Abstract
The decomposition of plant litter in soil is a dynamic process during which substrate chemistry and microbial controls interact. We more clearly quantify these controls with a revised version of the Guild-based Decomposition Model (GDM) in which we used a reverse Michaelis-Menten approach to simulate short-term (112 days) decomposition of roots from four genotypes of Zea mays that differed primarily in lignin chemistry. A co-metabolic relationship between the degradation of lignin and holocellulose (cellulose+hemicellulose) fractions of litter showed that the reduction in decay rate with increasing lignin concentration (LCI) was related to the level of arabinan substitutions in arabinoxylan chains (i.e., arabinan to xylan or A∶X ratio) and the extent to which hemicellulose chains are cross-linked with lignin in plant cell walls. This pattern was consistent between genotypes and during progressive decomposition within each genotype. Moreover, decay rates were controlled by these cross-linkages from the start of decomposition. We also discovered it necessary to divide the Van Soest soluble (labile) fraction of litter C into two pools: one that rapidly decomposed and a second that was more persistent. Simulated microbial production was consistent with recent studies suggesting that more rapidly decomposing materials can generate greater amounts of potentially recalcitrant microbial products despite the rapid loss of litter mass. Sensitivity analyses failed to identify any model parameter that consistently explained a large proportion of model variation, suggesting that feedback controls between litter quality and microbial activity in the reverse Michaelis-Menten approach resulted in stable model behavior. Model extrapolations to an independent set of data, derived from the decomposition of 12 different genotypes of maize roots, averaged within <3% of observed respiration rates and total CO2 efflux over 112 days.
Introduction
Recent studies are challenging the ways that we have traditionally perceived and modeled decomposition. Decomposers were once thought to rapidly degrade labile fractions of plant litter, such as carbohydrates and proteins, leaving more recalcitrant compounds, like lignin, to become the foundation of stabilized soil organic matter [1], [2], [3]. However, microbial products rather than plant lignin actually may represent the larger fraction of SOM [4], [5], and lignin per se may not persist throughout decay [6]. Unfortunately, many mathematical models are based on the traditional view of decomposition. In part this is because the chemical composition of litter has often been evaluated by proximate carbon analysis, typically yielding three, qualitatively different pools of compounds: polar and nonpolar extractives, acid hydrolysable materials, and acid non-hydrolysable materials. These pools provide a convenient structural framework for modeling, but lack the resolution to address finer scale biochemical transformations revealed by more contemporary studies [4], [5], [6]. Mathematical models must change to reflect these observations.
One of the changes needed in traditional decomposition models is to explicitly simulate the activities of microorganisms. If microbial contributions to SOM are more substantial than lignin, then the litter compounds fueling microbial activity may be more important to C stabilization than the plant lignin pool. This point was suggested by Smith et al. [7] who noted that the proportion of initial litter C remaining over time (125 days) was directly related to the initial rate of decomposition prior to lignin decay. In other words, the incorporation of C into microbial biomass increased its overall persistence despite an initially higher fractional loss through respiration [2], [8]. A simple explanation is that the higher carbon use efficiency (CUE) of readily decayed materials, like sugars and proteins, transfers a larger fraction of substrate C to microbial biomass than lignin, and that these microbial products are more persistent [4], [5]. These results suggest that the early stage of litter decay when microbial activities are greatest should be an integral component of mathematical models.
A second change needed in decomposition models is how they respond to differences and interactions between the chemical constituents of decaying litter. Despite the questionable role of plant lignin in decomposition, both the initial lignin content and the lignocellulose index (LCI) of litter are often the best predictors of decay rate [1], [3]. The lignocellulose index is normally calculated as the ratio of the acid non-hydrolysable/(non-hydrolysable+hydrolysable) products of proximate C analysis (see above), and assumed to represent the lignin (non-hydrolysable) and holocellulose (hydrolysable) fractions of litter. Although this assumption may be nearly true for fresh litter, both microbial and lignin degradation products increasingly contribute to proximate C fractions during decay [5], [6]. Thus LCI and proximate C fractions are ambiguous metrics of litter chemistry and models using them to regulate decomposition conflate litter quality controls with the decomposition process. In reality, plant cell walls are usually the largest component of plant litter and have a specific biophysical structure of interconnected saccharide and phenolic polymers [9]. This arrangement partly explains the relatively consistent relationship between LCI and decay, but raises questions about the precise relationships between lignin and other litter constituents [10], [11], [12] needed to more accurately model substrate dynamics during decomposition.
Few empirical studies have examined litter chemistry during decay with sufficient resolution to improve models beyond the lignin or LCI controls normally calculated. However, Machinet et al. [11], [13], [14] examined the detailed litter chemistry of maize (Zea mays L.) roots for naturally occurring genotypes decomposing in laboratory incubations. These genotypes varied primarily in their lignin content. Long-term C losses (>200 days) were most often correlated with compounds associated with lignin and cross-linked between lignin and polysaccharides, whereas short-term (<200 days) losses were more closely related to soluble compounds and cell wall sugars [11]. Different loss rates of different sugars (e.g., glucans, xylans and arabinans) suggested that the relationship between lignin and decomposition was partly mediated by the specific composition of the polysaccharide fraction of the litter, probably because arabinan substitution in xylan chains interferes with the degradation of hemicellulose [9]. In other words, the sugar composition of cell walls appears to define the transition from short- to long-term patterns of decay, providing a mechanistic explanations for patterns in microbial activity, as well as the empirical relationships between LCI and decay most often used in decomposition models [15].
The overall goal of this study was to simulate the relationships between litter decay, microbial production and litter chemistry at the early stage of decomposition, using data collected by Machinet et al. [11], [13] to test and refine the Guild Decomposition Model (GDM) developed by Moorhead and Sinsabaugh [16]. Our two specific objectives were to simulate patterns of microbial productivity during decomposition as litter chemistry changed, and to simulate the relationship between measured changes in cell wall chemistry (sugars and lignin) and decay rate. We chose GDM because it calculates the decay of specific substrate pools as a combined function of both microbial activity and substrate characteristics, using the Michaelis-Menten equation of substrate saturation. The maximum velocity of decomposition (Vmax) is thus a function of microbial biomass [16], so that estimated decay rates reflect both the changing chemical composition of the decomposing litter and microbial productivity.
Methods
Our general approach was to revise GDM to use the empirical data collected by Machinet et al. [11], [13], [14] to derive parameters needed to simulate decomposition. Then, we analyzed the data from a 112-day laboratory study of four maize genotypes that differed from one another in initial litter chemistry [13], [14], to evaluate interactions between litter qualities likely controlling the decomposition process, and to obtain estimates for parameter values used to describe these processes. Next, we selected two key parameters for GDM, which preliminary analyses indicated varied with initial litter chemistry and had large effects on model behavior (below). We then optimized these parameters to produce the best possible fit between simulated and observed patterns of decomposition, with respect to CO2 efflux and chemical transformations in decaying litter. Best-fit parameter values were in turn compared to initial litter chemical characteristics to determine possible relationships. These relationships were then used to derive parameter values needed to simulate CO2 efflux during decomposition of maize roots for 12 additional genotypes [11].
Experimental data
The data used to drive the revised GDM model were obtained from Machinet et al. [13]. Briefly fine roots (diameter 2–3 mm) of four natural genotypes of maize (F2, F2bm1, F292 and F292bm3), which differed in their chemical composition (Table 1), were cut into 5 mm lengths, added to soil in laboratory microcosms and incubated at 15°C. Potassium-nitrate fertilizer was added to microcosms to insure no nitrogen limitation [17]. They monitored CO2-C efflux on days 3, 7, 10, 14, 21, 29, 36, 42, 51, 57, 70, 80, 87, 95 and 112. Chemical characteristics of litter were determined on days 0 (initial), 14, 36, 57 and 112, on roots manually removed from soils. Machinet et al. [11] used the same experimental methods for 12 additional maize genotypes, but examined only the initial litter chemistry and measured CO2 efflux during decomposition.
Table 1. Model parameters and state variables.
| Factor | Description | Valuesa (genotype) | Unit |
| k1max | Decay rate coefficient of substrate C1 | 0.1 | day−1 |
| k2max | Maximum decay rate coefficient of substrate C2 | 0.047b | day−1 |
| k3max | Maximum decay rate coefficient of substrate C3 | 0.001 | day−1 |
| e1 | Substrate C1 use efficiency | 0.4 | unitless |
| e2 | Substrate C2 use efficiency | 0.3 | unitless |
| g | Coefficient for basal respiration | 0.001 | day−1 |
| BCmax | Maximum Biomass: total C ratio | 0.05 | unitless |
| LCIT | LCI value at which C3 decay starts | 0.42 (F2) | unitless |
| 0.36 (F2bm1) | |||
| 0.46 (F292) | |||
| 0.45 (F292bm3) | |||
| LCImax | LCI value at which decay stops | 0.7 | unitless |
| KBj1 | Half-saturation coefficient of all Guilds for substrate C1 | 10 | mg C·kg−1 soil |
| KB22 | Half-saturation coefficient of Guild 2 for | 29 (F2) | mg C·kg−1 |
| substrate C2 | 33 (F2bm1) | soil | |
| 23 (F292) | |||
| 19 (F292bm3) | |||
| KB32 | Half-saturation coefficient of Guild 3 for | 300 | mg C·kg−1 |
| substrate C2 | soil | ||
| KB33 | Half-saturation coefficient of Guild 3 for substrate C3 | 500 | mg C·kg−1 soil |
| C1T | Non-decomposable fraction of C1 | 0.61b (F2) | unitless |
| 0.60 (F2bm1) | |||
| 0.71 (F292) | |||
| 0.90 (F292bm3) | |||
| C1 | Litter C in C1 pool (Van Soest soluble) | 364 (F2) | mg C·kg−1 |
| 419 (F2bm1) | soil | ||
| 361 (F292) | |||
| 288 (F292bm3) | |||
| C2 | Litter C in C2 pool (acid hydrolysable fraction) | 1238 (F2) | mg C kg−1 |
| 1166 (F2bm1) | soil | ||
| 1295 (F292) | |||
| 1387 (F292bm3) | |||
| C3 | Litter C in C3 pool (acid non-hydrolysable | 397 (F2) | mg C·kg−1 |
| fraction) | 415 (F2bm1) | soil | |
| 344 (F292) | |||
| 325 (F292bm3) | |||
| B1 | C in Guild 1 | 10 | mg C·kg−1 |
| soil | |||
| B2 | C in Guild 2 | 10 | mg C·kg−1 |
| soil | |||
| B3 | C in Guild 3 | 5 | mg C·kg−1 |
| soil |
Initial values for variables.
Empirically determined from experimental data.
The suite of litter chemical characteristics reported by Machinet et al. [11], [13] in root residues included C and N content, Van Soest soluble C (C-SOL) and N (N-SOL) contents, and cell wall polysaccharides: glucan (Glu), arabinan (Ara) and xylan (Xyl), which are the major polymer carbohydrates in graminaea, as well as galactan (Gal) and uronic acids contents (galacturonic (Ac Gal) and glucuronic (Ac Glu)). They also determined Klason lignin (KL) and the lignin monomers, guaiacyl (G) and syringyl (S), as well as ester-linked p-coumaric (pCA) and ferulic acids (FAester), and ether-linked ferulic acids (FAether). From these data we calculated the sum of cell wall sugars (∑Sug), arabinan∶xylan ratio (A∶X), carbon∶nitrogen ratio (C∶N), lignocellulose index (LCI = C3/[C2+C3])) with C1 ( = C-SOL), C2 ( = ∑Sug) and C3 ( = KL), and non-detergent fiber (NDF) content.
Machinet et al. [13] estimated mass loss of litter based on cumulative CO2 carbon efflux. Concentrations of litter chemical fractions at each date were multiplied by the estimated mass of remaining litter to estimate pool sizes during decomposition. Decay rate coefficients (ki) were calculated for chemical constituents C1, C2 and C3 for all 4 litter types over all 4 periods of observation (days 0–14, 14–36, 36–57 and 57–112), as the difference in the natural log of pool size between observation dates, divided by the time period.
Model revisions
This model was programmed in MATLAB (The MathWorks Inc., Natick, USA). We revised GDM to use Reverse Michaelis-Menten (RMM) functions to calculate decay rates as functions of microbial activity [18]: dCi/dt = ki·Ci·Bj/(KBji+Bj), where Ci is the amount of substrate i, Bj is the amount of microbial biomass in guild j, and KBji is the half-saturation coefficient of guild j for substrate i. Note that we altered the more common RMM approach by using biomass (B) in place of an enzyme (E) pool, which assumes a constant ratio of E∶B resulting from constitutive enzyme production [16], [18]. We limited access to substrate pools by guilds: guild 1 was assumed to access only soluble resources (C1); guild 2 was assumed to access both C1 and C2; guild 3 could access C1, C2 and C3 (Figure 1).
Figure 1. Carbon flow diagram for revised Guild Decomposition Model (GDM) simulating Zea mays root decomposition.
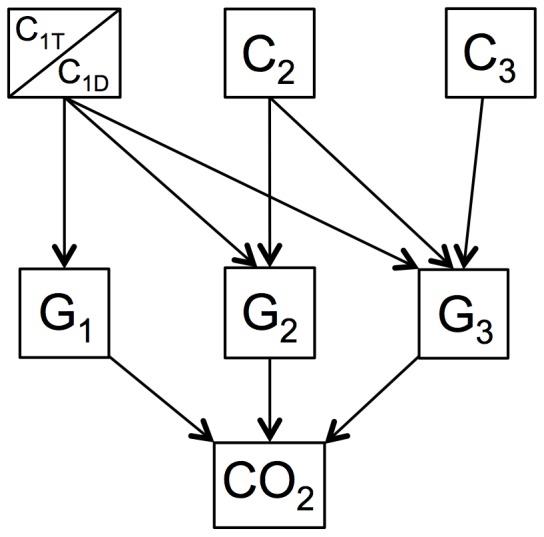
Litter pools are two Van Soest soluble fractions, a decomposable fraction (C1D) and a resistant fraction (C1T), acid hydrolysable (C2) and acid non-hydrolysable (C3) fractions, and microbial guilds of opportunists (G1), cellulolytic decomposers (G2) and lignolytic decomposers (G3).
A final revision to GDM was necessary to capture the dynamics of the soluble pool (C1), a fraction of which persisted throughout the study by Machinet et al. [13]. This persistent fraction (C1T) varied between genotypes and was considered to be non-decomposable during the time frame of the study (112 days).
Parameters and state variables
All parameters and state variables are reported in Table 1. Values of C1T, the persistent fraction of the initial C1 pool, were estimated from the average values of C1 over time for each genotype (Table 1). We selected a decay rate coefficient (k1) of 0.1 for the decomposable fraction of the C1 pool (C1D), due to its rapid loss.
The RMM approach made it possible to simplify the balance of C2 and C3 decay as functions of lignocellulose index (LCI) according to Moorhead et al. [19] (Text S1). In brief, the decay rate coefficients (k2 and k3) were described as linear functions of LCI: ki = mi·LCI+kimax, given empirically observed maximum values (kimax) and slopes (mi). This approach assumes that k3 = 0 at a threshold level of LCI = LCIT, wherein LCIT = 0.4 [20], which defines the point at which LCI changes from being solely determined by C2 decay (LCI≤LCIT) to also being determined by C3 decay (LCI>LCIT). The value of k2max (maximum coefficient of C2 decay) was estimated as the intercept of the linear regression of the observed decay rate coefficients for C2 (k2) against litter LCI for the four maize genotypes (Figure 2a; Table 1) over days 14–112, excluding values estimated over days 0–14 when we assumed that microorganisms had not begun to fully utilize pool C2. Values of k2 used during simulations were then estimated according to LCI of remaining litter. This revision to GDM provided a closer fit to observed patterns of C2 decay (Figure 2a; N = 12, R2 = 0.97, P<0.01) than Moorhead and Sinsabaugh [16]. The value of k3max was set to 0.001 because there was little evidence of C3 decomposition for any litter type during incubations [13].
Figure 2. Observed and simulated decay rate coefficients for holocellulose and araban∶xylan composition of remaining litter at residual values of lignocellulose index (LCI).
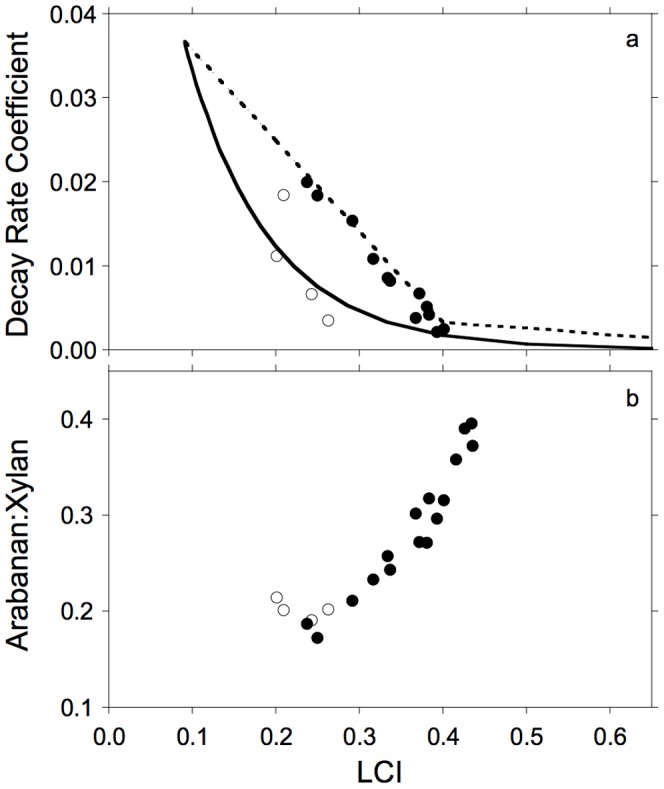
Relationships between observed and simulated: a. decay rate coefficients (k2) for litter pool C2 and lignocellulose index (LCI) of litter (solid line is simulated according to Moorhead and Sinsabaugh [16], dashed line is simulated according to Moorhead et al. [19]), b. relationship between arabinan∶xylan (A∶X) and LCI contents of decaying litter. Open circles are observations based on initial litter chemistry (day 0) and solid circles are from observations over time.
We selected identical values of KB11, KB12 and KB13, the half-saturation coefficients for the utilization of pool C1 by all three guilds, assuming all microorganisms have similar affinities for soluble substrates (Table 1). We initially set KB22 and KB32 at the same values, again assuming that organisms capable of using the resource would have similar half-saturation coefficients, but higher than those for C1, assuming that C2 was generally less decomposable than C1. We then selected an even higher value for KB33 assuming that C3 was even less decomposable than C2. We are unaware of any published values for these parameters, and so followed the rationale of Moorhead and Sinsabaugh [16] in choosing values reflecting relative access to substrates of different qualities.
GDM requires an estimate of initial decomposer biomass, which it divides among three distinct microbial pools: early opportunists (guild 1), subsequent decomposition specialists (guild 2) and a final group of lignin degraders (guild 3) (Figure 1). Machinet et al. [13] estimated that microorganisms colonizing maize roots contributed 0–11% of the initial litter carbon content, and other studies suggest that microbial biomass rarely exceeds 2–3% of soil organic matter [21], [22]. We set the initial microbial biomass pool at 1.25% (25 mgC·kg−1 soil) of total litter mass, assuming 10 mgC·kg−1 soil for guilds 1 and 2, and 5 mgC kg−1 soil for guild 3 [16].
We estimated microbial production as the difference between the quantity of carbon released from decaying substrates and the amount mineralized through both growth- and maintenance-associated respiration. We assumed that this difference was immobilized in microbial biomass. GDM also calculates microbial turnover necessary to keep total microbial C less than 5% of the total system's organic C (microorganisms+substrates; Table 1). We estimated carbon use efficiency (CUE) as the difference between the amounts of C released from decaying substrates and mineralized through respiration, divided by the amount of C released by decomposition [23].
Optimizing parameter values
The values of two key model parameters that showed correlations with initial chemistry, LCIT, and KB22, were optimized for the four maize genotypes [13] using the fmincon function of the MATLAB Optimization Toolbox (The MathWorks, Inc., Natick, USA), which applies a sequential quadratic programming algorithm. The objective function was the sum of the root mean square errors calculated between the experimental and simulated CO2 efflux rates, cumulative amount of carbon mineralized over 112-days and chemical evolution in the C1 and C2 pools during decomposition, normalized by the means of the observations.
We determined the best-fit estimates of parameters KB22 and LCIT that most closely matched simulations to observed patterns of CO2 efflux and both C1 and C2 pools over time (Table 1). We selected these parameters because the dynamics of pool C2 were most closely related to cumulative CO2 efflux and thus, estimated mass loss.
Sensitivity analysis
To evaluate the sensitivity of the model to parameter estimates (Table 1), we randomly varied model parameters e1, e2, k1max, BCmax and KB11 within ±10% of their initial values (Table 1), 100 times for each litter type. We then calculated the differences between simulated and observed values of C1, C2, cumulative CO2, and respiration rates on each date of observation [13] as a relative value = (observation-simulation)/observation. We summed these relative differences for each type of observation over all days of observation (i.e., through day 112), which produced a composite measure of the relative differences between observations and model output for each maize genotype. ANCOVA evaluated the contributions of variations in parameter values to variations in the relative differences between model output and observations by litter type. The type II sums of squares from ANCOVA were interpreted to represent the relative contribution of each parameter to model behavior.
Model extrapolation
The last set of simulations estimated decomposition for the 12 additional maize genotypes examined by Machinet et al. [13]. Relationships between values of C1T and best-fit values of LCIT and KB22 (Table 1) and the initial characteristics of litter chemistry were estimated for the four litter types described by Machinet et al. [13]: LCIT = −0.36·KL/AX+0.70 (N = 4; R2 = 0.923; P≤0.05); C1T = 0.02 · ∑Sug – 0.64 (N = 4; R2 = 0.998; P≤0.05); and KB22 = 53.22 · KL/AX −13.65 (N = 4; R2 = 0.975; P≤0.05). These relationships were then used to estimate parameter values of C1T, LCIT and KB22 for simulations with each of the 12 additional genotypes reported by Machinet et al. [11], based on their initial chemistry. Principal components analyses evaluated relationships between the relative differences in observed and simulated values of respiration rates and cumulative CO2 efflux, and initial litter chemistry characteristics for all 12 litters to determine if more detailed litter chemical characteristics than currently used in the model could provide additional insights to litter quality controls on decomposition.
Results
Decomposition dynamics
We discovered a close correspondence between the arabinan∶xylan ratio (A∶X) and lignocellulose index (LCI) of the residue (Figure 2b) in the empirical data [11]. Between days 14 and 112, a linear regression of AX over LCI yielded an R2 = 0.91 (N = 12, P≤0.01); we omitted days 0–14 because we expected decomposition to be limited by microbial activity rather than substrate [16], [18]. Perhaps this relationship also explains why both AX and KL were related to best-fit model parameters LCIT and KB22.
Rates of CO2 efflux for all four litter types rapidly increased to peak values within 14–21 days followed by gradual declines (Figure 3); cumulative CO2 efflux rose rapidly during the first 36 days and then more slowly until day 112. For 3 of the 4 genotypes, the C1 pool declined rapidly from day 1 to day 14, and then remained relatively constant during the rest of the incubation (Figure 4). For genotype F292bm3, the pool of C1 remained essentially unchanged throughout the study. The C2 pools of all four litters declined throughout the incubations, accounting for most of the litter mass loss over time (Figure 4). The C3 fraction of the remaining litter showed a slight increase over the first 14 days of incubation (ca. 5–10%) for both F2 genotypes (not shown), but remained roughly constant for the two F292 genotypes [13].
Figure 3. Observed and simulated respiration rates and cumulative CO2 efflux during decomposition of Zea mays root litter.
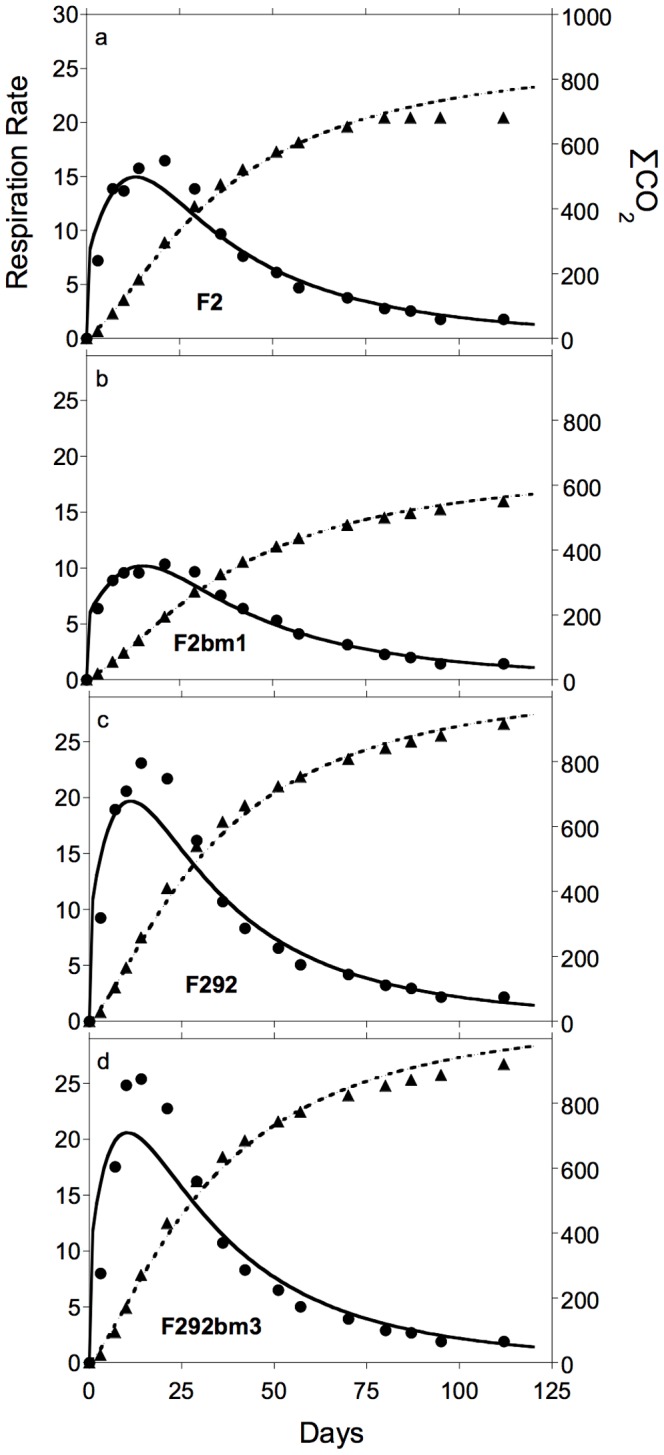
Observed (symbols) and simulated (lines) patterns of respiration rates (solid lines and circles, mgC·kg soil−1·d−1) and cumulative CO2 efflux (dashed lines and triangles, mgC·kg soil−1) for maize mutants, a. F2, b. F2bm1, c. F292 and d. F292bm3.
Figure 4. Observed and simulated mass remaining for soluble (C1) and acid hydrolysable (C2) chemical fractions of litter during decomposition of Zea mays roots.
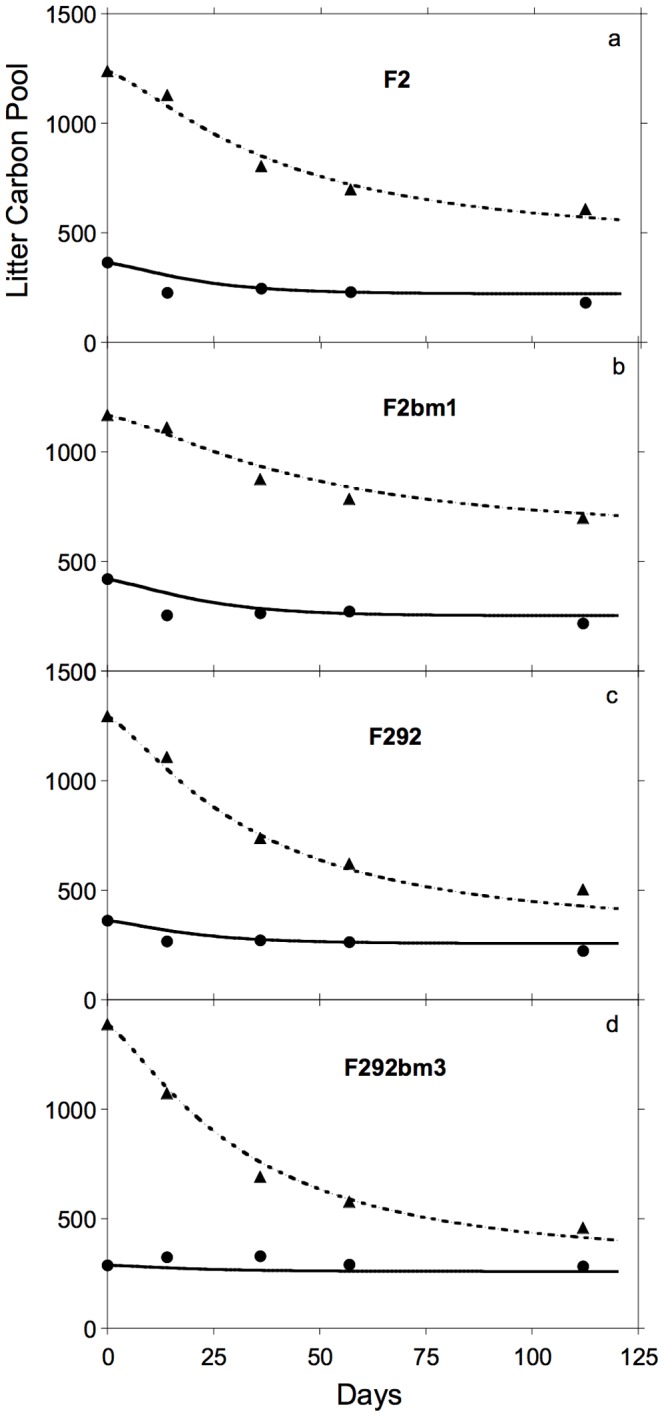
Observed (symbols) and simulated (lines) patterns of mass remaining for soluble C1 pool (solid lines and circles, mgC·kg soil−1) and acid hydrolysable C2 pool (dashed lines and triangles, mgC·kg soil−1) during decomposition of litter from maize mutants, a. F2, b. F2bm1, c. F292 and d. F292bm3.
Microbial production
Microbial biomass rapidly increased to peak values within 20–40 days, varying among genotypes (Figure 5a), declining most rapidly for those genotypes supporting the most rapid initial growth with the highest decay rates (F292 and F292bm3). Microbial turnover rates (Figure 5b), which should correlate with the generation of microbial products such as cell walls, were similar in shape but had lower peak values and lagged behind the patterns of rising and falling respiration rates (Figure 3). Genotypes with higher turnover rates also showed the greatest declines in biomass by day 112. Litter LCI increased over time in all litter types, coincident with declining biomass and turnover rates (Figure 5c), increasing most rapidly for litters decaying most rapidly (Figure 3). CUE declined over time for all litter types (Figure 5d), coincident with increasing LCI (Figure 5c), but started at higher values in litter types with larger pools of labile C1 (i.e., C1D), which had a higher C-assimilation efficiency (ei) than C2 (Table 1).
Figure 5. Simulated values of microbial biomass and turnover rate, remaining litter lignocellulose index (LCI), and carbon use efficiency (CUE), during decomposition of Zea mays root litter.
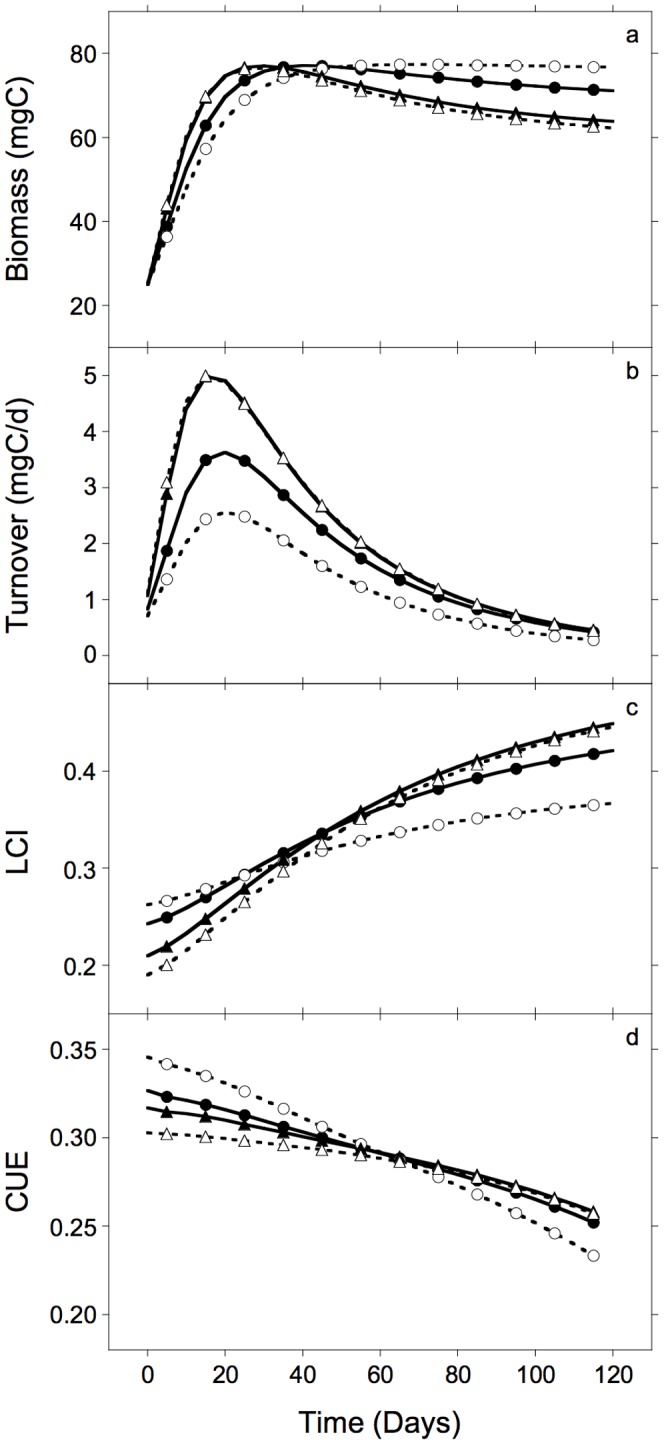
Simulated values of a. total microbial biomass, b. daily microbial turnover rate, c. remaining litter lignocellulose index (LCI), and d. realized carbon use efficiency (CUE), over time, for litter genotypes F2 (filled circles), F2bm1 (open circles), F292 (filled triangles) and F292bm3 (open triangles).
The total microbial production (C-immobilized into biomass including microbial turnover) by day 112 varied between litter types: 248, 195, 295 and 295 mgC for genotypes F2, F2bm1, F292 and F292bm3, respectively. These values were negatively related to initial litter KL and KL/AX contents, as well as final biomass and best-fit values of KB22. They were positively related to total cumulative CO2 efflux by day 112 and best-fit values of LCIT (all N = 4, P≤0.05).
Model test
Our initial model parameter set, including best-fit estimates of KB22 and LCIT, simulated rates of CO2 efflux closely matching observations (all N = 15; omitting day 0), with R2 values ranging from 0.87 for genotype F2bm3 to 0.98 for genotype F2bm1 (Fig. 3). Simulated rates peaked before observations for all litter types and at slightly lower values. However, simulated peak rates were within 10% of observed peaks for both F2 genotypes and 20% for both F292 genotypes. Simulated values of cumulative CO2 efflux (to day 112) were even closer to observations, with R2 values in excess of 0.99 for all genotypes (Figure 3). All simulations were within 5% of observed cumulative CO2 efflux values. Simulated values of C2 seldom differed by more than 5% of the observed values, with all R2≥0.99 (Figure 3). Finally, our model did not estimate any loss in pool C3 during simulations and has no mechanism to generate an increase in this pool's size (Figure 1).
Sensitivity analysis
The relative contributions of individual parameters to explaining variations in model behaviors differed by litter type and observation (Table 2). For example, the efficiency of C assimilation from substrate C1 (e1) rarely explained more than 1% of the variation in model behavior for any litter type. Moreover, no single parameter made a substantial contribution (≥10%) to explaining the variation in any model behavior when all four genotypes were pooled (not shown). When litter types were examined separately, variations in k1max and KB11 often made their largest contributions to the same output variables for the same litter types. For example both parameters made substantial contributions (≥10%) to variations in C1 for litter types F2, F2bm1 and F292bm3; cumulative CO2 efflux for litter types F2, F2bm1 and F292; respiration rates for litter types F2bm1 and F292; and overall model fit for litter type F2bm1. Neither parameter made a substantial contribution to variation in C2 for any litter type. In contrast, parameters e2 and BCmax often made their largest contributions to model behaviors for litter types when k1max and KB11 did not. For example, e2 and BCmax made substantial contributions to variations in C2 for litter types F2, F2bm1 and F292bm3; cumulative CO2 efflux for litters F292 and F292bm1; respiration rates for F2, F292 and F292bm3; and overall model fit for F2.
Table 2. Results of sensitivity analysis quantifying the relative contributions (%) of random variations in model parameters (column headings) to resulting variations in model behaviors (row labels), based on sums of squares from ANOVA relating model output to parameters.
| Source | Genotype | e1 | e2 | k1max | BCmax | KB11 |
| C1-Carbon | F2 | 0.2* | 1.3* | 47.5** | 3.9* | 30.6** |
| C1-Carbon | F2bm1 | 2.0* | 0.5* | 63.7** | 0.7* | 38.9** |
| C1-Carbon | F292 | 0.1* | 1.7 | 3.9* | 42.5** | 1.2 |
| C1-Carbon | F292bm3 | 0.0 | 2.3* | 65.6** | 7.0* | 22.1** |
| C1-Carbon | All Litters | 0.2 | 0.0 | 2.1* | 0.1 | 6.3* |
| C2-Carbon | F2 | 1.1* | 38.4** | 0.2* | 53.9** | 0.1 |
| C2-Carbon | F2bm1 | 1.2 | 13.2** | 0.1* | 71.4** | 0.0 |
| C2-Carbon | F292 | 0.0 | 3.5* | 0.8* | 73.0** | 0.1 |
| C2-Carbon | F292bm3 | 0.0 | 40.8** | 0.0 | 40.9** | 0.0 |
| C2-Carbon | All Litters | 1.0* | 1.6* | 0.2 | 3.3* | 0.4 |
| Cumulative CO2 | F2 | 0.2* | 1.2* | 48.1** | 3.8* | 30.1** |
| Cumulative CO2 | F2bm1 | 0.4* | 0.1 | 64.4** | 0.2* | 40.3** |
| Cumulative CO2 | F292 | 0.7* | 11.7** | 35.7** | 12.3** | 38.1** |
| Cumulative CO2 | F292bm3 | 0.5* | 48.8** | 6.6* | 21.2** | 4.5* |
| Cumulative CO2 | All Litters | 0.2 | 0.0 | 0.6 | 0.3 | 1.6* |
| Respiration Rate | F2 | 1.0 | 28.7** | 9.6* | 17.8** | 7.6* |
| Respiration Rate | F2bm1 | 0.3 | 0.1 | 48.5** | 0.3 | 30.2** |
| Respiration Rate | F292 | 2.1* | 40.8** | 20.6** | 11.4** | 14.0** |
| Respiration Rate | F292bm3 | 0.2* | 45.3** | 1.8* | 34.7** | 0.5* |
| Respiration Rate | All Litters | 0.0 | 0.2 | 0.0 | 0.4 | 0.1 |
| Overall Fit | F2 | 3.0* | 34.8** | 4.0* | 31.9** | 2.1* |
| Overall Fit | F2bm1 | 0.6* | 0.2 | 57.9** | 0.6* | 36.3** |
| Overall Fit | F292 | 2.6* | 46.5** | 4.6* | 16.8* | 7.9* |
| Overall Fit | F292bm3 | 0.3* | 49.2** | 0.9* | 31.3* | 0.7* |
| Overall Fit | All Litters | 0.0 | 0.2 | 0.6 | 0.4 | 0.6 |
| Maximum | 30.4 | 49.2 | 65.6 | 73.0 | 40.3 | |
| Count>10% | 0 | 11 | 9 | 12 | 9 |
*significant parameter contributions (P≤0.05) to variation in model behavior.
**contributions of parameters representing at least 10% of the explained variation in model behavior (P≥0.05).
Model extrapolation
When we simulated decomposition of the 12 different maize genotypes [11], we found that overall simulated rates of respiration were strongly related to observations (N = 180, R2 = 0.69, P≤0.01), and that simulated rates of respiration averaged 1±17% lower than observed rates (data not shown). However, the relative differences ([observations-simulations]/observations) varied over time (Text S2). The first two axes of the principal components analysis explained 54% of the variation in the relative differences between observed and simulated respiration and litter chemical characteristics (Figure 6a). Differences in rates between days 10–21 were more strongly related to axis 2, along with litter LCI, KL/AX and ∑Sug. In contrast, differences in respiration for most days ≥50 were more strongly associated with axis 1, along with several chemical characteristics, the strongest being arabinans, AX, galactose, NDF and pCA (Figure 6a).
Figure 6. Principal components analysis of variations in initial litter chemistry characteristics and relative differences between observed and simulated rates of microbial respiration and cumulative CO2 efflux during decomposition of Zea mays roots.
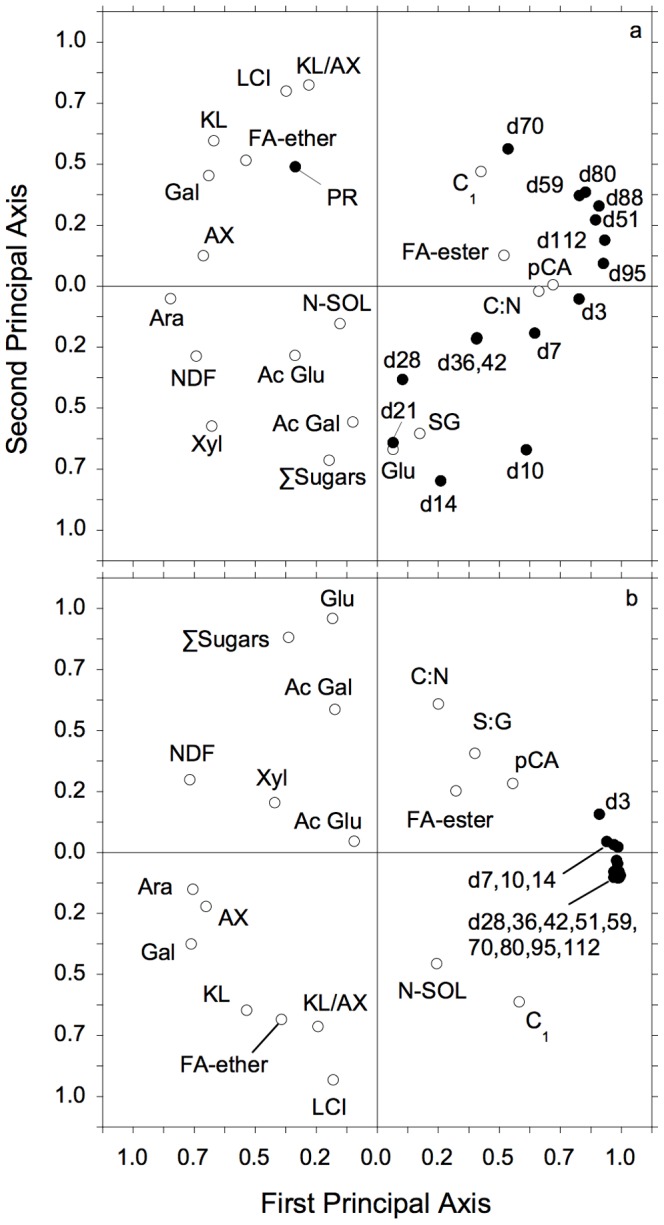
Results of principal components analysis of variations in initial litter chemistry characteristics and relative differences = (observations – simulations)/observations from simulated decomposition of 12 novel maize mutants [11] for: a. rates of respiration on days 3–112 (e.g., d3 = rate on day 3) and peak respiration (PR), and b. cumulative CO2 efflux by days 3–112 (parameter definitions given in text).
In contrast to the daily respiration rates, there was no significant relationship between simulated and observed peak respiration rates (means = 17.5±1.8 and 21.4±9.9 mgC·kg soil−1·d−1, respectively; not shown). The PCA showed that relative differences between peak rates (PR) were more closely related to axis 2 and opposite those of rates between days 10–21 (Figure 6a), with significant positive correlations with KL and KL/AX.
Simulated values of cumulative CO2 efflux were also strongly related to observations (N = 180, R2 = 0.923, P≤0.01), and averaged only 3±18% greater than observations. The first two axes of the PCA explained 69% of the variation in the relative differences between observed and simulated cumulative CO2 efflux and initial litter chemistry characteristics (Figure 6b). All values for CO2 efflux were tightly clustered and closely associated with the first axis. Litter chemical characteristics, NDF, arabinans, galactose and AX, were also closely related to the first axis. The relative differences between observations and simulations at day 36 (d36) showed a significant, positive correlation with the initial soluble content of litter (C1 = C-SOL) and negative correlation with galactan (GAL) content. Differences by day 59 were also related to C1 and galactan. Although there were no significant differences between simulations and observations at day 112, these variations were significantly related to several aspects of initial litter chemistry, including C1.
Estimates of microbial production by day 112 for these 12 genotypes ranged 203–303 mgC (mean = 270±25 mgC), and were negatively correlated with KL/AX and final microbial biomass, as well as initial LCI. Production was positively related to total cumulative CO2 efflux by day 112 and best-fit values of LCIT, as well as initial litter concentrations of Glu, ∑Sug, and C2 (all N = 12, P≤0.05). A stepwise regression explained nearly all variation in microbial production as a function of total CO2 efflux and initial concentrations of xylans (Xyl) and guaiacyl (G) in litter (N = 12, R2 = 0.998, P≤0.01).
Discussion
Lignin-cellulose interaction
Simulations provided a close match to observed patterns of holocellulose (C2) decay (Figure 4). This pool represented the largest fraction of litter (Table 1), also explaining why simulations closely fit patterns of CO2 efflux (Figure 3). Long-term decomposition (months to years) has often been negatively correlated to initial lignin content of litter [3], partly because lignin decays slowly and partly because some products of decomposition increase the size of the non-hydrolysable pool often interpreted as lignin [6]. However, Machinet et al. [11] also demonstrated the effects of biochemical connections between cell wall polysaccharides and lignin on the pattern of cumulative CO2 efflux from decomposing maize roots. The fit between k2 and LCI (Figure 2a) reveals an interaction between C2 and C3 in early stages of litter decay [19], well before C3 begins to decline [13], [20]. In other words, there is unlikely to be qualitatively separate pools of lignin-shielded and unshielded holocellulose, as is sometimes implied [1], [3]. Instead, biochemical linkages between cellulose, hemicellulose and lignin components of cell walls influence patterns of decomposition throughout the process.
A brief explanation of these relationships is that the structural composition of hemicellulose is a primary chain consisting mainly of xylans with branching arabinan side chains that interact with other cell wall polymers. The level of arabinoxylan substitution (represented by A∶X) increases with the progressive enzymatic degradation of plant material, during both digestion in the rumen [9], [24] and decomposition in soils [12], [13], as the more exposed elements of xylan chains are more readily hydrolyzed than those near arabinan branches. Gunnarsson et al. [25] found that initial xylan and arabinan concentrations in litter were as important as the total amount of hemicellulose in describing C mineralization over the first 9 days of laboratory incubations, and that arabinan was the single most important factor. These results were consistent with those of Machinet et al. [11], who identified initial arabinan content as an early predictor of C mineralization (days 3–7), followed by AX (days 7–14). In addition, hydroxycinnamic acids (ferulic and p-coumaric acids) play a key role in cross-linking arabinoxylans with lignin and this cohesive network also hampers decomposition [10], [11], [12]. These cross-linkages explain the negative effect of lignin on holocellulose decay long reported in the literature [1], [3] and also why k2 decreased as AX increased early in decomposition (Figure 2).
Our model revision approximated this complex control with a straightforward relationship between LCI and k2 (Figure 2) that included interactions between C2 and C3 (based on empirical sugar and lignin determinations). Moreover, key model parameters LCIT and KB22 were most closely related to initial KL/AX ratios of litter, emphasizing the interaction between holocellulose and lignin throughout the decomposition process.
Soluble dynamics
Data from Machinet et al. [13] also revealed that the soluble C1 pool included a fraction (C1T) that persisted throughout the study (Figure 4). Although the soluble fraction of litter is usually considered to be highly decomposable, the persistence of a relatively large soluble pool is common in both soils and decaying litter [26], [27]. The soluble pool usually contains a hydrophilic fraction including sugars and amino acids that are readily used by microorganisms [28], [29], and a more hydrophobic portion, including soluble polyphenols (e.g., tannins) that are less rapidly utilized [26], [30]. This differential use of compounds in the C1 pool by decomposer microorganisms changes the pool's overall quality and decomposability with time [1]. Although the soluble pool is replenished with degradation products from non-soluble substrates [1], [31], products of C2 hydrolysis would enter the more labile fraction of the soluble pool, which cycles much more rapidly than the more persistent fraction [28], [29]. In addition, Machinet et al. [13] found no decrease in the C3 pool over time (not shown), so that it's degradation products could not have increased the more persistent fraction of the C1 pool (C1T). Simulating the persistence of a sizeable pool of C1 with GDM required dividing the pool into labile and persistent fractions (Figure 1), with the labile fraction (C1D) rapidly declining during decay and the persistent C1T pool remaining intact (Figure 4). Moorhead and Sinsabaugh [16] found that simply routing the products of C2 degradation through the whole C1 pool GDM could not explain the size of this composite pool.
Respiration patterns
Simulations tended to overestimate rates of CO2 efflux between days 0–14, and underestimate rates between days 14–36 (Figure 3). A positive relationship between the most labile components of litter and early respiration (hours to weeks) or decay rate is commonly reported [32], [33], and most models, including GDM, assume a greater decay rate for soluble substrates [15], [16]. In terms of substrate dynamics, GDM tended to underestimate early losses (day 14) of C1 and overestimate C2 losses (Figure 4), suggesting that values of k1max should be slightly increased and KB1i (i = 1,2,3) reduced (Table 1), to stimulate decay of C1D and possibly initial CO2 efflux. However, GDM underestimated respiration to a greater extent for litter types with lower amounts of C1D, i.e., with higher C1T (F292 and F292bm3). We found that C1T was most closely related to the total sugar content of cell walls (∑Sug; N = 4; R2 = 0.998; P≤0.05) and that xylan, arabinan and glucan concentrations were highly correlated with each other (not shown). Further resolution of the chemical composition of the soluble fraction (C1D and C1T) and its dynamics during decomposition are needed to improve the mathematical descriptions of these relationships.
Microbial production
Moorhead and Sinsabaugh [16] argued that litter decay is initially limited by microbial action, because there is a time lag in the colonization of fresh litter by decomposer microorganisms (Figure 2). Whether this lag was a numerical (biomass) or functional (physiological) response in the study by Machinet et al. [13] is unknown because they did not monitor microbial biomass. Therefore, the most speculative part of this study was our simulation of microbial dynamics. Nonetheless, the patterns and magnitudes of simulated microbial biomass and turnover rates were consistent with observed patterns of respiration (Figures 3, 5b), reported limits to microbial biomass concentration in soils [21], [22], and changes in litter chemistry likely controlling microbial activities (Figures 2, 4 and 5c; [1]) as well as CUE (Figure 5d) [23]. Parameter estimates for C-assimilation efficiencies (ei) for various substrate pools, coefficient of microbial basal respiration rate (g), and maximum biomass∶total system C ratio (BCmax), most directly affected the relationships between substrate decomposition and biomass dynamics (Table 1). However, variations in any or all of these parameter values generate similar patterns with respect to different litter chemistry, although amplitudes and temporal regimes vary [16].
Our primary reason for simulating biomass dynamics was to determine if litter quality affected microbial production consistent with the idea that more decomposable chemical fractions of litter not only decay more rapidly but also generate more microbial products likely to enter stable soil organic matter pools [4], [5]. In fact, simulated microbial production was significantly and negatively correlated to KL and KL/AX, which are inversely related to decomposability, but production showed no significant positive relationship to any simple measure of litter quality. However, it was positively related to the sum of the two most decomposable carbon pools, i.e., C2+C1D, (N = 4, rho = 0.982, P≤0.05). The contributions of the relatively larger pools of C1D in litter types F2 and F2bm1 to simulated production were small compared to the larger pools of C2 in the other litters, despite the lower C assimilation efficiency of C2 versus C1 (Table 1). Thus our results were consistent with the observations of Smith et al. [7] and others who found that higher initial rates of C incorporation into biomass coincided with higher losses of litter through respiration [2], [8].
Sensitivity analysis
The most interesting result of our sensitivity analysis was the lack of any simple, overall interpretation. No single parameter consistently explained>10% of the observed variability in any model behavior (Table 2). Usually a parameter important to explaining one model behavior, such as the contributions of k1max to simulated C1 dynamics or cumulative CO2 efflux (Table 2), made little contribution to other model outputs. The largest discrepancies between simulations and observations were in early respiration and C1 loss (discussed above). Of the tested parameters, e2, the C assimilation efficiency for C2, appeared to be most important to respiration rates, which seems reasonable because C2 was the largest substrate pool and provided most of the C respired (Table 1, Figure 4). As for the dynamics of the C1 pool, parameter k1max appeared to be most important, followed by KB11; thus parameters controlling the degradation rate of this pool explained differences between simulations and observations (Figure 4). These results are consistent with our earlier conclusion that greater resolution of the C1 pool composition and dynamics might provide a better understanding of decomposition.
Extrapolations
We simulated the decomposition of 12 additional maize genotypes in the second set of incubations conducted by Machinet et al. [11] based on the assumption that the relationships between key parameters, LCIT, KB22 and C1T, and litter chemical characteristics were consistent with those for the four genotypes examined by Machinet et al. [13]. The assumption seemed reasonable because all 16 genotypes were naturally occurring varieties of Zea mays, and likely to be more similar in chemistry and tissue architecture than unrelated species more commonly used in comparative decomposition experiments [6].
In general, simulations were within a few percent of observed values of respiration rates and cumulative CO2 efflux over the entire period of incubation. Analyses of these differences between simulations and observations provided relatively little additional insight to patterns of cumulative CO2 efflux (Figure 6b). The importance of the initial C1 pool (as both C1T and C1D estimates) to cumulative CO2 efflux through time again emphasized the importance of initial decay rate to longer-term patterns [7], [8]. The relationships between CO2 efflux on day 112 and initial arabinan and p-coumaric acid concentrations and AX suggest that the cross-linkages among hemicellulose and lignin became increasingly important with progressive decay. The tight cluster of cumulative CO2 efflux along the first axis of our PCA also underscored the importance of cross-linkages among cell wall constituents (e.g., arabinan, AX, NDF and p-coumaric acid) to litter decay (Figure 6b), consistent with Machinet et al. [11].
The differences between simulated and observed respiration rates were more variable, suggesting temporally shifting controls on decomposition. Peak rates and those on days 14–28 were related to initial KL and KL/AX, as well as glucan and S∶G (Figure 6a). In contrast, rates on days>42 were more closely associated with arabinan, AX, p-coumaric acid and ester-linked ferrulic acids, perhaps because polysaccharide-ester linked ferulic acids can form ether-links with lignin [34], [35] and the syringyl units of lignin can be esterified by p-coumaric acids, which is typical of grass cell walls [11]. However, the biggest differences between simulated and observed rates were on days 36–42, which were most closely related to KL, ether-linked ferrulic acids and galactan (Figure 6b). The frequent importance of KL and AX (or their chemical constituents) to these patterns was surprising, because KL (in LCI) was used to estimate k2, and KL/AX to estimate LCIT and KB22 (previously discussed) used in simulations. Clearly, simple linear relationships were insufficient to capture the subtleties of these controls. The relationships between respiration and galactan and C∶N ratio (Figure 6b) hint at a microbial control [14], in part because galactan is sometimes used as an index to microbial contributions [36], but it is also a hemicellulosic sugar, along with arabinan, rhamnan, and xylan [37].
Conclusions
We found that the level of arabinan substitution in xylan chains (AX) was an important control in early stages of decomposition, and was also linearly related to LCI calculated on the basis of fine scale cell wall chemistry. These relationships provide a plausible, mechanistic explanation for earlier, empirical descriptions of LCI effects on decomposition as a result of biochemical cross-linkages between polysaccharides and lignin. Thus lignin and LCI serve as convenient, negative proxies for the decomposability of litter even at the start of decay. However, additional research is needed to determine the chemical composition and dynamics of the non-hydrolysable product of proximate C analysis that is typically termed “lignin” if we are to discover the mechanistic relationships between LCI and latter stages of decomposition.
We also found that dividing the soluble pool of litter (C1) into separate persistent (C1T) and labile (C1D) pools, was necessary to accurately simulate the dynamics of the composite soluble pool during early decomposition because not all soluble compounds are equally decomposable. The finer scale chemical composition and dynamics of the soluble component of litter is needed to determine the possible sources and fates of these compounds.
These relationships between simulated patterns of litter decay, litter chemistry (LCI, AX, C1T and C1D) and microbial productivity were consistent with the notion that the more rapid utilization of substrates with high carbon use efficiency generates greater amounts of microbial products that can contribute to stable soil organic matter pools than more persistent substrates with lower C-assimilation, like lignin. Our assumption that lignin decay provides little to no net C-acquisition by microorganisms is also consistent with recent observations that little lignin C enters stable SOC pools.
Finally, our extrapolations with litter types that differed in initial chemistry demonstrated that relationships we found between key model parameters, LCIT, KB22 and C1T, and decomposition were robust across a large range of maize litter types, highlighting the importance of access (KB22) to the largest, rapidly decaying pool of substrate (C2) by microorganisms able to use this resource (G2), as well as negative controls imposed by the less-accessible substrate pools (KL, C1T). Moreover, differences between simulations and observations indicated that temporal controls on decay rates shifted from relative substrate pool sizes (both accessible and persistent) dominating at the start to factors related to cross-linkages between structural polysaccharides and lignin with progressive decomposition.
In closing, our results suggest that interactions between decomposer microorganisms and litter quality characteristics at the earlier stages of decomposition may provide more insights to soil organic C stabilization than later stages dominated by the more persistent chemical characteristics of the cell wall, if indeed microbial products comprise a large fraction of stable soil C pools.
Supporting Information
Data used to test and refine model. Observed respiration rates and chemical composition of residues of maize roots during decomposition [13].
(DOCX)
Differences over time between observed and simulated respiration rates and cumulative CO2 efflux from the decomposition of Zea mays roots. Relative differences between observations and simulations of litter decomposition from 12 novel maize genotypes [11] for: a. respiration rates (mgC·kg soil−1·d−1) over time (means±95% confidence intervals, all N = 12), b. cumulative CO2 efflux (mgC·kg soil−1) over time (means±95% confidence intervals, all N = 12).
(TIFF)
Lignocellulose controls. Relationships between decay rate coefficients for polysaccharides (C2) and polyphenolics (C3) as functions of lignocellulose index (LCI = C3/[C2+C3])).
(DOCX)
Sensitivity analysis. Patterns of differences between observed and modeled patterns of respiration associated with decomposition of decaying maize roots.
(DOCX)
Data Availability
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. The data used in this study were included in previous publications, raising ethical questions about linking them to this publication. Nonetheless, they were used and presented differently in our study. Thus we include two supplemental tables of data that we used to test and refine our model. This should permit readers to duplicate our results. Also, all data used in this study are available upon request from author Isabelle Bertrand, INRA, UMR614 Fractionnement des AgroRessources et Environnement, F-51100 Reims, France. email: isabelle.bertrand@reims.inra.fr.
Funding Statement
This study was supported by United States National Science Foundation grants DEB-0946257 and DEB-0918718, and Institut National de la Recherche Agronomique. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Berg B, McClaugherty C (2008) Plant litter: Decomposition, humus formation, carbon sequestration, Second edition. New York: Springer. 338 p. [Google Scholar]
- 2. McMahon SK, Williams MA, Bottomley PJ, Myrold DD (2005) Dynamics of microbial communities during decomposition of carbon-13 labeled ryegrass fractions in soil. Soil Sci Soc Am J 69: 1238–1247. [Google Scholar]
- 3. Melillo JM, Aber JD, Muratore JF (1982) Nitrogen and lignin control of hardwood leaf litter decomposition dynamics. Ecology 63: 621–626. [Google Scholar]
- 4. Wickings K, Grandy AS, Reed SC, Cleveland CC (2012) The origin of litter chemical complexity during decomposition. Ecol Lett 15: 1180–1188. [DOI] [PubMed] [Google Scholar]
- 5. Cotrufo MF, Wallenstein MD, Boot CM, Denef K, Paul E (2013) The microbial efficiency-matrix stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: do labile plant inputs form stable soil organic matter? Glob Change Biol 19: 988–995. [DOI] [PubMed] [Google Scholar]
- 6. Preston CM, Nault JR, Trofymow JA (2009) Chemical changes during 6 years of decomposition of 11 litters in some Canadian forest sites. Part 2. 13C abundance, solid-state 13C NMR spectroscopy and the meaning of “Lignin”. Ecosystems 12: 1078–1102. [Google Scholar]
- 7. Smith JL, Bell JM, Bolton H Jr, Bailey VL (2007) The initial rate of C substrate utilization and longer-term soil C storage. Biol Fertil Soils 44: 315–320. [Google Scholar]
- 8. Blagodatskaya EV, Blagodatsky SA, Anderson T –H, Kuzyakov Y (2009) Contrasting effects of glucose, living roots and maize straw on microbial growth kinetics and substrate availability in soil. Eur J Soil Sci 60: 186–197. [Google Scholar]
- 9. Chesson A (1988) Ligninopolysaccharide complexes of the plant cell wall and their effect on microbial degradation in the rumen. Anim Feed Sci Tech 21: 219–228. [Google Scholar]
- 10. Grabber JH, Mertens DR, Kim H, Funk C, Lu FC, et al. (2008) Cell wall fermentation kinetics are impacted more by lignin content and ferulate cross-linking than by lignin composition. J Sci Food Agr 89: 122–129. [Google Scholar]
- 11. Machinet GE, Bertrand I, Barriere Y, B Chabbert B, Recous S (2011) Impact of plant cell wall network on biodegradation in soil: Role of lignin composition and phenolic acids in roots from 16 maize genotypes. Soil Biol Biochem 43: 1544–1552. [Google Scholar]
- 12. Amin BAZ, Chabbert B, Moorhead D, Bertrand I (2014) Impact of fine litter chemistry on lignocellulolytic enzyme efficiency during decomposition of maize leaf and root in soil. Biogeochemistry 117: 169–183. [Google Scholar]
- 13. Machinet GE, Bertrand I, Chabbert B, Recous S (2009) Decomposition in soil and chemical changes of maize roots with genetic variations affecting cell wall quality. Eur J Soil Sci 60: 176–185. [Google Scholar]
- 14. Machinet GE, Bertrand I, Chabbert B, Watteau F, Villemin G, et al. (2009) Soil biodegradation of maize root residues: Interaction between chemical characteristics and the presence of colonizing micro-organisms. Soil Biol Biochem 41: 1253–1261. [Google Scholar]
- 15. Manzoni S, Porporato A (2009) Soil carbon and nitrogen mineralization: theory and models across scales. Soil Biol Biochem 41: 1355–1379. [Google Scholar]
- 16. Moorhead DL, Sinsabaugh RL (2006) A theoretical model of litter decay and microbial interaction. Ecol Monogr 76: 151–174. [Google Scholar]
- 17. Recous S, Robin D, Darwis D, Mary B (1995) Soil inorganic N availability: Effect on maize residue decomposition. Soil Biol Biochem 27: 1529–1538. [Google Scholar]
- 18. Schimel JP, Weintraub MN (2003) The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: A theoretical model. Soil Biol Biochem 35: 549–563. [Google Scholar]
- 19. Moorhead DL, Lashermes G, Sinsabaugh RL, Weintraub MN (2013) Calculating co-metabolic costs of lignin decay and their impacts on carbon use efficiency. Soil Biol Biochem 66: 17–19. [Google Scholar]
- 20. Herman J, Moorhead D, Berg B (2008) The relationship between rates of lignin and cellulose decay in aboveground forest litter. Soil Biol Biochem 40: 2620–2626. [Google Scholar]
- 21. Anderson T-H, Domsch KH (1989) Ratios of microbial biomass carbon to total organic carbon in arable soils. Soil Biol Biochem 21: 471–479. [Google Scholar]
- 22. Wardle DA (1998) Controls of temporal variability of the soil microbial biomass: A global-scale synthesis. Soil Biol Biochem 30: 1627–1637. [Google Scholar]
- 23. Sinsabaugh RL, Manzoni S, Moorhead DL, Richter A (2013) Carbon use efficiency of microbial communities: stoichiometry, methodology and modeling. Ecol Lett 16: 930–939. [DOI] [PubMed] [Google Scholar]
- 24. Vailhé MAB, Provan GJ, Scobbie L, Chesson A, Maillot MP, et al. (2000) Effect of phenolic structures on the degradability of cell walls isolated from newly extended apical internode of tall fescue (Festuca arundinacea Schreb.). J Agr Food Chem 48: 618–623. [DOI] [PubMed] [Google Scholar]
- 25. Gunnarsson S, Marstorp H, Dahlin AS, Witter E (2008) Influence of non-cellulose structural carbohydrate composition on plant material decomposition in soil. Biol Fertil Soils 45: 27–36. [Google Scholar]
- 26. Cleveland CC, Neff JC, Townsend AR, Hood E (2004) Composition, dynamics, and fate of leached dissolved organic matter in terrestrial ecosystems: Results from a decomposition experiment. Ecosystems 7: 275–285. [Google Scholar]
- 27. Bengtson P, Bengtsson G (2007) Rapid turnover of DOC in temperate forests accounts for increased CO2 production at elevated temperatures. Ecol Lett 10: 783–790. [DOI] [PubMed] [Google Scholar]
- 28. Kalbitz K, Schmerwits J, Schwesig D, Matzner E (2003) Biodegradation of soil-derived dissolved organic matter as related to its properties. Geoderma 113: 273–291. [Google Scholar]
- 29. Bertrand I, Prevot M, Chabbert B (2009) Soil decomposition of wheat internodes of different maturity stages: Relative impact of the soluble and structural fractions. Bioresource Technol 100: 155–163. [DOI] [PubMed] [Google Scholar]
- 30. Nishimura S, Maie N, Baba M, Sudo T, Sugiura T, et al. (2012) Changes in the quality of chromophoric dissolved organic matter leached from senescent leaf litter during the early decomposition. J Environ Qual 41: 823–833. [DOI] [PubMed] [Google Scholar]
- 31. Sinsabaugh RL (1994) Enzymatic analysis of microbial pattern and process. Biol Fertil Soils 17: 69–74. [Google Scholar]
- 32. Lagomarsino A, Moscatelli MC, De Angelis P, Grego S (2006) Labile substrates quality as the main driving force of microbial mineralization activity in a poplar plantation soil under elevated CO2 and nitrogen fertilization. Sci Total Environ 372: 256–265. [DOI] [PubMed] [Google Scholar]
- 33. Hernandez DL, Hobbie SE (2010) The effects of substrate composition, quantity, and diversity on microbial activity. Plant Soil 335: 397–411. [Google Scholar]
- 34.Jung HG, Deetz DA (1993) Cell wall lignification and degradability. In: Jung HG, Buxton DR, Hatfield RD, Ralph J, editors. Forage cell wall structure and digestibility. Madison: ASA-CSSA-SSSA. Pp. 315–346. [Google Scholar]
- 35. Jacquet G, Pollet B, Lapierre C (1995) New ether-linked ferulic acid-coniferyl alcohol dimers identified in grass straws. J Agr Food Chem 43: 2746–2751. [Google Scholar]
- 36. Oades JM (1984) Soil organic matter and structural stability: mechanisms and implications for management. Plant Soil 76: 319–337. [Google Scholar]
- 37. Merila P, Malmivaara-Lamsa M, Spetz P, Stark S, Vierikko K, et al. (2010) Soil organic matter quality as a link between microbial community structure and vegetation composition along a successional gradient in a boreal forest. Appl Soil Ecol 46: 259–267. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data used to test and refine model. Observed respiration rates and chemical composition of residues of maize roots during decomposition [13].
(DOCX)
Differences over time between observed and simulated respiration rates and cumulative CO2 efflux from the decomposition of Zea mays roots. Relative differences between observations and simulations of litter decomposition from 12 novel maize genotypes [11] for: a. respiration rates (mgC·kg soil−1·d−1) over time (means±95% confidence intervals, all N = 12), b. cumulative CO2 efflux (mgC·kg soil−1) over time (means±95% confidence intervals, all N = 12).
(TIFF)
Lignocellulose controls. Relationships between decay rate coefficients for polysaccharides (C2) and polyphenolics (C3) as functions of lignocellulose index (LCI = C3/[C2+C3])).
(DOCX)
Sensitivity analysis. Patterns of differences between observed and modeled patterns of respiration associated with decomposition of decaying maize roots.
(DOCX)
Data Availability Statement
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. The data used in this study were included in previous publications, raising ethical questions about linking them to this publication. Nonetheless, they were used and presented differently in our study. Thus we include two supplemental tables of data that we used to test and refine our model. This should permit readers to duplicate our results. Also, all data used in this study are available upon request from author Isabelle Bertrand, INRA, UMR614 Fractionnement des AgroRessources et Environnement, F-51100 Reims, France. email: isabelle.bertrand@reims.inra.fr.


