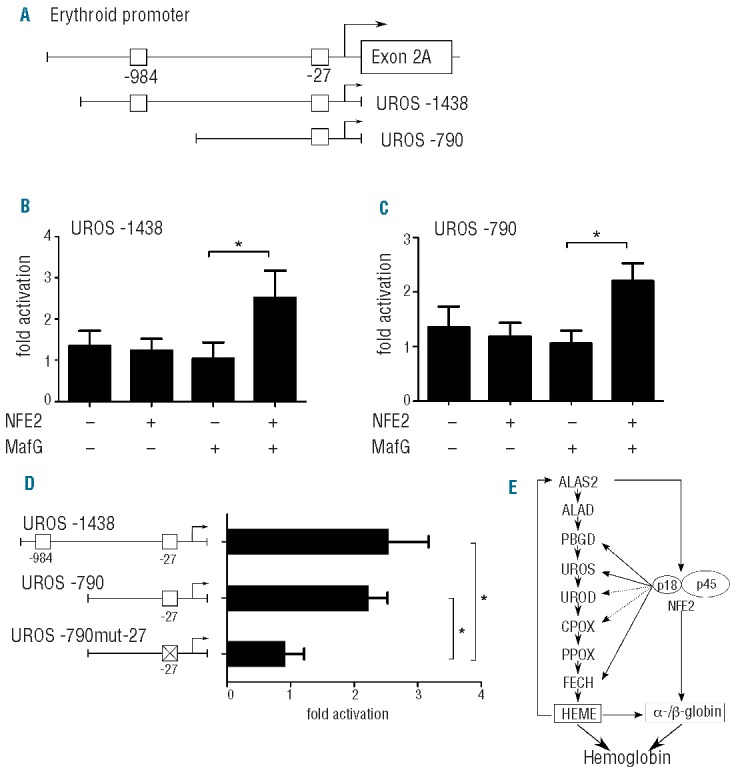
(A) Schematic representation of the UROS erythroid promoter and sequences inserted into reporter gene constructs. The constructs include bp -1438 to bp 105 (UROS -1438) and bp 790 to bp 105 (UROS -790), respectively, of the erythroid promoter. NFE2 sites predicted by in silico analysis and investigated by ChIP are indicated by open boxes. (B and C) The constructs were transfected into HEK293T cells together with expression plasmids for MafG, NFE2 or both as indicated. 24 h after transfection, luciferase activity was determined. Results were normalized for transfection efficiency by co-transfection of a Tk-driven Renilla luciferase reporter gene vector and for background luminescence of the empty pGL3 luciferase vector as previously reported.5 Values are given as fold activation with NFE2 and MafG compared to an internal calibrator consisting of the reporter constructs transfected in the presence of MafG expression vectors alone, which was set to 1. Results represent the mean and standard deviation of at least three independent experiments, each performed in duplicate. *P<0.05 one-way Mann-Whitney test. (D) By site-directed mutagenesis, a reporter construct containing a mutated NFE2 site at bp -27 (UROS-790mut-27) was generated. The constructs were co-transfected into HEK293T cells together with expression plasmids for NFE2 and MafG. Bar graphs represent the mean and standard deviation of at least three independent experiments, each performed in duplicate. *P<0.05 one-way Mann-Whitney test. (E) Proposed model of regulation of heme synthesis by NFE2.
Taken together, our results demonstrate that NFE2 modulates the heme biosynthesis pathway in erythroid cells at multiple steps and point to the existence of a novel regulatory pathway (Figure 2E). A critical step during erythropoiesis is the upregulation of the erythroid isoform of the first heme biosynthetic enzyme, 5-aminolevulinic acid synthase 2 (ALAS2),8 followed by a sequential upregulation of the remaining enzymes. Mouse erythroleukemia cells transduced with an antisense RNA against ALAS2 show decreased levels of NFE2,9 suggesting that ALAS2 expression positively regulates NFE2 expression. Thus, the initial induction of erythroid heme synthesis by activation of ALAS2 will result in increased NFE2 activity. Furthermore, hemin induction increases the activity of NFE2.10,11 NFE2 co-operates with other erythroid transcription factors that regulate the expression of heme biosynthetic enzymes, such as EKLF12 and GATA1.13 Together, this will lead to transcriptional activation of successive enzymes in the pathway, with NFE2 contributing particularly to the activation of PBGD3 and FECH,4 and, as shown here, of UROS and potentially to UROD and CPOX (Figure 2E). For hemoglobin synthesis, globin proteins and heme molecules are needed at a fixed stoichiometric ratio. NFE2 is a crucial regulator of β-globin transcription in erythroid cells.2 Regulation of both pathways by a common transcription factor such as NFE2 provides an attractive possibility of fine-tuning hemoglobin synthesis and avoiding both an excess production of individual components and an accumulation of possibly toxic metabolites.
Based on our data, we propose that NFE2 regulates the transcription of several downstream enzymes of the heme biosynthesis pathway after the initial induction by activation of ALAS2. In this way, NFE2 may establish a positive feedback loop (Figure 2E) that secures a constant supply of both heme and globin for the assembly of hemoglobin and ensures co-ordinated levels of synthesis of both proteins during erythroid differentiation.
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Ingley E, Tilbrook PA, Klinken SP. New insights into the regulation of erythroid cells. IUBMB Life. 2004;56(4):177–84 [DOI] [PubMed] [Google Scholar]
- 2.Johnson KD, Grass JA, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, et al. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc Natl Acad Sci USA. 2002;99(18):11760–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mignotte V, Eleouet JF, Raich N, Romeo PH. Cis- and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc Natl Acad Sci USA. 1989;86(17):6548–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tugores A, Magness ST, Brenner DA. A single promoter directs both housekeeping and erythroid preferential expression of the human ferrochelatase gene. J Biol Chem. 1994;269(49):30789–97 [PubMed] [Google Scholar]
- 5.Wehrle J, Seeger TS, Schwemmers S, Pfeifer D, Bulashevska A, Pahl HL. Transcription factor nuclear factor erythroid-2 mediates expression of the cytokine interleukin 8, a known predictor of inferior outcome in patients with myeloproliferative neoplasms. Haematologica. 2013;98(7):1073–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roelz R, Pilz IH, Mutschler M, Pahl HL. Of mice and men: human RNA polymerase III promoter U6 is more efficient than its murine homologue for shRNA expression from a lentiviral vector in both human and murine progenitor cells. Exp Hematol. 2010;38(9):792–7 [DOI] [PubMed] [Google Scholar]
- 7.Sassa S. Sequential induction of heme pathway enzymes during erythroid differentiation of mouse Friend leukemia virus-infected cells. J Exp Med. 1976;143(2):305–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadlon TJ, Dell’Oso T, Surinya KH, May BK. Regulation of erythroid 5-aminolevulinate synthase expression during erythropoiesis. Int J Biochem Cell Biol. 1999;31(10):1153–67 [DOI] [PubMed] [Google Scholar]
- 9.Meguro K, Igarashi K, Yamamoto M, Fujita H, Sassa S. The role of the erythroid-specific delta-aminolevulinate synthase gene expression in erythroid heme synthesis. Blood. 1995;86(3):940–8 [PubMed] [Google Scholar]
- 10.Palma JF, Gao X, Lin CH, Wu S, Solomon WB. Iron protoporphyrin IX (hemin) but not tin or zinc protoporphyrin IX can stimulate gene expression in K562 cells from enhancer elements containing binding sites for NFE2. Blood. 1994;84(4):1288–97 [PubMed] [Google Scholar]
- 11.Solomon WB, Lin CH, Palma J, Gao XY, Wu S. Suppression of a cellular differentiation program by phorbol esters coincides with inhibition of binding of a cell-specific transcription factor (NFE2) to an enhancer element required for expression of an erythroid-specific gene. J Biol Chem. 1993;268(7):5089–96 [PubMed] [Google Scholar]
- 12.Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107(8):3359–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katsumura KR, DeVilbiss AW, Pope NJ, Johnson KD, Bresnick EH. Transcriptional mechanisms underlying hemoglobin synthesis. Cold Spring Harb Perspect Med. 2013;3(9):a015412. [DOI] [PMC free article] [PubMed] [Google Scholar]


