Abstract
Purpose
Early pregnancy in women by the age of 20 is known to have a profound effect on reduction of lifelong breast cancer risk as compared to their nulliparous counterparts. Additional pregnancies further enhance the protection against breast cancer development. Nationwide trend of delayed pregnancy may contribute to the recently reported increase in the incidence of advanced breast cancer among young women in this country. The underlying mechanism for the parity-associated reduction of breast cancer risk is not clearly understood. The purpose of the current study is to use whole-genome DNA methylation profiling to explore a potential association between parity and epigenetic changes in breast tissue from women with early parity and nulliparity.
Methods
Breast tissue was collected from age-matched cancer-free women with early parity (age < 20; n = 15) or nulliparity (n = 13). The methyl-CpG binding domain-based capture (MBDCap)-sequencing technology was used for whole-genome DNA methylation profiling. Potential parity-associated hypermethylated genes were further verified by locus-specific pyrosequencing, using an expanded cohort of parous (n = 19) and nulliparous (n = 16) women that included the initial samples used in the global analysis.
Results
Our study identified 6 genes that are hypermethylated in the parous group (p < 0.05). Pyrosequencing confirmed parity-associated hypermethylation at multiple CpG islands of the FOXA1 gene, which encodes a pioneer factor that facilitates chromatin binding of estrogen receptor α (ERα).
Conclusions
Our work identifies several potential methylation biomarkers for parity-associated breast cancer risk assessment. In addition, the results are consistent with the notion that parity-associated epigenetic silencing of FOXA1 contributes to long-term attenuation of the estrogenic impact on breast cancer development.
Keywords: parity, FOXA1, DNA methylation, breast cancer risk reduction
INTRODUCTION
Besides the BRCA1/2 breast cancer susceptibility genes, several endocrine-related factors are known to significantly influence the risk for breast cancer. In particular, women who have their first full-term pregnancy before the age of 20 have approximately half of the risk for developing breast cancer as compared to their nulliparous counterparts, and additional pregnancies further accentuate the parity-associated protection against breast cancer [1]. This lifelong risk-reducing effect of early parity is in contrast to the risk-enhancing effect of late pregnancy after age 35 [2–4]. A number of studies in human and rodents have demonstrated the strong and lifelong protective role of early pregnancy on breast cancer [5–7]. While the exact underlying mechanism by which early parity reduces lifelong breast cancer incidence is not known, it is most likely due to enduring biological changes in the breast tissue. Several models have been proposed based on parity-associated changes in breast epithelial cells and its surrounding stromal compartments [1, 8]. For example, it has been hypothesized that early parity-induced differentiation of mammary epithelial cells could render them more resistant to oncogenesis [9]. In a similar vein, early parity may decrease the pools of mammary stem/progenitor cells, thus decreasing the number of putative cell of origin for breast tumors [10, 11]. As early parity predominantly reduces risk of estrogen receptor α (ER)-positive breast cancer [12], it is also conceivable that attenuation of the estrogen-related pathways could contribute to parity-mediated risk reduction [13]. In addition, the roles of hormones and hormone-sensing cells in the early parity-based protection on breast cancer has been recently documented [14]. Better understanding of the cellular and molecular basis for this longstanding biological phenomenon will go a long way in risk assessment and breast cancer prevention.
ERα is a site-specific transcription factor that plays a key role in normal breast ductal development and luminal breast cancer [15]. Traditionally, estrogen-stimulated ERα alone was thought to be sufficient to bind to the cognate estrogen-responsive enhancers for hormone-stimulated transcriptional activation. However, whole genome-based studies in breast cancer cells indicate that FOXA1, another site-specific transcription factor important in normal and breast cancer development [16], tends to co-occupy ERα-bound transcriptional enhancers [17, 18]. Importantly, it has been demonstrated that FOXA1 serves as a pioneer factor that facilitates ERα binding to compacted chromatin DNA. Without FOXA1, ERα cannot bind to the corresponding enhancers even in the presence of estrogens, and consequently, estrogen-mediated transcription and breast cancer cell proliferation are abrogated. More recent studies further underscore the clinical relevance of the FOXA1-ERα-DNA interaction complex [19, 20]. For example, it was shown that FOXA1 is capable of reprogramming of ER chromatin binding and this FOXA1-mediated event correlates with the clinical outcome in breast cancer [20, 21]. Furthermore, breast cancer risk-associated single nucleotide polymorphisms (SNPs) are found in the FOXA1/ERα-occupied enhancers, which alter FOXA1 chromatin binding and expression of FOXA1/ERα downstream target genes [22]. These findings highlight the biological and pathological impact of the cooperative chromatin binding of FOXA1-ERα on breast cancer development and progression.
In the current study, we sought to uncover DNA epigenetic signatures that are associated with breast tissue of women with early parity. By combining genome-wide MBDCap-sequencing and locus-specific pyrosequencing, our work identified distinct CpG islands in the FOXA1 gene that are preferentially hypermethylated in the early-parity group. Our findings draw attention to sustained FOXA1 gene silencing and reduced estrogen actions as a potential contributing factor to early parity-associated reduction of breast cancer risk.
MATERIALS AND METHODS
Clinical Specimens
Cancer-free breast tissue was procured from women undergoing cosmetic reduction mammoplasty, following a protocol approved by the Institutional Review Board at the University of Texas Health Science Center at San Antonio. All donors signed a written consent form authorizing the use of the specimens for breast cancer-related laboratory investigations. The participants also filled out a questionnaire concerning personal medical history and health- and diet-related behaviors. Supplemental Table 1 provides more detailed information about the age-matched parous and nulliparous groups used in the current study.
DNA Isolation and MBDCap-seq
MBDCap libraries for sequencing were prepared following standard protocols from Illumina (San Diego, CA). Briefly, genomic DNA was prepared from frozen tissue by using QiaAmp DNA extraction kit (Qiagen, USA) and subsequently fragmented by sonication to reach an average size of 250 base pairs (bp). Methylated DNA fragments were captured and eluted by using MethylMiner Methylated DNA Enrichment Kit (Invitrogen, USA), following standard protocol from the manufacturer. MBDCap libraries were sequenced using the Illumina Genome Analyzer II (GA II) per manufacturer’s instructions. Sequencing was performed up to 25 cycles for mapping to the human genome reference sequence. Image analysis and base calling were performed using the standard Illumina software.
Bioinformatic analysis of MBDCap-seq data
Reads (up to 50 bp) were mapped to the human reference genome (hg18) using the BWA algorithm, with up to two base-pair mismatches. The uniquely mapped reads were used for additional linear normalization and differential methylation analysis as previously described [23].
The methylation level was calculated by accumulating the number of reads. NRead,i is the number of normalized reads at the ith bin; URead,i is the number of uniquely mapped reads at the ith bin, and NU is the number of total uniquely mapped reads. “INT” rounds the element to the nearest integers toward minus infinity, “^” means the power operator.
A region of methylation level was represented by the average of the normalized unique reads. Comparison of group A and B (G = A or B), the average methylation level (AVGR,G) was calculated separately at two groups in a given region R (which includes m bin size, and start at the sth bin). The number of sample is SA for group A, and SB for group B. AVGR,G means the average methylation level of group G at the R region. MR,G is the methylation levels of each sample of group G at the R region.
Pyrosequencing assay
The pyrosequencing target sequence and location in each selected CpG site for FOXA1 were listed in Supplemental Figure 1. Analysis of each site involved one forward and one biotin-labeled reverse PCR primer, as well as one sequencing primer for pyrosequencing. Genomic DNA was bisulfite-converted using EZ-96 DNA Methylation kit (ZYMO Research, PN D5001). Twenty ng of bisulfite-converted DNA was used for PCR amplification and the products were verified for size and overall quality by gel electrophoresis. The PCR conditions were as follows: 95°C for 5 min, followed by 50 cycles of 95°C for 1 min, 60°C for 1 min, 72°C for 1 min, then 72°C for 7 min using the Taq DNA polymerase (Applied Biosystem, N12338).
For pyrosequencing, 10 µL of the PCR product was mixed with streptavidin-coated agarose beads (GE Healthcare UK) and processed through the Qiagen Vacuum Prep Tool for purification of the biotinylated single-stand DNA fragments. PyroMark Gold Q96 CDT Reagents were used for sequencing reactions. The automated pyrosequencing instrument, PyroMark Q96 MD (Qiagen), was employed for the sequencing-by-synthesis method to detect the methylation status of each CpG site in a specific region using the sequencing primer at 500 nM. Quantification of each CpG site was performed using the methylation Software PyroMark CpG 1.0 software. The built-in internal quality control for bisulfite treatment and non-specific background was set at 6.5%. Methylated DNA (Universal DNA with methylated enzyme treatment) served as the positive control.
Statistical analysis
Welch t-test and the analysis of variance (ANOVA) were applied to determine whether the percentages of CpG islands were different across groups in various factors of interest. Welch t-test was used to examine whether two groups in a factor were significantly different from each other. It compared the mean values of two groups with the assumption that the variances were unequal in the groups. P-value less than 0.05 suggested that there was a statistically difference in two groups. ANOVA was conducted to analyze group differences when there were more than two groups in a factor (ethnicity in the current study). Variance in a factor was partitioned into two components: variance between groups and variance within groups. F-test was used in ANOVA to examine two components by comparing the ratio of variance between groups and variance within groups to a critical value. When P-value is less than 0.05, the result implies that at least one group in the factor is different from the others.
RESULTS AND DISCUSSION
MBDCap-seq, a high throughput technology for surveying genome-wide DNA methylation patterns, combines capturing of methylated genomic DNA by the methyl-CpG-binding domain (MBD) of MeCP2 with next-generation sequencing. The robust procedure allows comprehensive and unbiased detection of DNA methylation across the entire genome. In the current study, we applied this technology to compare the methylation status in the breast tissue of women with early parity versus nulliparity. To this end, we collected breast tissue from two age-matched cohorts of women who underwent elective reduction mammoplasty (Supplemental Table 1). The first group (n = 15; average age of 37.2 years old) consisted of those who had the first full-term pregnancy by the age of 19 years old. The second cohort (n = 13; average age of 37.8 years old) was from women who had no history of childbearing. The same age frames of these two cohorts allowed us to exclude any age-related epigenetic differences.
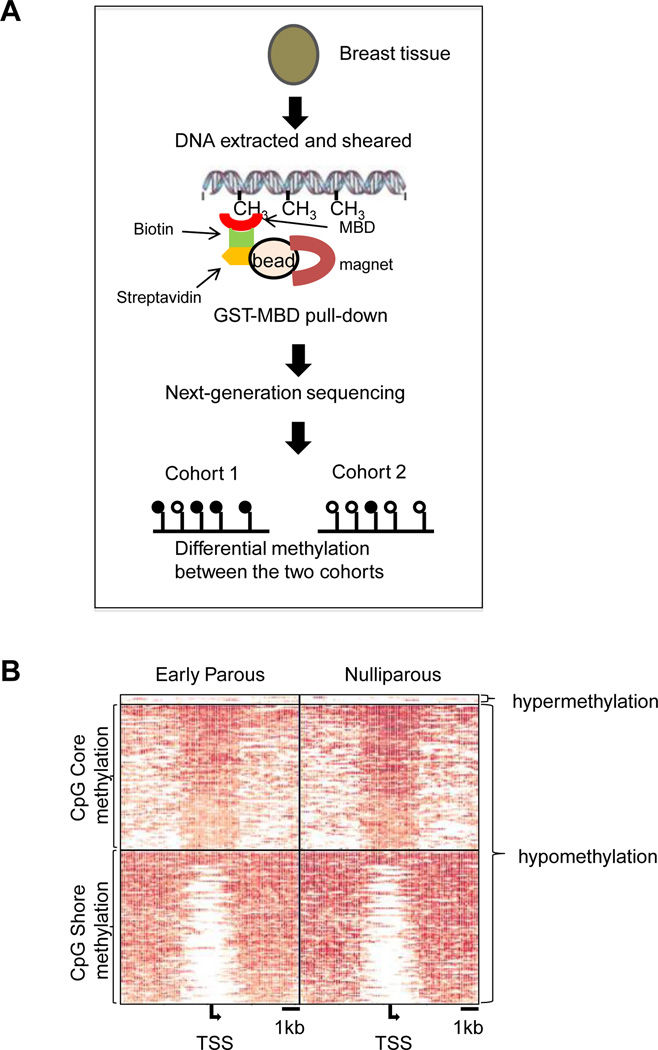
Genomic DNA was isolated from the frozen breast tissue. Methylated DNA fragments were bound to the GST-MBD resin, eluted, and subjected to MBDCap-seq by the Illumina HiSeq 2000 sequencing system (Figure 1A). A total of 42 billion bp from the 28 clinical samples were processed, and 75% of the sequenced fragments were mapped to unique genome locations (Supplementary Table 2). As DNA methylation tends to be found in GC-rich regions, a minimum of 10 million unique reads is usually considered sufficient to cover adequate sequence depth for whole-genome methylation profiling [23]. Upon data normalization, we conducted a pairwise comparison within an 8 kb window to identify differentially methylated CpG island loci in the early parous and nulliparous samples.
Figure 1. Methylation profiling of early parous and nulliparous women.
(A). Schematic outline of the experimental procedure of MBDCap-seq. DNA from the tissue samples were subjected to MBD2 capture. After library construction and next-generation sequencing, data were analyzed to identify differentially methylated loci. (B). Methylation levels of loci, 4 kb upstream and downstream from the transcription start site of the differentiated methylated genes in early parous and nulliparous samples.
We analyzed a total of 13,081 known promoter CpG islands and found that only 6 genes (0.046%) were significantly hypermethylated in parous samples relative to the nulliparous ones (Figure 1B). They are BCAR3, FOXA1, FOXB2, GRIK5, MGST3, and OLIG3 (Table 1). Two of the parity-associated hypermethylated genes exhibited methylation of the CpG core (BCAR3 and OLIG3), whereas the rest showed CpG shore methylation (right, left or both sides). In contrast, a far greater number of genes (211 or 1.62%) were hypomethylated in the parous samples (Figure 1B, Supplemental Table 2). Of the hypomethylated genes, 104 exhibited CpG core methylation, whereas 107 showed CpG shore methylation. No known breast tumor suppressor genes were identified from the hypomethylated genes.
Table 1.
Potential hypermethylated genes in the early parous group
| Gene symbol |
Gene ID |
Chr.# | Region start |
Region end | Fold change |
p-value |
|---|---|---|---|---|---|---|
| FOXA1 | 3169 | 14 | 37130240 | 37138240 | −0.47 | 0.0356 |
| GRIK5 | 2901 | 6 | 101949581 | 101957581 | −0.47 | 0.047 |
| OLIG3 | 167826 | 6 | 137853224 | 137861224 | −0.57 | 0.0055 |
| MGST3 | 4259 | 1 | 163863073 | 163871073 | −0.6 | 0.0462 |
| BCAR3 | 8412 | 1 | 93915973 | 93923973 | −0.71 | 0.0336 |
| FOXB2 | 442425 | 9 | 78820390 | 78828390 | −0.73 | 0.0446 |
Among the 6 hypermethylated genes in the parous group, FOXA1 encodes a site-specific transcription factor that acts as a pioneer factor to promote and dictate chromatin binding of ERα [17, 18]. Its amplification is associated with development of multiple cancer types including breast cancer [24]. The role of FOXB2, another FOX family gene, in cancer is not known. BCAR3 encodes a signaling molecule that plays an important role in breast cancer invasion and resistance to anti-estrogen therapies [25, 26]. Therefore, it is conceivable that epigenetic changes at these loci could affect hormonal responsiveness of breast epithelial cells and their propensity for tumorigenesis.
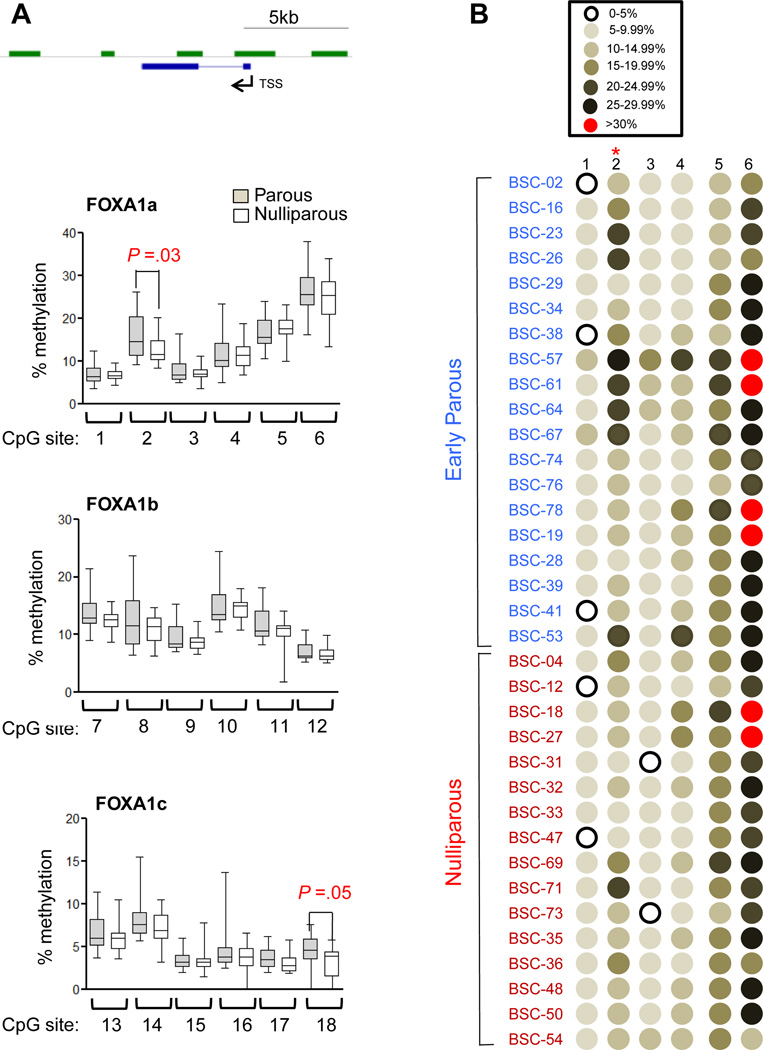
To confirm the MBDCap-seq data, we conducted pyrosequencing analysis of the FOXA1, FOXB2, and BCAR3 loci, using the original cohorts for the MBDCap-seq plus 4 additional early parous and 3 nulliparous samples subsequently procured (Supplemental Table 1). MBD-seq and pyrosequencing are two different and mutually complementary methods for detecting DNA methylation. While MBD-seq assesses genome-wide DNA methylation in a relatively low resolution, pyrosequencing is particuarly suitable for determining DNA methylation level at the single base-pair resolution in a relatively small region. Pyrosequencing primers were designed for the specific CpG islands that displayed parity-associated hypermethylation as revealed by MBDCap-seq (Supplemental Figure 1–3). The methylation status of the individual CpG sites at these gene loci are presented in Figure 2 (FOXA1) and Supplemental Figure 4 (FOXB2 and BCAR3). Among the 18 CpG sites interrogated at the FOXA1 locus, sites 2 and 18 were found to be significantly hypermethylated in the parous group (P < 0.05, Figure 2), thus confirming the genome-wide MBDCap-seq data. In contrast, no statistically significant, parity-associated difference was detected by pyrosequencing at the FOXB2 or BCAR3 loci, although several CpG sites at these two gene loci displayed a trend of hypermethylation in the parous group (Supplemental Figure 4). More sample analysis will be needed to resolve this discrepancy between the two methods.
Figure 2. Pyrosequencing validation of the parity-associated hypymethylation at FOXA1.
(A). Graphical representation of the % methylation at various CpG sites in the FOXA1 gene by using 3 sets of primers (FOXA1a, FOXA1b and FOXA1c). See Supplemental Figure 1 for the genomic locations of the CpG sites. The boxes cover sample values from the 25 percentiles (in the bottom) to 75 percentiles (in the top), the whiskers extending from the boxes stretch to the minimum and the maximum values of the samples. Welch t-tests were used to determine the p-values between two groups. (B). Graphic representation of the methylation pattern in the first 6 CpG sites analyzed for the FOXA1 locus. The color denotation is shown on the top.
As higher levels of methylation are usually associated with silenced transcription, our result raises the distinct possibility that parity-associated epigenetic silencing of FOXA1 may result in attenuated ER function and reduced risk of tumorigenesis for the corresponding breast epithelial cells. One caveat in the current work was our inability of directly measuring the FOXA1 mRNA levels in the same breast tissue samples used for the methylation study, which was likely due to the sub-optimal preservation of the clinical samples. We are also aware of the possibility that epigenetic changes and ER functions could be influenced by multiple known risk factors including menopausal status, body weight index (BMI), comorbidity with metabolism-related disease, and ethnicity, etc. However, using the current cohort of relatively small sample size, we did not find any significant correlation between the methylation pattern at the two CpG sites (#2 and #18) of FOXA1 and the other confounding factors (Supplemental Table 3). Lastly, the size of the cohorts used in the current study is relatively small. It is therefore important to validate the findings in future with larger pools of clinical samples. Upon confirmation, the hypermethylation sites identified in the current study could serve as quantifiable metrics for assessing parity-associated breast cancer risk.
Delayed pregnancy is a nationwide trend among women of childbearing age. This could be one of the factors contributing to the recently reported increase in the incidence of advanced breast cancer among young women (age 25–39) in this country over the past three decades [27]. Conversely, early and multi-parity among Hispanic women could partly account for the relatively low breast cancer incidence in this population (www.cancer.org). In fact, tissue procurement in our pilot study was facilitated by the relatively high percentage of women with early/multi-parity in San Antonio and South Texas. The average age of first-time mothers in our current cohort is 21.6 years, as compared to national average of 25 years. In addition, close to 40% of first births in our cohort are to mothers under age 20, whereas only 21% are so nationwide. Establishing a causal relationship between FOX1A methylation and breast cancer risk in future work may could inform targeted ways of mimicking the protective effect of early parity among late-parous and nulliparous women.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the NIH grants to R.L. (CA161349) and T.H.M.H (U54 CA113001), Y.H. (CA118578), and a Department of Defense Breast Cancer Research grant to R.L (W81XWH-14-1-0129). We also thank the generous support by the Cancer Therapy and Research Center at University of Texas Health Science Center at San Antonio (P30CA054174). The genomic data are available at NIH Gene Expression Omnibus site (accession number pending).
Footnotes
AUTHOR CONTRIBUTION
R.L. managed the project, R.L., T.H.M.H., P.R., Y.H., and S.G. designed the experiments, S.G., F.G., J.L., C-M.W., and H.W. carried out the experiments, T.H.M.H., F.G., and C-L.L. analyzed the data, R.L. and S.G. wrote the manuscript.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFRRENEES
- 1.Britt K, Ashworth A, Smalley M. Pregnancy and the risk of breast cancer. Endo-Rel Cancer. 2007;14:907–933. doi: 10.1677/ERC-07-0137. [DOI] [PubMed] [Google Scholar]
- 2.Schedin P. Pregnancy-associated breast cancer and metastasis. Nat Rev Cancer. 2006;6:281–291. doi: 10.1038/nrc1839. [DOI] [PubMed] [Google Scholar]
- 3.Polyak K. Pregnancy and breast cancer: the other side of the coin. Cancer Cell. 2006;9:151–153. doi: 10.1016/j.ccr.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 4.Haricharan S, Dong J, Hein S, Reddy JP, Du Z, Toneff M, Holloway K, Hilsenbeck SG, Huang S, Atkinson R, et al. Mechanism and preclinical prevention of increased breast cancer risk caused by pregnancy. eLife. 2013;2:e00996. doi: 10.7554/eLife.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacMahon B, Cole P, Lin TM, Lowe CR, Mirra AP, Ravnihar B, Salber EJ, Valaoras VG, Yuasa S. Age at first birth and breast cancer risk. Bull World Health Organ. 1970;43:209–221. [PMC free article] [PubMed] [Google Scholar]
- 6.Albrektsen G, Heuch I, Hansen S, Kvale G. Breast cancer risk by age at birth, time since birth and time intervals between births: exploring interaction effects. Br J Cancer. 2005;92:167–175. doi: 10.1038/sj.bjc.6602302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt K, Ashworth A, Smalley M. Pregnancy and the risk of breast cancer. Endocr Relat Cancer. 2007;14:907–933. doi: 10.1677/ERC-07-0137. [DOI] [PubMed] [Google Scholar]
- 8.Russo J, Balogh GA, Heulings R, Mailo DA, Moral R, Russo PA, Sheriff F, Vanegas J, Russo IH. Molecular basis of pregnancy-induced breast cancer protection. Eur J Cancer Prev. 2006;15:306–342. doi: 10.1097/00008469-200608000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Russo J, Moral R, Balogh GA, Mailo D, Russo IH. The protective role of pregnancy in breast cancer. Breast Cancer Res. 2005;7:131–142. doi: 10.1186/bcr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silvaraman L, Medina D. Hormone-induced protection against breast cancer. J Mammary Gland Biol Neoplasia. 2002;7:77–92. doi: 10.1023/a:1015774524076. [DOI] [PubMed] [Google Scholar]
- 11.Siwko SK, Dong J, Lewis MT, Liu H, Hilsenbeck SG, Li Y. Evidence that an early pregnancy causes a persistent decrease in the number of functional mammary epithelial stem cells--implications for pregnancy-induced protection against breast cancer. Stem Cells. 2008;26:3205–3209. doi: 10.1634/stemcells.2008-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colditz GA, Rosner BA, Chen WY, Holmes MD, Hankinson EE. Risk factors for breast cancer according to estrogen and progesterone receptor status. J Natl Cancer Inst. 2004;96:218–228. doi: 10.1093/jnci/djh025. [DOI] [PubMed] [Google Scholar]
- 13.Thordarson G, Jin E, Guzman RC, Swanson SM, Nandi S, Talamantes F. Refractoriness to mammary tumorigenesis in parous rats: is it caused by persistent changes in the hormonal environment or permanent biochemical alterations in the mammary epithelia? Carcinogenesis. 1995;16:2847–2853. doi: 10.1093/carcin/16.11.2847. [DOI] [PubMed] [Google Scholar]
- 14.Meier-Abt F, Milani E, Roloff T, Brinkhaus H, Duss S, Meyer DS, Klebba I, Balwierz PJ, van Nimwegen E, Bentires-Alj M. Parity induces differentiation and reduces Wnt/Notch signaling ratio and proliferation potential of basal stem/progenitor cells isolated from mouse mammary epithelium. Breast Cancer Res. 2013;15:R36. doi: 10.1186/bcr3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernardo GM, Keri RA FOXA1. a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32:113–130. doi: 10.1042/BSR20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll JS, Liu S, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Augello MA, Hickey TE, Knudsen KE. FOXA1 master of steroid receptor function in cancer. EMBO J. 2011;30:3885–3894. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watters RJ, Benos PV, Oesterreich S. To bind or not to bind--FoxA1 determines estrogen receptor action in breast cancer progression. Breast Cancer Res. 2012;14:312. doi: 10.1186/bcr3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2010;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, Moore JH, Lupien M. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang TT, Gonzales CB, Gu F, Hsu YT, Jadhav RR, Wang CM, Redding SW, Tseng CE, Lee CC, Thompson IM, et al. Epigenetic deregulation of the anaplastic lymphoma kinase gene modulates mesenchymal characteristics of oral squamous cell carcinomas. Carcinogenesis. 2013;34:1717–1727. doi: 10.1093/carcin/bgt112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh M, Igarashi M, Fukuda H, Nakagama H. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 25.van Agthoven T, Sieuwerts AM, Meijer-van Gelder ME, Look MP, Smid M, Veldscholte J, Sleijfer S, Foekens JA, Dorssers LC. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol. 2009;27:542–549. doi: 10.1200/JCO.2008.17.1462. [DOI] [PubMed] [Google Scholar]
- 26.Wilson AL, Schrecengost RS, Guerrero MS, Thomas KS, Bouton AH. Breast cancer antiestrogen resistance 3 (BCAR3) promotes cell motility by regulating actin cytoskeletal and adhesion remodeling in invasive breast cancer cells. PloS One. 2013;8:e65678. doi: 10.1371/journal.pone.0065678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson RH, Chien FL, Bleyer A. Incidence rate of breast cancer in young women--reply. JAMA. 2013;309:2435–2436. doi: 10.1001/jama.2013.6464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.